Abstract
Androgen receptor (AR), a hormonal transcription factor, plays important roles during prostate cancer progression and is a key target for therapeutic interventions. While androgen-deprivation therapies are initially successful in regressing prostate tumors, the disease ultimately comes back as castration-resistant prostate cancer (CRPC) or at the late stage as neuroendocrine prostate cancer (NEPC). CRPC remains largely dependent on hyperactive AR signaling in the milieu of low androgen, while NEPC is negative of AR expression but positive of many AR-repressed genes. Recent technological advances in genome-wide analysis of transcription factor binding sites have revealed an unprecedented set of AR target genes. In addition to its well-known function in activating gene expression, AR is increasingly known to also act as a transcriptional repressor. Here, we review the molecular mechanisms by which AR represses gene expression. We also summarize AR-repressed genes that are aberrantly upregulated in CRPC and NEPC and represent promising targets for therapeutic intervention.
Keywords: androgen receptor, CRPC, EZH2, histone modifications, NEPC, targeted therapy, transcriptional repressor
INTRODUCTION
Androgen receptor (AR) is a member of the superfamily of hormonal nuclear receptors.1 Unliganded AR is secured in the cytoplasm by heat-shock proteins.2 Once exposed to its cognate ligand, the male hormone androgen, AR, becomes activated, separates from heat-shock proteins, and translocates to the nucleus, where it binds to the chromatin at androgen response elements (ARE) to initiate its transcriptional program. This hormone-stimulated AR signaling is important for proper tissue differentiation and homeostasis during prostate development and function. AR signaling, however, is hijacked in prostate tumors, turning into a powerhouse for continuous cancer progression. Thus, the mainstay treatment for metastatic prostate cancer (PCa) is androgen deprivation therapy (ADT).1 However, despite initial regression of the androgen-dependent tumors (ADPC) on ADT, the disease inevitably comes back in a more aggressive form termed castration-resistant prostate cancer (CRPC).3 Importantly, although CRPC is resistant to androgen depletion, it continues to depend on AR signaling. Thus, higher affinity, second-generation AR pathway inhibitors, such as enzalutamide and abiraterone, are useful to further delay CRPC progression.4,5 Nevertheless, CRPC sustains, ultimately turning into an incurable disease. The search for the mechanisms underlying CRPC progression and treatment resistance will benefit patients who have already exhausted all currently available treatment measures. At the center of such efforts is a comprehensive understanding of AR function.
AR is best known as a transcription activator, with prostate-specific antigen (PSA) (KLK3) being a prototype AR-induced gene. AR engages multiple coactivators and chromatin modifiers, which are assembled into pro-transcriptional complexes. These complexes facilitate RNA polymerase II recruitment to the transcription start site (TSS) of AR target genes, which are defined by direct AR binding to AREs at their regulator elements.6 AR recruitment to such chromatin is facilitated by pioneering factor forkhead-box A1 (FOXA1), which unwinds chromatin and makes it accessible to AR.7,8 Other transcription factors, such as GATA-binding protein 2 (GATA2) and homeobox B13 (HOXB13), have also been reported to contribute to chromatin accessibility by AR.9,10,11 AR binding to its regulatory elements is followed by the recruitment of coactivators CBP (CREB binding protein) and SRC-1 (steroid receptor coactivator 1), among others, that initiate transcription of target genes.6,12 Many of the AR-induced genes are involved in cellular processes such as hormonal responses, cell cycle, and lipid metabolism.7,13,14
Evidence has recently emerged, suggesting that AR can also function as a transcriptional repressor. As AR primarily binds to distal enhancers that are often more than 10 kb away from the target promoter, the identification of direct AR target genes has been impeded.15,16 With the advent of ChIP-based genome-wide location analysis assays, recent studies have discovered a plethora of AR target genes, many of which are repressed by androgen, suggesting AR as a transcriptional repressor.17,18 Here, we review the molecular mechanisms, by which AR directly inhibits transcription, highlight AR-repressed genes that have been identified thus far, and evaluate their roles during PCa progression and castration resistance.
AR AS A TRANSCRIPTIONAL REPRESSOR
We define AR as a transcriptional repressor only when it binds to DNA at enhancers and/or promoters to directly inhibit genes expression. Genes that are repressed by AR through indirect mechanisms are not considered in this review. We will start out by discussing literatures reporting molecular mechanisms underlying the role of AR as a transcriptional repressor, which are schematically presented in Figure 1.
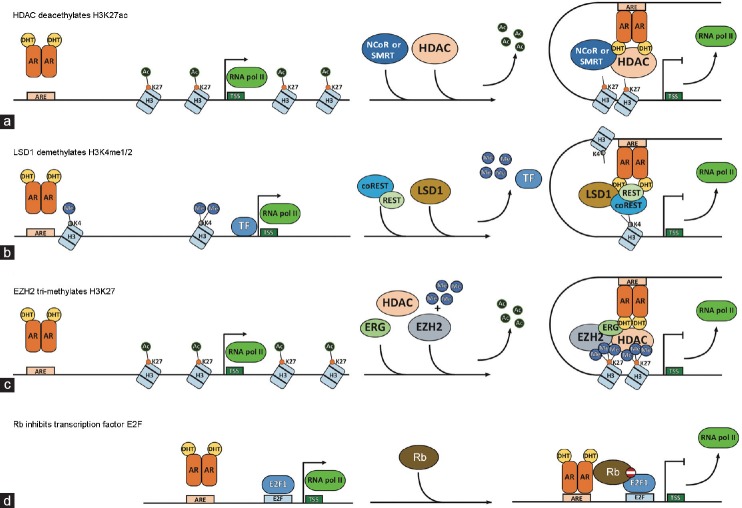
Figure 1.
Mechanism of AR-regulated gene repression. Ligand (DHT)-bound AR dimerizes and translocates to the nucleus, where it binds to androgen response elements (ARE) at enhancers of target genes. (a) DHT-AR binding to ARE at target enhancers recruits repressive complex that consists of NCoR/SMRT and HDAC. DNA looping brings AR-repressive complex to proximity with target gene promoter, where HDAC deacetylates H3K27ac, causing chromatin inaccessibility and transcription repression. (b) Gene enhancers are enriched for H3K4me1/2 and gene promoters are enriched for H3K4me3. DHT-AR binding to ARE at target enhancers recruits REST/coREST and LSD1 complex, which demethylates H3K4me1/2 leading to enhancer deactivation. DNA looping further exposes target promoters to LSD1, which catabolizes H3K4me3 demethylation and promoter inactivation. (c) DHT-AR accumulation at target regulatory elements recruits ERG, EZH2, and HDAC repressive complex. HDAC removes acyl groups from H3K27ac, priming it for subsequent trimethylation by EZH2, resulting in epigenetic silencing. (d) AR mediates gene repression through direct binding to Rb. Thus, AR stabilizes Rb interaction with nearby E2F1 transcription activator, inhibiting its transcriptional activity. AR may also itself interact with E2F1 to remove it from chromatin, enabling the recruitment of E2F4 transcriptional repressor. AR: androgen receptor; NCoR: nuclear receptor corepressor; SMRT: silencing mediator of retinoid and thyroid receptors; REST: repressive element 1 silencing transaction. HDAC: histone deacetylase; LSD1: lysine-specific demethylase 1; ERG: ETS-related gene; EZH2: enhancer of zeste homolog 2.
HDAC and histone deacetylation
Most nuclear receptors, including AR, can both repress or activate target genes under the instrumentation of essential regulatory cofactors. Gene repression depends on common nuclear receptor corepressors, such as nuclear receptor corepressor (NCoR) and its homolog silencing mediator of retinoid and thyroid receptors (SMRT).19 NCoR and SMRT are structurally similar; both interact with nuclear receptors through C-terminal domains, while their N-terminal domains recruit histone deacetylases through either direct binding to histone deacetylase 3 (HDAC3), HDAC4, and HDAC5 or indirect interaction with HDAC1 and HDAC2 through Sin3 adaptor protein.19 Unoccupied HDACs are enzymatically inactive and require stimulation by protein–protein interaction with cofactors. Both SMRT and NCoR carry a conserved N-terminal sequence known as deacetylates activating domain (DAD), which interacts with HDACs, stabilizing their enzymatically active form.20 Activated HDACs then remove neutral acyls from amino groups on lysine residues of histones 3 and 4, causing positive charge on them. This increases histone's adherence to negatively charged DNA and, thus, facilitates a tight chromatin compaction, which limits DNA accessibility for gene transcription.21
NCoR has been shown to not only compete with nuclear transcriptional coactivators cAMP-response element (CRE)-binding protein (CREB)-binding protein (CBP) and steroid receptor co-activator-1 (SRC-1) for AR binding but also recruit and activate HDACs at AR target genes, which facilitate local chromatin compaction.22 In another study, SMRT was shown to mediate transcriptional repression of AR target genes through recruitment of HDACs.23 Moreover, AR itself has been reported to directly bind to cytoplasmic HDAC7 and transport it into the nuclei, wherein HDAC7 facilitates AR-driven target gene repression.24
LSD1 and lysine demethylation
Local histones undergo methylation at various lysine residues, which affects nucleosome accessibility to transcription factors. Three forms of lysine methylation exist: mono-, di-, and trimethylation. Methylated forms of histone 3 lysine 4 (H3K4me1, me2, me3) correlate with transcriptional activation.25,26 Specifically, H3K4me3 is tightly associated with active gene promoters and H3K4me1 with gene enhancers, while H3K4me2 is described to be present at both promoters and enhancers.25,26,27,28 Demethylation of activated chromatin represents a potent mechanism for genes repression. Histone-demethylating enzyme, lysine-specific demethylase 1 (LSD1), has been identified as a member of transcription-repressive complex that blocks expression of neuron-specific genes.28,29 During gene repression, LSD1 is recruited by repressive element 1 silencing transaction (REST)/coREST repressive complex to the target DNA site.30 When lysine's side chain amino group becomes mono- or dimethylated, LSD1 reverses lysine methylation through reaction of amine oxidation. The LSD1-catalyzed demethylation is limited to mono- and dimethylated H3K4 substrates because of the biochemical specificity of this reaction.28 H3K4me3, on the other hand, is metabolized by a group of histone demethylases that contain JmjC domain, which exploits Fe(II)-containing protein cofactor to remove a single methylation group from H3K4me3.31 The resulting H3K4me2 can further be demethylated by LSD1.
AR has been shown to repress certain target genes, including itself, through engaging LSD1-mediated histone demethylation.17 Ligand-bound AR coenriches with LSD1 at the ARE-containing enhancer element of the AR gene. This enhancer brings a corepressive complex to the gene promoter through DNA looping, where LSD1 metabolizes demethylation of H3K4me2. The decrease of overall level of histone methylation at the target gene impedes the recruitment of RNA pol II and hence decreases transcription.17 A more recent work from the same group further demonstrated that coREST, an important LSD1-recruiting protein, cooccupies at the ARE-containing enhancers with LSD1.32 This study suggests that the ligand-activated AR recruits REST/coREST/LSD1-dependent repressive complex to the AR-regulated enhancers during target genes repression.32,33
Enhancer of zeste homolog 2 (EZH2) and histone methylation
Polycomb repression complex 2 (PRC2), consisted of core subunits suppressor of zeste 12 (SUZ12), embryonic ectoderm development (EED), and EZH2, plays important roles in maintaining embryonic stem cell identity through epigenetic silencing of a large cohort of developmental regulators.34 EZH2 is the enzymatic subunit of PRC2 that catalyzes histone H3 lysine 27 trimethylation (H3K27me3), a reported mark for repressive histone environment.35,36 We have recently reported that EZH2 also plays a polycomb- and methylation-independent role in gene activation, one of its targets being AR, potentiating its function in PCa.37 EZH2 is among the most upregulated genes in aggressive PCa and its expression is inversely correlated with PCa clinical outcomes.38 EZH2 behaves as a bona fide oncogene as its overexpression promotes prostate tumorigenesis while its depletion leads to cellular senescence and halts tumor progression.39
EZH2 has recently been shown to coordinate with AR on transcriptional repression. Through ChIP-seq analysis of the key transcription factors and histone marks, our group identified an integrative transcriptional network involving AR, EZH2, and ETS-related gene-1 (ERG1) in PCa cells.16 ERG belongs to the ETS family of transcription factors and is frequently fused with the 5’ untranslated region of the TMPRSS2 gene in PCa.40 We showed that ERG cooccupies with AR on many ARE-containing genomic loci to inhibit gene expression, which may be mediated, at least partially, by direct induction of EZH2 expression and recruitment to the target sites for epigenetic silencing. Further, integrating genome-wide AR localization data with androgen-induced gene expression data, we have identified a large number of genes whose expression is directly inhibited by AR but can be rescued through EZH2 inhibition.18,41,42 Collectively, our data establish that EZH2 facilitates AR-transcriptional repression through catalyzing H3K27me3.
An additional report from an independent research group confirms the importance of AR-centered coregulatory network, which wires in ERG and EZH2, in the gene repressive program.43 Chng and colleagues further identified the collaboration between HDAC2/3 and EZH2, as both enzymes were shown to be recruited to the AR target genes by AR-ERG corepressive complex. HDACs probably cooperate with EZH2 through the removal of acyl groups H3K27ac, which primes H3K27 for subsequent methylation by EZH2.
Retinoblastoma protein (Rb) and E2F family transcription factors
A recent study demonstrated that AR contributes to direct repression of the genes involved in DNA replication in PCa through cooperation with Rb.44 Gao and colleagues revealed that acute stimulation of AR rapidly suppresses expression of the proliferative genes, such as Minichromosome complex maintenance component 7 (MCM7), FANCI, and lamin B1 (LMNB1) in VCaP cells. Interestingly, most of these AR-repressed regulators of DNA replication contain within their promoter ARE and E2F motifs. E2F transcription factors are controlled by Rb, a prominent oncosuppressor, which inhibits E2F transcriptional activity through direct protein–protein interaction.45,46 AR was shown to physically interact with Rb and recruit it to the target genomic loci that locate in proximity to E2F motif. Hence, AR stabilizes the inhibiting interaction between Rb and E2F, abrogating the expression of target genes.
The family of E2F transcription factors includes activator E2F1 and repressor E2F4, which cooperate to facilitate proper transition through cell cycle.46,47 AR has been reported to regulate switching between E2F transcription factors.48 They demonstrated that AR directly represses expression of cyclin B1 in PCa stromal cells through switching E2F family transcription factors.48 In cyclin B1 promoter, ARE is localized nearby to E2F response element. When AR occupies the ARE site after ligand stimulation, it directly binds to E2F1 and removes it from the E2F response element, which becomes quickly occupied by transcriptional repressor E2F4. This AR-driven enrichment of E2F4 at gene promoter stalks transcription of cyclin B1, causing inhibited proliferation of PCa stromal cells.48
AR-REPRESSED GENES AND THEIR ROLES IN PROSTATE CANCER
Since the discovery that PCa depends on androgen signaling1 and generalization of ADT as a standard of care for PCa patients,49 research community continuously attempts to understand the transcriptional program governed by AR in PCa. Thus far, the major effort has been primarily focused on AR-activated genes and their roles in PCa development.49,50,51 In this section, we will review the current reports on key AR-repressed genes and their roles in PCa (Table 1). We define genes as direct targets of AR only if their regulatory DNA elements contain AREs that are bound by AR.
Table 1.
Androgen receptor-repressed genes and their roles in prostate cancer
| Gene name | Reference | AR repressive mechanism | Function |
|---|---|---|---|
| AR-repressed oncosuppressors | |||
| DEPTOR | Kanno et al. 201553 | AR binds to ARE located at the gene’s 4th intron. AR recruits histone-deacetylating repressive complex | DEPTOR, an endogenous mTOR inhibitor, binds to mTORC1 and mTORC2 protein kinase complexes and inhibits their enzymatic activities |
| CDHE | Liu et al. 200856 | AR binds to ARE at CDHE promoter and recruits HDAC1 based repressive complex | E-cadherin (CDHE) is an epithelial cell surface molecule, which stabilizes cell–cell adhesion. CDHE inhibits EMT and prevents metastatic dissemination |
| VCL | Chng et al. 201243 | AR cooperates with ERG to recruit repressive HDACs to VCL enhancer region | Vinculin (VCL), a cytoskeleton protein important for stabilization of the cell-to-cell and cell-to-matrix contacts. Loss of VCL promotes metastases |
| CC3/NOV | Fong et al. 2017;42 Wu et al. 201441 | AR binds to CC3/NOV promoter and recruits EZH2 that metabolizes repressive trimethylation of H3K27 | NOV inhibits prostate cancer growth. NOV binds to the N-terminus of AR and sequesters it from nuclear translocation |
| AR-repressed oncogenes | |||
| CCND1 | Holter et al. 2002;62 Lanzino et al. 201061 | AR binds to ARE at CCND1 cis regulatory element. AR then recruits repressive DAX1, NCoR, HDAC. | CCND1 regulates cells cycle. CCND1 forms a complex with CDK kinases that phosphorylate Rb, causing its degradation |
| CCNB1 | Li et al. 201248 | ARE is localized at the gene promoter in proximity to E2F response element. Recruited AR binds and removes E2F1 transcription activator from CCNB1 promoter. The emptied E2F-response element is then occupied by repressive E2F4 | CCNB1 plays an important role for a proper progress through G2/M cell cycle check point |
| hTERT | Moehren et al. 200871 | AR represses hTERT through direct binding to the gene promoter. However, the exact repressive complex remains to be identified | TERT prevents cell senescence and facilitates uncontrolled proliferation |
| MET | Verras et al. 200775 | AR binds to MET enhancer and competes with transcription activator Sp1 | HGF/SF receptor (c-MET) is a receptor tyrosine kinase. c-MET facilitates cell cycle progress, tumor cells migration, tumor angiogenesis and survival |
| MUC1 | Rajabi et al. 2011112 | AR directly inhibits MUC1 expression through binding to ARE in the gene promoter. The repressive complex has not been tested yet | MUC1 is overexpressed in aggressive PCa. MUC1 is a transmembrane receptor, which transduces molecular signal through NF-kB, beta-catenin, and STAT1/3 |
| CHRM1 | Mannan Baig et al. 2017;113 Prescott et al. 2007114 | AR binds to CHRM1 promoter and directly represses gene transcription even in the absence of androgen. The mechanism remains to be described | Stimulation of acetyl choline muscarinic receptor (CHRM1) increases prostate cells survival and proliferation. Targeting of CHRM1 decreases PCa growth. CHRM1 signaling overlaps with PIP3-AKT-CaM pathway |
| BRN2 (POU3F2) | Bishop et al. 201784 | AR binds to ARE in the gene enhancer. AR is suggested to interfere with SOX2, which activates BRN2 transcription | BRN2 is a neural transcription factor. It drives NEPC transcriptional program in cooperation with SOX2. BRN2 cooperates with SOX2 to activate neuronal genes transcription |
| SOX2 | Kregel et al. 2013;83 Mu et al. 201781 | AR was shown to bind to SOX2 enhancer and inhibit its transcription in AR-overexpressing PC3 cells. The AR-recruited complex then enriches local histone for repressive H3K27me3 | SOX2 activates NE differentiation in enzalutamide-resistant PCa cells |
| PEG10 | Akamatsu et al. 201585 | AR is recruited to PEG10 promoter, where it stabilizes repressive Rb binding to E2F transcription activator | PEG10 is normally expressed in placenta. NEPC shows overexpression of PEG10, which drives cancer cells proliferation. PEG10 silencing upregulates cell cycle checkpoint mediators, CDKN1A (p21) and CBKN1B (p27) |
| AR | Cai et al. 201117 | AR binds to its own enhancer at intron 2 and recruits LSD1 | AR self-repression represents a mechanism of feedback loop. Thus, AR targeting causes increased AR expression |
| NR3C1(GR) | Arora et al. 2013;116 Xie et al. 2015115 | AR binds to GR enhancer region and directly represses its transcription. The exact mechanism remains to be identified | GR mediates bypass for AR pathway targeting in advanced CRPC, as GR shares common target genes with AR |
| NCOA2 (TIF2) | Agoulnik et al. 2006117 | AR enriches at gene promoter and distal enhancer at intron 8. Direct AR binding to these elements promotes gene repression. The repressive complex is yet unknown | TIF2 is a member of p160 family of coactivators, related to SRC-1. TIF2 is overexpressed in aggressive CRPC, where it works as AR-coactivator |
| PADI2 | Wang et al. 201789 | ARE is located at PADI2 promoter. AR direct binding causes gene repression, evident through decreased RNA pol II occupancy | PADI2, is a cytoplasmic enzyme that citrullinates AR protecting it from degradation. Also, PADI2 citrullinates H3R26 at AR binding sites, stabilizing AR interaction with chomatin |
AR: androgen receptor; DEPTOR: DEP containing mTOR interacting protein; CCND1: cyclin D1; CCNB1: cyclin B1; TERT: telomerase reverse transcriptase; HGF/SF: hepatocyte growth factor/scatter factor; MUC1: mucin 1; GR: glucocorticoid receptor; PADI2: peptidylarginine deiminase 2; SRC-1: steroid receptor coactivator-1; TIF2: transcription intermediary complex 2; PEG10: paternally expressed 10 imprinted gene; PCa: prostate cancer; ARE: androgen response elements; CRPC: castration-resistant prostate cancer; SOX2: sex-determining region Y-box2; BRN2: Bruno-like 2; LSD1: lysine-specific demethylase 1
AR-repressed oncosuppressors
AR signaling regulates normal prostate tissue development and homeostasis, but becomes hijacked during PCa oncogenesis. Thus, androgen deprivation causes cell cycle arrest in PCa cell culture.52 The communication between AR and mammalian target of rapamycin (mTOR), a molecular master regulator that governs DNA replication during cell cycle, has been addressed in a recent report.53 AR maintains mTOR signaling through direct repression of oncosuppressor DEP domain-containing mTOR interacting protein (DEPTOR), an endogenous mTOR inhibitor. DEPTOR neutralizes mTORC1 and mTORC2 enzymatic activities through direct interaction. In androgen-dependent LNCAP and VCAP cells, ligand-activated AR enriches to the enhancer ARE region located in DEPTOR intron 4 and recruits repressive complex including HDACs. Meanwhile, in AR-negative CRPC cell line, such as PC-3, treatment with androgens fails to reduce DEPTOR expression.53 Nevertheless, DEPTOR remains repressed in CRPC cells, possibly through an alternative mechanism. Along this line, a recent report demonstrated that, in androgen-independent CRPC tumors, DEPTOR becomes increasingly ubiquitinated and thus degraded because of the upregulated SAG E3 ubiquitin ligase.54
Epithelial–mesenchymal transition (EMT) stands behind metastatic dissemination, a hallmark of advanced PCa. During EMT, cancer cells lose their epithelial markers, primarily E-cadherin and keratins and upregulate mesenchymal N-cadherin and vimentin.55 AR may contribute to EMT in PCa via negatively regulating expression of the genes encoding epithelial markers. Liu and colleagues have shown that AR directly represses E-cadherin. Agonist-stimulated AR binds to ARE located in E-cadherin regulatory sequence and recruits corepressor HDAC1, which initiates repressive histone deacetylation.56 In addition, AR controls the expression of vinculin, a cytoskeleton protein that cooperates with E-cadherin in order to stabilize cell–cell adhesion, thus preventing EMT. AR binds to ARE in the intronic enhancer together with cofactor ERG. AR complexed with ERG brings HDAC and EZH2 holoenzymes, which coordinate exchange of H3K27ac for H3K27me3 and repress vinculin expression.43 Nevertheless, tumor specimens collected from ADT-treated patients show increased expression of mesenchymal markers.57 Moreover, in androgen-independent cells that lack endogenous AR, overexpression of ectopic AR inhibits EMT-driven cell invasiveness.58 This phenomenon can be explained by the cellular context in these cells that overrides AR-downstream effects. For example, androgen-independent PCa cells upregulate Zinc finger E-box-binding protein 1 (ZEB1), a master EMT inducer, which contributes to ADT resistance.59 ZEB2, another master transcription factor for EMT markers, is also upregulated in these cells and can be indirectly repressed by ectopic AR.60
Normally, AR is integrated in a tightly organized feedback loop, which stops constitutive AR signaling. However, dysregulation of the feedback response in PCa can cause prolonged overactivation of AR, which in turn enforces tumor growth. Our group has previously identified that AR directly represses the transcription of nephroblastoma overexpressed (NOV), a tumor suppressor arresting cytoplasmic AR from nuclear translocation and inhibiting PCa cells proliferation, migration, and growth.42 We found that AR directly binds to NOV enhancer, recruiting there a corepressive EZH2 complex.41 Ligand-activated AR in androgen-dependent PCa cells promotes rapid tumor growth partially through the suppression of NOV and thus the negative feedback loop it mediates. Surprisingly, NOV remains repressed in CRPC and enzalutamide-resistant PCa, probably due to additional mechanisms such as EZH2 upregulation. Although AR targeting might de-repress distinct oncosuppressors in androgen-dependent PCa, which are manifested by initial regress of tumor growth, the disease eventually reemerges in the form of aggressive, AR-independent CRPC, armed with a range of molecular mechanisms to keep oncosuppressors inhibited.
AR-repressed oncogenes
AR has also been shown to directly repress many oncogenes, which regulate DNA synthesis and cell cycle progression.17 Accordingly, AR inhibits CCND1, a cyclin D1-encoding gene, through direct binding to its proximal cis enhancer element,61 where it recruits a corepressor complex, consisted of DAX1, NCoR, and HDACs.62,63 Oncogenic cyclin D1 activates cyclin-dependent kinases CDK4 and CDK6, which phosphorylate Rb, causing its degradation. Eradication of phosphorylated Rb unleashes E2F transcriptional activity, necessary for cell cycle progression and proliferation.64 In PCa, increased cyclin D1 expression has been shown to promote resistance to chemotherapy.65 A recent report has also confirmed an important role of cyclin D1 in enzalutamide resistance in CRPC patients.66
Cyclin B1 is another example of AR-repressed oncogene that drives cell cycle progression. The promoter of the cyclin B1 encoding gene, CCNB1, carries both ARE and E2F response elements in proximity one to another. AR recruitment facilitates the switching between E2F activator (E2F1) and E2F repressor (E2F4) and, therefore, inhibits transcription of cyclin B1.48 Cyclin B1 promotes cell cycle progression through activation of CDK1, which mediates proper progress through G2/M checkpoint.67 Extensive molecular analyses of clinical specimens and of CRPC murine models suggest that increased expression of cyclin-dependent genes positively correlates with incidence of therapy-resistant CRPC tumors.68,69,70
Continuous cell proliferation requires a sustained telomerase (TERT) activity, which is generally induced during oncogenesis. In androgen-dependent PCa cells, AR represses human telomerase reverse transcriptase (hTERT) through direct binding to its gene promoter.71 The nature of the repressive complex recruited by AR to hTERT remains to be identified. However, it has been reported that mutant AR T877A loses control over hTERT expression, suggesting that in CRPC, which accumulates AR mutants, tumor-repressive function of AR becomes compromised. Moreover, a recent study has demonstrated that AR is downregulated in hTERThigh PCa cells, which exhibit cancer stem cell-like properties, in comparison to their hTERTlow counterparts.72 An increased expression of hTERT in aggressive PCa tumors leads to evasion from cell cycle checkpoint control, prompting hTERT targeting for therapeutic evaluation in ADT-resistant prostate cancer.73
Uncontrolled proliferation in cancer cells commonly relies on constitutively active cellular stimulations by ligands such as cytokines and growth factor. Hepatocyte growth factor/scatter factor (HGF/SF) receptor with tyrosine kinase activity, also known as c-MET, is a well-known oncogene.74 AR represses MET through direct binding to the ARE element located near the gene promoter. Recruited AR outcompetes transcriptional activator Sp1, whose cognate binding site locates in proximity to ARE. AR-dependent inhibition of Sp1 binding blocks c-MET expression in androgen-responsive PCa cells.75 In fact, androgen ablation correlates with increased expression of c-MET during CRPC progression. Small-molecule inhibitors specific to c-MET have shown a promising antiproliferative effect for CRPC in preclinical studies.76 Moreover, anti-c-MET therapeutic agents are currently undergoing clinical trials and have demonstrated preliminary anti-tumor effects in patients with metastatic CRPC.77
As neuroendocrine prostate cancer (NEPC) features an entire loss of AR expression,78,79 and androgen deprivation is known to enable NE differentiation in androgen-sensitive PCa cells,80 a question arises as to whether AR is capable to repress the expression of NE-driving oncogenes. A recent report has demonstrated that evolution of ADT resistance in PCa coincides with cancer cell transformation from epithelial to neuroendocrine phenotype, driven by lineage plasticity regulated by reprogramming transcription factor sex-determining region Y-box2 (SOX2).81 Wang et al.82 studying Pten-null murine PCa model have shown that Sox2-positive luminal PCa cells markedly expand during tumor growth and further increase on castration. AR has been shown to repress SOX2 expression through direct binding to SOX2 cis regulatory element.83 The exact repressive complex remains unclear. However, AR binding is found to correlate with enrichment for repressive H3K27me3 histone marks at the SOX2 promoter, suggesting the involvement of epigenetic regulation.
In addition to SOX2, AR has recently been shown to directly inhibit the expression of Bruno-like 2 (BRN2, encoded by POU3F2)84 and paternally expressed 10 (PEG10),85 which are key regulators of NEPC progression. BRN2 is a target gene for SOX2 transcription factor, and one of the proposed mechanisms, by which AR represses BRN2, is through interference with SOX2 for DNA binding at POU3F2 enhancer. BRN2 has been described as a central and clinically relevant driver of NEPC, as BRN2 is upregulated during NE differentiation and overexpression of BRN2 alone is sufficient to drive resistance to enzalutamide in PCa. On the other hand, PEG10 is a retrotransposon-derived gene that is required for cell proliferation and survival and normally expressed during placental development. PEG10, similarly to other cell cycle regulators, is transcribed by E2F1 transcription factor. AR directly represses PEG10 through recruitment and stabilization of inhibitory Rb binding to E2F1.44 NE transformation, driven by AR loss, correlates with upregulation of PEG10.85 Accordingly, clinical data confirmed an association between the expressions of PEG10 and NEPC markers. In AR-independent tumors, PEG10 promotes tumor progression at several levels. First, it facilitates a continuous stimulation for expression of proliferative genes, such as cell cycle regulators. Second, PEG10 cooperates with the transforming growth factor-beta (TGF-β) pathway in order to promote NEPC cells invasiveness.85 Normally, PEG10 is tightly controlled by AR, Rb, and TP53; however, the loss of all three molecules in NEPC upregulates PEG10, leading to an accelerated progression of the lethal disease.
AR-mediated gene repression and its therapeutic implications
AR-mediated gene repression plays important roles in PCa treatment, response, resistance, and progression. During PCa progression to CRPC under ADT, a number of mechanisms have been discovered that bypass androgen dependence, the most frequent one of which is AR hyperactivation through either AR amplification and mutations,86 or AR cofactors, such as TRIM24, TRIM28, HOTAIR, PADI2, and others.87,88,89 In these tumors, AR continues to induce essential oncogenes and repress tumor suppressor genes, contributing to disease progression in the milieu of low androgen. These CRPC tumors have also been shown to be able to reprogram intracellular molecular signaling in a way to bypass AR dependency.66,78,90,91,92,93
With recent use of high-affinity AR antagonist enzalutamide and androgen synthesis inhibitor abiraterone in the clinic, increasing studies have reported the essential roles of AR-repressed genes in treatment resistance. These drugs appear to immediately contribute to CRPC progression through elimination of the repressive arm of AR-dependent regulation of oncogenes. For example, AR-repressed genes cyclins B1 and D1 are both important for cancer cell expedition through cell cycle.48,61 Overexpression of cell cycle regulators CDK4/6, which are activated by cyclin D1, is sufficient to promote enzalutamide resistance, and cotargeting of CDK4/6 can be a promising therapeutic strategy in therapy-resistant CRPC.94 Clinical characterization of CRPC patients after treatment with enzalutamide and/or abiraterone revealed that cyclin D1 and TGF-β pathways are upregulated in therapy-resistant tumors.66 In a recent study, we demonstrated that Forkhead Box A1 (FOXA1) loss induces TGF-β signaling, which can be targeted by transforming growth factor beta type I receptor (TGFBR1) inhibitors.37 Further, we and others have demonstrated in preclinical models that concurrent TGF-β targeting enhances response to enzalutamide and delays the onset of resistance.95,96 In addition, data from our group (unpublished) and others have revealed chemokine (CXC motif) receptor 7 (CXCR7), an AR-repressed gene, is upregulated during ADT and contributes to enzalutamide resistance.97 Another exemplary AR-repressed oncogene is c-MET, which mediates tumor metastasis and has been the center for drug development for the treatment of CRPC.75,76,77 Finally, during CRPC progression, prostate cancer reactivates telomerase TERT.72 Wild-type AR directly represses TERT expression, and TERT targeting potentiates antitumor efficacy of enzalutamide.71,98
The major clinical post-ADT challenges are stemming from emergence of CRPC with aggressive phenotypes, such as neuroendocrine NEPC78 or double-negative DNPC, which lacks both NEPC- and androgen-dependent CRPC-defining markers.90 The loss of molecular targets in advance CRPC leaves clinicians with very limited therapeutic armamentarium,3,4,99,100,101 rendering NEPC an increasingly abundant lethal disease. This disease can be partially attributed to the restoration of a number of AR-repressed genes on AR elimination in NEPC following strong ADT.81,83,84,85 For example, AR has been shown to directly repress transcription of SOX2, a reprogramming transcription factor that drives PCa lineage plasticity into enzalutamide-resistant CRPC and NEPC.81 Moreover, a recent report revealed that AR directly represses a master neural transcription factor BRN2, which promotes enzalutamide-resistant NEPC phenotype in patients.84 Restored expression of PEG10, another AR-repressed gene, has also been shown to promote progression of NEPC.85 Collectively, these evidences reinforce the protective role of AR through gene repression in delaying aggressive CRPC and NEPC progression.
AR regulates differentiation of embryonic epithelial cells in urogenital sinus into matured organ during prostate development and maintains homeostatic integrity of prostate tissue in adults.102,103,104,105 Considering this physiologic role of AR, it is acceptable that, under certain context, AR might, at least partially, restrain aggressive prostate cancer progression. Therapeutic approaches using supraphysiologic levels of testosterone to treat CRPC have been proposed. This idea is appealing considering ADT-related adverse effects, including those caused by testosterone-deficiency, such as anemia, depression, fatigue, and metabolic dysfunctions. In contrary, testosterone administration delivers the opposite, positive effects to patient life quality, advocating for reevaluation of testosterone administration to PCa patients. Despite the lack of controlled clinical studies, some retrospective analyses of previously published data related to therapeutic administration of testosterone suggest that testosterone can mitigate ADT-associated adverse effects without exacerbating PCa.106,107,108,109 Moreover, preliminary preclinical studies report that testosterone or testosterone metabolites slow down CRPC tumor growth.110,111 Mechanistically, this can be supported by the important role of AR in suppressing many oncogenes and nonprostatic transcription factors. Intermittent ADT and supraphysiologic testosterone administration may lead to a balance in killing tumor cells and yet maintaining the prostate lineage. Clearly, more rigorous clinical studies are necessary in order to distil a cohort of CRPC patients that would successfully respond to AR-stimulating therapy.
CONCLUSION
Androgen deprivation and AR-targeted therapies remain as the mainstay treatment for metastatic PCa. The initial survival benefit from utilizing currently approved therapeutic means is undermined by imminent relapse with CRPC. We have already identified a number of mechanisms, such as AR amplification, AR alternative splicing, AR crossing activation with other signaling pathways, or AR bypass, that propagate resistance in CRPC patients to AR-targeted therapies. While the significance of AR-induced gene expression in PCa progression has been extensively reported, the importance of AR-driven gene repression is currently unveiling with the wide application of ADT followed by drug resistance and NEPC progression. In this review, we attempted to critically analyze the current literature on AR-repressed genes and their regulation and function during PCa progression. As a transcriptional repressor, ligand-activated AR binds to the enhancers and/or promoter elements of target genes and mediates assembly of the repressive complexes, including HDACs, LSD1, and EZH2. The resulting histone modifications create repressive environment at the local chromatin, which makes the regulatory elements inaccessible and the gene repressed.
ADT or AR-targeted therapies de-repress both oncosuppressor and oncogenes that are normally inhibited by AR. CRPC appears to adapt and keep oncosuppressors continuously silenced by bypass mechanisms. On the other hand, prosurvival and proliferative oncogenes become activated, driving CRPC to achieve expedited tumor growth and therapy resistance. Accordingly, pharmacological targeting of AR-repressed genes such as c-MET shows promising results in a number of clinical trials for advanced CRPC.77 AR-repressed genes may be important targets for therapeutic intervention in the postenzalutamide/abiraterone era.
AUTHOR CONTRIBUTIONS
GG and JY designed the study, wrote, and reviewed the manuscript. WQG read and commented on the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
ACKNOWLEDGMENTS
This work was supported in part by the U.S. the Research Scholar Award RSG-12-085-01 (to JY) from the American Cancer Society, the National Institutes of Health R01CA172384 (to JY), the Northwestern Prostate SPORE (P50 CA180995, to JY), Department of Defense, Impact Award #W81XWH-17-1-0405 (to JY), and the NIH/NCI training grant T32 CA009560 (to GG).
REFERENCES
- 1.Huggins C. Prostatic cancer treated by orchiectomy; the five year results. J Am Med Assoc. 1946;131:576–81. doi: 10.1001/jama.1946.02870240008003. [DOI] [PubMed] [Google Scholar]
- 2.Azad AA, Zoubeidi A, Gleave ME, Chi KN. Targeting heat shock proteins in metastatic castration-resistant prostate cancer. Nat Rev Urol. 2015;12:26–36. doi: 10.1038/nrurol.2014.320. [DOI] [PubMed] [Google Scholar]
- 3.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–11. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 5.Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–92. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- 6.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–10. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 7.Jin HJ, Zhao JC, Wu L, Kim J, Yu J. Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun. 2014;5:3972. doi: 10.1038/ncomms4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao JC, Fong KW, Jin HJ, Yang YA, Kim J, et al. FOXA1 acts upstream of GATA2 and AR in hormonal regulation of gene expression. Oncogene. 2016;35:4335–44. doi: 10.1038/onc.2015.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene. 2009;28:3847–56. doi: 10.1038/onc.2009.243. [DOI] [PubMed] [Google Scholar]
- 11.He B, Lanz RB, Fiskus W, Geng C, Yi P, et al. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc Natl Acad Sci U S A. 2014;111:18261–6. doi: 10.1073/pnas.1421415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foley C, Mitsiades N. Moving beyond the androgen receptor (AR): targeting AR-interacting proteins to treat prostate cancer. Horm Cancer. 2016;7:84–103. doi: 10.1007/s12672-015-0239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Li W, Zhang Y, Yuan X, Xu K, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–4. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai C, He HH, Chen S, Coleman I, Wang H, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20:457–71. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao JC, Yu J, Runkle C, Wu L, Hu M, et al. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome Res. 2012;22:322–31. doi: 10.1101/gr.131508.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones PL, Shi YB. N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors. Curr Top Microbiol Immunol. 2003;274:237–68. doi: 10.1007/978-3-642-55747-7_9. [DOI] [PubMed] [Google Scholar]
- 20.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Kawate H, Ohnaka K, Nawata H, Takayanagi R. Nuclear compartmentalization of N-CoR and its interactions with steroid receptors. Mol Cell Biol. 2006;26:6633–55. doi: 10.1128/MCB.01534-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao G, Chen LY, Zhang A, Godavarthy A, Xia F, et al. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J Biol Chem. 2003;278:5052–61. doi: 10.1074/jbc.M206374200. [DOI] [PubMed] [Google Scholar]
- 24.Karvonen U, Jänne OA, Palvimo JJ. Androgen receptor regulates nuclear trafficking and nuclear domain residency of corepressor HDAC7 in a ligand-dependent fashion. Exp Cell Res. 2006;312:3165–83. doi: 10.1016/j.yexcr.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–7. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 26.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–65. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 30.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 31.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 32.Cai C, He HH, Gao S, Chen S, Yu Z, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9:1618–27. doi: 10.1016/j.celrep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svensson C, Ceder J, Iglesias-Gato D, Chuan YC, Pang ST, et al. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 2014;42:999–1015. doi: 10.1093/nar/gkt921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 35.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Lee Y, Lu X, Song B, Fong KW, et al. Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 2018;25:2808–20.e4. doi: 10.1016/j.celrep.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Yu J, Rhodes DR, Tomlins SA, Cao X, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–63. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 40.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 41.Wu L, Runkle C1, Jin HJ, Yu J1, Li J, et al. CCN3/NOV gene expression in human prostate cancer is directly suppressed by the androgen receptor. Oncogene. 2014;33:504–13. doi: 10.1038/onc.2012.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fong KW, Zhao JC, Kim J, Li S, Yang YA, et al. Polycomb-mediated disruption of an androgen receptor feedback loop drives castration-resistant prostate cancer. Cancer Res. 2017;77:412–22. doi: 10.1158/0008-5472.CAN-16-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chng KR, Chang CW, Tan SK, Yang C, Hong SZ, et al. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012;31:2810–23. doi: 10.1038/emboj.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao S, Gao Y, He HH, Han D, Han W, et al. Androgen receptor tumor suppressor function is mediated by recruitment of retinoblastoma protein. Cell Rep. 2016;17:966–76. doi: 10.1016/j.celrep.2016.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–82. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crosby ME, Almasan A. Opposing roles of E2Fs in cell proliferation and death. Cancer Biol Ther. 2004;3:1208–11. doi: 10.4161/cbt.3.12.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Zhang DY, Ren Q, Ye F, Zhao X, et al. Regulation of a novel androgen receptor target gene, the cyclin B1 gene, through androgen-dependent E2F family member switching. Mol Cell Biol. 2012;32:2454–66. doi: 10.1128/MCB.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong YN, Ferraldeschi R, Attard G, de Bono J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nat Rev Clin Oncol. 2014;11:365–76. doi: 10.1038/nrclinonc.2014.72. [DOI] [PubMed] [Google Scholar]
- 50.Jariwala U, Prescott J, Jia L, Barski A, Pregizer S, et al. Identification of novel androgen receptor target genes in prostate cancer. Mol Cancer. 2007;6:39. doi: 10.1186/1476-4598-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, et al. Cell-and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–17. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J Biol Chem. 1998;273:20213–22. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 53.Kanno Y, Zhao S, Yamashita N, Yanai K, Nemoto K, et al. Androgen receptor functions as a negative transcriptional regulator of DEPTOR, mTOR inhibitor. J Toxicol Sci. 2015;40:753–8. doi: 10.2131/jts.40.753. [DOI] [PubMed] [Google Scholar]
- 54.Tan M, Xu J, Siddiqui J, Feng F, Sun Y. Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis. Mol Cancer. 2016;15:81. doi: 10.1186/s12943-016-0567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamouille S, XuJ, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu YN, Liu Y, Lee HJ, Hsu YH, Chen JH. Activated androgen receptor downregulates E-cadherin gene expression and promotes tumor metastasis. Mol Cell Biol. 2008;28:7096–108. doi: 10.1128/MCB.00449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y, Wang BE, Leong KG, Yue P, Li L, et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: implications for androgen-deprivation therapy. Cancer Res. 2012;72:527–36. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- 58.Huo C, Kao YH, Chuu CP. Androgen receptor inhibits epithelial-mesenchymal transition, migration, and invasion of PC-3 prostate cancer cells. Cancer Lett. 2015;369:103–11. doi: 10.1016/j.canlet.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Li P, Wang J, Chu M, Zhang K, Yang R, et al. Zeb1 promotes androgen independence of prostate cancer via induction of stem cell-like properties. Exp Biol Med (Maywood) 2014;239:813–22. doi: 10.1177/1535370214538727. [DOI] [PubMed] [Google Scholar]
- 60.Jacob S, Nayak S, Fernandes G, Barai RS, Menon S, et al. Androgen receptor as a regulator of ZEB2 expression and its implications in epithelial-to-mesenchymal transition in prostate cancer. Endocr Relat Cancer. 2014;21:473–86. doi: 10.1530/ERC-13-0514. [DOI] [PubMed] [Google Scholar]
- 61.Lanzino M, Sisci D, Morelli C, Garofalo C, Catalano S, et al. Inhibition of cyclin D1 expression by androgen receptor in breast cancer cells-identification of a novel androgen response element. Nucleic Acids Res. 2010;38:5351–65. doi: 10.1093/nar/gkq278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holter E, Kotaja N, Mäkela S, Strauss L, Kietz S, et al. Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX-1. Mol Endocrinol. 2002;16:515–28. doi: 10.1210/mend.16.3.0804. [DOI] [PubMed] [Google Scholar]
- 63.Agoulnik IU, Krause WC, Bingman WE 3rd, Rahman HT, Amrikachi M, et al. Repressors of androgen and progesterone receptor action. J Biol Chem. 2003;278:31136–48. doi: 10.1074/jbc.M305153200. [DOI] [PubMed] [Google Scholar]
- 64.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 65.Casimiro MC, Di Sante G, Ju X, Li Z, Chen K, et al. Cyclin D1 promotes androgen-dependent DNA damage repair in prostate cancer cells. Cancer Res. 2016;76:329–38. doi: 10.1158/0008-5472.CAN-15-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pal SK, Patel J, He M, Foulk B, Kraft K, et al. Identification of mechanisms of resistance to treatment with abiraterone acetate or enzalutamide in patients with castration-resistant prostate cancer (CRPC) Cancer. 2018;124:1216–24. doi: 10.1002/cncr.31161. [DOI] [PubMed] [Google Scholar]
- 67.Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, et al. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–37. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 68.Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nucl Recept Signal. 2008;6:e001. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–25. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 70.Maddison LA, Huss WJ, Barrios RM, Greenberg NM. Differential expression of cell cycle regulatory molecules and evidence for a “cyclin switch” during progression of prostate cancer. Prostate. 2004;58:335–44. doi: 10.1002/pros.10341. [DOI] [PubMed] [Google Scholar]
- 71.Moehren U, Papaioannou M, Reeb CA, Grasselli A, Nanni S, et al. Wild-type but not mutant androgen receptor inhibits expression of the hTERT telomerase subunit: a novel role of AR mutation for prostate cancer development. FASEB J. 2008;22:1258–67. doi: 10.1096/fj.07-9360com. [DOI] [PubMed] [Google Scholar]
- 72.Zhang K, Guo Y, Wang X, Zhao H, Ji Z, et al. WNT/beta-catenin directs self-renewal symmetric cell division of hTERT(high) prostate cancer stem cells. Cancer Res. 2017;77:2534–47. doi: 10.1158/0008-5472.CAN-16-1887. [DOI] [PubMed] [Google Scholar]
- 73.Biroccio A, Leonetti C. Telomerase as a new target for the treatment of hormone-refractory prostate cancer. Endocr Relat Cancer. 2004;11:407–21. doi: 10.1677/erc.1.00764. [DOI] [PubMed] [Google Scholar]
- 74.Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3(1 Suppl):S7–19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verras M, Lee J, Xue H, Li TH, Wang Y, et al. The androgen receptor negatively regulates the expression of c-Met: implications for a novel mechanism of prostate cancer progression. Cancer Res. 2007;67:967–75. doi: 10.1158/0008-5472.CAN-06-3552. [DOI] [PubMed] [Google Scholar]
- 76.Tu WH, Zhu C, Clark C, Christensen JG, Sun Z. Efficacy of c-Met inhibitor for advanced prostate cancer. BMC Cancer. 2010;10:556. doi: 10.1186/1471-2407-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varkaris A, Corn PG, Gaur S, Dayyani F, Logothetis CJ, et al. The role of HGF/c-Met signaling in prostate cancer progression and c-Met inhibitors in clinical trials. Expert Opin Investig Drugs. 2011;20:1677–84. doi: 10.1517/13543784.2011.631523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lipianskaya J, Cohen A, Chen CJ, Hsia E, Squires J, et al. Androgen-deprivation therapy-induced aggressive prostate cancer with neuroendocrine differentiation. Asian J Androl. 2014;16:541–4. doi: 10.4103/1008-682X.123669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan TC, Veeramani S, Lin FF, Kondrikou D, Zelivianski S, et al. Androgen deprivation induces human prostate epithelial neuroendocrine differentiation of androgen-sensitive LNCaP cells. Endocr Relat Cancer. 2006;13:151–67. doi: 10.1677/erc.1.01043. [DOI] [PubMed] [Google Scholar]
- 81.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–8. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang J, Zhu HH, Chu M, Liu Y, Zhang C, et al. Symmetrical and asymmetrical division analysis provides evidence for a hierarchy of prostate epithelial cell lineages. Nat Commun. 2014;5:4758. doi: 10.1038/ncomms5758. [DOI] [PubMed] [Google Scholar]
- 83.Kregel S, Kiriluk KJ, Rosen AM, Cai Y, Reyes EE, et al. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PLoS One. 2013;8:e53701. doi: 10.1371/journal.pone.0053701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7:54–71. doi: 10.1158/2159-8290.CD-15-1263. [DOI] [PubMed] [Google Scholar]
- 85.Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 2015;12:922–36. doi: 10.1016/j.celrep.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 86.Jernberg E, Bergh A, Wikström P. Clinical relevance of androgen receptor alterations in prostate cancer. Endocr Connect. 2017;6:R146–61. doi: 10.1530/EC-17-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fong KW, Zhao JC, Song B, Zheng B, Yu J. TRIM28 protects TRIM24 from SPOP-mediated degradation and promotes prostate cancer progression. Nat Commun. 2018;9:5007. doi: 10.1038/s41467-018-07475-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, et al. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13:209–21. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Song G, Zhang X, Feng T, Pan J, et al. PADI2-mediated citrullination promotes prostate cancer progression. Cancer Res. 2017;77:5755–68. doi: 10.1158/0008-5472.CAN-17-0150. [DOI] [PubMed] [Google Scholar]
- 90.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–89.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brennen WN, Isaacs JT. Cellular origin of androgen receptor pathway-independent prostate cancer and implications for therapy. Cancer Cell. 2017;32:399–401. doi: 10.1016/j.ccell.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 92.Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, et al. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci U S A. 2015;112:E6544–52. doi: 10.1073/pnas.1518007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sheahan AV, Ellis L. Epigenetic reprogramming: a key mechanism driving therapeutic resistance. Urol Oncol. 2018;36:375–9. doi: 10.1016/j.urolonc.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 94.Han GC, Hwang J, Mullane SA. Clinical and genomic resistance to second generation androgen blockade in paired biopsies of metastatic castration-resistant prostate cancer. Cancer Res. 2017;77(13 Suppl):2905. doi: 10.1200/PO.17.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song B, Park SH, Zhao JC, Fong KW, Li S, et al. Targeting FOXA1-mediated repression of TGF-beta signaling suppresses castration-resistant prostate cancer progression. J Clin Invest. 2018 doi: 10.1172/JCI122367. Doi: 10.1172/JCI122367. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paller C, Pu H, Begemann DE, Wade CA, Hensley PJ, et al. TGF-beta receptor I inhibitor enhances response to enzalutamide in a pre-clinical model of advanced prostate cancer. Prostate. 2019;79:31–43. doi: 10.1002/pros.23708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo Y, Azad AK, Karanika S, Basourakos SP, Zuo X, et al. Enzalutamide and CXCR7 inhibitor combination treatment suppresses cell growth and angiogenic signaling in castration-resistant prostate cancer models. Int J Cancer. 2018;142:2163–74. doi: 10.1002/ijc.31237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gecgel KK, Muduroglu M, Erdogan S. Inhibition of telomerase potentiates enzalutamide efficiency of androgen-sensitive human prostate cancer cells. J BUON. 2017;22:1570–6. [PubMed] [Google Scholar]
- 99.Beltran H, Tagawa ST, Park K, MacDonald T, Milowsky MI, et al. Challenges in recognizing treatment-related neuroendocrine prostate cancer. J Clin Oncol. 2012;30:e386–9. doi: 10.1200/JCO.2011.41.5166. [DOI] [PubMed] [Google Scholar]
- 100.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–33. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Attard G, Reid AH, A’Hern R, Parker C, Oommen NB, et al. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J Clin Oncol. 2009;27:3742–8. doi: 10.1200/JCO.2008.20.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou Y, Bolton EC, Jones JO. Androgens and androgen receptor signaling in prostate tumorigenesis. J Mol Endocrinol. 2015;54:R15–29. doi: 10.1530/JME-14-0203. [DOI] [PubMed] [Google Scholar]
- 103.Wen S, Chang HC, Tian J, Shang Z, Niu Y, et al. Stromal androgen receptor roles in the development of normal prostate, benign prostate hyperplasia, and prostate cancer. Am J Pathol. 2015;185:293–301. doi: 10.1016/j.ajpath.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cooke PS, Young P, Cunha GR. Androgen receptor expression in developing male reproductive organs. Endocrinology. 1991;128:2867–73. doi: 10.1210/endo-128-6-2867. [DOI] [PubMed] [Google Scholar]
- 105.Wilson JD, George FW, Griffin JE. The hormonal control of sexual development. Science. 1981;211:1278–84. doi: 10.1126/science.7010602. [DOI] [PubMed] [Google Scholar]
- 106.Khera M. Testosterone replacement in men with treated and untreated prostate cancer. Sex Med Rev. 2013;1:143–9. doi: 10.1002/smrj.16. [DOI] [PubMed] [Google Scholar]
- 107.Kaplan AL, Hu JC, Morgentaler A, Mulhall JP, Schulman CC, et al. Testosterone therapy in men with prostate cancer. Eur Urol. 2016;69:894–903. doi: 10.1016/j.eururo.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morgentaler A. Testosterone therapy in men with prostate cancer: scientific and ethical considerations. J Urol. 2013;189(1 Suppl):S26–33. doi: 10.1016/j.juro.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 109.Kacker R, Hult M, San Francisco IF, Conners WP, Rojas PA, et al. Can testosterone therapy be offered to men on active surveillance for prostate cancer? Preliminary results. Asian J Androl. 2016;18:16–20. doi: 10.4103/1008-682X.160270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Song W, Soni V, Soni S, Khera M. Testosterone inhibits the growth of prostate cancer xenografts in nude mice. BMC Cancer. 2017;17:635. doi: 10.1186/s12885-017-3569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bremmer F, Jarry H, Unterkircher V, Kaulfuss S, Burfeind P, et al. Testosterone metabolites inhibit proliferation of castration- and therapy-resistant prostate cancer. Oncotarget. 2018;9:16951–61. doi: 10.18632/oncotarget.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rajabi H, Joshi MD, Jin C, Ahmad R, Kufe D. Androgen receptor regulates expression of the MUC1-C oncoprotein in human prostate cancer cells. Prostate. 2011;71:1299–308. doi: 10.1002/pros.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mannan Baig A, Khan NA, Effendi V, Rana Z, Ahmad HR, et al. Differential receptor dependencies: expression and significance of muscarinic M1 receptors in the biology of prostate cancer. Anticancer Drugs. 2017;28:75–87. doi: 10.1097/CAD.0000000000000432. [DOI] [PubMed] [Google Scholar]
- 114.Prescott J, Jariwala U, Jia L, Cogan JP, Barski A, et al. Androgen receptor-mediated repression of novel target genes. Prostate. 2007;67:1371–83. doi: 10.1002/pros.20623. [DOI] [PubMed] [Google Scholar]
- 115.Xie N, Cheng H, Lin D, Liu L, Yang O, et al. The expression of glucocorticoid receptor is negatively regulated by active androgen receptor signaling in prostate tumors. Int J Cancer. 2015;136:E27–38. doi: 10.1002/ijc.29147. [DOI] [PubMed] [Google Scholar]
- 116.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155:1309–22. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman WE, 3rd, et al. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res. 2006;66:10594–602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]