Abstract
Germ-line (micronuclear) genes in hypotrichous ciliates are interrupted by numerous, short, noncoding, AT-rich segments called internal eliminated segments, or IESs. IESs divide a gene into macronuclear destined segments, or MDSs. IESs are excised from micronuclear genes, and the MDSs are spliced when a micronuclear genome is processed into a macronuclear genome after cell mating. In the micronuclear version of the actin I gene intramolecular recombination between IESs during evolution has put MDSs into a scrambled disorder in some but not all hypotrichs. Studies using rDNA sequences to define phylogenetic relationships among eight hypotrichs suggests that evolution of the micronuclear actin I gene proceeds by successive addition of IESs in earlier diverging species, without MDS scrambling. Continued addition of IESs and recombination among IESs in later diverging species produced actin I genes with scrambled MDSs. Subsequent to MDS scrambling, additional IESs were inserted into the more recently evolved species. Thus, IES insertions and gene scrambling occur in a progressive manner during species evolution to produce micronuclear actin I genes of increasing structural complexity.
Genes in micronuclear (germ-line) DNA of hypotrichous ciliates are interrupted by multiple, short, AT-rich, noncoding segments called internal eliminated segments, or IESs. IESs divide a gene into macronuclear destined segments, or MDSs. In some genes intramolecular recombination between IESs during evolution has disordered MDSs—i.e., MDSs have become scrambled. During development of a micronucleus into a new macronucleus after cell mating, IESs are spliced out of genes and MDSs are rearranged and ligated in the orthodox order to produce a transcribable gene (1). After MDS ligation the genes are excised from chromosomal DNA as individual, short DNA molecules, and all of the remaining DNA (spacer DNA), which accounts for ≈95% of the nucleotide sequence complexity in the micronuclear genome, is destroyed. Generally, one gene is present per molecule, but a small percentage of molecules may contain two genes (2) or even three genes (D.M.P., J. D. Prescott, and R. M. Prescott, unpublished observation).
The significance of IESs in micronuclear genes is not understood, although it has been suggested that they facilitate evolution of new genes; recombination among IESs, which results in shuffling of MDSs, could conceivably create new, intergenic MDS combinations. Thus, the presence of scrambled MDSs in contemporary hypotrichs is a manifestation of intragenic MDS shuffling caught in evolutionary time. In addition, IES position within a gene can change (shift), which alters the sequence composition and size of MDSs in that gene (3), increasing further the versatility of MDS shuffling. At the very least, IES shifting and MDS scrambling by IES recombination, as reaffirmed in the data reported here, attest to an extraordinarily dynamic nature of the germ-line genome in hypotrichs over evolutionary time.
This paper discusses the origin and evolution of IESs and intragenic MDS shuffling, or gene scrambling. We have determined the MDS structure of the actin I gene in the micronuclear genome of five hypotrichs. We have compared them among themselves and with the MDS structure of the actin I gene previously published for three other hypotrichs. These comparisons imply a pathway for the origin and evolution of IESs and for MDS scrambling—a pathway that also coincides closely with the evolutionary pathway for these organisms as defined by the nucleotide sequences of rDNAs.
Materials and Methods
Organisms.
Oxytricha sp. (Misty) was newly isolated from a pond in Sarasota, Florida. Stylonychia lemnae was isolated from Teller Lake in Boulder, Colorado. Stylonychia pustulata was isolated from the Roaring Fork River, Aspen, Colorado. All three were grown on Chlorogonium as described (4). Engelmanniella mobilis was a gift from W. Foissner (Universität Salzburg, Salzburg, Austria). It was cultured on a mixture of unidentified bacteria in rice grain cultures. Urostyla grandis was isolated from a pond on the campus of the University of Colorado, Boulder. It was grown on Tetrahymena thermophila.
DNA Preparation.
Macronuclei and micronuclei were isolated from 10–20 ml of packed cells as described (4). DNA was purified by phenol/chloroform extractions. Micronuclear DNA was filtered through a Sephacryl 1000s column (Amersham Pharmacia), which removed most of the contaminating macronuclear DNA. After sephacryl 1000s filtration, micronuclear DNA from U. grandis was treated with 2 units of Bal31 exonuclease (New England Biolabs) in 50 μl of buffer for one hour at 30°C to eliminate any residual macronuclear DNA (5). Sephacryl 1000s-treated micronuclear DNA of E. mobilis was freed of traces of macronuclear DNA contamination by gel electrophoresis, taking advantage of the much more rapid migration of macronuclear DNA. For PCR of the micronuclear actin I gene of Oxytricha sp. (Misty), S. lemnae, and S. pustulata advantage was taken of the inversion of MDS 2 and the downstream location of MDS 1 to design primer pairs that yielded a PCR product only with micronuclear DNA (see Figs. 1 and 3 for MDS locations).
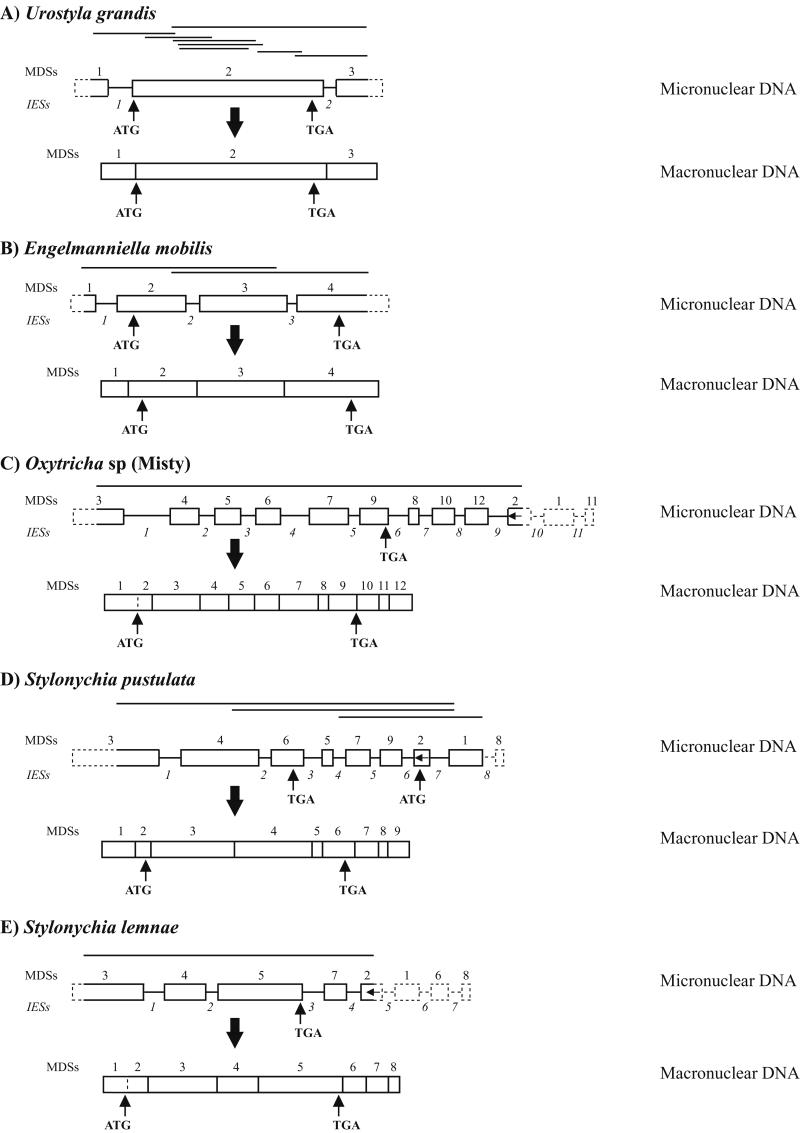
Figure 1.
Structure of micronuclear and macronuclear actin I genes in U. grandis (A), E. mobilis (B), Oxytricha sp. (Misty) (C), S. pustulata (D), and S. lemnae (E). Micronuclear DNA PCR products are indicated by lines above each micronuclear gene. MDSs are blocks and IESs are lines between blocks. Unsequenced regions are indicated by dashed blocks and dashed lines.
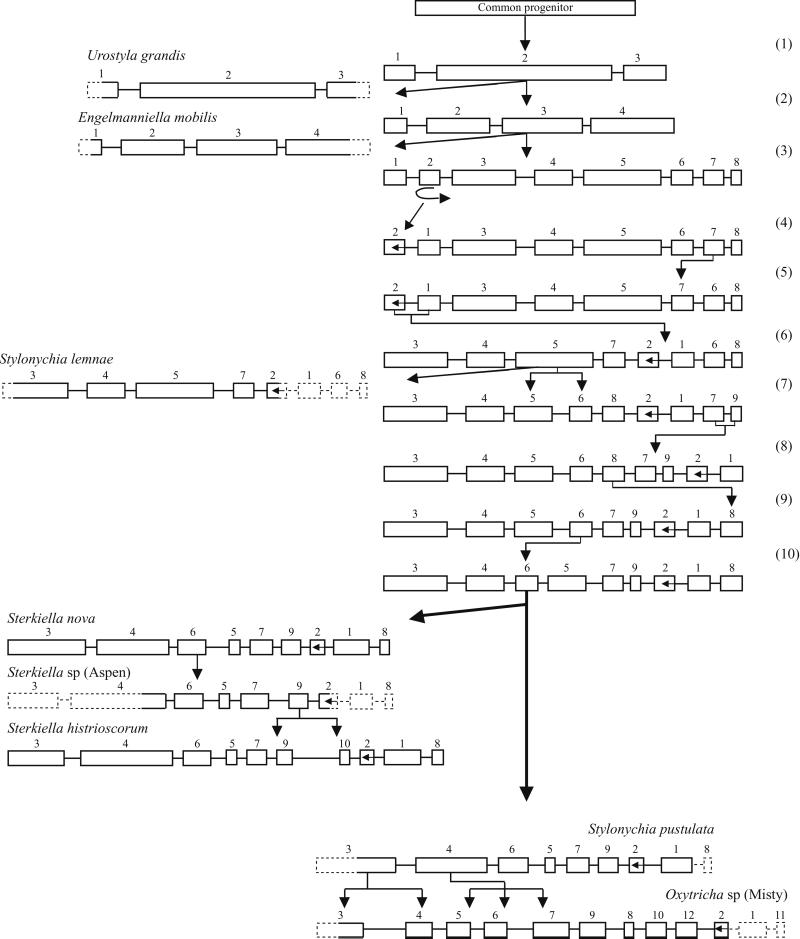
Figure 3.
Evolutionary scheme for the insertion of IESs and the scrambling of MDSs in the micronuclear actin I gene in eight hypotrichs. MDSs are blocks, and IESs are lines between blocks. Regions not sequenced in micronuclear DNA are dashed blocks and lines.
Macronuclear Actin I Sequences.
The actin I sequence for U. grandis was obtained from the GenBank database (accession no. AF188160). Descriptions of the actin I sequence for E. mobilis, S. lemnae, S. pustulata, and Oxytricha sp. (Misty) and completion of the macronuclear actin I sequence for Sterkiella sp. (Aspen) [formerly, Oxytricha sp. (Aspen) (6)] will be published elsewhere and have been deposited under accession nos. AY044837, AY046534, AY044838, AY044839, and U63566, respectively.
Micronuclear Actin I Sequences.
These were determined from PCR products generated with primers designed from macronuclear sequences. PCR products were cloned into pGEM-T Easy Vector (Promega) and sequenced by the departmental sequencing facility, using M13F and M13R sequencing primers and species-specific internal primers.
Results
Phylogenetic Trees.
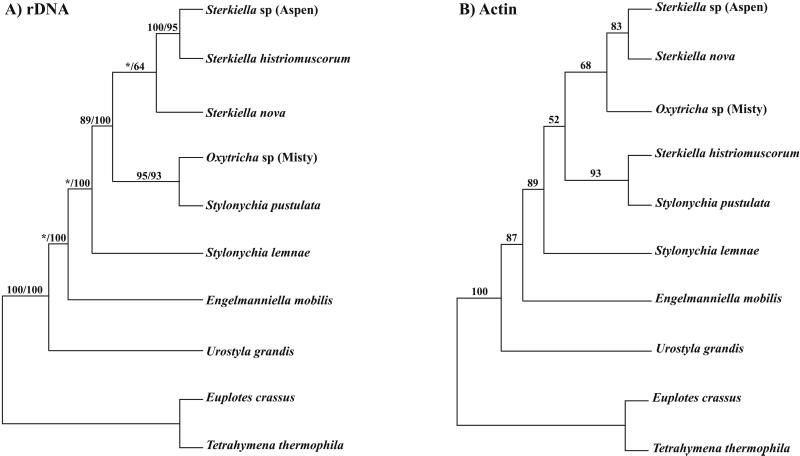
Parsimony analysis on both the rDNA and the amino acid sequence of the actin I gene was applied by using an exhaustive search criteria in the program PAUP 4.0 beta 8 (7). Support for nodes on parsimony trees was established by using bootstrap resampling (% of 1,000 replicates). The quartet puzzling method was applied to only the rDNA data set. This method employs maximum-likelihood tree reconstruction to all possible quartets that can be formed from all sequences that serve as starting points to reconstruct a set of optimal n-taxon trees (8). This method has been shown to be equally or better able to reconstruct the true tree than neighbor-joining methods (8). The values (Fig. 2) represent the percentage of times that a particular cluster was found among the 1,000 intermediate trees [quartet puzzling steps (QPS) values].
Figure 2.
Maximum parsimony trees based on the sequence of rDNA (A; ssrDNA–ITSI–5.8S rDNA–ITS2–1400 bp of the large subunit lsrDNA; the first number is the bootstrap value and the second is for quartet puzzling) and the sequence of amino acids in the actin I gene (B).
U. grandis.
Eight overlapping PCR products of the micronuclear actin I gene were obtained with primers designed from the macronuclear actin I sequence by using a single preparation of Bal31-treated micronuclear DNA (Fig. 1A). The structure of the micronuclear version of the actin I gene was assembled from these sequences as shown in Fig. 1A. Two IESs are present. IES 1 (102 bp; 91% AT) is located in the 5′ leader. IES 2 (28 bp; 96% AT) is located in the 3′ trailer. The two IESs divide the micronuclear precursor into three MDSs. MDS 1 contains the 5′ leader. The sequence of the first 62 bp of MDS 1 was not obtained for the micronuclear gene because a PCR product could not be generated from this AT-rich region (80% AT). The 3′ end of MDS 1, where it joins IES 1, contains the sequence TTACTTAT. The 5′ end of MDS 2 begins with TTACTTAT. The AT at the 3′ end of this repeat in MDS 2 is followed by a G in MDS 2, forming the ATG start codon.
MDS 2 contains the entire actin I coding region (except the AT of the ATG start) and ends in the 3′ trailer 19 bp downstream of the TGA stop codon. MDS 3 contains the rest of the 3′ trailer. The sequence of the last 66 bp of the 3′ trailer in the micronuclear precursor could not be determined by PCR because of its AT richness (77% AT). It is unlikely that the first 62 bp in MDS 1 and/or the last 66 bp in MDS 3 are interrupted by an additional IES(s). Among the 13 micronuclear genes sequenced in their entirety in the Sterkiella/Stylonychia group no IESs have been found within the several hundred base pairs at the ends of the micronuclear precursors. The ORF consists of 1131 bp, including the TGA, encoding a putative protein of 376 aa. The nucleotide sequence of the micronuclear ORF differs from the macronuclear ORF sequence in two positions; neither results in a difference in amino acid sequence.
E. mobilis.
Two overlapping PCR products of the micronuclear actin I gene were generated with primers designed from the macronuclear sequence by using gel-purified micronuclear DNA. The assembled micronuclear sequence (Fig. 1B) lacks 40 bp of MDS 1 at the 5′ end and 97 bp of MDS 4 at the 3′ end. Three IESs are present: IES 1 (85 bp; 81% AT) is in the 5′ leader, and IES 2 (37 bp; 86% AT) and IES 3 (12 bp; 100% AT) are in the ORF. The three IESs are flanked by repeat sequences of ATA, GAAAAGA, and AATGC, respectively. The four MDSs are in the orthodox order. The ORF (1131 bp) extends from MDS 2 to MDS 4 and encodes a putative protein of 376 aa. The micronuclear ORF differs from the macronuclear ORF in one nucleotide, which does not change an amino acid.
Oxytricha sp. (Misty).
We considered that the micronuclear actin I gene in this organism might have a scrambled pattern like Sterkiella nova (formerly, Oxytricha nova), Sterkiella histriomuscorum (formerly, Oxytricha trifallax), and Sterkiella sp. (Aspen). MDS 2 in the micronuclear actin I gene in these three organisms is inverted (see Fig. 3). Primers were designed from the macronuclear sequence of Oxytricha sp. (Misty) for putative MDSs 3 and 2. These yielded a PCR product containing ≈90% of the micronuclear actin I gene (Fig. 1C). It contains nine IESs, which divides the PCR sequence into eight complete MDSs and two partial MDSs, MDSs 3 and 2. Two hundred and fifteen bp at the 5′ end of MDS 3 are missing from the PCR product. Two hundred and twenty-five bp corresponding to the 5′ part of MDS 2 (inverted) and all of MDSs 1 and 11 are missing from the right end of the micronuclear gene. The presence of MDS 11 (14 bp) is defined by the macronuclear segment that is missing from the PCR sequence between MDSs 10 and 12 in micronuclear DNA in Fig. 1C. We postulate the existence of an IES of undetermined size (IES 10 in Fig. 1C) that creates MDS 1 and 2, as it does in the scrambled actin I genes in S. nova, S. histriomuscorum, and Sterkiella sp. (Aspen). The position of MDS 11 has not been established, but is assumed to follow MDS 1. It seems unlikely that any additional IESs are present, although MDS 1 could be split by an IES. In any case, ≈90% of the micronuclear gene is accounted for by the PCR product, which allows a detailed comparison with the actin I gene structures of the other hypotrichs. With these assumptions about MDSs 1 and 11 and IES 10 and 11, the micronuclear actin I gene in Oxytricha sp. (Misty) consists of 11 IESs and 12 MDSs. MDS 1 contains the entire 5′ noncoding leader. The stop codon (TGA) is at the end of MDS 9, and MDSs 10, 11, and 12 contain the 3′ noncoding trailer.
Stylonychia pustulata.
Pairs of primers were designed from the macronuclear actin I sequence based on the assumption that the micronuclear version would be scrambled in a pattern similar to the pattern in Sterkiella nova, S. histriomuscorum, and Sterkiella sp. (Aspen) (Fig. 3). The primers encompassed the region from putative MDS 3 to MDS 1, defining the micronuclear gene structure in Fig. 1D. The pattern is the same as in S. nova and Sterkiella sp. (Aspen), but the PCR product lacks 255 bp at the 5′ end of MDS 3 and all of MDS 8 (8 bp). Thus, the micronuclear gene of S. pustulata consists of nine MDSs and eight IESs, with MDS 2 inverted. The ATG is in MDS 2, and the TGA is in MDS 6.
Stylonychia lemnae.
Again, it was assumed that the micronuclear actin I gene of S. lemnae would have a scrambled pattern similar to S. nova, S. histriomuscorum, and Sterkiella sp. (Aspen). A pair of primers designed to span the region from MDS 3 to inverted MDS 2 yielded a micronuclear-specific product (Fig. 1E). The sequence of the PCR product defines the MDS pattern 3-4-5-7-2. The existence of MDSs 1, 6, and 8 can be deduced from the sequence of the micronuclear PCR product and the macronuclear sequence, but their positions are assumed based on the micronuclear actin I gene structure in S. nova, S. histriomuscorum, and Sterkiella sp. (Aspen). Fifty-two bp are missing from the 5′ end of MDS 3 in the micronuclear PCR product (the 5′ end of MDS 3 is defined in the macronuclear molecule because the immediately adjoining 3′ end of MDS 2 [inverted] is known from the sequence of the micronuclear PCR product). The 5′ end of MDS 2 is undefined. The sequence of MDS 6 is defined as the segment between MDSs 5 and 7 in macronuclear DNA. The sequence of MDS 8 is defined as the region in the macronuclear molecule from the 3′ end of MDS 7 to the telomere. In short, the micronuclear actin I gene in S. lemnae consists of eight MDSs and seven IESs. It is possible, but unlikely, that MDSs 1, 6, and/or 8, missing from the micronuclear PCR product, are split by an additional IES(s). Similarly, it is possible, but unlikely, that the missing portions of MDSs 2 and 3 contain an IES(s).
Discussion
With the sequencing of the micronuclear actin I gene in five new organisms the total number in which most of the structure of the micronuclear gene is known is eight. The complete sequences of the micronuclear actin I genes in S. nova and S. histriomuscorum, and a partial sequence for Sterkiella sp. (Aspen) were published earlier (3, 9). Our purpose is to compare the arrangements of IESs and MDSs in the micronuclear actin I gene in these eight organisms to make deductions about the origin and evolution of IESs and the evolution of MDS scrambling.
As a starting point, maximum parsimony trees based on rDNA sequences (E.A.H., K.M.M., D.J.H., and D.M.P., unpublished observations) and amino acid sequences of actin I for the eight organisms were constructed (Fig. 2 A and B). The rDNA sequences extend from the beginning of the small subunit (ss)-rDNA, through ITS1, 5.8S rDNA, ITS2, and 1400 bases of the large subunit (ls)-rDNA. The two trees define identical phylogenetic relationships for the early divergence of U. grandis, followed by divergence of E. mobilis, S. lemnae, S. pustulata, and Sterkiella sp. (Aspen). They differ with respect to the later divergence of S. nova, Oxytricha sp. (Misty), and S. histriomuscorum. Trees based on rDNA are generally considered to define phylogenetic relationships more accurately than trees based on actin I.
Comparative analysis of the eight micronuclear actin I genes has a significant bearing on two crucial issues: (i) the insertion of IESs into micronuclear DNA during hypotrich evolution; and (ii) the evolutionary timing and mechanism of gene scrambling in hypotrichs.
The micronuclear gene structures for the eight organisms are arranged in Fig. 3 according to the phylogenetic tree defined by their rDNA sequences. The tree is rooted by a hypothetical progenitor that lacks IESs in its actin I gene. In the earliest branch U. grandis has acquired two IESs, and the three MDSs are nonscrambled [step (1) in Fig. 3]. U. grandis is followed by E. mobilis [step (2) in Fig. 3], whose actin I gene contains three IESs and four MDSs. None of the three IESs in E. mobilis match either of the two IESs in U. grandis in position, size, or sequence. Nevertheless, one or even two of the IESs in E. mobilis may have derived from the same IES insertion event as one or the two IESs in U. grandis because IESs shift position along DNA by stepwise mutations, accumulate mutations at a high rate, and change in length during evolution. The closest correspondence in position is between IES 1 in the two organisms; IES 1 interrupts the 5′ leader six bases upstream of the ATG in U. grandis and 37 bp upstream of the ATG in E. mobilis; this IES is 102 bp and 83 bp in the two organisms, respectively, and no sequence identity beyond what is expected for AT-rich DNA (91 and 81% AT, respectively) is observed between these two IESs. Nevertheless, they may represent a single original IES that has shifted position along the DNA and accumulated many mutations. It is also possible that the IESs in U. grandis and E. mobilis were inserted as separate, independent events after divergence of the two evolutionary lines.
Subsequent to the branching of E. mobilis, four more IESs were inserted to give a total of eight MDSs, one of which is MDS 2 [step (3) in Fig. 3]. In this theoretical intermediate, MDS 2 was inverted by recombination between the two IESs that flank MDS 2 [step (4) in Fig. 3]. Next, two MDS translocation events produced the pattern seen in S. lemnae [steps (5) and (6) in Fig. 3]. This pattern was further elaborated by insertion of one more IES [step (7) in Fig. 3] and by three translocations of MDSs [steps (8), (9), and (10) in Fig. 3], producing the pattern 3-4-6-5-7-9-2-1-8 in a common, theoretical progenitor of S. pustulata and Oxytricha sp. (Misty) on the right branch in Fig. 3 and of the three Sterkiella organisms on the left branch in Fig. 3. The pattern of the theoretical progenitor is maintained in S. pustulata, S. nova, and Sterkiella sp. (Aspen). Insertion of three more IESs into this pattern, creating three more nonscrambled MDSs, produced the pattern in Oxytricha sp. (Misty) (right branch at the bottom of Fig. 3). Insertion of one IES in the pattern shared by S. nova and Sterkiella sp. (Aspen) yielded the additional, nonscrambled MDS in S. histriomuscorum (left branch at the bottom of Fig. 3).
The scheme in Fig. 3 describing evolution of actin I gene structure in eight organisms could undoubtedly be refined and elaborated by inclusion of additional hypotrichs perhaps even detecting one or more of the theoretical intermediates in Fig. 3. However, the basic tenets of this IES/MDS evolutionary scheme are validated by the close match of the scheme with the evolutionary tree defined by rDNA sequences (Fig. 2A). We are led to conclude that IESs are added successively in the evolution of hypotrich genes. Whether they are ever deleted during evolution is uncertain; there are no observations that suggest IES deletion. Accumulation of IESs in a gene creates the possibility for intragenic recombination among IESs, resulting in the scrambling of MDSs. What might propel such recombinations isn't known. Recombination has not always occurred; the βTP gene in S. histriomuscorum contains six IESs, but the seven MDSs have remained in the orthodox order (10). In parallel with recombinations that scramble MDSs, new IESs continue to be inserted into existing MDSs, increasing the number of MDSs—e.g., Oxytricha sp. (Misty).
Although we believe that we have begun to unravel the origin and evolution of IESs and MDS scrambling, we do not understand the significance of these phenomena in the evolution and genetic function of the organisms. Possibly, IESs create opportunities to shuffle segments (MDSs) intergenically by recombination, thereby facilitating evolution of new genes. MDS scrambling may be no more than a manifestation of such MDS shuffling in the evolution of new genes.
Acknowledgments
We are grateful to Gayle Prescott for typing several versions of the manuscript. This work is supported by National Institute of General Medical Sciences Research Grant R01 GM 56161 and National Science Foundation Research Grant MCB-9974680 (to D.M.P.). D.J.H. was partially supported by the Hughes Undergraduate Biological Science Education Initiative, the University of Colorado Undergraduate Research Opportunities Program, and the University of Colorado Cancer Center. K.E.O. was partially supported by the University of Colorado Undergraduate Research Opportunities Program. K.M.M. was supported by start-up funds from the University of Waterloo.
Abbreviations
- IES
internal eliminated segment
- MDS
macronuclear destined segment
Footnotes
References
- 1.Prescott D M. Nat Rev Genet. 2000;1:191–198. doi: 10.1038/35042057. [DOI] [PubMed] [Google Scholar]
- 2.Seegmiller A, Williams K R, Herrick G. Dev Genet. 1997;20:348–357. doi: 10.1002/(SICI)1520-6408(1997)20:4<348::AID-DVG6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 3.DuBois M, Prescott D M. Proc Natl Acad Sci USA. 1995;92:3888–3892. doi: 10.1073/pnas.92.9.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prescott D M, Greslin A F. Dev Genet. 1992;13:66–74. doi: 10.1002/dvg.1020130111. [DOI] [PubMed] [Google Scholar]
- 5.Mitcham J L, Lynn A J, Prescott D M. Genes Dev. 1992;6:788–800. doi: 10.1101/gad.6.5.788. [DOI] [PubMed] [Google Scholar]
- 6.Foissner W, Berger H. Acta Protozool. 1999;38:215–248. [Google Scholar]
- 7.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 8.Strimmer K, von Haeseler A. Mol Cell Biol. 1996;13:964–969. [Google Scholar]
- 9.DuBois M L, Prescott D M. Mol Cell Biol. 1997;17:326–337. doi: 10.1128/mcb.17.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prescott D M, DuBois M L. J Eukaryotic Microbiol. 1996;43:432–441. doi: 10.1111/j.1550-7408.1996.tb04502.x. [DOI] [PubMed] [Google Scholar]