Short abstract
Background
Carboxymethyl lysine is an advanced glycation end product of interest as a potential biomarker of cardiovascular and other diseases. Available methods involve ELISA, with potential interference, or isotope dilution mass spectrometry (IDMS), with low-throughput sample preparation.
Methods
A high-throughput sample preparation method based on 96-well plates was developed. Protein-bound carboxymethyl lysine and lysine were quantified by IDMS using reversed phase chromatography coupled to a high-resolution accurate mass Orbitrap Exactive mass spectrometer. The carboxymethyl lysine concentration (normalized to lysine concentration) was measured in 1714 plasma samples from the British Regional Heart Study (BRHS).
Results
For carboxymethyl lysine, the lower limit of quantification (LLOQ) was estimated at 0.16 μM and the assay was linear between 0.25 and 10 μM. For lysine, the LLOQ was estimated at 3.79 mM, and the assay was linear between 2.5 and 100 mM. The intra-assay coefficient of variation was 17.2% for carboxymethyl lysine, 9.3% for lysine and 10.5% for normalized carboxymethyl lysine. The inter-assay coefficient of variation was 18.1% for carboxymethyl lysine, 14.8 for lysine and 16.2% for normalized carboxymethyl lysine. The median and inter-quartile range of all study samples in each batch were monitored. A mean carboxymethyl lysine concentration of 2.7 μM (IQR 2.0–3.2 μM, range 0.2–17.4 μM) and a mean normalized carboxymethyl lysine concentration of 69 μM/M lysine (IQR 54–76 μM/M, range 19–453 μM/M) were measured in the BRHS.
Conclusion
This high-throughput sample preparation method makes it possible to analyse large cohorts required to determine the potential of carboxymethyl lysine as a biomarker.
Keywords: Carboxymethyl lysine, high-resolution accurate mass Orbitrap mass spectrometry, Orbitrap, isotope dilution mass spectrometry
Introduction
Carboxymethyl lysine (CML) is an advanced glycation end product (AGE), produced in vivo, particularly under hyperglycaemic conditions, and available from the diet.1,2 Increased CML concentrations are associated with cardiovascular disease, diabetic nephropathy and retinopathy, chronic kidney disease and others.2–4 CML has been proposed as a potential biomarker of cardiovascular disease; however, conflicting results have been found.4–6 Enzyme linked immunosorbent assays (ELISAs) are available but suffer from steric hindrance of the antigen and interference from endogenous anti-AGE antibodies.1,2 Isotope dilution mass spectrometry (IDMS) methods are available for quantification of protein-bound CML, and samples require chemical reduction, protein denaturation, hydrolysis and drying prior to IDMS analysis. Published sample preparation methods are individual tube-based, which have limited throughput.1,5 Therefore, we developed a high-throughput sample preparation method using 96-well plates.
Published IDMS methods for CML quantification rely on multiple reaction monitoring (MRM) detection using triple quadrupole mass spectrometers (MS).7 In MRM, the first quadrupole is optimized to select for the parent ion of interest (CML) based on the mass to charge ratio (m/z). A collision cell fragments the parent ions into product ions,7,8 while the third quadrupole is optimized to select for specific product ions: the quantifier (for quantification) and the qualifier (for verification the identity).7–9 As triple quadrupole detectors provide excellent sensitivity and specificity, even in complex biofluids, they are widely used in clinical chemistry laboratories (e.g. toxicology, endocrinology and new born screening).7–9
There is increasing interest in the use of high-resolution accurate mass (HRAM) MS for absolute quantification of ions, including for routine clinical analysis and clinical research.10,11 HRAM MS relies on superior mass accuracy (typically sub 3 ppm), which allows excellent ion selectivity, provided appropriate mass-extraction windows (based on the theoretical m/z of the ion of interest) are chosen.10–13 HRAM MS analysis is commonly run in full scan mode, enabling the detection of all ionized compounds, without the need to optimize quadrupoles and collision energies for individual ions.12 The quantitative performance of HRAM MS now equals that of triple quadrupole mass spectrometry: in terms of sensitivity, mass accuracy, selectivity, although this does depend on the conditions, parameters and the metabolite of interest used.10–12,14 Some of the major advantages of HRAM MS are that data can be reanalysed retrospectively to investigate further biomarkers and that those biomarkers can be more easily identified (based on molecular formula).10–12,14 HRAM MS has been successfully used to quantify a number of small molecule groups: over 50 metabolites (including amino acids);14 amino acids (within 3 min);15 drugs and drug metabolites;16 circulating steroids11 and plasma metanephrines.11
We therefore optimized an HRAM IDMS method to quantify protein-bound CML and lysine and their deuterated internal standards using an Orbitrap Exactive mass spectrometer. To account for variation in plasma total protein concentration and variation introduced during sample preparation, the CML concentration was normalized to the lysine concentration.5,6 The three measures reported are CML (μM), lysine (μM) and normalized CML (μM per M lysine).
Methods
Sample preparation
Serum collected from one healthy volunteer, stored at –80°C in multiple aliquots, was used as a quality control (QC) sample. The Glasgow University Ethics Committee provided ethical approval for collection of anonymized samples for QC (Project number 200140133). Unthawed fasting EDTA plasma samples (n = 1714) from the 30th year re-examination of the British Regional Heart Study (BRHS),17,18 stored at –80°C, were randomized to 21 batches. Approval for collection was obtained from the local research ethics committees of the 24 towns where participants were recruited. All participants provided written informed consent to participate in the study.18 The study is consistent with the World Medical Association Declaration of Helsinki. The BRHS is a prospective study which recruited 7735 men between 1978 and 1980 from 24 British towns. At the 30-year re-examination, samples were collected from 1722 men between 2010 and 2012, with the men then being aged 71 to 92 years.17
A high-throughput 96-well deep-well plate method of sample preparation based on previously published methods was developed.1,4–6 Sodium tetraborate (Sigma, Dorset, UK), sodium borohydride (Alfa Aesar, Lancashire, UK), trichloroacetic acid (Sigma, Dorset, UK), hydrochloric acid (Sigma, Dorset, UK) were used for sample preparation. Plasma samples were defrosted for 90 min and centrifuged at 20,000 × g for 5 min. Ten microlitres of plasma were added to 300 μL sodium tetraborate (0.2 M)/borohydride (0.1 M) buffer in a 96-well deep-well polypropylene plate (Thermo Fisher Scientific, Hemel Hempstead, UK). The samples were chemically reduced overnight at 4°C to prevent further production of CML (or other advanced glycation end products) during subsequent hydrolysis.1 The protein was denatured in 20% trichloroacetic acid, and the pellet was washed in 20% trichloroacetic acid. The protein pellet was then hydrolysed at 110°C in 600 μL 6 M hydrochloric acid for 24 h, using a ceramic bead-bath. After hydrolysis, the samples were dried to completion at 95°C (approximately 24 h). Immediately prior to analysis, the samples were spiked with 10 μL of 20 μM CML-d4 (Toronto Research Chemicals, Ontario, Canada, 98% pure) and 10 μL of 150 mM universally 13C labelled L-lysine:2HCl) (Cambridge Isotope Laboratories Inc., MA, USA, 98% pure) as internal standards (ISs) and reconstituted in 270 μL of 5 mM nonafluoropentanoic acid (NFPA) (Sigma, Dorset, UK) as an ion-pairing agent.
CML (Toronto Research Chemicals, Ontario, Canada, 96% pure) and lysine (Sigma, Dorset, UK) were used to prepare calibrator samples: made up in water and then mixed with IS and NFPA. A seven-point calibration curve (CML: 0, 0.25, 0.5, 1, 2, 5, 10 μM and lysine: 0, 2.5, 5, 10, 20, 50, 100 mM) was used to quantify both CML and lysine relative to their ISs. The concentration ranges were chosen based on the concentrations previously reported using IDMS quantification.1,6 Previous studies demonstrated that acid hydrolysis of calibration solutions did not alter peak area; therefore, calibrator samples were not hydrolysed.1
Chromatography and HRAM mass spectrometry
Chromatography was carried out on an UltiMate 3000 RSLC system (Thermo Fisher Scientific, Hemel Hempstead, UK) using an ACQUITY UPLC BEH C18 column (100 mm × 2.1 mm, 1.7 μm column, Waters, Wilmslow) with VanGuard pre-column (Waters, Wilmslow, UK). Mobile phase A was 5 mM NFPA (Sigma, Dorset, UK) in HPLC grade water (Fisher, Loughborough, UK). Mobile phase B was HPLC grade acetonitrile (Fisher, Loughborough, UK). The column was maintained at 50°C, and samples were eluted with a linear gradient over 9.0 min at a flow rate of 0.3 ml/min. Starting conditions were 90% mobile phase A, decreasing to 20% between 0.1 to 4.6 min; this was held between 4.6 and 6.1 min, then increased to 90% at 6.2 min and held until 9.0 min to re-equilibrate the column. The injection volume was 5 μL, and samples were maintained at 5°C prior to injection. For HRAM MS, an Orbitrap Exactive (Thermo Fisher Scientific, Hemel Hempstead, UK) was operated in high-resolution full scan positive mode, at a scan range of 120–250 m/z, a probe temperature of 150°C and capillary temperature 275°C. The mass resolution was 50,000, providing a mass accuracy of less than 1 ppm. A mass calibration was performed prior to each batch using Pierce LTQ Velos positive ion calibration solution (Thermo Fisher Scientific, Hemel Hempstead, UK). TraceFinder 3.3 (Thermo Fisher Scientific, Hemel Hempstead, UK) was used to automatically detect peaks within expected retention time and mass extraction windows.
Protein-bound CML and lysine concentrations were calculated based on integrated areas relative to those of the ISs. The normalized CML concentration (μM/M lysine) was calculated from the measured CML and lysine concentrations observed (CML/lysine × 1,000,000). This allows for variation in total protein concentration to be accounted for, analogous to reporting HbA1c in relation to haemoglobin concentration.19 It also allows variation in hydrolysis to be accounted for. Once the method was optimized, 1714 samples from the BRHS were analysed.
Results and discussion
Isolation and detection
Mean Pearson correlation coefficients, mean response factors (gradients) and mean y-intercepts for protein-bound CML and lysine calibration were calculated (Table 1).
Table 1.
Figures demonstrating assay performance and median concentration observed in BRHS plasma samples.
| CML | Lys | CML (normalized) | |
|---|---|---|---|
| Calibration | |||
| Pearson correlation coefficient (r2) (SD) | 0.9994 (0.003) | 0.9993 (0.0008) | NA |
| Gradient (SD) | 0.0223 (0.0102) | 0.0016 (0.0007) | NA |
| Intercept (SD) | 0.0020 (0.0008) | 1.146 (1.18986) | NA |
| Water-based QC | |||
| Measured concentration (μM) | 0.29 | 2,377 | NA |
| Inter-assay CV (%) | 10.1 | 18.3 | NA |
| Estimated LOB (μM) | 0.13 | 136 | NA |
| Estimated LOD (μM) | 0.12 | 1250 | NA |
| Estimated LOQ (μM) | 0.16 | 3789 | NA |
| Chosen LOQ | 0.25 | 2500 | NA |
| Serum based QC | |||
| Intrasample CV | 2.7 | 2.1 | 3.9 |
| Intra-assay CV | 17.2 | 9.3 | 10.5 |
| Inter-assay CV | 18.1 | 14.8 | 16.2 |
| BHRS plasma samples | |||
| Median measured Concentration (μM) | 2.5 | 39,773.5 | 65 |
| Interquartile range (μM) | 2.0 to 3.2 | 36,109 to 43,210.6 | 54 to 76 |
| Estimated reference range (μM) | 1.1 to 5.6 | 26,182 to 57,677 | 34 to 123 |
CML: carboxymethyl lysine; LOQ: limit of quantification; LOB: limit of blank; LOD: limit of detection.
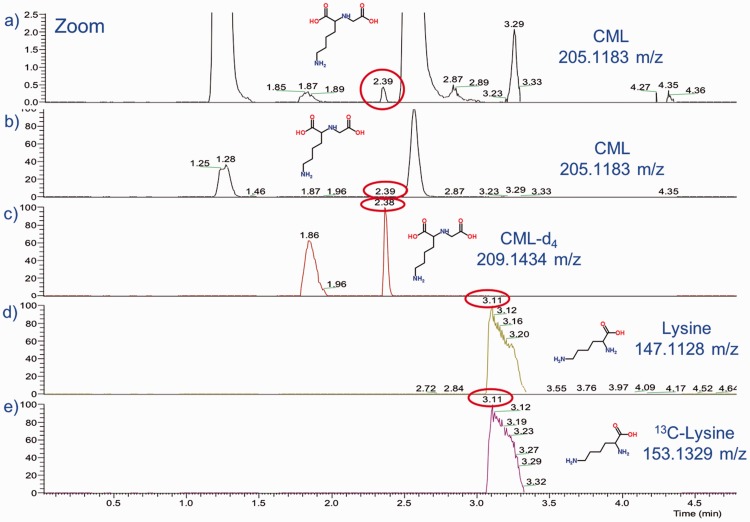
Separation of CML from the closely eluting peak, suspected to be valylserine or serylvaline, was good in most (1614 [97%]) samples (Figure 1). Valylserine and serylvaline are dipeptides composed of valine and serine with the same molecular formula (C8H16N2O4) and m/z as CML and are likely to be produced during acid hydrolysis. Since this is an isobaric interference, reduction of the mass extraction window cannot eliminate this interferent. The CML and CML-d4 appear to have isomerized, resulting in two peaks with the same m/z at two different retention times (1.86 and 2.38 min). The peak with the latter retention time was chosen for integration due to better peak shape.
Figure 1.
Extracted-ion-chromatograms (0–5 min): (a) Close-up of CML (and other metabolites) in serum with m/z of 205.1183 and retention time of 2.39 min (note splitting of the CML peak [at 1.87 and 2.39 min] and closely eluting peak thought to be valylserine or serylvaline; (b) CML (and other metabolites) in serum with m/z of 205.1183 and retention time of 2.39 min; (c) deuterated CML in serum with m/z of 209.1343 and retention time of 2.38 min (note splitting at 1.86 and 2.38 min); (d) lysine in serum with m/z of 147.1128 and retention time of 3.11 min; (e) universally labelled 13C-lysine in serum with m/z of 153.1329 and retention time of 3.11 min.
CML: carboxymethyl lysine.
Limits of blank, detection and quantification
In IDMS, the lower limit of detection (LLOD) and quantification (LLOQ) are usually determined by calculating the signal to noise ratio (SNR) in a spectrum, with an SNR of 3 being used as an LLOD and an SNR of 10 (with accuracy of 80–120% and <20% imprecision) being used as an LLOQ.8 Due to the signal processing of the Orbitrap Exactive, with baseline removal inherent in Orbitrap data acquisition, there is generally no noise in the extracted ion chromatogram.11 Therefore, the SNR for all peaks was infinity, and SNR cannot be used to estimate LLOD and LLOQ. Instead, the LLOD and LLOQ were estimated based on the slope and SD of the y-intercept of the calibration curve.20
In the lowest water-based calibrator sample (0.25 μM), the mean CML concentration (over 21 batches) was 0.29 μM (<20% bias), with an inter-assay coefficient of variation (CV) of 10.1% (<20% variation). In the water-based calibrators, the results were linear between 0.25 μM and 10 μM. The limit of blank (LOB) for the zero-calibrator sample was estimated as 0.13 μM based on an average concentration of 0.04 μM and a standard deviation of 0.06 (LOB = mean + 1.645 SD).21 The LLOD was estimated as 0.12 μM, by multiplying the SD of the y-intercept by 3.3 and dividing by the slope of the calibration curve.20 The LLOQ was estimated as 0.16 μM, by multiplying the SD of the y-intercept by 10 and dividing by the slope of the calibration curve.20 The concentration of the lowest water-based calibrator (0.25 μM) was chosen as the LLOQ, in order to avoid extrapolation,22 since CML is endogenous to serum samples. Only one plasma sample was observed with a CML concentration below the LLOQ of 0.25 μM. Thirteen samples were observed with a CML concentration of over 10 μM (ranging between 11.9 and 17.4 μM). Since there was no evidence of detector saturation with the closely eluting isobaric contaminant peak present at approximately 1000 times higher concentrations (based on peak area), the linearity can be extrapolated.
In the lowest water-based calibrator sample, the mean lysine concentration (over 21 batches) was 2377 μM (<5% bias), with an inter-assay CV of 18.3% (<20%).22 In the water-based calibrators, the results were linear between 2500 μM and 100,000 μM. The LOB for the zero-calibrator sample was estimated as 136 μM based on an average concentration of –539 μM and an SD of 410 (LOB = mean + 1.645 SD).21 The LLOD was estimated as 1250 μM, by multiplying the SD of the y-intercept by 3.3 and dividing by the slope of the calibration curve.20 The LLOQ was also estimated as 3789 μM, by multiplying the SD of the y-intercept by 10 and dividing by the slope of the calibration curve.20 The concentration of the lowest water-based calibrator, 2500 μM, was chosen as the LLOQ, in order to avoid extrapolation,22 since lysine is endogenous to serum samples. No serum samples were observed with a lysine concentration below 2500 μM (all ≥ 8,343 μM). No serum samples were observed with a lysine concentration over 100,000 μM (all ≤ 58,231 μM).
Serum-based QC
Six serum QC samples were prepared and re-injected 16 times each. The mean intra-assay, intra-sample CVs were 2.7% for CML, 2.1 for lysine and 3.9% for normalized CML. This demonstrates that repeated analysis of the same sample preparation is robust. The CVs for normalized CML are increased, since the variability of both the CML measurement and the lysine measurement is contributing to the overall variability.
The intra-assay CV (based on 30 freshly prepared and reconstituted samples, each injected only once) was 17.2% for CML, 9.3% for lysine and 10.5% for CML normalized to lysine. The CV for normalized CML is lower than that of directly measured CML in serum samples, as normalization accounts for variation incorporated during individual sample preparation.
The overall inter-assay CV was 18.1% for CML, 14.8% for lysine and 16.2% for CML: lysine. The inter-assay CVs for CML (directly measured and normalized) are outside the target CV of 15% recommended by Food and Drug Administration and other guidelines for validation of bioanalytical methods within regulated environments.22 In non-regulatory environments, a CV of 20–25% is a commonly used target.23 The variation may have been introduced during high-throughput sample preparation, particularly during hydrolysis or during HRAM MS analysis.
It is recommended that the normalized CML concentration is used for clinical research studies, as it accounts for variation in sample preparation and in blood total protein concentrations. This is analogous to reporting HbA1C in relation to haemoglobin concentration.19
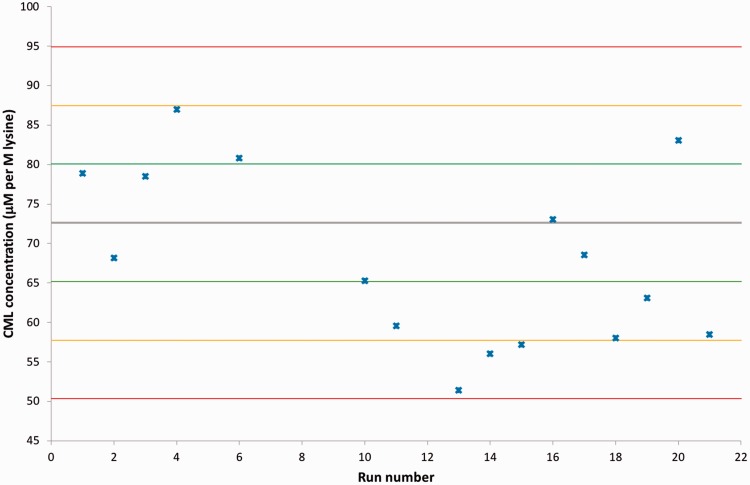
To assess the variability of the HRAM MS analysis and the sample preparation, a serum-based QC was prepared and run with every plate. The normalized CML concentration obtained was within QC limits, according to the Westguard Multi-rules, for all but one batch, and all but three within 2 SD of the mean of 12 run-in samples (Figure 2). Unfortunately, in some runs, this serum QC sample was contaminated – either from an unknown contaminant eluting over a wide retention range or from co-elution of the isobaric peak (suspected to be valylserine or serylvaline) (Figure 1).
Figure 2.
Levy-Jennings plot displaying the variability of the measured concentration of CML (normalized to the measured lysine concentration) in the quality control serum samples run with every batch. The mean normalized CML concentration derived from previous analysis of 12 quality control serum samples is referenced as the grey line. The green, yellow and red lines reference the mean ± 1 2 and 3 SD, respectively. Note for some runs co-elution with an isobaric interferant or contamination of the QC sample meant that CML concentration could not be measured.
CML: carboxymethyl lysine.
Batch-to-batch variation
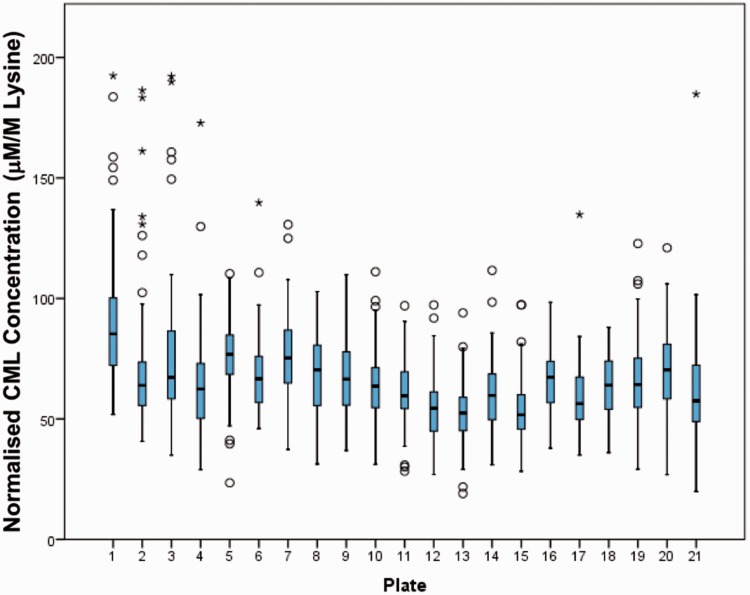
Since no external or commercial QC material exists, the overall mean, median and interquartile range (IQR) was monitored for each batch of 95 samples to check for batch effects. The batch-to-batch values are expected to vary, since each batch includes different samples; however, no obvious trends were observed in normalized lysine concentration (Figure 3).
Figure 3.
Box plots showing median (line), box (interquartile range) and whiskers (<1.5 × IQR) for normalized CML concentration in BRHS plasma samples run in each batch, arranged by plate number (n = 95 per batch). The circles represent outliers (<3 × IQR) and the stars represent extreme outliers (>3 × IQR). No obvious trends are observed from batch to batch; samples were randomized before sample preparation. Normalized CML concentrations of ≥ 200 mM/M lysine were excluded from the figure for clarity.
CML: carboxymethyl lysine.
To attempt to investigate the stability of the dried hydrolysed samples, 12 samples which were stored for three months at –80°C were reconstituted with NFPA and analysed by HRAM MS. The mean CML, lysine and normalized CML concentrations were not significantly different. To check for unwanted trends in the data over time, the box plots were arranged by date run on the LC-MS, by date of sample preparation and by delay between sample preparation and LCMS run. No obvious trends were observed (data not shown).
EDTA plasma versus serum samples
Paired samples of serum (serum separator vacutainers) or plasma (K+EDTA vacutainers) from seven healthy volunteers were run in triplicate (Table 2). The concentrations observed were not statistically significantly different for all three analytes. CML concentrations were also found to be similar in plasma versus serum samples in a previous study.4
Table 2.
Comparison of results from paired serum and EDTA plasma samples (seven paired samples run in triplicate). Results of paired t-test demonstrated no significant (ns) difference between the two sample types.
| CML (μM) | Lys (μM) | CML (μM/M lys) | |
|---|---|---|---|
| Serum (n = 7) (mean [SD]) | 2.543 (0.44) | 44,881 (3646) | 57 (8.0) |
| EDTA plasma (n = 7) (mean [SD]) | 2.509 (0.37) | 44,675 (2506) | 56 (6.5) |
| P (paired t-test) | 0.665 (ns) | 0.740 (ns) | 0.697 (ns) |
EDTA: ethylenediaminetetraacetic acid; CML: carboxymethyl lysine.
Throughput
We developed a 96-well deep-well method of sample preparation based on previously published methods individual tube methods.1 The hands-on preparation time (not including incubations) is reduced more than five-fold compared with the estimated sample preparation time reported for individual tube methods1 (Table 3). It also uses lower volumes of reagents. MS analysis time is slightly increased: 9-min run compared with the 7.5-min run published.4 However, analysis of a 96-well plate can still be completed within an overnight run.
Table 3.
Comparison of hands-on sample preparation time using the 96-well versus individual tube method and comparison of HRAM MS analysis time for 1000 samples.
| Per 1000 samples | Traditional method (hr) | HT method (h) |
|---|---|---|
| Sample preparation (hands-on) | 150 | 26 |
| Chromatography and detection | 125 | 150 |
Comparison with other IDMS methods
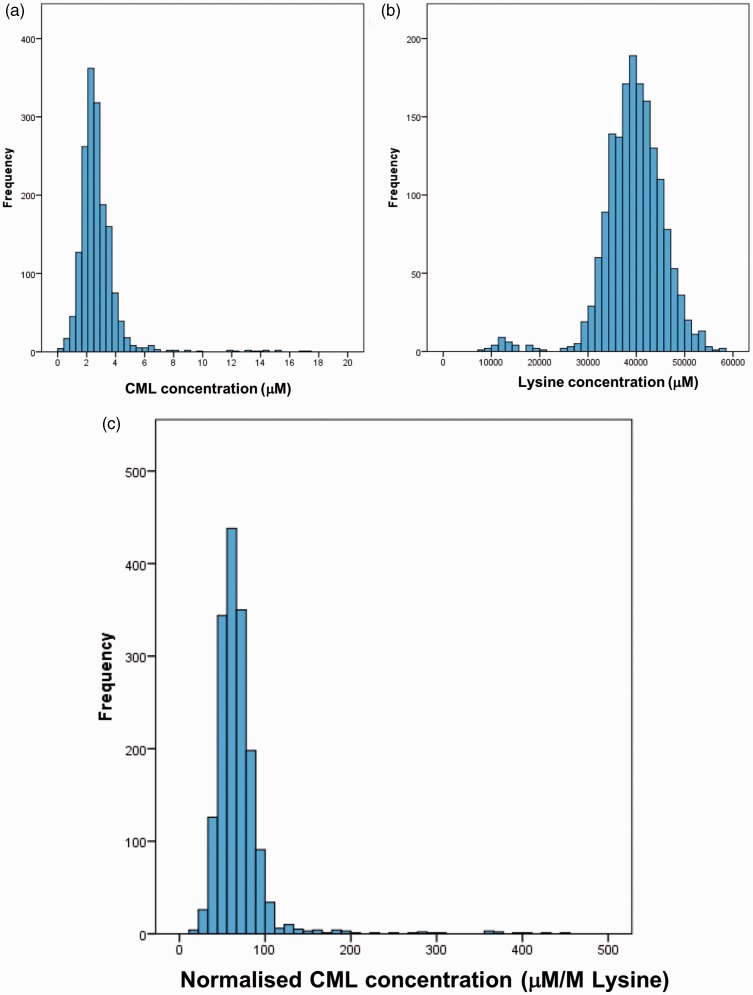
The CML concentration was measurable in 1664 samples from the BRHS. For 50 samples (3%), the CML peak co-eluted with the isobaric peak (suspected to be valylserine or serylvaline) or there was broad contamination. The CML and normalized CML concentrations were positively skewed and results for lysine were negatively skewed (Figure 4).
Figure 4.
Histogram of (a) CML concentration (μM); (b) lysine concentration (μM); (c) normalized CML concentration (μM/M lysine) in 1664 BRHS EDTA plasma samples.
CML: carboxymethyl lysine.
A mean of 2.7 μM (full range: 0.2–17.4 μM) with an SD of 1.4 μM and a median of 2.5 μM (IQR: 2.0–3.2 μM) was found for CML concentration in the BHRS samples (n = 1664). A reference range of 1.1 to 5.6 μM was calculated (mean ± 1.96 SD, after log transformation). It should be noted that the samples obtained at the 30th year re-examination of the BRHS were from white European males aged 71 to 92 years and that up to 444 of the participants had a diagnosis of CVD.17
The mean CML concentration of 2.7 μM measured is similar to the mean of 2.8 μM (SD 0.4 μM), previously reported for 10 healthy controls1 (Table 4). The BRHS median CML concentration of 2.5 μM is also similar to the median concentration of 2.9 μM (range 1.7 to 4.4 μM) observed in 31 individuals with type 1 diabetes mellitus with normal renal function.5
Table 4.
Comparison of CML and normalized CML concentration observed by HRAM MS with high-throughput sample preparation vs. LC-MS/MS with individual tube-based sample preparation.
| Mean (±SD) or median (IQR) in health | Mean (±SD) or median (IQR) in disease | ||||
|---|---|---|---|---|---|
| Reference | Sample type | CML (μM) | Population information | CML (μM) | Population information |
| This study | Fasting EDTA plasma | 2.7 (±1.4)2.5 (2.0 to 3.2) | 1664 Caucasian Europeans aged 71–92, 444 with CVD | NA | NA |
| Gaens et al.24 | EDTA plasma and serum | 1.61 (±0.38) | 738 Individuals in the Dutch Hoorn Study | 1.77 (±0.45) | 532 Individuals in the Dutch CODAM study |
| Teerlink et al.1 | Plasma | 2.8 (±0.4) | 10 Healthy individuals | 7.26 (±1.36); 8.01 (±3.8) | 17 Individuals on haemodialysis; 9 individuals on peritoneal dialysis |
| Liew-A-Fa et al.5 | Plasma | 2.9 (1.7 to 4.4) | 31 Individuals with T1DM and normal eGFR (>80 mL/min) | 4.9 (2.0 to 12.6) | 29 Individuals with T1DM and decreased eGFR (<80 mL/min) |
|
Normalized CML (μM/M lysine) |
Normalized CML (μM/M lysine) |
||||
| This study | Fasting EDTA plasma | 69 (±34)65 (54 to 76) | 1664 Caucasian Europeans aged 71–92, 444 with CVD | NA | NA |
| Anwar et al.25 | Fasting EDTA | 158 (±26) | 21 Healthy children | 190 (±38) | 27 Children with autism spectrum disorder |
| Maessen et al.26 | Serum | 68 (56–76) | 18 Sedentary individuals | 80 (73 to 89) | 18 Athletes |
| De Courten et al.27 | Serum | NA | NA | 77.6 (±14) | 20 Overweight or obese individuals |
| Linssen et al.28 | Serum | 31.6 (27.4 to 37.3) | >200 Individuals in top tertile for diastolic function | 33.6 (28.5 to 38.7) | >200 Individuals in bottom tertile for diastolic function |
| Hanssen et al.29 | Plasma | 52.1 (46.1 to 59.6) | ∼70 Individuals in bottom tertile for AGE score | 80.4 (72.1 to 91.7) | ∼70 Individuals in top tertile for AGE score |
| Gopal et al.30 | EDTA plasma | 82.9 (±19.3) | 44 Ex-smokers | 61.6 (±15.6) | 88 Individuals with COPD |
| Hanssen et al.6 | Fasting EDTA plasma | 34 (29 to 39) | 733 Individuals without prior CVD | 33 (27 to 38) | 558 Individuals with prior CVD |
| Hull et al.4 | Fasting serum and plasma | Medians 132 to 140 | Single pool of 10 healthy individuals measured with different sample processing | NA | NA |
| Ga Van Eupen et al.31 | Fasting EDTA plasma | 92.5 (±15.7) | 169 Individuals without Diabetes | 104.6 (±19.4) | 165 Individuals with T1DM |
| Rabbani et al.32 | Plasma | NA | NA | 52 (±14) | 52 Individuals with T2DM and microalbuminuria |
| Engelen et al.33 | Plasma | NA | NA | 49.9 (±11.9) | 125 Individuals with T2DM and microalbuminuria |
| Thornalley et al.34 | Plasma | 21 (±5) | Five healthy controls | NA | NA |
AGE: Advanced glycation end-product; COPD: chronic obstructive pulmonary disease; CVD: cardiovascular disease; EDTA: ethylenediaminetetraacetic acid; eGFR: estimated glomerular filtration rate; IQR: interquartile range; NA: not applicable; SD: standard deviation; T1DM: type 1 diabetes mellitus; T2DM: type 2 diabetes mellitus; CML: carboxymethyl lysine.
The mean lysine concentration in the BHRS samples was 39,490.5 μM, with an SD of 6268.6 μM, after hydrolysis. The median was 39,773.5 μM (IQR: 36,109.0–43,210.6). A reference range of 26,182 to 57,677 μM was calculated (mean ± 1.96 SD, after log transformation). To our knowledge, no reference ranges have been reported for lysine concentration in serum or plasma hydrolysate, as this is not a physiologically relevant measure. The lysine concentration correlated with the albumin concentration (r2 = 0.24), demonstrating that variation in protein concentration and variation in sample preparation contribute to the variation in lysine concentration.
A mean normalized CML concentration of 69 μM/M lysine (range: 19–453; SD 34) was reported in the BRHS. The median was 65 μM/M lysine (IQR: 54–76). A reference range of 34 to 123 μM/M was estimated from the log-transformed data (mean ± 1.96 SD).
The median normalized CML concentration of 65 μM/M lysine measured is similar to the median of 68 μM/M lysine, previously reported for 18 sedentary individuals26 (Table 4). It is also broadly similar to the medians of 51 μM/M lysine and 80 μM/M lysine reported for 70 individuals in the top and bottom tertile of AGE score, respectively.29
The mean of 69 μM/M lysine observed in the BRHS samples was far lower than the medians reported by Hull et al.: 132 to 140 μM/M lysine4 (Table 4). They measured lysine concentration using IDMS; however, they did not report the raw CML or lysine concentrations observed. Their range is based on repeated analysis of a pooled sample from 10 healthy volunteers analysed under different preanalytical conditions, none of which were found to significantly affect the CML concentration.4 It is also far lower than the mean of 158 μM/M lysine reported in 21 healthy children25 and lower than the mean of 83 μM/M lysine reported in 44 ex-smokers30 and of 93 μM/M lysine reported in 169 individuals without diabetes.31
The median normalized CML concentration is about double that reported by Hanssen et al.6: 34 μM/M lysine (IQR: 29 to 39) based on 558 individuals without prior CVD. They derivatized CML with 1-butanol: HCl, as an alternative to using NFPA as an ion-pairing agent, before IDMS analysis. The sample was split after hydrolysis and the lysine concentration was measured separately, again using IDMS. Hanssen et al. did not report the raw CML or lysine concentrations observed. Perhaps incomplete derivatization or differences in sample population account for the difference in ranges observed between our study and theirs. It is also approximately double that reported for over 200 individuals in the top tertile for diastolic function28 and more than double that reported in five healthy individuals.34
Conclusion
CML is a challenging AGE to measure, as it is being detected in the presence of other amino acids (including lysine) present at 1000 times higher concentrations in the sample. However, the results suggest that this is a robust method for the quantification of CML (normalized to lysine), despite reducing the hands-on time (and reagent volumes used) for sample preparation substantially. There are no gold standard methods available for comparison at present, only ELISAs and other IDMS methods. Our method appears to compare relatively well to other IDMS-based methods. At present, we recommend this method for research use only. Further work is required in clinical research to determine whether CML is indeed a useful biomarker. The increased throughput provided by this sample preparation method will aid this endeavour. If CML is a useful biomarker, then it will be appropriate to evaluate whether the improved mass accuracy of an ultra high-resolution instrument is better for sensitivity and selectivity than a triple quadrupole instrument for this analyte.
Acknowledgements
We thank Erin Manson, Ana Montiero and Sara-Jane Duffus (University of Glasgow) for technical assistance.
Declaration of conflicting interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: NJR, SW, KB, GW and PW report no conflicts of interest. NS has consulted for AstraZeneca, Bristol-Myers Squibb, Amgen, Sanofi and Boehringer Ingelheim.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: CML analysis: the BRHS, Glasgow Polyomics Facility and NR were supported by the Dunhill Medical Trust (R419/0515), BHF Programme Grant (RG/13/1630528), the Welcome Trust (grant nos. 097821/Z/11/Z and 105614/Z/14/Z) and the Glasgow Molecular Pathology NODE (funded by the MRC and the EPSRC (MR/N005813/1, project code 69042/1)), respectively.
Ethical approval
Ethical approval for the BRHS was obtained from the LRECs of the 24 towns where participants were recruited.
Guarantor
NJR.
Contributorship
PW and NS researched literature and conceived the study. GW gained ethical approval and funding and contributed to patient recruitment and statistical analysis. NJR, SW and KB were involved in method development. NJR wrote the first draft of the manuscript. All authors reviewed and edited the manuscript for important intellectual content and approved the final version of the manuscript.
References
- 1.Teerlink T, Barto R, Ten Brink HJ, et al. Measurement of N-(carboxymethyl)lysine and N-(carboxyethyl)lysine in human plasma protein by stable-isotope-dilution tandem mass spectrometry. Clin Chem 2004; 50: 1222–1228. [DOI] [PubMed] [Google Scholar]
- 2.Delgado-Andrade C. Carboxymethyl-lysine: thirty years of investigation in the field of AGE formation. Food Funct 2016; 7: 46–57. [DOI] [PubMed] [Google Scholar]
- 3.Ni JYuan X, Gu J, et al. Plasma protein pentosidine and carboxymethyllysine, biomarkers for age-related macular degeneration. Mol Cell Proteomics 2009; 8: 1921–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hull GLJ, Woodside JV, Ames JM, et al. Validation study to compare effects of processing protocols on measured Nε-(carboxymethyl) lysine and Nε-(carboxyethyl) lysine in blood. J Clin Biochem Nutr 2013; 53: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieuw-A-Fa MLMVan Hinsbergh V, Teerlink T, et al. Increased levels of N -(carboxymethyl)lysine and N -(carboxyethyl)lysine in type 1 diabetic patients with impaired renal function: correlation with markers of endothelial dysfunction. Nephrol Dial Transplant 2004; 19: 631–636. [DOI] [PubMed] [Google Scholar]
- 6.Hanssen NMJEngelen L, Ferreira I, et al. Plasma levels of advanced glycation endproducts Nϵ -(carboxymethyl)lysine, Nϵ -(carboxyethyl)lysine, and pentosidine are not independently associated with cardiovascular disease in individuals with or without type 2 diabetes: The Hoorn and CODAM Studies. J Clin Endocrinol Metab 2013; 98: E1369–E1373. [DOI] [PubMed] [Google Scholar]
- 7.Adaway JE, Keevil BG, Owen LJ. Liquid chromatography tandem mass spectrometry in the clinical laboratory. Ann Clin Biochem 2015; 52: 18–38. [DOI] [PubMed] [Google Scholar]
- 8.Honour JW. Development and validation of a quantitative assay based on tandem mass spectrometry. Ann Clin Biochem 2011; 48: 97–111. [DOI] [PubMed] [Google Scholar]
- 9.Clarke W, Rhea JM, Molinaro R. Challenges in implementing clinical liquid chromatography-tandem mass spectrometry methods – seeing the light at the end of the tunnel. J Mass Spectrom 2013; 48: 755–767. [DOI] [PubMed] [Google Scholar]
- 10.Rochat B. From targeted quantification to untargeted metabolomics: why LC-high-resolution-MS will become a key instrument in clinical labs. TrAC Trends Anal Chem 2016; 84: 151–164. [Google Scholar]
- 11.Grund B, Marvin L, Rochat B. Quantitative performance of a quadrupole-Orbitrap-MS in targeted LC–MS determinations of small molecules. J Pharm Biomed Anal 2016; 124: 48–56. [DOI] [PubMed] [Google Scholar]
- 12.Glauser GGrund B, Gassner A, et al. Validation of the mass-extraction-window for quantitative methods using liquid chromatography high resolution mass spectrometry. Anal Chem 2016; 88: 3264–3271. [DOI] [PubMed] [Google Scholar]
- 13.Eliuk S, Makarov A. Evolution of Orbitrap mass spectrometry instrumentation. Annu Rev Anal Chem (Palo Alto Calif) 2015; 8: 61–80. [DOI] [PubMed] [Google Scholar]
- 14.Gertsman I, Gangoiti JA, Barshop BA. Validation of a dual LC–HRMS platform for clinical metabolic diagnosis in serum, bridging quantitative analysis and untargeted metabolomics. Metabolomics 2014; 10: 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemkov T, D’Alessandro A, Hansen KC. Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole Orbitrap mass spectrometry. Amino Acids 2015; 47: 2345–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer MR, Maurer HH. Current applications of high-resolution mass spectrometry in drug metabolism studies. Anal Bioanal Chem 2012; 403: 1221–1231. [DOI] [PubMed] [Google Scholar]
- 17.Wannamethee SGWelsh P, Papacosta O, et al. Circulating soluble receptor for advanced glycation end product: cross-sectional associations with cardiac markers and subclinical vascular disease in older men with and without diabetes. Atherosclerosis 2017; 264: 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennon LTRamsay S, Papacosta O, et al. Cohort profile update: the British Regional Heart Study 1978–2014: 35 years follow-up of cardiovascular disease and ageing. Int J Epidemiol 2015; 44: 826–826g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeppsson J-OKobold U, Barr J et al. Approved IFCC reference method for the measurement of HbA1c in human blood. Clin Chem Lab Med 2002; 40: 78–89. [DOI] [PubMed] [Google Scholar]
- 20.ICH Expert Working Group. ICH Harmonised Tripartite Guideline. Validation of analytical procedures: text and methodology Q2(R1). Federal Register 1997; 62: 1–2. [Google Scholar]
- 21.Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 2008; 29 Suppl 1: S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 22.Center for Drug Evaluation and Research. Food and Drug Administration. Bioanalytical Method Validation: Guidance for Industry Report, May 2018. https://www.fda.gov/downloads/Drugs/Guidance/ucm070107.pdf (accessed 21 February 2019). [Google Scholar]
- 23.Dubbelman A-CCuyckens F, Dillen L, et al. Mass spectrometric recommendations for Quan/Qual analysis using liquid-chromatography coupled to quadrupole time-of-flight mass spectrometry. Anal Chim Acta 2018; 1020: 62–75. [DOI] [PubMed] [Google Scholar]
- 24.Gaens K HJFerreira I, Van de Warrenburg MP, et al. Protein-bound plasma Nε-(carboxymethyl)lysine is inversely associated with central obesity and inflammation and significantly explain a part of the central obesity-related increase in inflammation: The Hoorn and CODAM studies. Arterioscler Thromb Vasc Biol 2015; 35(12): 2707–2713. doi:10.1161/ATVBAHA.115.306106 [DOI] [PubMed] [Google Scholar]
- 25.Anwar AAbruzzo P, Pasha S, et al. Advanced glycation endproducts, dityrosine and arginine transporter dysfunction in autism – a source of biomarkers for clinical diagnosis. Mol Autism 2018; 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maessen MFHSchalkwijk CG, Verheggen RJHM, et al. A comparison of dicarbonyl stress and advanced glycation endproducts in lifelong endurance athletes vs. sedentary controls. J Sci Med Sport 2017; 20: 921–926. [DOI] [PubMed] [Google Scholar]
- 27.De Courten Bde Courten MP, Soldatoa G, et al. Diet low in advanced glycation end products increases insulin sensitivity in healthy overweight individuals: a double-blind, randomized, crossover trial 1-3. Am J Clin Nutr 2016; 103: 1426–1459. [DOI] [PubMed] [Google Scholar]
- 28.Linssen PBCHenry RM, Schalkwijk CG, et al. Serum advanced glycation endproducts are associated with left ventricular dysfunction in normal glucose metabolism but not in type 2 diabetes: The Hoorn Study. Diabetes Vasc Dis. Res 2016; 13(4): 278–285. doi:10.1177/1479164116640680 [DOI] [PubMed]
- 29.Hanssen NMJBeulens JW, van Dieren S, et al. Plasma advanced glycation end products are associated with incident cardiovascular events in individuals with type 2 diabetes: a case-cohort study with a median follow-up of 10 years (EPIC-NL). Diabetes 2015; 64: 257–265. [DOI] [PubMed] [Google Scholar]
- 30.Gopal PReynaert NL, Scheijen JL, et al. Plasma advanced glycation end-products and skin autofluorescence are increased in COPD. Eur Respir J 2014; 43: 430–438. [DOI] [PubMed] [Google Scholar]
- 31.Ga Van Eupen MSchram MT, Colhoun HM, et al. Plasma levels of advanced glycation endproducts are associated with type 1 diabetes and coronary artery calcification. Cardiovasc Diabetol 2013; 12: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabbani NAdaikalakoteswari A, Rossing K, et al. Effect of Irbesartan treatment on plasma and urinary markers of protein damage in patients with type 2 diabetes and microalbuminuria. Amino Acids 2012; 42(5): 1627–1639. doi:10.1007/s00726-011-0857-7 [DOI] [PubMed]
- 33.Engelen LPersson F, Ferreira I, et al. Irbesartan treatment does not influence plasma levels of the advanced glycation end products N e (1-carboxymethyl)lysine and N e (1-carboxyethyl)lysine in patients with type 2 diabetes and microalbuminuria. A randomized controlled trial. Nephrol Dial Transpl 2011; 26: 3573–3577. [DOI] [PubMed] [Google Scholar]
- 34.Thornalley PJBattah S, Ahmed N, et al. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem J 2003; 375: 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]