Abstract
Asparagine-linked (N-linked) glycosylation is one of the most common protein modification reactions in eukaryotic cells, occurring upon the majority of proteins that enter the secretory pathway. X-ray crystal structures of the single subunit OSTs from eubacterial and archaebacterial organisms revealed the location of donor and acceptor substrate binding sites and provided the basis for a catalytic mechanism. Cryoelectron microscopy structures of the octameric yeast OST provided substantial insight into the organization and assembly of the multisubunit oligosaccharyltransferases. Furthermore, the cryoelectron microscopy structure of a complex consisting of a mammalian OST complex, the protein translocation channel and a translating ribosome revealed new insight into the mechanism of cotranslational glycosylation.
Keywords: cotranslational glycosylation, lipid-linked oligosaccharide, N-glycosylation, oligosaccharyltransferase, OST structure
Introduction
Oligosaccharyltransferase (OST) catalyzes N-linked glycosylation of proteins by transferring a preassembled oligosaccharide from a lipid-linked oligosaccharide (LLO) donor onto consensus acceptor sites (sequons, (NXT/S/C; X≠P)) in cell surface proteins in bacterial organisms or in nascent polypeptides within the ER lumen of eukaryotic organisms. N-glycosylation of proteins is a critical protein modification reaction in eukaryotes, as N-linked glycans are important for glycoprotein folding in the ER, intracellular traffic, stability and function, cell–cell communication and normal development (Helenius and Aebi 2004; Hebert and Molinari 2007). Defects in N-linked glycosylation, including those caused by mutations in OST subunit genes, cause a family of inherited diseases known as congenital disorders of glycosylation (CDG, as reviewed by Freeze (2013); Hennet and Cabalzar (2015). Current understanding of the OST has significantly increased during the past decade due to information that has been obtained by X-ray crystallography and single particle cryoelectron microscopy analysis of bacterial and eukaryotic oligosaccharyltransferases.
Structures of PglB and AglB illuminate the catalytic mechanism of the OST
The eubacterial OST (PglB) and archaebacterial OST (AglB) and the homologous catalytic subunits of eukaryotic OST complexes (STT3 proteins) are comprised of 13 transmembrane (TM) spans followed by a C-terminal extracellular or ER-lumenal domain. The X-ray crystal structure of the extracellular C-terminal domain of an archaebacterial OST (A. fulgidus AglB) (Igura et al. 2008) provided the first information about the location of conserved sequence motifs in the OST (Table I). Analysis of the structure of the C-terminal domain of AglB led to the identification of the DK motif which is located near the universally conserved WWD motif in the AglB structure (Igura et al. 2008). The WWD motif had been shown to be essential for OST activity in the yeast (Yan and Lennarz 2002b) and bacterial model systems (Wacker et al. 2002; Yan and Lennarz 2002b).
Table I.
Conserved sequence motifs involved in substrate recognition and catalysis
| Organism | C. lari | A. fulgidus | S. cerevisiae | H. sapiens | H. sapiens |
|---|---|---|---|---|---|
| Protein | PglBa | AglBa | STT3a | STT3A | STT3B |
| Sequon | D/EXNXT/S | NXT/S | NXT/S | NXT/S/C | NXT/S/C |
| Sequon bindingb | |||||
| –2 position | R331 | L379 | D362 | D363 | D417 |
| −1 position | T54 | G45 | E45 | E47 | E101 |
| WWD motif | |||||
| +2 position | W463 | W550 | W516 | W525 | W604 |
| +2 position | W464 | W551 | W517 | W526 | W605 |
| +2 position | D465 | D552 | D518 | D527 | D606 |
| DK/MI motif | |||||
| +2 position | I572 | K618 | K586 | K595 | K674 |
| Active site | |||||
| Mn++, Asn | D56 | D47 | D47 | D49 | D103 |
| Mn++ | D154 | D161 | D166 | D167 | D221 |
| Mn++ | D156 | H163 | E168 | E169 | E351 |
| Mn++, Asn | E319 | E360 | E350 | E351 | E405 |
| LLO binding | |||||
| PPi | Y196 | W215 | W208 | W209 | W263 |
| PPi | R375 | R426 | R404 | R405 | R459 |
| NAc | Y468 | H555 | Y521 | Y530 | Y609 |
| DGGK loop | D481GGK | N568PFQ | D534NNT | D543NNT | D662NNT |
aResidues and motifs in italics have been shown to be important for oligosaccharyltransferase activity in one or more of the following publications: PglB (Wacker et al. 2002; Lizak et al. 2011; Gerber et al. 2013; Barre et al. 2017; Napiorkowska et al. 2017); AglB (Matsumoto et al. 2013); yeast STT3 (Yan and Lennarz 2002b; Chavan et al. 2003; Li et al. 2005; Igura et al. 2008; Wild et al. 2018).
bThe positions shown (−2, −1 and +2) are relative to the asparagine residue in a glycosylation site. PglB, AglB and STT3 residues involved in binding the asparagine residue in the sequon are listed in the active site section of this table.
A breakthrough in the understanding of the structure and catalytic mechanism of the OST was achieved upon co-crystallization of an intact eubacterial OST (C. lari PglB) with a hexapeptide containing a eubacterial acceptor sequon (D/EXNXT/S) in the peptide-binding site (Lizak et al. 2011). The PglB structure consists of 13 TM spans followed by a lumenal domain that is structurally homologous to the C-terminal domain of AglB. Residues in the WWD motif form hydrogen bonds to the hydroxyamino acid residue at the +2 position of the sequon, while I572 in the MI motif, which is the eubacterial equivalent of the DK motif, contacts the methyl group of threonine in the acceptor peptide (Table I). The MI/DK motif interaction explains the higher affinity of bacterial and eukaryotic OSTs for peptides with threonine at the +2 position relative to serine (Breuer et al. 2001; Chen et al. 2007; Gerber et al. 2013). The side chain of asparagine in the sequon projects through a porthole formed upon assembly of extracellular loop 5 (EL5), a conformationally mobile segment of the OST that is responsible for binding both the acceptor and donor substrates (Lizak et al. 2011; Napiorkowska et al. 2017). An acidic residue cluster (D56, D154, D156 and E319, see Table I) comprised of residues from three extracellular loops (EL1, EL2 and EL5) coordinate the active site divalent metal ion (Mn++) and activate the amide nitrogen of asparagine for formation of the N–C bond to the oligosaccharide (Lizak et al. 2011).
Unlike the smaller archaebacterial or eukaryotic sequons (NXT/S), an acidic residue (D or E) at the −2 position in the eubacterial sequon is necessary for acceptor substrate recognition and catalysis (Kowarik et al. 2006; Chen et al. 2007; Gerber et al. 2013). An arginine residue (R331) in PglB makes contact with the −2 position of the acceptor peptide. Critical roles for the acidic residue cluster and R331 in acceptor peptide-binding and product formation have been experimentally verified (Lizak et al. 2011; Gerber et al. 2013).
An X-ray crystal structure of intact AglB in the absence of an acceptor or donor substrate revealed two noteworthy differences (Matsumoto et al. 2013). Unlike the PglB-acceptor peptide structure (Lizak et al. 2011), where the N-terminal end of EL5 was disordered, EL5 was ordered in the AglB apo-structure (Matsumoto et al. 2013). Secondly, packing of the membrane embedded domain of AglB differed from PglB with respect to the location of TM8 and TM9. The difference in the fold of the membrane embedded region of AglB relative to PglB is not explained by the absence of the acceptor peptide as an AglB structure obtained in the presence of a covalently tethered acceptor peptide showed the same packing of the TM spans (Matsumoto et al. 2017). The presence of the tethered acceptor peptide and the lack of a donor oligosaccharide led to a N-terminally disordered conformation of EL5 that was similar to EL5 in the PglB structure. OST activity assays utilizing covalently tethered substrates allowed the contribution of peptide affinity to be experimentally separated from the contribution of sequon residues to acceptor activity. While sequon tethering eliminated the requirement for a hydroxyamino acid at the +2 position, the substitution of glutamine for asparagine reduced transfer activity by 85-fold rather than the 200,000-fold observed for an untethered DQQAT peptide by PglB (Lizak et al. 2013). Although proline remained disallowed at the X position in a sequon (N-X-T/S) even when the acceptor peptide was tethered, all other amino acids at the X position yielded similar transfer rates when tethered unlike free acceptor peptides where the X position impacts transfer rate (Matsumoto et al. 2017).
The donor substrates for N-glycosylation are polysaccharides linked to a polyisoprenoid carrier by phosphate or pyrophosphate. The in vivo LLO donors are not water-soluble as the polyisoprenoid carriers consist of eleven to twenty 5-carbon isoprenyl units. Chemical synthesis of donor substrates and competitive inhibitors that contain two or three isoprenyl units (geranyl-PP or farnesyl-PP instead of undecaprenyl-PP) provided the critical reagents for the identification of the LLO binding site in the OST (Napiorkowska et al. 2017). PglB was crystallized in the presence of a water-soluble nonhydrolyzable donor substrate and the acceptor peptide (Napiorkowska et al. 2017). When the acceptor peptide and donor substrate analog are present, EL5 is completely ordered, clamping both substrates into the enzyme active site. The polyprenol anchor is located in a hydrophobic groove that is in part formed by EL5, TM6 and TM11. An aromatic residue (Y196 in PglB) and a conserved basic residue (R375) contact the pyrophosphate group of the donor substrate while Y468 contacts the N-acetyl group on the first saccharide residue (Table I). The DGGK loop, which was previously shown to be essential for PglB activity (Jaffee and Imperiali 2011; Barre et al. 2017), makes additional contacts to the acceptor substrate and the first saccharide unit in the LLO donor. Entry of the physiological heptasaccharide donor into the LLO binding site of PglB cannot occur when the N-terminal end of EL5 is in the ordered conformation consistent with the disordered nature of this section of EL5 in the PglB-acceptor peptide co-crystal structure. The C-terminal end of EL5 would have to detach from the active site following catalysis to allow glycopeptide product release from the active site.
Structure guided alignment of the AglB and PglB indicated that the critical residues and sequence motifs involved in acceptor peptide recognition, LLO binding and activation of the amide nitrogen for N–C bond formation are conserved (Table I).
Structure of the yeast oligosaccharyltransferase: conservation of catalytic mechanism
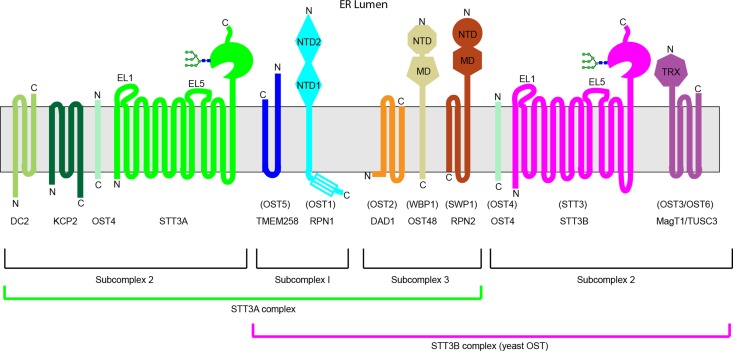
Remarkable technical and computational improvements in cryoelectron microscopy allowed two groups to solve high-resolution structures of the yeast oligosaccharyltransferase in the apo-conformation (Bai et al. 2018; Wild et al. 2018). Unlike the single subunit bacterial OST discussed above, the majority of eukaryotic organisms assemble OST complexes that contain between four and nine non-identical subunits. The yeast OST is composed of eight subunits that can be grouped into subcomplex 1 (OST1 and OST5), subcomplex 2 (STT3, OST4 and OST3/OST6) and subcomplex 3 (WBP1, SWP1 and OST2) as indicated in Figure 1. The evidence for three subcomplexes was initially based upon genetic and biochemical evidence (e.g., te Heesen et al. 1993; Karaoglu et al. 1997; Spirig et al. 1997; Mueller et al. 2015) as summarized in an earlier review (Kelleher and Gilmore 2006). In addition to the 13 TM spans of STT3, there are 15 additional TM spans provided by the other seven subunits.
Fig. 1.
Subunit composition, membrane topology and domain organization of the STT3A and STT3B complexes. Multisubunit OSTs are composed of three subcomplexes. Proteins are shown using their mammalian names, with yeast protein names shown in parentheses. Yeast lack homologs of DC2 and KCP2. The C-terminal four-helix bundle of RPN1 and the N-terminal domain of RPN2 are not present in the yeast homologs (Ost1 and Swp1, respectively). NTD, MD and TRX designate N-terminal domains, middle domains and the thioredoxin domain, and are based upon the cryo-EM structure of the yeast OST (Bai et al. 2018; Wild et al. 2018) and the structures of OST6 and TUSC3 (Schulz et al. 2009; Mohorko et al. 2014). The diagrams for STT3A and STT3B depict the location of EL1, EL5 and a well-ordered N-glycan that is important for STT3 function.
The resolution of the 3D reconstruction was excellent for both structures (3.3 Å (Wild et al. 2018) and 3.5 Å (Bai et al. 2018)), allowing the assignment of 26 TM spans and atomic resolution structures for the lumenal domains of all subunits except OST3. TM9 and EL5 of STT3 were disordered due to the absence of substrates. The 13 TM spans of yeast STT3 are arranged in a fold that is more similar to AglB than to PglB based upon the locations of TM8 and TM9 relative to the other TM spans (Bai et al. 2018; Wild et al. 2018).
A comparison of the yeast STT3 and eubacterial PglB structures revealed extensive conservation of the residues involved in donor and acceptor substrate binding and catalysis (Table I). The importance of several key catalytic residues in the yeast OST was confirmed by testing whether yeast expressing stt3 mutations could survive in the absence of a single subunit OST from L. major (Wild et al. 2018). Other residues that had been shown to be critical for yeast OST function had been identified previously based upon sequence conservation (Yan and Lennarz 2002b; Chavan et al. 2003; Li et al. 2005) or by analysis of the AglB structure (Igura et al. 2008). One noteworthy difference between PglB and STT3 is in the acceptor peptide-binding site. Like AglB, STT3 proteins lack the basic residue that aligns with R331 in PglB to form the salt bridge with the −2 position of the extended eubacterial sequon (Table I). The corresponding residue in yeast STT3 is D362, which by virtue of having a shorter, albeit charged, side chain poses no restriction on amino acids that are permitted at the −2 position relative to the eukaryotic sequon. Although the peptide-binding pocket of eukaryotic STT3 proteins have a conserved glutamate residue near the modeled location of the −1 residue of an acceptor peptide (E45 in yeast STT3, Table I) there is no indication that sequons with a basic residue at the −1 position are preferred substrates.
Bacterial and eukaryotic OSTs can transfer truncated oligosaccharides that contain one or two saccharide residues (Lee and Coward 1992; Tai and Imperiali 2001; Napiorkowska et al. 2017). For example, PglB can utilize a LLO donor that consists of a single GlcNAc residue (Napiorkowska et al. 2017). Transfer of truncated oligosaccharides by the OST is explained by the presence of active site residues that directly contact the polyisoprenoid, the pyrophosphate group and the acetylated 2-amino sugar that is the first saccharide residue in the LLO donor (Table I). Nonetheless, the transfer rates for truncated oligosaccharides are reduced compared to the natural LLO donor, and this results in the synthesis of glycoproteins that have fewer N-linked glycans in cells that are defective in LLO assembly. In eukaryotic cells, assembly of the polysaccharide of LLO is catalyzed by the glycosyltransferases in the ALG pathway (asparagine-linked glycosylation). Due to competition between truncated and fully-assembled LLO donors for the OST active site, cells with a partial defect in an ALG pathway enzyme will synthesize glycoproteins that contain mixtures of normal and truncated N-linked glycans. A novel N-linked tetrasaccharide (Neu5Ac-α2,6Galβ1,4-GlcNAcβ1,4 GlcNAc) that is derived by Golgi processing of N-linked GlcNAc2 has been detected in serum and in fibroblast lysates from patients with ALG1-CDG, PMM2-CDG and MPI-CDG (Zhang et al. 2016).
Structure of the yeast OST: subunit structures and interactions
The structures of the lumenal domain of yeast OST6 and a human homolog (TUSC3) were solved previously (Schulz et al. 2009; Mohorko et al. 2014). These OST subunits are oxidoreductases with a thioredoxin fold. Interestingly, the thioredoxin domain and first TM span of OST3 were not resolved in either cryo-EM structure indicating that OST3 is conformationally flexible with respect to STT3 (Bai et al. 2018; Wild et al. 2018). The active site CXXC motif in these subunits (OST3/OST6 or MagT1/TUSC3) is important for glycosylation of proteins by yeast OST (Schulz and Aebi 2009; Schulz et al. 2009) and by the mammalian STT3B complex (Cherepanova et al. 2014; Cherepanova and Gilmore 2016) which is homologous to the yeast OST (Figure 1). It has been proposed that the CXXC motif in OST3 family members can form a mixed disulfide with an unfolded protein to enhance STT3/STT3B access to acceptor sequons (Schulz et al. 2009; Cherepanova et al. 2014; Mohorko et al. 2014). Rotational flexibility of the lumenal domain of these oxidoreductases with respect to the STT3 catalytic site might facilitate the presentation of acceptor sites to STT3 or STT3B.
The role of the non-catalytic OST subunits with large lumenal domains (OST1, WBP1 and SWP1) in OST function was unclear despite previous publications that reported roles for these proteins, or their mammalian homologs, in donor or acceptor substrate binding or catalysis based upon chemical modification experiments (Pathak et al. 1995; Bause et al. 1997) or photolabeling (Yan and Lennarz 2002a). As these reports predated the identification of STT3 as the catalytic subunit of the OST, there is good reason to question whether these earlier reports remain informative. Ribophorin I (RPN1), the mammalian homolog of OST1, was proposed to act as a chaperone to escort specific substrates to STT3A or STT3B (Wilson and High 2007; Wilson et al. 2008).
The lumenal domains of WBP1, SWP1 and OST1 were sufficiently well resolved in the cryo-EM structures to allow their comparison to previously solved proteins (Bai et al. 2018; Wild et al. 2018). While both groups identified the same structural homologs, they reached rather different conclusions about whether the homologies were informative. Bai and colleagues (Bai et al. 2018) concluded that the structural homologs of WBP1 and SWP1 provided support for a role in recruiting LLO to the STT3 active site. However as noted by Wild et al. (Wild et al. 2018), the N-terminal domain of Swp1 (Figure 1, Figure 2A) is attached to the membrane by a long linker, placing the globular SWP1 domain well outside the LLO binding pocket. OST1, which does not contain a domain similar to a chaperone or a saccharide binding protein (Wild et al. 2018), is not located in the vicinity of the acceptor peptide-binding site (Figure 2A) casting doubt on the hypothesis that RPN1 mediates selective entry of substrates into the OST active site. Instead, Wild and colleagues proposed that OST1, WBP1 and SWP1 might be involved in recruitment of accessory factors that interact with nascent glycoproteins in the ER lumen. In mammalian cells, an ER localized lectin (malectin) that recognizes the GlcNAc2Man9Glc2 trimming intermediate of an N-linked oligosaccharide (Schallus et al. 2008, 2010), is a substoichometric subunit of the OST that binds directly to RPN1 (Qin et al. 2012; Shrimal et al. 2017).
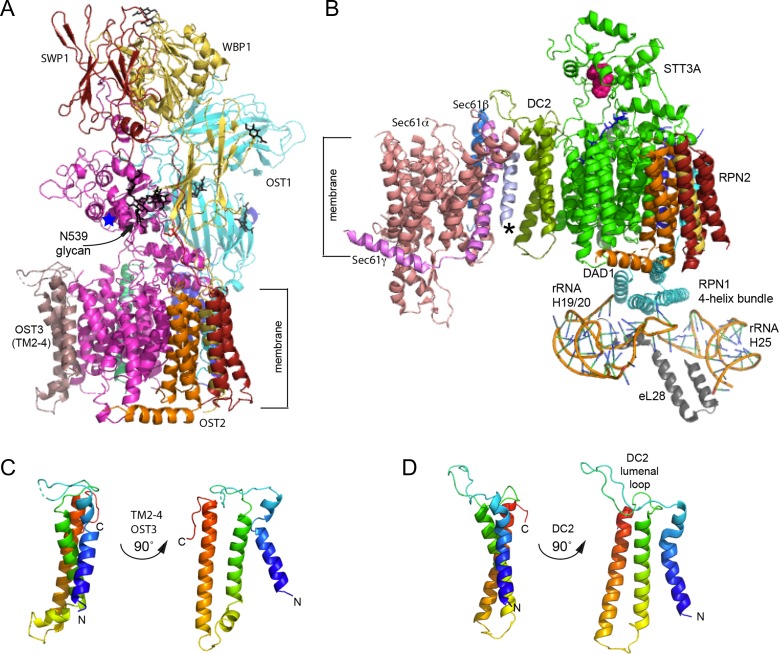
Fig. 2.
Structure of the eukaryotic oligosaccharyltransferases. (A) High-resolution structure of the yeast OST viewed from the plane of the membrane with the ER lumenal domain at the top. Subunits are color coded as in Figure 1 and are labeled. The blue star is positioned near the WWD motif. N-glycans are shown as black sticks. (B) Interaction between the STT3A complex and the RNC-Sec61 complex shown in the same orientation as in panel A. OST subunits are color coded as in Figure 1. The lumenal loop of DC2 interacts with the C-terminal tails of Sec61β (marine) and Sec61γ (light magenta). Lumenal domains of RPN1, RPN2, OST48 and TMEM258 are not shown. The C-terminal four-helix bundle of RPN1 interacts with 60 S ribosomal subunit protein eL28 and 28 S rRNA helices H25 and H19/20. The WWD motif in STT3A is shown as magenta spheres. An unidentified TM span near Sec61 is designated by an asterisk. (C, D) Structural homology between TMs 2-4 of OST3 (C) and DC2 (D) as viewed from the plane of the membrane. N and C-termini are labeled, where N of OST3 corresponds to residue S216. The figure was made with PYMOL v2.1software and PDB files 6FTI (B) and 6EZN (A, C).
The interactions between STT3 and the seven accessory subunits can be categorized as follows: (a) protein–protein interactions between TM spans, (b) protein–protein interactions between hydrophobic patches on lumenal domains, (c) phospholipid mediated interactions between nearby TM spans, and (d) N-glycan dependent interactions between lumenal domains (Bai et al. 2018; Wild et al. 2018). Rather than review all of the observed interactions, we highlight one or two from each category.
Three (OST2, OST4 and OST5) of the eight yeast subunits are relatively small integral membrane proteins without extensive lumenal or cytoplasmic domains. The TM spans of OST2 bind to TM5, TM7 and TM8 of STT3 to recruit subcomplex 3. Subcomplex 3 is stabilized by evolutionarily conserved (Wild et al. 2018) contacts between the lumenal domains of SWP1 and WBP1 and by several well ordered phospholipids that fill gaps between the TM spans of SWP1, OST2 and STT3 (Bai et al. 2018; Wild et al. 2018). Contacts between the C-terminal tail of STT3 and the N-terminal lumenal domains of WBP1 and SWP1 are the most extensive lumenal contacts between STT3 and subcomplex 3 (Bai et al. 2018; Wild et al. 2018). STT3–subcomplex 1 interactions within the membrane bilayer are primarily mediated by well-ordered phospholipids (Bai et al. 2018; Wild et al. 2018). Evolutionarily conserved hydrophobic patches on the lumenal surface of STT3 and OST1 mediate protein–protein contacts.
STT3, WBP1 and OST1 are glycoproteins that contain a total of seven N-linked glycans based upon previous analysis (Kelleher and Gilmore 1994; Li et al. 2005). The cryo-EM structures of the yeast OST were of sufficient resolution to visualize either three (Bai et al. 2018) or five of these glycans (Wild et al. 2018). As N-glycans are conformationally flexible, the well-resolved portion of protein-linked N-glycans is typically limited to one or two saccharide residues. A remarkable exception is the glycan linked to N539 of yeast STT3 that is sufficiently well ordered that GlcNAc2Man5-6 can fit into the observed density (Figure 2A). Hydrogen bonds between mannose residues and Wbp1 and Swp1 are responsible for the ordered structure of the N539 glycan, raising the possibility that the glycan helps stabilize the interaction between STT3 and subcomplex 3.
Potential roles for the conserved STT3 glycan
N539 in yeast STT3 is located in a protein sequence (D534NNTWN539NT) that contains a pair of sequons and initiates with the eukaryotic equivalent of the DGGK loop (D534NNT, Table I). This sequence is highly conserved in fungal and metazoan STT3 sequences (Table II). Glycoproteomic analysis of murine proteins (Zielinska et al. 2010) indicates that the glycosylated asparagine is located in the second acceptor site (e.g., N548 in STT3A, N624 in STT3B) as in yeast STT3. Interestingly the double sequon motif (NXTZNXT/S, where Z can be any residue and X can be any residue except proline) is also conserved in most multisubunit protist STT3s (Table II) that express a Wbp1 homolog, but is not present in STT3 proteins from organisms that express single subunit OSTs (e.g., L. major (Kelleher et al. 2007; Nasab et al. 2008). In S. cerevisiae, the stt3 N539Q mutation is lethal when this allele is the sole source of STT3 (Li et al. 2005). Importantly, when expressed along with wild type STT3, the stt3 N539Q protein co-precipitates with WBP1 and OST1 indicating that the non-glycosylated variant is neither unstable nor defective in OST complex assembly. It is not known whether the stt3 N539Q complexes contain SWP1. Thus, N539 appears to be essential for OST activity, presumably due to the presence of the N-glycan, as the stt3 T541A mutant is temperature sensitive and displays severe growth and N-glycosylation defects at the permissive temperature (Li et al. 2005).
Table II.
N-glycosylation acceptor sites that include the “DGGK” loop
| Organism | Phylum | Sequence | OST subunitsa |
|---|---|---|---|
| STT3 | |||
| S. cerevisiae | Fungi | DNNTWNNT | 8 |
| H. sapiens (A) | Metazoa | DNNTWNNT | 9 |
| H. sapiens (B) | Metazoa | DNNTWNNS | 8 |
| D. melanogaster (A) | Metazoa | DNNTWNNT | 9 |
| D. melanogaster (B) | Metazoa | DNNTWNNT | 8 |
| O. sativa (A) | Viridiplantae | DNNTWNNT | 9 |
| O. sativa (B) | Viridiplantae | DNNTWNNT | 8 |
| T. vaginalis | Parabasalia | DGNTNNFT | 4 |
| E. histolytica | Amoebozoa | DNNTWNNS | 4 |
| T. thermophila | Alveolata | DNNTWNNT | 5 |
| P. falciparum | Alveolata | DNNTWNPE | 4 |
| C. parvum | Alveolata | DNNTWNNT | 5 |
| T. brucei | Euglenozoa | DGNTWNHE | 1 |
| L. major | Euglenozoa | DGNTWNHE | 1 |
| G. lamblia | Diplomonadida | DGLTRSTY | 1 |
| PglB | |||
| C. lari | Proteobacteria | DGGKHLGK | 1 |
| AglB | |||
| A. fulgidus | Euryarchaeota | NPFQAGIG | 1 |
| M. jannaschii | Euryarchaeota | DGGSQNSP | 1 |
| P. furiosus | Euryarchaeota | DGGHARDR | 1 |
| S. solfataricus | Crenarchaeotab | ENNTLNGT | 1 |
| P. calidifontis | Crenarchaeotab | DNSTINAT | 1 |
| M. yellowstonensis | Crenarchaeotab | ENNTLNDT | 1 |
| Bathyarchaeota sp.c | Bathyarchaeotab | DNGTINMT | 1 |
| N. viennensis | Thaumarchaeotab | DNATINQT | 1 |
| N. maritimus | Thaumarchaeotab | DNSTILDH | 1 |
| Odinarchaeota sp.c | ASGARD | DNGTFNQT | 1 |
| Thorarchaeota sp.c | ASGARD | DNATFNAT | 1 |
| N. equitans | Nanoarchaeota | DGGNYYPY | 1 |
aSubunit compositions of OST complexes that have not been purified are based upon protein database searches. Four subunit complexes consist of STT3, WBP1, OST1 and OST2 while five subunit OST complexes in addition contain OST3. It is not known whether OST4 or OST5 are also present in the four or five subunit OST complexes as these small hydrophobic proteins can be overlooked in protein database searches.
bCrenarchaeota, Bathyarchaeota and Thaumarchaeota are part of the TACK superphylum.
cProtein sequences derived from DNA sequencing of uncultured organisms.
The N539 glycan in STT3 is located in a central cavity in the yeast OST that extends from the active site of STT3 to the WBP1 and SWP1 subunits (Bai et al. 2018; Wild et al. 2018). This cavity is large enough to accommodate the oligosaccharide (GlcNAc2Man9Glc3) on the fully assembled LLO donor utilized by most fungal and metazoan OSTs. The ability of eukaryotic OSTs to discriminate between fully-assembled LLO and assembly intermediates varies between organisms, with a single subunit OST (T. cruzi) showing lower selectivity than a four subunit OST that contains WBP1 as a subunit (T. vaginalis or E. histolytica) which in turn showed lower specificity than the eight subunit yeast OST that contains both SWP1 and WBP1 subunits (Kelleher et al. 2007).
Wild and colleagues (Wild et al. 2018) made the intriguing proposal that the N539 glycan may contribute directly to the recognition of LLOs that contain a terminal α−1,2 linked glucose residue by forming hydrogen bonds to the donor oligosaccharide. A role for the N539 glycan in LLO recognition would be one possible explanation for the critical function of this glycan in the yeast STT3 (Li et al. 2005). Interestingly, P. falciparum STT3 does not retain the sequon corresponding to N539TT, and utilizes a truncated LLO donor (Dol-PP-GlcNAc2) in vivo (Bushkin et al. 2010).
Remarkably, the double sequon motif (DNXTZNXT/S) is present in AglB sequences from the ASGARD and TACK superphylums of Archaea (Table II) but not in AglB sequences from species in the Euryarchaeota superphylum. Eukaryotes are thought to be most closely related to the ASGARD and TACK superphylums (Castelle and Banfield 2018). Nonetheless, conservation of these potential glycosylation sites would be unexpected unless the extended DGGK loop containing a sequon is important in most eukaryotes and a subset of Archaea. It is not known whether the DNXTZNXT site is glycosylated in the AglB protein.
Analysis of the lipid-linked oligosaccharide of the Crenoarchaeota, Pyrobaculum calidifontis revealed a decasaccharide linked to dolichol pyrophosphate (Taguchi et al. 2016). Structural analysis of a protein-linked oligosaccharide from this organism indicated that the oligosaccharide resembles a eukaryotic high mannose glycan in having a biantenary structure consisting of nine mannose residues linked to two di-N-acetylated glucose residues linked to asparagine. Although we appreciate that this suggestion is speculative, an interaction between the LLO donor and a protein-linked oligosaccharide on STT3/AglB could be involved in LLO recognition.
Interaction of the OST with the protein translocation channel
Multicellular plant and metazoan genomes encode two STT3 proteins (STT3A and STT3B) that are assembled into separate OST complexes (Figure 1) that contain either eight or nine subunits (Kelleher et al. 2003; Cherepanova et al. 2014; Shrimal et al. 2017). Subcomplex 1 and subcomplex 3, which contain five of the six shared subunits, are present in both complexes. STT3A complexes contain DC2 and KCP2, while STT3B complexes contain an oxidoreductase subunit (either MagT1 or TUSC3), hence the STT3B complex is homologous to the yeast OST. Based upon gene expression data (NCBI, EST database), STT3A and STT3B are expressed in all human tissues, and to our knowledge are expressed in all mammalian cell lines. Depletion of STT3A or STT3B by siRNA knockdown, or by CRISPR/Cas9 mediated gene disruption revealed that the STT3A and STT3B complexes have distinct roles in N-linked glycosylation (Ruiz-Canada et al. 2009; Cherepanova and Gilmore 2016). The STT3A complex is associated with the protein translocation channel (Sec61 complex) and mediates cotranslational glycosylation of nascent polypeptides using an N-terminal to C-terminal scanning mechanism (Ruiz-Canada et al. 2009; Shrimal and Gilmore 2013). The STT3B complex can post-translocationally glycosylate acceptor sites that have been skipped by STT3A including extreme C-terminal sites (Ruiz-Canada et al. 2009; Shrimal and Gilmore 2013; Shrimal et al. 2013).
Biochemical and cell biological insight into the interaction between the STT3A complex and the protein translocation channel was obtained by analyzing HEK293 cells that lack STT3A, DC2 or KCP2 (Shrimal et al. 2017). The DC2 deficient STT3A complexes were unable to form 80S ribosome-SEC61-STT3A complexes as monitored by blue native gel electrophoresis and have a glycosylation defect that resembles STT3A deficient cells (Shrimal et al. 2017). Cells that lack KCP2 have a less severe glycosylation defect.
A low-resolution structure of an active protein translocation channel was obtained by cryoelectron tomography of pancreatic microsomal membranes (Pfeffer et al. 2014). In addition to the SEC61-bound 80 S ribosome, additional protein density corresponding to the TRAP complex and the OST complex was observed (Pfeffer et al. 2014, 2017). However, it was not known which OST complex (STT3A or STT3B) was visualized, nor was the resolution sufficient to define the interaction site between the OST and the SEC61 complex. To address these questions, microsomal membranes from the STT3A null or STT3B null HEK293 cells were analyzed by cryoelectron tomography (Braunger et al. 2018). Elimination of STT3A, but not STT3B, prevented the association between the OST and the ribosome-bound SEC61 complex, providing complementary evidence that the STT3A complex interacts directly with the translocation channel in the ER.
Structure of a mammalian STT3A complex bound to the protein translocation channel
A breakthrough in understanding the mechanism of cotranslational protein glycosylation was achieved by obtaining a high-resolution cryoelectron microscopy structure of an active protein translocation channel isolated from canine microsomes programmed in vitro to synthesize a truncated derivative of the membrane glycoprotein opsin. These in vitro assembled complexes consisted of the ribosome-nascent chain (RNC) complex, the SEC61 complex and the STT3A complex (Braunger et al. 2018). The TRAP complex was also present, but poorly resolved due to disorder or low occupancy.
The resolution of the RNC-Sec61-STT3A complex was not as high as the yeast OST complex, particularly in the membrane-distal lumenal domains of RPN1, RPN2 and OST48. Nonetheless, all of the OST subunits except KCP2 could be unequivocally identified based upon the organization of the TM spans (Figure 2B). Unlike the yeast OST structure, density for pyrophosphate was readily detected in the LLO binding site of STT3A. TM9 was not disordered and the C-terminal end of EL5 was ordered consistent with the presence of the LLO donor. The acceptor sites in the opsin nascent chain had been glycosylated prior to solubilization of the membrane. The 3-D reconstruction lacks density for the nascent chain in the acceptor peptide-binding pocket in STT3A, so it was not possible to visualize the path taken by the nascent chain between the lumenal face of Sec61 and the peptide-binding site in STT3A. The presence of STT3A-bound LLO suggests that LLO binding can precede acceptor substrate binding (Karaoglu et al. 2001), thereby placing the OST in a primed state awaiting the arrival of an acceptor substrate.
The most significant information that was provided by the cryo-EM structure was the visualization of the interaction site between the STT3A complex and the RNC-SEC61 complex. The DC2 protein mediates the primary interaction between the STT3A complex and the SEC61 complex (Figure 2B), with DC2 adjacent to TM1 of Sec61α, and the C-terminal tails of Sec61β and Sec61γ (Braunger et al. 2018). Although not resolved in the cryo-EM structure, the KCP2 protein contributes to the Sec61 interaction, as KCP2 interacts with DC2 and is needed for optimal glycosylation of STT3A-dependent protein substrates (Shrimal et al. 2017). DC2 occupies the same position in the STT3A complex that is occupied by the yeast OST3 protein (Figure 2A and B), hence DC2 and KCP2 are part of subcomplex 2 (Figure 1). Segments of DC2 that were shown to be necessary for incorporation into the STT3A complex, or for the interaction with the Sec61complex (Shrimal et al. 2017), contact STT3A and the Sec61 complex respectively in the cryo-EM structure (Braunger et al. 2018).
A second interaction that contributes to the stability of the RNC-SEC61-STT3A complex is between a four helix bundle that corresponds to the C-terminal cytoplasmic tail of RPN1 and the 28 S rRNA and ribosomal protein eL28 (Figure 2B, (Braunger et al. 2018)). RPN1 and RPN2 were initially identified as ribosome binding proteins (Kreibich et al. 1978) long before they were shown to be OST subunits (Kelleher et al. 1992). Fab fragments derived from an antibody raised against the C-terminus of RPN1 sterically block protein translocation by preventing ribosome targeting to microsomal membranes (Yu et al. 1990) consistent with the interaction between RPN1 and the large ribosmal subunit. OST1, the yeast RPN1 homolog, lacks the C-terminal segment that forms the four helix bundle. The RPN1-60S subunit interaction apppears not to be conserved in all metazoa as protein database searches indicate that insect genomes from several orders (Diptera, Coleoptera and Lepidoptera) encode C-terminally truncated forms of RPN1 that lack the four helix bundle, yet these genomes encode STT3A, DC2 and KCP2. We conclude that the RPN1-60S subunit contact stabilizes the RNC-Sec61-STT3A complex, but is not a universally conserved requirement for the interaction between STT3A and Sec61.
Docking of the yeast OST structure into the previously published cryoelectron tomagraphy structures of the native protein translocation channel (Pfeffer et al. 2014, 2017) allowed Wild and colleauges to localize mammalian OST subunits in the observed EM density and to conclude that the STT3A–Sec61 interaction is mediated by DC2 (Wild et al. 2018). The same docking approach led Bai and colleagues to conclude that the OST3 protein mediates the interaction of the yeast OST with the Sec61 complex (Bai et al. 2018). Although a complex between the yeast OST and an 80 S ribosome was assembled from purified components for cryoelectron microscopy (Harada et al. 2009), the EM structure had insufficient resolution to identify the knob-like interaction sites, which at the time were assumed to be cytosolic tails of the yeast OST subunits.
In addition to the positive determinants that promote the STT3A–SEC61 interaction, there may be negative determinants that disfavor the STT3B–SEC61 interaction. One potential negative determinant is the N-terminal extension on STT3B relative to STT3A. This segment, due to its proximity to the C-terminal domain of RPN1, might interfere with the interaction between RPN1 and the 60 S ribosomal subunit (Braunger et al. 2018). The N-terminal extension on STT3B appears to be limited to vertebrate organisms, so this is not a universal mechanism to prevent an STT3B interaction with the SEC61 complex. The lumenal domain and first transmembrane span of the oxidoreductases (MagT1/TUSC3) are proposed to interfere with the Sec61–STT3B interaction as these segments are not present in DC2 (Braunger et al. 2018).
Specificity of multisubunit OST complex assembly
Until the yeast and mammalian OST structures were solved, it was not known how the complex specific accessory subunits (DC2, KCP2 and MagT1/TUSC3) interact with STT3A or STT3B. Sequence alignment between DC2 and TMs 2-4 of OST3 revealed short blocks of sequence identity interspersed with short blocks of non-homology (Wild et al. 2018). Moreover, DC2 and TMs 2-4 of OST3 have a similar three-dimensional structure (Braunger et al. 2018) as depicted in Figure 2C and D. Thus, the DC2 protein can be viewed as an N-terminally truncated derivative of OST3. Given the homology between DC2 and OST3, and their shared binding surface on STT3 proteins (TM10-13 of STT3A or yeast STT3), what accounts for the specificity of the interaction of DC2 with STT3A and MagT1 with STT3B? Sequence alignments revealed that TMs 10-13 of STT3A and STT3B show lower sequence identity than other regions of the proteins and highlight an expansion of the TM12/13 loop in STT3B relative to STT3A (Braunger et al. 2018). Residues that are conserved between DC2 and OST3 may be responsible for the similarity in the protein fold, while non-homologous segments likely specify the interactions with STT3A, STT3B and the SEC61 complex. Notably, the lumenal loop (L1/2) of DC2 is larger than the corresponding loop in OST3 (L2/3) and adopts a markedly different conformation to promote the interaction with the Sec61 complex (Figure 2B). Differences in the structure (Figure 2C and D) and sequence of the C-terminal tails of OST3 and DC2 also influence the specificity of OST complex formation as an STT3A interaction motif was mapped to the C-terminus of DC2 (Shrimal et al. 2017).
Distinct mechanisms for substrate recogniton by the STT3A and STT3B complexes: implications for N-linked glycosylation in eukaryotess that lack the STT3A complex
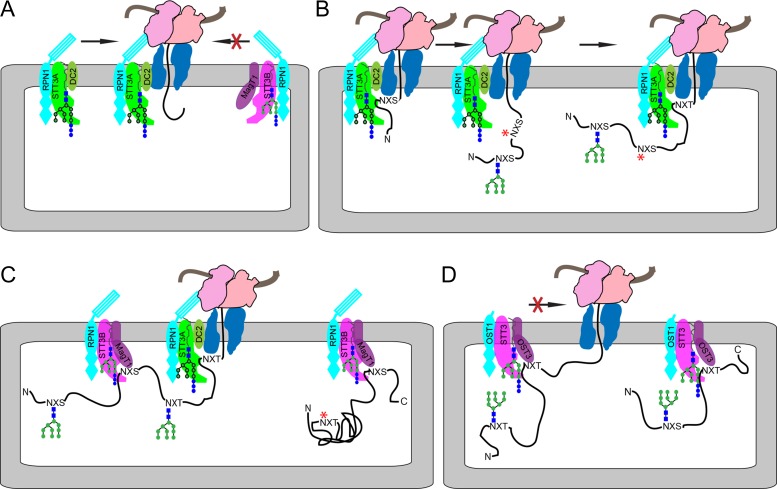
The combination of positive and negative determinants are proposed to regulate the association between the OST and the Sec61 complexes (Figure 3A). Nascent glycoproteins are glycosylated in a highly synchronized manner (Chen et al. 1995) consistent with a defined distance between the peptidyltransferase site on the ribosome and the STT3A active site (Whitley et al. 1996; Nilsson et al. 2003). The STT3A complex (Figure 3B) utilizes an N-terminal to C-terminal cotranslational scanning mechanism to locate acceptor sequons (Shrimal and Gilmore 2013). Internal glycosylation sites that have been skipped by STT3A can be glycosylated by STT3B within the time-frame of a brief pulse-labeling experiment (Ruiz-Canada et al. 2009). Hence, the STT3B complex can mediate cotranslational glycosylation by a mechanism that does not involve a direct interaction with the Sec61 complex (Figure 3C). Posttranslocational glycosylation of extreme C-terminal sites in proteins by STT3B does not occur by a scanning mechanism as indicated by the more rapid kinetics of glycosylation of NXT sites than NXS sites (Shrimal et al. 2013). Importantly, skipped sequons in folded protein domains are no longer substrates for posttranslocational glycosylation (Figure 3C). We propose that cooperation between the STT3A and STT3B complexes enhances N-glycosylation efficiency and led to the expansion of the N-glycoproteome to include acceptor sites that are not efficiently modified by the STT3B complex.
Fig. 3.
Oligosaccharyltransferase in metazoan and fungal organisms. (A) Formation of complexes between the oligosaccharyltransferase and the RNC-Sec61 complex is controlled by the presence of positive determinants in the STT3A complex and negative determinants in the STT3B complex. For simplicity, the diagrams do not show RPN2, OST48, DAD1, TMEM258, KCP2 and OST4. (B) STT3A cotranslationaly glycosylates proteins using a scanning mechanism. A skipped sequon is indicated by a red asterisk. (C) STT3B can mediate cotranslational or posttranslocational glycosylation of acceptor sites that were skipped by STT3A before the protein folds. (D) By analogy to the mammalian STT3B complex we propose that the yeast OST glycosylates substrates by a mechanism that does not rely on a direct interaction with the Sec61-RNC complex. For simplicity, we do not depict glycoproteins that are translocated by the heptameric Sec complex or the Ssh1p complex.
Given that the yeast OST lacks DC2, KCP2 and the RPN1 tail, we favor the hypothesis that the yeast OST does not interact directly with the yeast Sec61 complex (Figure 3D). If OST3 was responsible for proposed interaction between the yeast OST and the Sec61 complex (Yan and Lennarz 2005; Bai et al. 2018), one would predict that the mammalian STT3B complex would interact with the Sec61 complex via MagT1 or TUSC3. However, the STT3B–SEC61 interaction was not detected by cryoelectron tomography in STT3A null cells (Braunger et al. 2018) nor by native gel electrophoresis when mammalian cells lack both DC2 and KCP2 (Shrimal et al. 2017). We propose that organisms that lack the STT3A complex mediate glycosylation by a mechanism that does not rely upon formation of a complex between the OST and the protein translocation channel (Figure 3B), but instead acceptor sites are recognized by a non-scanning mechanism that is facilitated by OST3 (Schulz and Aebi 2009; Schulz et al. 2009; Mohorko et al. 2014).
Future goals and outstanding questions
In our opinion, an important goal for the future will be to determine whether protein N-glycosylation in yeast utilizes a translocation-channel coupled mechanism despite the absence of the STT3A complex. Resolving this issue may shed light on the mechanism of N-glycosylation in other eukaryotes that lack an STT3A complex including protist organisms and species in the genus Caenorhabditis. Another objective of future research will be to obtain a better understanding of the role of the non-catalytic subnunits that have large lumenal domains (OST1/RPN1, WBP1/OST48 and SWP1/RPN2). It will be particularly important to determine whether WBP1 and SWP1 have a role in LLO recognition either directly via interacting with the LLO donor or indirectly by stablizing the conformation of the conserved STT3 glycan.
Funding
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number GM43768.
Conflict of interest statement
None declared.
Abbreviations
CDG, congenital disorders of glycosylation; EL5, extracellular loop 5; LLO, lipid linked oligosaccharide; OST, oligosaccharyltransferase; RNC, ribosome-nascent chain complex; TM, transmembrane.
References
- Bai L, Wang T, Zhao G, Kovach A, Li H. 2018. The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature. 555:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre Y, Nothaft H, Thomas C, Liu X, Li J, Ng KKS, Szymanski CM. 2017. A conserved DGGK motif is essential for the function of the PglB oligosaccharyltransferase from Campylobacter jejuni. Glycobiology. 27:978–989. [DOI] [PubMed] [Google Scholar]
- Bause E, Wesemann M, Bartoschek A, Breuer W. 1997. Epoxyethylglycyl peptides as inhibitors of oligosaccharyltransferase: Double-labeling of the active site. Biochem J. 322:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunger K, Pfeffer S, Shrimal S, Gilmore R, Berninghausen O, Mandon EC, Becker T, Forster F, Beckmann R. 2018. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science. 360:215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer W, Klein RA, Hardt B, Bartoschek A, Bause E. 2001. Oligosaccharyltransferase is highly specific for the hydroxy amino acid in Asn-Xaa-Thr/Ser. FEBS Lett. 501:106–110. [DOI] [PubMed] [Google Scholar]
- Bushkin GG, Ratner DM, Cui J, Banerjee S, Duraisingh MT, Jennings CV, Dvorin JD, Gubbels MJ, Robertson SD, Steffen M et al. 2010. Suggestive evidence for Darwinian Selection against asparagine-linked glycans of Plasmodium falciparum and Toxoplasma gondii. Eukaryot Cell. 9:228–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, Banfield JF. 2018. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 172:1181–1197. [DOI] [PubMed] [Google Scholar]
- Chavan M, Rekowicz M, Lennarz W. 2003. Insight into functional aspects of Stt3p, a subunit of the oligosaccharyl transferase. Evidence for interaction of the N-terminal domain of Stt3p with the protein kinase C cascade. J Biol Chem. 278:51441–51447. [DOI] [PubMed] [Google Scholar]
- Chen MM, Glover KJ, Imperiali B. 2007. From peptide to protein: Comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry (Mosc). 46:5579–5585. [DOI] [PubMed] [Google Scholar]
- Chen W, Helenius J, Braakman I, Helenius A. 1995. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci USA. 92:6229–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova NA, Gilmore R. 2016. Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Sci Rep. 6:20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova NA, Shrimal S, Gilmore R. 2014. Oxidoreductase activity is necessary for N-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J Cell Biol. 206:525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH. 2013. Understanding human glycosylation disorders: Biochemistry leads the charge. J Biol Chem. 288:6936–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S, Lizak C, Michaud G, Bucher M, Darbre T, Aebi M, Reymond JL, Locher KP. 2013. Mechanism of bacterial oligosaccharyltransferase: In vitro quantification of sequon binding and catalysis. J Biol Chem. 288:8849–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Li H, Lennarz WJ. 2009. Oligosaccharyltransferase directly binds to ribosome at a location near the translocon-binding site. Proc Natl Acad Sci U S A. 106:6945–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. 2007. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol Rev. 87:1377–1408. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 73:1019–1049. [DOI] [PubMed] [Google Scholar]
- Hennet T, Cabalzar J. 2015. Congenital disorders of glycosylation: A concise chart of glycocalyx dysfunction. Trends Biochem Sci. 40:377–384. [DOI] [PubMed] [Google Scholar]
- Igura M, Maita N, Kamishikiryo J, Yamada M, Obita T, Maenaka K, Kohda D. 2008. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27:234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffee MB, Imperiali B. 2011. Exploiting topological constraints to reveal buried sequence motifs in the membrane-bound N-linked oligosaccharyl transferases. Biochemistry (Mosc). 50:7557–7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. 1997. The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J Biol Chem. 272:32513–32520. [DOI] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. 2001. Allosteric regulation provides a molecular mechanism for preferential utilization of the fully assembled dolichol-linked oligosaccharide by the yeast oligosaccharyltransferase. Biochem. 40:12193–12206. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Banerjee S, Cura AJ, Samuelson J, Gilmore R. 2007. Dolichol-linked oligosaccharide selection by the oligosaccharyltransferase in protist and fungal organisms. J Cell Biol. 177:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. 1994. The Saccharomyces cerevisiae oligosaccharyltransferase is a protein complex composed of Wbp1p, Swp1p, and four additional polypeptides. J Biol Chem. 269:12908–12917. [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. 2006. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 16:47–62. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R. 2003. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol Cell. 12:101–111. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Kreibich G, Gilmore R. 1992. Oligosaccharyltransferase activity is associated with a protein complex composed of ribophorins I and II and a 48 kd protein. Cell. 69:55–65. [DOI] [PubMed] [Google Scholar]
- Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF et al. 2006. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25:1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich G, Czako-Graham M, Grebenau R, Mok W, Rodriguez-Boulan E, Sabatini DD. 1978. Characterization of the ribosomal binding site in rat liver rough microsomes: Ribophorins I and II, two integral membrane proteins related to ribosome binding. J Supramol Struct. 8:279–302. [DOI] [PubMed] [Google Scholar]
- Lee J, Coward JK. 1992. Enzyme catalyzed glycosylation of peptides using a synthetic lipid disaccharide substrate. J Org Chem. 57:4126–4135. [Google Scholar]
- Li G, Yan Q, Nita-Lazar A, Haltiwanger RS, Lennarz WJ. 2005. Studies on the N-glycosylation of the subunits of oligosaccharyl transferase in Saccharomyces cerevisiae. J Biol Chem. 280:1864–1871. [DOI] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Michaud G, Schubert M, Fan YY, Bucher M, Darbre T, Aebi M, Reymond JL, Locher KP. 2013. Unexpected reactivity and mechanism of carboxamide activation in bacterial N-linked protein glycosylation. Nat Commun. 4:2627. [DOI] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Numao S, Aebi M, Locher KP. 2011. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 474:350–355. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Shimada A, Nyirenda J, Igura M, Kawano Y, Kohda D. 2013. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci U S A. 110:17868–17873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Taguchi Y, Shimada A, Igura M, Kohda D. 2017. Tethering an N-glycosylation sequon-containing peptide creates a catalytically competent oligosaccharyltransferase complex. Biochemistry (Mosc). 56:602–611. [DOI] [PubMed] [Google Scholar]
- Mohorko E, Owen RL, Malojcic G, Brozzo MS, Aebi M, Glockshuber R. 2014. Structural basis of substrate specificity of human oligosaccharyl transferase subunit N33/Tusc3 and its role in regulating protein N-glycosylation. Structure. 22:590–601. [DOI] [PubMed] [Google Scholar]
- Mueller S, Wahlander A, Selevsek N, Otto C, Ngwa EM, Poljak K, Frey AD, Aebi M, Gauss R. 2015. Protein degradation corrects for imbalanced subunit stoichiometry in OST complex assembly. Mol Biol Cell. 26:2596–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napiorkowska M, Boilevin J, Sovdat T, Darbre T, Reymond JL, Aebi M, Locher KP. 2017. Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat Struct Mol Biol. 24:1100–1106. [DOI] [PubMed] [Google Scholar]
- Nasab FP, Schulz BL, Gamarro F, Parodi AJ, Aebi M. 2008. All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol Biol Cell. 19:3758–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, Von Heijne G, Johnson AE. 2003. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol. 161:715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R, Hendrickson TL, Imperiali B. 1995. Sulfhydryl modification of the yeast Wbp1p inhibits oligosaccharyl transferase activity. Biochemistry (Mosc). 34:4179–4185. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Forster F. 2014. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat Commun. 5:3072. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Schaffer M, Ng BG, Albert S, Plitzko JM, Baumeister W, Zimmermann R, Freeze HH, Engel BD et al. 2017. Dissecting the molecular organization of the translocon-associated protein complex. Nat Commun. 8:14516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin SY, Hu D, Matsumoto K, Takeda K, Matsumoto N, Yamaguchi Y, Yamamoto K. 2012. Malectin forms a complex with ribophorin I for enhanced association with misfolded glycoproteins. J Biol Chem. 287:38080–38089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R. 2009. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 136:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallus T, Feher K, Sternberg U, Rybin V, Muhle-Goll C. 2010. Analysis of the specific interactions between the lectin domain of malectin and diglucosides. Glycobiology. 20:1010–1020. [DOI] [PubMed] [Google Scholar]
- Schallus T, Jaeckh C, Feher K, Palma AS, Liu Y, Simpson JC, Mackeen M, Stier G, Gibson TJ, Feizi T et al. 2008. Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol Biol Cell. 19:3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz BL, Aebi M. 2009. Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol Cell Proteomics. 8:357–364. [DOI] [PubMed] [Google Scholar]
- Schulz BL, Stirnimann CU, Grimshaw JP, Brozzo MS, Fritsch F, Mohorko E, Capitani G, Glockshuber R, Grutter MG, Aebi M. 2009. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc Natl Acad Sci USA. 106:11061–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Cherepanova NA, Gilmore R. 2017. DC2 and KCP2 mediate the interaction between the oligosaccharyltransferase and the ER translocon. J Cell Biol. 216:3625–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Gilmore R. 2013. Glycosylation of closely spaced acceptor sites in human glycoproteins. J Cell Sci. 126:5513–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Trueman SF, Gilmore R. 2013. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J Cell Biol. 201:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig U, Glavas M, Bodmer D, Reiss G, Burda P, Lippuner V, te Heesen S, Aebi M. 1997. The STT3 protein is a component of the yeast oligosaccharyltransferase complex. Mol Gen Genet. 256:628–637. [DOI] [PubMed] [Google Scholar]
- Taguchi Y, Fujinami D, Kohda D. 2016. Comparative analysis of archaeal lipid-linked oligosaccharides that serve as oligosaccharide donors for Asn glycosylation. J Biol Chem. 291:11042–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai VW, Imperiali B. 2001. Substrate specificity of the glycosyl donor for oligosaccharyl transferase. J Org Chem. 66:6217–6228. [DOI] [PubMed] [Google Scholar]
- te Heesen S, Knauer R, Lehle L, Aebi M. 1993. Yeast Wbp1p and Swp1p form a protein complex essential for oligosaccharyl transferase activity. EMBO J. 12:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW et al. 2002. N-linked glycosylation in C. jejuni and its functional transfer to E. coli. Science. 298:1790–1793. [DOI] [PubMed] [Google Scholar]
- Whitley P, Nilsson IM, von Heijne G. 1996. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J Biol Chem. 271:6241–6244. [DOI] [PubMed] [Google Scholar]
- Wild R, Kowal J, Eyring J, Ngwa EM, Aebi M, Locher KP. 2018. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science. 359:545–550. [DOI] [PubMed] [Google Scholar]
- Wilson CM, High S. 2007. Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J Cell Sci. 120:648–657. [DOI] [PubMed] [Google Scholar]
- Wilson CM, Roebuck Q, High S. 2008. Ribophorin I regulates substrate delivery to the oligosaccharyltransferase core. Proc Natl Acad Sci U S A. 105:9534–9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Lennarz WJ. 2002. a. Studies on the function of oligosaccharyl transferase subunits: A glycosylatable photoprobe binds to the luminal domain of Ost1p. Proc Natl Acad Sci USA. 99:15994–15999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Lennarz WJ. 2002. b. Studies on the function of the oligosaccharyltransferase subunits: Stt3p is directly involved in the glycosylation process. J Biol Chem. 277:47692–47700. [DOI] [PubMed] [Google Scholar]
- Yan A, Lennarz WJ. 2005. Two oligosaccharyl transferase complexes exist in yeast and associate with two different translocons. Glycobiology. 15:1407–1415. [DOI] [PubMed] [Google Scholar]
- Yu Y, Sabatini DD, Kreibich G. 1990. Antiribophorin antibodies inhibit the targeting to the ER membrane of ribosomes containing secretory polypeptides. J Cell Biol. 111:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, James PM, Ng BG, Li X, Xia B, Rong J, Asif G, Raymond K, Jones MA, Hegde M et al. 2016. A novel N-tetrasaccharide in patients with congenital disorders of glycosylation, including asparagine-linked glycosylation Protein 1, phosphomannomutase 2, and mannose phosphate isomerase deficiencies. Clin Chem. 62:208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wisniewski JR, Mann M. 2010. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 141:897–907. [DOI] [PubMed] [Google Scholar]