Abstract
Background
Current research has shown that microRNAs (miRNAs) play vital roles in plant response to stress caused by heavy metals such as aluminum, arsenic, cadmium (Cd), and mercury. Cd has become one of the most hazardous pollutants in the environment. Maize can be a potential model to study phytoremediation of Cd-contaminated soil owing to its large biomass production. However, little is known about miRNAs as a response to Cd stress in maize.
Results
To investigate the role of miRNAs in response to Cd stress, roots of seedlings of the inbred maize lines B73 and Mo17 were collected and treated with 200 mg/L CdCl2·2.5 H2O over different exposure times. Enzyme activities of superoxide dismutase and peroxidase were measured to confirm Cd stress. The expression of six candidate miRNAs and their targets were validated using quantitative real-time PCR (qRT-PCR) technology. In addition, the expression of Zma-miR171b was assessed using in situ hybridization.
Conclusions
Our results showed that miRNAs and their respective target genes were differentially expressed in maize seedling roots exposed to Cd stress. This research produced new insights into the molecular mechanism of miRNAs responsive to Cd stress in plants and sheds light on the latent roles of miRNAs in plants exposed to heavy metal stresses.
Electronic supplementary material
The online version of this article (10.1186/s12867-019-0131-1) contains supplementary material, which is available to authorized users.
Keywords: miRNAs, Cadmium stress, Maize (Zea mays), Target gene
Background
Contamination of soils with heavy metals has become a prevalent topic in ecology owing to their widespread release from industry, agriculture, and other human activities [1]. Cadmium (Cd) adversely affects various physiological and biochemical processes such as nitrogen metabolism, photosynthesis, mineral utilization, carbohydrate metabolism, and water uptake, thereby inhibiting plant growth and development [2, 3]. Cd pollution has substantially increased over the least four decades worldwide, which is a matter of concern for human health [4]. Therefore, research on the molecular mechanisms underlying Cd pollution in plants is urgently required.
Recently, several studies investigated the molecular mechanisms association with Cd tolerance and toxicity in plants [2, 3]. Exceptional microRNAs (miRNA) expression was identified in tissues and cells of stressed plants, and the involvement of miRNAs in Cd-stress responses was suggested [5–7]. In rapeseed under Cd stress, the miRNA bna-miR393 was specifically expressed in leaves, bna-miR156a and bna-miR167a/c were expressed in roots and leaves, respectively, and bna-miR164b and bna-miR394a/b/c were expressed in all tissues [5]. Similarly, differential expression of numerous non-conserved and conserved miRNAs was reported to be induced by Cd stress in Brassica napus roots [5]. In the same study, 84 conserved and non-conserved miRNAs families with differential expression were identified in shoots and roots under Cd stress [5]. In rice, 19 Cd-responsive miRNAs and their targets including transcription factors and stress-related proteins were identified in seedlings using a microarray assay [6]. In addition, to assess Cd-responsive miRNAs and their associated genes in radish (Raphanus sativus), small-RNA libraries were constructed from plants under Cd stress and from controls. A total of 23 miRNA families with notable differential expression (15 previously known and 8 novel miRNA families) was identified in Cd-stressed plants, and their targets were then predicted using a degradome analysis [7]. Numerous potential heavy Cd-responsive miRNA candidates were identified in rice, B. napus, radish, and many other plants [5–7]. However, information on the molecular mechanism of Cd stress resistance in maize is limited. Therefore, we performed a characterization of Cd stress-mediated miRNAs and their target genes in roots of B73 and Mo17 maize under Cd stress and in negative controls. We identified target genes associated with Cd-regulated candidate miRNAs in maize seedling roots. The candidate miRNAs that respond to Cd stress will be useful for further improving Cd tolerance in maize.
Results
Expression pattern of conserved miRNAs in plant under different heavy metal stress
Recent studies showed that miRNAs play vital roles in regulating the expression of stress response genes following stress caused by various heavy metals. In this study, we assessed the expression patterns of miRNAs in plant exposed to different heavy metals, including aluminum (Al), arsenic (As), cadmium (Cd), and mercury (Hg) [6, 8–11]. Significant differential expression of miRNAs due to heavy metal stress was observed, such as down-regulation of common miRNAs (miR159, miR160, miR166, miR319, and miR396) and up-regulation of miRNA miR393. Other miRNAs also showed differential expression in response to heavy metal stress: miR156, miR159, miR160, miR166, miR167, miR171, miR319, miR390, miR394, miR396, and miR408 were down-regulated, and miR393 was up-regulated under Cd stress (Fig. 1). A conservation analysis of those miRNAs showed that miR156/miR157, miR159/319, miR166, and miR170/miR171 were higher conserved in all organisms, followed by miR160, miR396, miR167, and miR390. The miRNAs miR398 and miR394 were less conserved in all organisms (Additional file 1: Table S4). Based on the results of significant differential expression and conservation analyses of these miRNAs under Cd stress, miR156, miR166, miR167, miR171, and miR393 were selected as candidates for differentially expressed miRNAs responsive to Cd stress. In addition, we analyzed the expression profiles of these miRNAs and their target genes in the inbred maize lines B73 and Mo17 using qRT-PCR.
Fig. 1.

Altered expression of miRNAs in plants in response to heavy metal stress. Green indicates down-regulation, and red indicates up-regulation. Quantitative data of miRNA abundance in diverse plant species were collected; green indicates down-regulated miRNAs, red indicates up-regulated miRNAs
Alterations in the development of maize leaves and root system in response to Cd stress
To confirm the appropriate treatment time for validation of candidate Cd-regulated miRNAs, we analyzed the enzymatic activities of superoxide dismutase (SOD) and peroxidase (POD) in Cd-treated maize roots and leaves after treatment times of 6–96 h. We found that SOD activity increased after 12, 24, and 48 h of the Cd-treatment, and particularly so after 12 h, in both Mo17 and B73. POD activity showed the opposite pattern in both phenotypes. After 48 h, SOD and POD activity gradually decreased to a constant level. SOD and POD activity was higher in roots than in leaves, in both phenotypes (Additional file 2: Figure S1). These results suggest that certain Cd-tolerant mechanisms were activated to maintain the normal physiological and metabolic activities shortly after the Cd treatment. Subsequently, RNA was isolated from the B73 and Mo17 treatments and control groups.
Cd-regulated miRNA expression levels
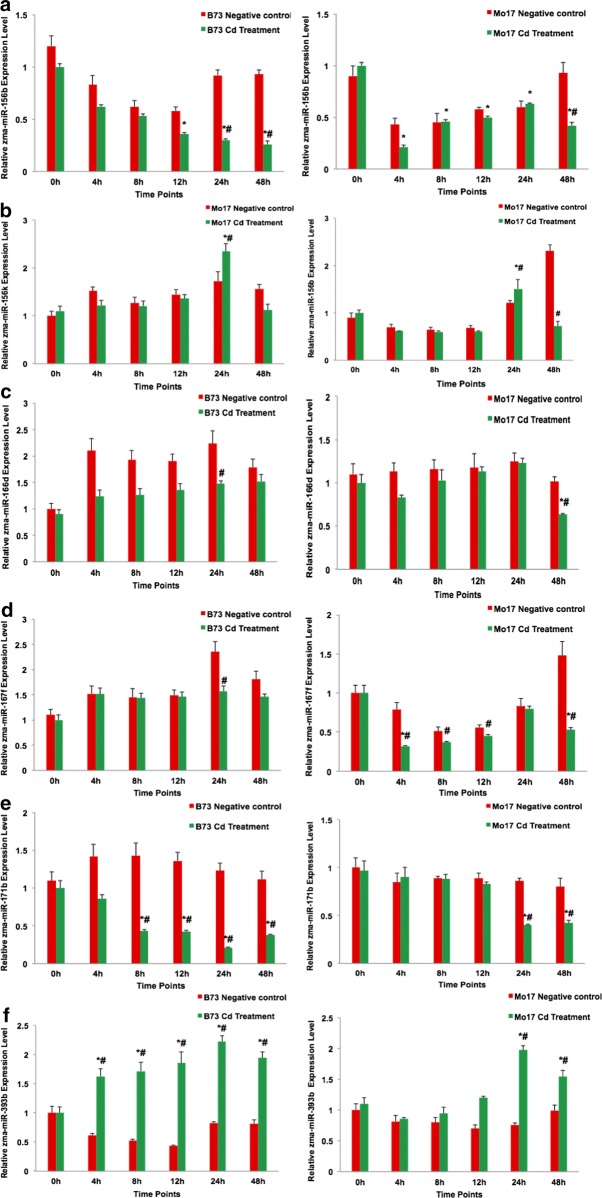
To assess the functions of these candidate miRNAs selected according to altered expression levels of conserved miRNAs in plants under Cd stress, we analyzed their expression profiles using qRT-PCR. Zma-miR156b, Zma-miR156k, Zma-miR166d, Zma-miR167f, Zma-miR171b, and Zma-miR393b were expressed in maize roots of both maize lines (Fig. 2). Zma-miR156b showed a trend of down-regulation in roots after the Cd treatment in B73, compared with the control, whereas no significant changes were observed in Mo17 (Fig. 2a). Zma-miR156k showed no significant alterations in B73 and Mo17 (Fig. 2b). Zma-miR166d was more down-regulated in B73 than in Mo17 roots under Cd stress (Fig. 2c), and Zma-miR167f was more down-regulated in Mo17 than in B73 (Fig. 2d). Zma-miR171b was down-regulated in both maize lines (Fig. 2e), in contrast to miR393b (Fig. 2f).
Fig. 2.
Validation of candidate microRNAs related to Cd stress using qRT-PCR. Left: B73, right; Mo17. Red and green vertical bars indicate the negative control (CK) and the Cd treatment, respectively. The letters A to F indicate the expression levels of Zma-miR156b, Zma-miR156k, Zma-miR166d, Zma-miR167f, Zma-miR171b, and Zma-miR393b. Shown are the mean ± SEM. *p < 0.05 Cd exposure time vs. 0 h; #p < 0.05 treatment vs. negative control
qRT-PCR analysis of target genes related to candidate microRNAs in response to Cd stress
The results suggested that the activities of miRNA-mRNA complexes could be altered by stress. To identify target genes of candidate miRNAs regulated by Cd stress, the target genes were predicted using WMD3 and PsRNA target website using the MGC database (ZmB73_v5b, Table. S5). Most of the target genes association with the candidate miRNAs belonged to transcription factor families, e.g., Zma-miRNA156 targeting the SBP transcription factor family, Zma-miR166 acting on the basic-leucine zipper (bZIP) transcription factors, Zma-miR167 regulating auxin response factors (ARFs), GRAS transcription factors were predicted targets of Zma-miR171, and F-box proteins were predicted targets of Zma-miR393. The relative expression level of target genes seemed negatively correlated with that of the respective miRNAs (Fig. 3). The target genes of Zma-miR156b (SBP-box), Zma-miR166d (bZIP), and Zma-miR171b (GRAS) were significantly up-regulated during the dynamic development of roots in B73, compared to in Mo17 (Fig. 3a, c, e). In contrast, the target gene of Zma-miR393b (F-box-like protein) was observed to be down-regulated in B73 than in Mo17 samples (Fig. 3f). However, the target genes of Zma-miR156k (SBP-box) and Zma-miR167f (ARF) showed no significant up-regulation in either maize line (Fig. 3b, d). In conclusion, candidate miRNAs such as Zma-miR156b, Zma-miR166d, Zma-miR171b, and Zma-miR393b may negatively regulate their respective target genes in maize under Cd stress, which likely involves transcriptional and post-transcriptional regulatory networks.
Fig. 3.
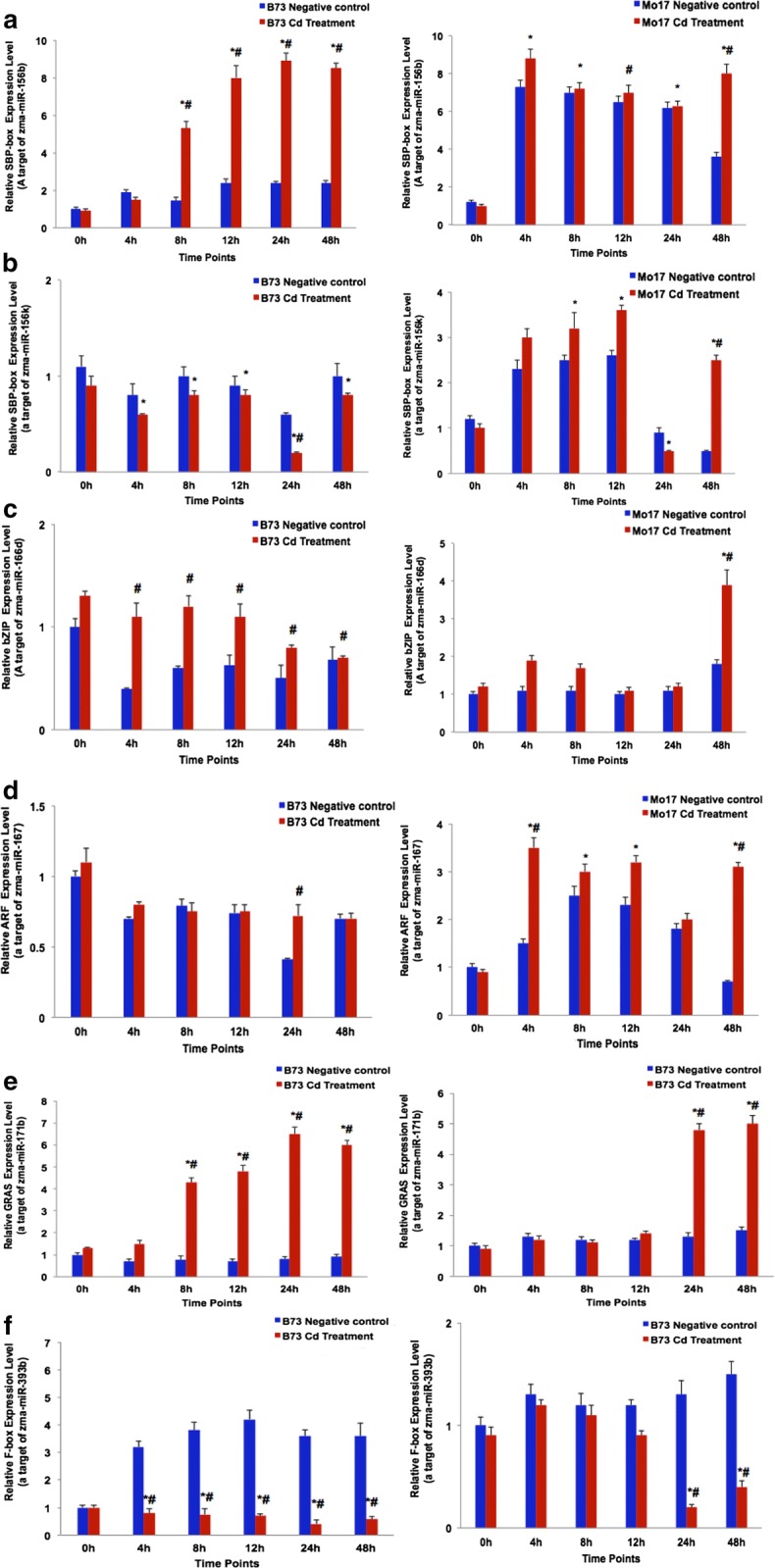
Validation of target genes of candidate microRNAs related to Cd stress using qRT-PCR technology. Negative controls (CK) sample and Cd-treated plants. The color nodes indicate the expression of fold change. Letters from A to F indicate expression levels of SBP-box, SBP-box, bZIP, ARF, GRAS, and F-box-like protein, respectively. Error bars indicate the standard error calculated from three biological replicates. Shown are the mean ± SEM. *p < 0.05 treatment time vs. 0 h; #p < 0.05 treatment vs. negative control
Expression of Zma-miR171b identified by in situ hybridization
Based on the results obtained from the qRT-PCR of candidate miRNAs and their respective targets genes, the expression of Zma-miR171b was selected and further analyzed using in situ hybridization (ISH). Black-blue positive hybrid signals of Zma-miR171b were detected throughout at 0, 4, 12, and 24 h of treatment in maize roots of B73 and Mo17. Expression of Zma-miR171b was down-regulated following Cd stress at different stages in B73 roots, compared to that in the control, and reached a minimum at 48 h; similar trends were observed in Mo17, but expression levels of Zma-miR171b were consistently higher in B73 than in Mo17 (Fig. 4). Zma-miR171b was down-regulated in both phenotypes, thus B73 likely accumulated GRAS gene products earlier in response to Cd stress than Mo17 (Figs. 2e, 3e).
Fig. 4.
Change of relative quantity of Zma-miR171b in B73 and Mo17 using ISH. The microscopic results were quantified as black-blue (ISH) staining as a positive signal in per unit area (n = 9). Error bars show the standard error calculated from three biological replicates. Shown are the means per section ± SEM. *p < 0.05 treatment time vs. 0 h; #p < 0.05 treatment vs. negative control (NC)
Gene ontology annotation and analysis of target genes
Gene ontology (GO) analyses were performed to gain understanding of the function of target genes. Target genes that were specifically enriched in the regulation of biological processes showed transcription regulation activity and binding activity; however, those target genes were decreased in metabolism processes, and their catalytic activities were also reduced (Additional file 2: Figure S2).
Discussion
miRNAs are one of the major types of endogenous, small, non-protein-coding single-stranded RNA, which act as regulators of plant growth and development at the translation and post-transcription levels by silencing gene expression [12, 13]. Previous studies demonstrated that miRNAs acts as regulators in plants exposed to abiotic stressors [14]. Moreover, many miRNAs were found to be important for plant responses to heavy metal stress using small RNA profiling [15, 16]. Recent studies identified numerous heavy metals such as Hg, Al, As, and Cd to regulate miRNAs in plants [6, 8–11]. However, we found that heavy metals regulated miRNAs varied from different plants and metal species. As shown in Fig. 1, some miRNA families (miR396, miR390, miR171, miR166, miR162, and miR159) were down-regulated, whereas miR395 and miR393 were up-regulated by most heavy metals. The expression of miR396, miR390, and miR159 was consistently down-regulated, and miR393 was up-regulated under exposure to four heavy metals. Ding et al. demonstrated that miR390, miR171, miR168, miR166, and miR156 responded to Cd stress in rice [17], and Zhou et al. found that expression of miR408, miR398, miR397, and miR396 was associated with Cd exposure in Brassica [5]. In addition, miR393 and miR171 were found to be affected by Cd stress through negatively regulating their target genes in Oryza sativa [18], Medicago truncatula [5], and B. napus [19]. In the current study, we demonstrated that Zma-miR156b, Zma-miR166d, Zma-miR171b, and Zma-miR393b were affected by Cd exposure in maize roots, using qRT-PCR-based analyses.
Recently, identification of target genes of metal-regulated miRNAs showed that most of these miRNAs encoded key regulatory proteins or TFs that were associated with diverse metabolic pathways [20–22]. In our study, several candidate miRNAs such as Zma-miR156b, Zma-miR166d, Zma-miR167f, Zma-miR171b, and Zma-miR393b were found to target the genes of TF families (SBP-box, bZIP, ARFs, GRAS), and F-box-like protein, most of which have been confirmed to play vital roles in the response to Cd stress (Additional file 1: Table S5, Fig. 4). Previous studies confirmed that SBP-box transcription factors were involved in a broad range of developmental and stress response processes targeted by miR156 in Arabidopsis [23, 24]. In addition, the SBP-box gene family was characterized as metal-containing transcription factors regulating copper homeostasis in Arabidopsis [25]. In the present study, two miRNAs belonged to the miRNA156 family (Zma-miR156b and Zma-miR156k) targeting SBP-box transcription factors. It may be inferred that miR156 may be an important regulator in Cd2+ homeostasis in maize roots by targeting SBP-box transcription factors. Moreover, for the bZIP family, bZIP62, ThbZIP1, and BjCdR15 were reported to be associated with Cd stress. Of those, the gene encoding bZIP62 was induced by Cd exposure in soybean roots, whereas ThbZIP1 showed increased expression in Tamarix hispida [26, 27]. In the current study, we found that bZIP transcription factors were regulated by Zma-miRNA166d. We suggest that Zma-miR166 acted in Cd2+ homeostasis in maize roots by targeting bZIP transcription factors. Furthermore, previous studies showed that some miRNAs responded to heavy metal stress through regulating phytohormone biosynthesis and signaling responses. For instance, As stress in B. juncea led to an increased miR167 abundance [11]. The auxin response factors ARF6 and ARF8 targeted by miR167 were found positively correlated with adventitious rooting and development of anthers and ovules via auxin homeostasis in Arabidopsis [28]. Moreover, ARF proteins may activate or repress the transcription of primary auxin response genes through binding to auxin-responsive elements [29]. F-box proteins, including transport inhibitor response 1 (TIR1) and auxin-signaling F-box proteins (AFB1, AFB2, and AFB3) can perceive auxins in plants [30–33]. In addition, TIR1 encoded an E3 ubiquitin ligase containing F-box subunit were targeted by miR393 [34] which interacts with Aux/IAA proteins as an auxin receptor, thereby resulting in Aux/IAA ubiquitination and subsequent degradation in auxin signal pathway [35]. In our study, we demonstrated that Zma-miR393b and Zma-miR167f targeted auxin signaling F-box proteins and that ARFs transcription factors responded to the Cd2+ treatment.
Conclusion
In conclusion, this is the first study to characterize Cd-stress induced miRNAs and their target genes in maize roots. Six previously known miRNAs and their respective target genes were found to respond to Cd stress. The expression patterns of these differentially regulated miRNAs and association targets were shown to be affected by Cd stress in the inbred maize lines B73 and Mo17. Most of the target transcripts belonged to transcription factors that were predicted to be functionally involved in different pathways that respond to Cd stress. These results provide new insights for the functional characterization of miRNAs and their targets in plant responses to heavy metal stress.
Methods
Plant material and experimental design
The inbred maize line B73 was previously reported to be more tolerant to heavy metal stress than Mo17 [36], particularly regarding Cd stress. In this study, seeds collected from Mo17 and B73 were washed using distilled water and germinated at 26 °C in the dark for 3 days. Germinated seeds were grown in a growth chamber under the following conditions: a photoperiod of 14 h light/10 h dark and relative humidity of 70% at 26 °C. After 20 days, the seedlings were transplanted to a plastic container with a modified half-strength Hoagland nutrient medium (see Additional file 1: Table S1). Subsequently, the plants were randomly assigned to a Cd stress treatment (200 mg/L CdCl2·2.5 H2O) and a control (Cd-free).
Enzymatic activity assays of POD and SOD
Maize seedlings of the treatment group and the control were collected after exposed to 200 mg/L CdCl2·2.5H2O for 0, 4, 8, 12, 24, 48, 72, or 96 h. POD and SOD activities were determined according to the methods described by Beyer [37] and Kim [38], respectively. All measurements were performed three times using three biological replicates.
RNA isolation from maize roots
Based on the results of the POD and SOD enzymatic activity assays, total RNA was isolated from Cd-free and Cd-exposed roots of B73 and Mo17 after 0, 4, 8, 12, 24, and 48 h, using Trizol reagent (Life, US) following the manufacturer’s instructions. Small RNA was isolated from each sample using the mirVana™miRNA Isolation Kit (Ambion, US) according to the manufacturer’s instructions.
Validation of candidate miRNAs using real-time PCR
Total miRNA was isolated from B73 and Mo17 using a Universal Plant microRNA Extraction Kit (Spin-column, BioTeke, China). Reverse transcription reactions of miRNAs were performed, and PCRs were performed in triplicate, according to Jian et al. [39]. Mitochondrial 5S RNA was used as a control, and specific primers were for real-time PCR (Additional file 1: Table S2). The absolute amount of each miRNA was calculated according to a standard curve using the 2−∆∆CT method [40].
ISH assays for Zma-miR171b
For ISH, miRCURY 5′-DIG and 3′-DIG-labeled locked nucleic acid (LNA)-miRNAs were utilized (Zma-miR171b) as detection probes. The sequence of the probe Zma-miR171b was 5′-DIG-UUGAGCCGUGCCAAUAUCAC-DIG-3′. Sections of 30 µm thickness were performed using LNA probes, and all steps were carried out as described by Urbanek et al. [41].
Target gene prediction of candidate miRNAs in the Cd2+ treatment
To predict the accuracy of target genes, the online tools WMD3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi?) and psRNA (http://plantgrn.noble.org/psRNATarget/) were used with default settings [42, 43].
qRT-PCR confirmation of target genes
Specific qRT-PCR primers were designed for target genes using the software Primer Premier (version 5.0); the primers are shown in Additional file 1: Table S3. The amplification programs were performed according to the standard protocol of the ABI7500 system and were conducted in triplicate, according to the method of Jian et al. [39]. The cycle threshold (Ct) of each tested gene were averaged for triplicate reactions, and the values were normalized according to the Ct of the control products of the Actin1 gene. Statistical analysis was performed using the 2−∆∆CT method.
GO annotation of putative target genes responsive to Cd stress
To annotate target genes we used singular enrichment analysis by examining the AgriGO database [44]. Using a customized or pre-calculated background, enriched GO terms were removed after statistical testing. GO annotation comprised biological processes, cellular components, and component functions of the putative sequences.
Additional files
Additional file 1: Figure S1. Effect of Cd stress on SOD and POD enzyme activities in B73 and Mo17. The graphs (A) and (B) depicted the average change of superoxide dismutase (SOD) activities and contents of peroxidase (POD) in maize inbred line B73 and Mo17 leaf and roots respectively. The data are presented as the mean number per section ± SEM. *p<0.05 experimental vs. 0 h; #p<0.05 root vs. leaf. Figure S2. GO annotation of target genes of the candidate miRNAs. The Y -axis is the percentage of targeted genes mapped by the term, and represents the abundance of the GO term. The X -axis is the definition of GO terms.
Additional file 2: Table S1. The detailed components of the nutrient solution used in this study. Table S2. List of primers for qRT-PCR analysis of candidate miRNAs responsive to Cd stress. Table S3. List of primers for qRT-PCR analysis of targets related to those candidate miRNAs under Cd treatment. Table S4. Conserved analysis of candidate miRNAs responsive to heavy metal in different species. Table S5. Conserved candidate microRNAs related to Cd stress and their targets identified in maize.
Authors’ contributions
JG and ML designed research and they contributed equally to this study; HP and WL performed research. WL and JG analyzed data and conducted qRT-PCR experiments; FC and JG wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its additional information files].
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
Normal Program supported this research for Science and Technology Research Program of Sichuan Municipal Education Commission (2018JY0170), Special Project of Science and Technology Innovation for Social Undertaking and People’s Livelihood Guarantee of CSTC (No. cstc2017shms-xdny80074).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Cd
cadmium
- Al
aluminum
- As
arsenic
- Hg
mercury
- SOD
superoxide dismutase
- POD
peroxidase
- miRNA
micro RNA
- qRT-PCR
quantitative real-time PCR
- ISH
in situ hybridization
- SEA
singular enrichment analysis
- GO
gene ontology
- TF
transcription factor
- ARF
auxin response factor
References
- 1.Valko M, Morris H, Cronin M. toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 2.Besson-Bard A, Gravot A, Richaud P, et al. Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol. 2009;149:1302–1315. doi: 10.1104/pp.108.133348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DalCorso G, Farinati S, Furini A. Regulatory networks of cadmium stress in plants. Plant Signal Behav. 2010;5:663–667. doi: 10.4161/psb.5.6.11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa S, Noriharu A, Murakami M, Wagatsuma T. Is Brassica juncea a suitable plant for phytoremediation of cadmium in soils with moderately low cadmium contamination?–Possibility of using other plant species for Cd-phytoextraction. Soil Sci Plant Nutr. 2006;52:32–42. doi: 10.1111/j.1747-0765.2006.00008.x. [DOI] [Google Scholar]
- 5.Zhao SZ, Jian BS, Zhi MY. Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J Exp Bot. 2012;63:4597–4613. doi: 10.1093/jxb/ers136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, Qu A, Gong S, et al. Molecular identification and analysis of Cd-responsive microRNAs in rice. J Agric Food Chem. 2013;61:11668–11675. doi: 10.1021/jf401359q. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Wang Y, Zhai L, et al. Genome-wide identification and characterization of cadmium-responsive microRNAs and their target genes in radish (Raphanus sativus L.) roots. J Exp Bot. 2013;64:4271–4287. doi: 10.1093/jxb/ert240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou ZS, Zeng HQ, Liu ZP, Yang ZM. Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 2012;35:86–99. doi: 10.1111/j.1365-3040.2011.02418.x. [DOI] [PubMed] [Google Scholar]
- 9.Valdéslópez O, Yang SS, Apariciofabre R, et al. MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 2010;187:805–818. doi: 10.1111/j.1469-8137.2010.03320.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Wang T, Zhao M, et al. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta. 2012;235:375–386. doi: 10.1007/s00425-011-1514-9. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava S, Srivastava AK, Suprasanna P, D’Souza SF. Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. J Exp Bot. 2013;64:303–315. doi: 10.1093/jxb/ers333. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 15.Sunkar R, Kapoor A, Zhu J-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down regulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y-F, Zhu C. The role of microRNAs in copper and cadmium homeostasis. Biochem Biophys Res Commun. 2009;386:6–10. doi: 10.1016/j.bbrc.2009.05.137. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y, Chen Z, Zhu C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa) J Exp Bot. 2011;62:3563–3573. doi: 10.1093/jxb/err046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Ying H, Yunxia L. Combined toxicity of copper and cadmium to six rice genotypes (Oryza sativa L.) J Environ Sci. 2009;21:647–653. doi: 10.1016/S1001-0742(08)62320-7. [DOI] [PubMed] [Google Scholar]
- 19.Huang SQ, Xiang AL, Che LL, et al. A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol J. 2010;8:887–899. doi: 10.1111/j.1467-7652.2010.00517.x. [DOI] [PubMed] [Google Scholar]
- 20.Van Ex F, Jacob Y, Martienssen RA. Multiple roles for small RNAs during plant reproduction. Curr Opin Plant Biol. 2011;14:588–593. doi: 10.1016/j.pbi.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noman A, Fahad S, Aqeel M, et al. miRNAs: major modulators for crop growth and development under abiotic stresses. Biotechol Lett. 2017;39(5):685–700. doi: 10.1007/s10529-017-2302-9. [DOI] [PubMed] [Google Scholar]
- 22.Noman A, Aqeel M. miRNA-based heavy metal homeostasis and plant growth. Environ Sci Pollut Res. 2017;24(11):10068–10082. doi: 10.1007/s11356-017-8593-5. [DOI] [PubMed] [Google Scholar]
- 23.Yu S, Galvão VC, Zhang Y-C, et al. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilon M, Cohu CM, Ravet K, et al. Essential transition metal homeostasis in plants. Curr Opin Plant Biol. 2009;12:347–357. doi: 10.1016/j.pbi.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Kropat J, Tottey S, Birkenbihl RP, et al. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. PNAS. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chmielowska-Bąk J, Lefèvre I, Lutts S, Deckert J. Short term signaling responses in roots of young soybean seedlings exposed to cadmium stress. J Plant Physiol. 2013;170:1585–1594. doi: 10.1016/j.jplph.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Wang W, Yin H, et al. Exogenous nitric oxide improves antioxidative capacity and reduces auxin degradation in roots of Medicago truncatula seedlings under cadmium stress. Plant Soil. 2010;326:321–330. doi: 10.1007/s11104-009-0011-4. [DOI] [Google Scholar]
- 28.Gutierrez L, Mongelard G, Floková K, et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell. 2012;24:2515–2527. doi: 10.1105/tpc.112.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. doi: 10.1023/A:1015207114117. [DOI] [PubMed] [Google Scholar]
- 30.Navarro L, Dunoyer P, Jay F, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Sci Signal. 2006;312:436. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 31.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 32.Dharmasiri N, Dharmasiri S, Weijers D, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z-H, Bao M-L, Sun Y-Z, et al. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol Biol. 2011;77:619–629. doi: 10.1007/s11103-011-9838-1. [DOI] [PubMed] [Google Scholar]
- 35.Si-Ammour A, Windels D, Arn-Bouldoires E, et al. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011;157:683–691. doi: 10.1104/pp.111.180083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxter IR, Gustin JL, Settles AM, Hoekenga OA. Ionomic Characterization of Maize Kernels in the Intermated B73 × Mo17 Population. Crop Sci. 2013;53:208–220. doi: 10.2135/cropsci2012.02.0135. [DOI] [Google Scholar]
- 37.Beyer WF, Fridovich I. Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal Biochem. 1987;161:559–566. doi: 10.1016/0003-2697(87)90489-1. [DOI] [PubMed] [Google Scholar]
- 38.Kim YH, Yoo YJ. Peroxidase production from carrot hairy root cell culture. Enzyme Microbial Technol. 1996;18:531–535. doi: 10.1016/0141-0229(95)00168-9. [DOI] [Google Scholar]
- 39.Gao J, Luo M, Zhang C, et al. A putative pathogen-resistant regulatory pathway between MicroRNAs and candidate target genes in maize. J Plant Biol. 2015;58(4):211–219. doi: 10.1007/s12374-014-0572-5. [DOI] [Google Scholar]
- 40.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 41.Urbanek MO, Nawrocka AU, Krzyzosiak WJ. Small RNA detection by in situ hybridization methods. Int J Mol Sci. 2015;16:13259–13286. doi: 10.3390/ijms160613259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(suppl 2):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 44.Du Z, Zhou X, Ling Y, et al. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(suppl 2):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Effect of Cd stress on SOD and POD enzyme activities in B73 and Mo17. The graphs (A) and (B) depicted the average change of superoxide dismutase (SOD) activities and contents of peroxidase (POD) in maize inbred line B73 and Mo17 leaf and roots respectively. The data are presented as the mean number per section ± SEM. *p<0.05 experimental vs. 0 h; #p<0.05 root vs. leaf. Figure S2. GO annotation of target genes of the candidate miRNAs. The Y -axis is the percentage of targeted genes mapped by the term, and represents the abundance of the GO term. The X -axis is the definition of GO terms.
Additional file 2: Table S1. The detailed components of the nutrient solution used in this study. Table S2. List of primers for qRT-PCR analysis of candidate miRNAs responsive to Cd stress. Table S3. List of primers for qRT-PCR analysis of targets related to those candidate miRNAs under Cd treatment. Table S4. Conserved analysis of candidate miRNAs responsive to heavy metal in different species. Table S5. Conserved candidate microRNAs related to Cd stress and their targets identified in maize.
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its additional information files].