Abstract
The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is critical for DNA repair via the nonhomologous end joining pathway. Previously, it was reported that bone marrow cells and spontaneously transformed fibroblasts from SCID (severe combined immunodeficiency) mice have defects in telomere maintenance. The genetically defective SCID mouse arose spontaneously from its parental strain CB17. One known genomic alteration in SCID mice is a truncation of the extreme carboxyl terminus of DNA-PKcs, but other as yet unidentified alterations may also exist. We have used a defined system, the DNA-PKcs knockout mouse, to investigate specifically the role DNA-PKcs specifically plays in telomere maintenance. We report that primary mouse embryonic fibroblasts (MEFs) and primary cultured kidney cells from 6–8 month-old DNA-PKcs-deficient mice accumulate a large number of telomere fusions, yet still retain wild-type telomere length. Thus, the phenotype of this defect separates the two-telomere related phenotypes, capping, and length maintenance. DNA-PKcs-deficient MEFs also exhibit elevated levels of chromosome fragments and breaks, which correlate with increased telomere fusions. Based on the high levels of telomere fusions observed in DNA-PKcs deficient cells, we conclude that DNA-PKcs plays an important capping role at the mammalian telomere.
Telomeres are composed of DNA/protein protective caps that prevent chromosome end to end fusions (1). A number of telomere-associated proteins have been identified from a wide variety of diverse species, and investigations into how these proteins function to maintain telomere homeostasis is underway (1). Telomere-associated proteins can bind telomeric DNA directly, such as TRF1 and TRF2 (2), or can localize to the telomere by means of interactions with telomere repeat binding proteins (1, 3–5). Interactions between telomere-associated proteins and telomeric DNA, along with telomere repeat synthesis by telomerase (6), are all critical for the maintenance of telomere length and capping function throughout development and the cell cycle.
Several genes known to play key roles in DNA repair have been shown to function in telomere maintenance (5, 7–12). The DNA-dependent protein kinase (DNA-PK) is a multicomponent complex consisting of the DNA-PK catalytic subunit (DNA-PKcs = ≈470 Kd) and the Ku heterodimer (Ku80 = ≈80 Kd and Ku70 = 70 Kd) (13). DNA-PKcs is a serine/threonine protein kinase containing a phosphoinositol 3-kinase (PI3-K) domain (13). Other PI3 kinase family members known to have telomere maintenance roles are the mammalian ataxia-telangiectasia mutated (ATM) protein and the yeast Tel1 and Mec1 proteins (11). DNA-PKcs is a multifunctional protein not only critical for the nonhomologous end joining (NHEJ) pathway, but also for V(D)J recombination and the innate immune response (13–17). Recently, DNA-PKcs was implicated in telomere maintenance by using severe combined immunodeficiency (SCID) mice and spontaneously transformed SCID cell lines. Telomere length in SCID mice was found to be 1.5–2 times longer than the telomeric DNA from other strains of mice (8) and spontaneously transformed cell lines from SCID mice were shown to accumulate telomere fusions (9). SCID mice are an inbred strain and cells from these animals express low levels of a C terminus truncated DNA-PKcs (18, 19). In addition to these defects in DNA-PKcs, SCID mice cells may contain other genomic alterations. When comparisons were made between SCID mice and DNA-PKcs null mice, many important similarities were found in defects related to the repair of double-strand breaks by means of NHEJ (13). However, significant phenotypic differences were also observed between SCID and DNA-PKcs null mice in T and B cell development (13, 15, 20) and the innate immune response, suggesting that the SCID defect is not completely equivalent to DNA-PKcs deficiency (17, 21).
In this report, we use DNA-PKcs null mice to determine the role of DNA-PKcs in telomere maintenance (15). The advantages of using DNA-PKcs null mice are: (i) this is a defined system with one known alteration; (ii) isogenic primary cells can be generated from wild-type, heterozygous, and homozygous deficient animals. We report that a deficiency in DNA-PKcs severely disrupts capping function, yet does not affect telomere length. In addition, telomerase activity is not affected by a DNA-PKcs deficiency. Thus, this is an example of a telomere maintenance protein, DNA-PKcs, which functions specifically in telomere capping not telomere length control.
Materials and Methods
DNA-PKcs Knockout Mice.
The targeting vector was constructed by substituting half of DNA-PKcs exon 3 and part of intron 3 with the PGK-neo gene as described (15). The DNA-PKcs−/− mice were obtained by intercrossing DNA-PKcs+/− mice.
Establishment of Primary Mouse Embryonic Fibroblasts (MEFs) and Primary Kidney Cells.
Primary MEFs were isolated from 13.5-day mouse embryos and harvested for analysis after eight to ten population doublings. Primary kidney cells were generated from 6–8-month mice and harvested for analysis after eight to ten population doublings. Independently isolated littermates of primary MEFs and primary kidney cells were generated from DNA-PKcs wild-type (+/+), heterozygous (+/−), and homozygous (−/−) genotype. Cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin and maintained at 37°C in a humidified atmosphere of 3% O2/10% CO2.
Genotyping of DNA-PKcs+/+, +/−, and −/− Mice.
To distinguish the endogenous from the targeted DNA-PKcs allele, PCR was performed with primers MD-32 (CTT GCA ACC GTT TTA GAG GTC TC) and MD-33 (GTT CTC TAA ACC ACA GCC TGA AG) for the endogenous allele, and MD-20 (TAT CCG GAA GTC GCT TAG CAT TG) and POL-8 (TTC ACA TAC ACC TTG TCT CCG ACG) for the targeted DNA-PKcs allele as described (15). These primer sets amplify a 450-bp and 360-bp fragment, respectively. All PCR products were analyzed by gel electrophoresis in 1.5% agarose.
Pulse-Field Gel Electrophoresis and TRF Analysis.
Cells were isolated and embedded in agarose plugs as described (27). DNA was digested with HinfI and RsaI (New England Biolabs) and electrophoresed on a 1% TBE agarose gel at 14°C, using a CHEF DR-II pulsed-field apparatus (Bio-Rad). Pulse-field electrophoresis was performed at 6 V/cm for 18 h at a ramp pulse of 0.2–13 s. Gels were transferred onto charged nylon membranes (Hybond N+, Amersham Pharmacia) as recommended by the supplier. Filters were hybridized with a labeled telomeric repeat (TTAGGG)3 probe and exposed to a PhosphoImager screen (Molecular Dynamics) for 4–10 h. The screen was scanned with a Storm 820 phosphor screen scanner (Molecular Dynamics).
Telomeric Repeat Amplification Protocol (TRAP) Assay.
Telomerase activity was measured by the TRAP (28) by using the TRAP-eze telomerase detection kit (Intergen, Purchase, NY). Cell lysate from 103 cells was used for each assay. The resulting PCR product and a 6-bp incremental ladder were electrophoresed on a 12.5% nondenaturing polyacrylamide gel and visualized by SYBR gold staining (Molecular probe).
Fluorescence in Situ Hybridization (FISH).
FISH analysis using a Cy3-labeled (CCCTAA)3 peptide nucleic acid probe was performed as described (29, 30). Cells were viewed with an Olympus (New Hyde Park, NY) BH2 microscope or a Zeiss Axioplan 2 imaging system equipped with a charge-coupled device (CCD) camera. Images were acquired by using CYTOVISION software (Applied Imaging, Santa Clara, CA) or ISIS software (Metasystems, Altlussheim, Germany). Quantitative analysis of telomere fluorescence was performed as described (30).
Results and Discussion
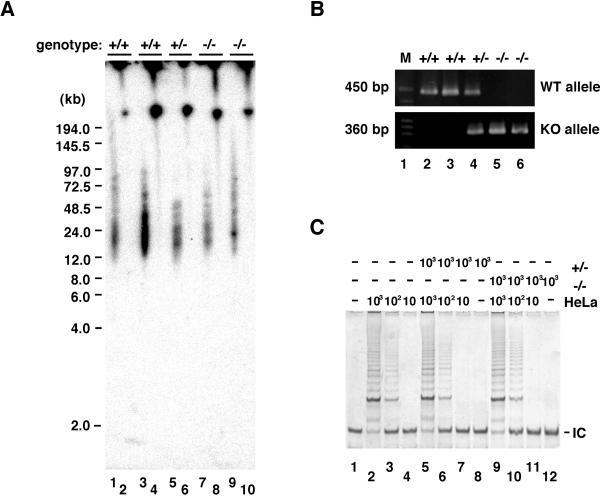
To determine the role of DNA-PKcs in telomere maintenance, DNA-PKcs+/− mice were mated to produce primary isogenic MEF lines of the three genotypes (DNA-PKcs+/+, DNA-PKcs+/−, and DNA-PKcs−/−). Genotyping of each primary MEF line was determined by PCR analysis (Fig. 1B). Two complementary methods were used to determine telomere length: Southern blot analysis and quantitative fluorescence in situ hybridization (qFISH). For Southern analysis, gel plugs were made of MEFs from each genotype and digested directly with HinfI and RsaI, and telomeric fragments were then separated by pulse-field gel electrophoresis and probed with a 32P-labeled telomeric oligo (T2AG3)3. We found that there was no significant change in telomere length regardless of MEF genotype (DNA-PKcs+/+, DNA-PKcs+/−, and DNA-PKcs−/−; Fig. 1A). Using the qFISH methodology, we determined telomere length of metaphase chromosome spreads prepared from early passage primary MEFs of the three possible genotypes. The frequency of distribution of telomere fluorescence (Fig. 2) was calculated, as well as the mean telomere length for each genotype (Table 1). In agreement with the Southern blot results, we found no significant change in telomere length associated with the loss of DNA-PKcs in MEFs.
Figure 1.
DNA-PKcs-deficient MEFs exhibit similar telomere lengths and telomerase activities when compared with wild-type MEFs. (A) Southern analysis of MEFs. Early passage MEFs from independently isolated littermates were prepared from wild-type (lanes 7 and 8), DNA-PKcs+/− (lanes 1, 2, 5, and 6), and DNA-PKcs−/− (lanes 3 and 4). Gel plugs containing genomic DNAs were digested with RsaI and HinfI (odd number lanes) or undigested (even number lanes), fractionated by pulse-field gel electrophoresis, and hybridized with the telomeric specific [TTAGGG]3 probe. The approximate sizes of the products (kb) are indicated based on molecular weight markers. The Southern hybridization signal observed with the [TTAGGG]3 probe under these conditions was sensitive to BAL-31 exonuclease digestion, suggesting that this is telomeric DNA (data not shown). (B) PCR analysis for genotyping. Using the specific primer pairs (see Materials and Methods), wild-type and targeted alleles were amplified as products of 450 bp and 360 bp, respectively. Lanes 2 and 4 show the DNA-PKcs+/− pattern, lane 3 shows the DNA-PKcs−/− pattern, and lane 5 shows the wild-type pattern. Lane 1 contains a size marker. (C) Telomerase activity in DNA-PKcs-deficient MEFs. TRAP assay was performed after 30 PCR cycles on cell extracts (10, 102, and 103 cells) prepared from DNA-PKcs−/− (lanes 1–3), DNA-PKcs+/− (lanes 4–6), and wild-type (lanes 7–9) MEFs. In lanes 10–12, a serial dilution of HeLa cell lysate was run as a positive control for quantitating relative telomerase activity levels. Lane 13 contains a negative control without cell lysate. IC denotes a standard internal control for PCR efficiency.
Figure 2.
The frequency distribution of telomere fluorescence in DNA-PKcs−/− cells reveals that telomere length does not vary significantly from wild type and heterozygotes. Data were collected from qFISH studies of metaphase chromosome spreads of the specified genotypes from independently isolated littermates. The x axis depicts the intensity of each signal as expressed in telomere fluorescence units (TFU; 1 TFU = 1 kb of telomeric repeats), and the y axis shows the frequency of telomeres of a given length.
Table 1.
Telomere length is not altered in DNA-PKcs-deficient MEFs
| DNA-PKcs genotype | Metaphases analyzed | Telomere fluorescence
(TFU, mean ± SD)
|
||
|---|---|---|---|---|
| p-arm | q-arm | All telomere | ||
| +/+ | 17 | 49.87 ± 14.2 | 61.87 ± 16.9 | 55.87 ± 16.8 |
| +/− | 23 | 53.82 ± 21.6 | 70.58 ± 26.1 | 62.13 ± 25.3 |
| +/− | 22 | 54.51 ± 22.3 | 63.02 ± 22.7 | 58.73 ± 23.2 |
| −/− | 26 | 56.00 ± 24.2 | 64.02 ± 25.5 | 59.96 ± 25.2 |
Telomere fluorescence intensity expressed as telomere fluorescence units (TFU), where 1 TFU corresponds to 1 kb of [TTAGGG] n sequence.
To determine whether the lack of telomere shortening was unique to this cell type, we determined telomere lengths in primary kidney cell lines established from 6–8 month-old wild-type, DNA-PKcs+/−, and DNA-PK−/− animals. As with the DNA-PKcs deficient MEF lines, primary kidney cells of animals from the three genotypes (DNA-PKcs+/+, DNA-PKcs+/−, and DNA-PKcs−/−) displayed no significant differences in telomere length (Fig. 3A).
Figure 3.
Telomere length does not change in DNA-PKcs−/− kidney cells compared with wild-type cells. (A) Southern analysis using a telomeric-specific probe of kidney cell genomic DNA from independently isolated littermates of each genotype. Gel plugs containing genomic DNA from kidney cells of 6–8-month-old mice were digested with RsaI and HinfI (odd number lanes) or undigested (even number lanes), fractionated by pulse field gel electrophoresis, and hybridized with the telomeric-specific [TTAGGG]3 probe. (B) PCR analysis for genotyping primary kidney cells. The endogenous and targeted alleles were distinguished by using a PCR primer specific for each allele. Lane 1 contains the size marker. Lanes 2 and 3, DNA-PKcs+/+; lane 4, DNA-PKcs+/−; lanes 5 and 6, DNA-PKcs−/−. (C) Telomerase activity is not present in DNA-PKcs+/− and DNA-PKcs−/− kidney cells. Lane 1, no extract as a negative control; lanes 2–4, the activity contained in serial dilutions of HeLa cell lysate; lanes 5–7 DNA-PKcs+/− kidney cell lysate with additions of serial dilutions of HeLa cell lysates; lane 8, DNA-PKcs+/− kidney cell lysate; lanes 9–11, DNA-PKcs−/− kidney cell lysates with serial dilution of HeLa cell lysate; lane 12, DNA-PKcs−/− kidney cell lysate. Numbers indicate approximate cell equivalents used in each assay. IC denotes a standard internal control for PCR efficiency.
Previously, it was reported that telomerase is active in MEFs (22). To determine whether lack of DNA-PKcs affects telomerase activity, we performed TRAP analysis of deficient lines. We found that loss of DNA-PKcs does not change the levels of telomerase activity in MEFs (Fig. 1C). We observed a robust telomerase activity, near that of HeLa cell levels, regardless of genotype (Fig. 1C). In addition, it has been reported that primary kidney cells have no detectable telomerase activity (23). In agreement with this, we also failed to observe telomerase activity in primary kidney cells (Fig. 3C, lanes 8 and 12). Through a series of mixing experiments with added HeLa cell extracts positive for telomerase activity, we were able to determine that the lack of telomerase activity in primary kidney cell extracts is not due to an inhibition factor of telomerase or TRAP (Fig. 3C, lanes 5–7 and 9–11).
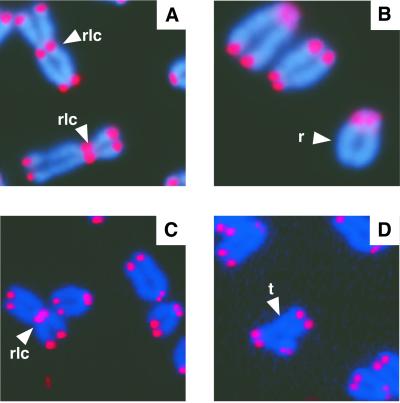
To determine whether DNA-PKcs functions to cap telomeres in mammals, we analyzed chromosome metaphase spreads from primary MEFs and primary kidney cell cultures from 6–8-month-old mice deficient for DNA-PKcs. High levels of telomere fusions were observed in metaphase chromosome spreads from DNA-PKcs−/− MEFs compared with wild-type MEFs (16.4–17% vs. 0%, respectively; Table 2, Fig. 4). A slight increase in telomere fusions was also observed in DNA-PKcs+/− MEFs with level between wild-type and DNA-PKcs null MEFs (Table 2, 1.5–2.9%). Primary kidney DNA-PKcs null metaphase cells showed even higher levels of telomere fusions when compared with null MEF cells (Table 3, 20.7–29.5%; Fig. 4). Similar to the DNA-PKcs+/− MEFs, heterozygous kidney cells accumulated telomere fusions to a level between wild-type and null cells (Table 3, 1.8–3.6%).
Table 2.
Telomere fusions accumulate in DNA-PKcs-deficient MEFs
| Genotypes* | Cell analyzed | Aneuploid (%) | Total fusions (% mean ± S.E.) | Fusions with TTAGGG repeats† (% mean ± S.E.) | Fragments (%) | Breaks (%)
|
|
|---|---|---|---|---|---|---|---|
| Chromosome | Chromatid | ||||||
| +/+ | 47 | 0 (0) | 0 | 0 | 1 (2.1) | 0 (0) | 0 (0) |
| +/+ | 55 (1.8) | 0 | 0 | 1 (1.8) | 0 (0) | 0 (0) | 0 (0) |
| +/− | 67 | 10 (14.9) | 2 (3.0 ± 2.1) | 1 (1.5 ± 1.5)‡ | 2 (3.0) | 0 (0) | 0 (0) |
| +/− | 68 | 12 (17.7) | 3 (4.5 ± 2.5) | 2 (2.9 ± 2.1)‡ | 1 (1.5) | 0 (0) | 0 (0) |
| +/− | 52 | 5 (9.6) | 1 (1.9 ± 1.9) | 1 (1.9 ± 1.9)‡ | 3 (5.8) | 0 (0) | 0 (0) |
| −/− | 61 | 20 (32.8) | 12 (19.7 ± 6.1) | 10 (16.4 ± 5.8)§ | 13 (21.3) | 8 (13.1) | 4 (6.6) |
| −/− | 53 | 16 (30.2) | 10 (18.9 ± 6.1) | 9 (17.0 ± 5.9)§ | 13 (24.5) | 7 (13.2) | 3 (5.7) |
Independently isolated littermates.
Number of fusions with telomeres at the fusion point. These values are included in the total fusions column.
Comparison between all lines of wild-type and +/− MEFs; P value = 0.34.
Comparison between all lines of wild-type and −/− MEFs; P value = 4.56 × 10−5.
Figure 4.
FISH analysis of metaphase chromosomes from DNA-PKcs null cells. Representative metaphase chromosome preparations from DNA-PKcs−/− MEFs (A and B) and DNA-PKcs−/− kidney cells (C and D) are shown. rlc, robertsonian fusion configurations; r, ring sister chromatid fusion; t, tri-radial fusion.
Table 3.
Telomere fusions accumulate in DNA-PKcs-deficient kidney cells
| Genotypes* | Cell analyzed | Aneuploid (%) | Total fusions (% mean ± S.E.) | Fusions with TTAGGG repeats† (% mean ± S.E.) | Fragments (%) | Breaks (%)
|
|
|---|---|---|---|---|---|---|---|
| Chromosome | Chromatid | ||||||
| +/+ | 50 | 1 (2.0) | 0 (0) | 0 (0) | 1 (2.0) | 0 (0) | 0 (0) |
| +/+ | 52 | 1 (1.9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| +/− | 56 | 9 (16.1) | 1 (1.8 ± 1.8) | 1 (1.8 ± 1.8)‡ | 5 (8.9) | 0 (0) | 1 (1.8) |
| +/− | 55 | 14 (25.5) | 2 (3.6 ± 2.5) | 2 (3.6 ± 2.5)‡ | 8 (14.5) | 1 (1.8) | 0 (0) |
| −/− | 58 | 21 (36.2) | 16 (27.6 ± 6.9) | 12 (20.7 ± 6.4)§ | 12 (20.7) | 3 (5.2) | 2 (3.5) |
| −/− | 61 | 30 (49.2) | 18 (29.5 ± 6.8) | 18 (29.5 ± 6.8)§ | 8 (13.1) | 3 (4.9) | 1 (1.6) |
Independently isolated littermates.
Number of fusions with telomeres at the fusion point. These values are included in the total fusions column.
Comparison between all lines of wild-type and +/− MEFs; P value = 0.28.
Comparison between all lines of wild-type and −/− MEFs; P value = 1.5 × 10−7.
qFISH analysis was performed in DNA-PKcs−/− cells to measure the telomere fluorescence units (TFUs) at fusion sites. The average TFU at telomeres of unfused chromosomes in DNA-PKcs−/− cells was 59.96 ± 25.2 (range of 34.76–85.16 kb; Table 1). At fusion sites involving two chromatids the average TFU was 83.6 ± 13.4 (range of 70.2–97 kb) and at two adjacent fusion sites involving four chromatids the average TFU was 157.7 ± 66.8 (range of 90.9–224.5 kb). Therefore, although qFISH provided a broad range of telomere length measurements, roughly twice the telomere fluorescence was observed at each fusion site, consistent with the majority of fusions being caused by loss of capping function, not from loss of telomeric DNA resulting in telomere to telomere end fusions.
As we previously reported with Ku-deficient MEFs (5), both primary MEF and kidney cultured cells deficient in DNA-PKcs exhibit a positive correlation between chromosome aberrations (increased chromosome fragments and breaks) and increased telomere fusions. In addition, we found that MEFs deficient in DNA-PKcs maintain normal G-stand overhangs (data not shown). The large accumulation of telomere fusions in DNA-PKcs-deficient primary MEFs and kidney cells indicates that DNA-PKcs likely plays an important telomere capping function, preventing end to end fusions.
We found that in the absence of DNA-PKcs, telomere length is maintained, yet capping is defective. This reveals that telomere length maintenance is a function separate from telomere capping function. In addition, these results demonstrate that analysis of only telomere length will not always establish whether a protein plays a role at the telomere. Thus, it is critical that capping function be examined in addition to telomere length before a particular genes role in telomere maintenance can be established. Previously, it was reported that telomere length in SCID mice is 1.5–2.0 times longer than telomeric DNA from other mouse strains (8). Because our results demonstrate that absence of DNA-PKcs does not cause changes in telomere length, the longer telomeres found in SCID mice must be due to a defect other than DNA-PKcs deficiency.
DNA-PKcs is a large polypeptide with multiple functions (13). We propose that DNA-PKcs might function at the telomere in several ways. One intriguing possibility is that DNA-PKcs may modify telomere maintenance proteins through phosphorylation. Alternatively, but not mutually exclusive, DNA-PKcs may play a structural role at the telomere. Recent work has shown that DNA-PKcs is localized to the mammalian telomere by chromatin immunoprecipitation (CHIP) analysis, providing further evidence that DNA-PKcs plays an important role at the mammalian telomere (24). DNA-PKcs may localize to the telomere by means of an interaction with Ku protein, because Ku and DNA-PKcs are thought to interact within the DNA–PK complex during the repair of DNA double-strand breaks and Ku is known to localize to the mammalian telomere (5, 10, 24–26). However, we previously demonstrated that Ku is capable of localizing to the telomere even in the complete absence of DNA-PKcs (10). Therefore, a complex between Ku and DNA-PKcs (the DNA–PK complex) is not required for Ku localization to the telomere and the accumulation of telomere fusions in DNA-PKcs-deficient cells is not simply an indirect effect of perturbing the association of Ku with the telomere. Establishing the exact role DNA-PKcs plays at the telomere will be crucial to furthering our understanding of telomere biology.
Acknowledgments
We especially thank Hsin-Ling Hsu for her valuable intellectual support during this work and Judith Campisi and Janice Pluth for extremely helpful comments about this manuscript. We thank Saira Mian for her expert statistical analysis. We are grateful to Hiroyuki Niida and Yoichi Shinkai for providing the human G9a cDNA. M.P.H. acknowledges the support of Professors David Brenner and Eric Hall and Grant SD#0029 from Radiological Society of North America. H.T. is a fellow of the Japan Society for the Promotion of Science. This work was supported by the U.S. Department of Energy under Contract DE-AC03–76SF00098 and National Institutes of Health Grants AG17709 and CA50519 (to D.J.C.).
Abbreviations
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- SCID
severe combined immunodeficiency
- MEF
mouse embryonic fibroblast
- qFISH
quantitative fluorescence in situ hybridization
- TRAP
telomeric repeat amplification protocol
References
- 1.Blackburn E H. Nature (London) 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer M R, Schnapp G, de Lange T. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S H, Kaminker P, Campisi J. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith S, de Lange T. Curr Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 5.Hsu H L, Gilley D, Galande S A, Hande M P, Allen B, Kim S H, Li G C, Campisi J, Kohwi-Shigematsu T, Chen D J. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greider C W. Cell. 1999;97:419–422. doi: 10.1016/s0092-8674(00)80750-3. [DOI] [PubMed] [Google Scholar]
- 7.d'Adda di Fagagna F, Hande M P, Tong W M, Lansdorp P M, Wang Z Q, Jackson S P. Nat Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- 8.Hande P, Slijepcevic P, Silver A, Bouffler S, van Buul P, Bryant P, Lansdorp P. Genomics. 1999;56:221–223. doi: 10.1006/geno.1998.5668. [DOI] [PubMed] [Google Scholar]
- 9.Bailey S M, Meyne J, Chen D J, Kurimasa A, Li G C, Lehnert B E, Goodwin E H. Proc Natl Acad Sci USA. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu H L, Gilley D, Blackburn E H, Chen D J. Proc Natl Acad Sci USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lansdorp P M. Mech Ageing Dev. 2000;118:23–34. doi: 10.1016/s0047-6374(00)00151-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X D, Kuster B, Mann M, Petrini J H, de Lange T. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 13.Smith G C, Jackson S P. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 14.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurimasa A, Ouyang H, Dong L J, Wang S, Li X, Cordon-Cardo C, Chen D J, Li G C. Proc Natl Acad Sci USA. 1999;96:1403–1408. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurimasa A, Kumano S, Boubnov N V, Story M D, Tung C S, Peterson S R, Chen D J. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu W, Gong X, Li Z, Takabayashi K, Ouyang H, Chen Y, Lois A, Chen D J, Li G C, Karin M, Raz E. Cell. 2000;103:909–918. doi: 10.1016/s0092-8674(00)00194-x. [DOI] [PubMed] [Google Scholar]
- 18.Danska J S, Holland D P, Mariathasan S, Williams K M, Guidos C J. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beamish H J, Jessberger R, Riballo E, Priestley A, Blunt T, Kysela B, Jeggo P A. Nucleic Acids Res. 2000;28:1506–1513. doi: 10.1093/nar/28.7.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taccioli G E, Amatucci A G, Beamish H J, Gell D, Xiang X H, Torres Arzayus M I, Priestley A, Jackson S P, Marshak Rothstein A, Jeggo P A, Herrera V L. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 21.Aderem A, Hume D A. Cell. 2000;103:993–996. doi: 10.1016/s0092-8674(00)00201-4. [DOI] [PubMed] [Google Scholar]
- 22.Blasco M A, Lee H W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 23.Coviello-McLaughlin G M, Prowse K R. Nucleic Acids Res. 1997;25:3051–3058. doi: 10.1093/nar/25.15.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.d'Adda di Fagagna F, Hande M P, Tong W, Roth D, Lansdorp P M, Wang Z, Jackson S P. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 25.Samper E, Goytisolo F A, Slijepcevic P, van Buul P P, Blasco M A. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song K, Jung D, Jung Y, Lee S G, Lee I. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 27.Birren B, Green E D, Klapholz S, Myers R M, Roskams J. Genome Analysis: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 28.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 29.Zijlmans J M, Martens U M, Poon S S, Raap A K, Tanke H J, Ward R K, Lansdorp P M. Proc Nat Acad Sci USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hande M P, Samper E, Lansdorp P, Blasco M A. J Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]