Abstract
Neuron-glial related cell adhesion molecule NrCAM is a newly identified negative regulator of spine density that genetically interacts with Semaphorin3F (Sema3F), and is implicated in autism spectrum disorders (ASD). To investigate a role for NrCAM in spine pruning during the critical adolescent period when networks are established, we generated novel conditional, inducible NrCAM mutant mice (Nex1Cre-ERT2: NrCAMflox/flox). We demonstrate that NrCAM functions cell autonomously during adolescence in pyramidal neurons to restrict spine density in the visual (V1) and medial frontal cortex (MFC). Guided by molecular modeling, we found that NrCAM promoted clustering of the Sema3F holoreceptor complex by interfacing with Neuropilin-2 (Npn2) and PDZ scaffold protein SAP102. NrCAM-induced receptor clustering stimulated the Rap-GAP activity of PlexinA3 (PlexA3) within the holoreceptor complex, which in turn, inhibited Rap1-GTPase and inactivated adhesive β1 integrins, essential for Sema3F-induced spine pruning. These results define a developmental function for NrCAM in Sema3F receptor signaling that limits dendritic spine density on cortical pyramidal neurons during adolescence.
Keywords: dendritic spine pruning, NrCAM, SAP102, Semaphorin3F
Introduction
Dendritic spines on pyramidal neurons harbor more than 90% of excitatory synapses in the mammalian brain (Shen and Cowan 2010). During postnatal development, spines are initially overproduced, eliminated in substantial numbers during adolescence, and then stabilized in adulthood (Huttenlocher 1979; McAllister 2007; Holtmaat and Svoboda 2009; Petanjek et al. 2011). These overlapping stages of spine and excitatory synapse maturation are essential for wiring and fine-tuning of neuronal circuits (Alvarez and Sabatini 2007). Although much is known about spine structural plasticity in the adult brain, less is understood about mechanisms that regulate dendritic spines during development. Altered regulation of such processes likely contributes to increased spine density in autism spectrum disorders (ASD) (Hutsler and Zhang 2010; Tang et al. 2014) and decreased spine density in schizophrenia and bipolar disorder (Glausier and Lewis 2013; Konopaske et al. 2014; Phillips and Pozzo-Miller 2015).
Neural cell adhesion molecule (CAM) NrCAM is a member of the L1 family of immunoglobulin (Ig)-class recognition molecules (NrCAM, L1, Close Homolog of L1 [CHL1], and Neurofascin), which have diverse roles in adhesion, axon growth, and synaptic targeting (Maness and Schachner 2007). Its importance is underscored by multiple studies in which genetic variants in the NrCAM gene have been associated with ASD (Pinto et al. 2010; Voineagu et al. 2011; Sakurai 2012). Furthermore, NrCAM null mice display autism-related behaviors of impaired sociability, reversal learning, and sensory processing (Moy et al. 2009; Demyanenko et al. 2011; Shigematsu et al. 2016). Studies with global knockout mice revealed a surprising new function for NrCAM as a negative regulator of dendritic spine density of cortical pyramidal neurons in a genetically interacting pathway with the repellent secreted ligand Sema3F (Demyanenko et al. 2014). Single knockout lines deleted for NrCAM, Sema3F, or the Sema3F receptor subunits Npn2 or PlexA3, all exhibit increased spine and excitatory synapse density on apical but not basal dendrites of pyramidal neurons in sensory cortical areas (Tran et al. 2009; Demyanenko et al. 2014). Like NrCAM, Npn2 (Melin et al. 2006; Wu et al. 2007; Weiss 2009; Hosseinpour et al. 2017), PlexA3 (D’Gama et al. 2015), and Sema3F (Butler et al. 2015) have been linked genetically to ASD.
A key question is whether NrCAM mediates spine pruning in cortical pyramidal neurons during the critical adolescent period, when an appropriate excitatory/inhibitory balance is established in cortical circuits. To investigate this, we generated a novel conditional mouse line in which NrCAM is deleted from pyramidal cells under tamoxifen-inducible control by Nex1Cre-ERT2 (Agarwal et al. 2012). It was of further interest to determine if NrCAM regulates spine density in the medial frontal cortex (MFC), where cortical and subcortical inputs integrate into circuits regulating ASD-relevant behaviors of sociability and cognitive flexibility (Douglas and Martin 2004; Pan and Gan 2008; Belmonte et al. 2010), in addition to the primary visual cortex (V1). Finally, we used molecular modeling and mutagenesis to investigate the mechanism by which NrCAM transduces Sema3F signaling leading to spine elimination.
We demonstrate for the first time that NrCAM functions cell autonomously in pyramidal neurons during early postnatal and adolescent development to limit spine density on apical dendrites in both the MFC and V1. Furthermore, we identify a novel mechanistic role for NrCAM in promoting oligomeric clustering of Sema3F receptor subunits through a molecular interface with Npn2, and by interaction with PDZ scaffold proteins such as SAP102. NrCAM-induced receptor clustering induces PlexA3’s intrinsic Rap-GTPase activating protein (GAP) activity, which in turn inhibits Rap1 GTPase and inactivates β1-integrins in the dendritic membrane. This NrCAM-dependent signaling pathway may promote spine detachment prior to elimination.
Materials and Methods
Generation of NrCAM Conditional Mutant Mice and Tamoxifen Induction
Murine ES cells (NrCAM clone EPD0479-4-E10) targeted to insert LoxP sites flanking NrCAM exons 11 and 12 were obtained from the NCRR-NIH supported KOMP Repository (www.komp.org) generated by the CSD consortium for the NIH funded Knockout Mouse Project (KOMP). Methods used for the targeted alleles have been published (Testa et al. 2004). Southern blotting of genomic DNA from the NrCAM E10 clone with a 5′ probe showed a novel band of correct size for the mutant allele (8 kb) compared with the WT allele (10 kb), indicating that targeting was correct. 3′ PCR analysis further validated the correct targeting. The NrCAM E10 clone was injected into C57Bl/6 N (Agouti) blastocysts, resulting in 5 pups that were strong chimeras. Germline transmission and removal of an internal Rosa26-flpe transgene and Frt-flanked neo cassette were obtained by breeding chimeras to FLPE deleter mice (C57Bl/6J-albino). Genotypes and selectable marker removal were assessed by PCR, and yielded 5 F1 offspring with the NrCAM floxed allele and Flpe removed. These NrCAM floxed mice were crossed with C57Bl/6 mice for colony expansion. For conditional expression of a fluorescent marker, NrCAM floxed mice were crossed to the Ai9 reporter line Rosa-CAG-(LoxP-Stop-LoxP-tdTomato)-WPRE (C57Bl/6 J) (Madisen et al. 2010).
NrCAM floxed mice were crossed to the NexCre-ERT2 line (C57Bl/6; from Klaus-Armin Nave) to induce recombination in postmitotic, postmigratory pyramidal neurons upon induction by treatment with tamoxifen as described (Agarwal et al. 2012). To induce expression of CreERT2 recombinase, daily doses of tamoxifen (100 mg/kg) were administered via intraperitoneal injection from P10 to P13, and mice analyzed at P21 and P80. For adult induction, 10 daily doses of tamoxifen were given starting at P55, and mice analyzed at P80. Other mice included NrCAM null mutants (Sakurai et al. 2001), which were on a C57Bl/6 background. All mice were handled according to the University of North Carolina Institutional Animal Care and Use Committee policies in accordance with NIH guidelines.
Immunoreagents
Monoclonal antibodies used were directed against: PSD95 (ThermoFisher Scientific), SAP102 (NeuroMab), and conformationally active β1 integrins (9EG7, BD Pharmingen). Polyclonal antibodies were: Npn-2 (R&D Systems), NR2B, PlexA3, and Rap1 (Santa Cruz Biotechnology). Normal goat, rabbit and mouse IgG, and AlexaFluor488, AlexaFluor565, and AlexaFluor647-conjugated secondary antibodies were from Jackson ImmunoResearch. Human IgG Fc was from Abcam, and Sema3F-Fc was from R&D Systems.
Immunostaining and Spine Analyses
Mice were anesthetized with 1.25% Avertin, perfused transcardially with PBS followed by 4% paraformaldehyde (PFA)/PBS, and brains were stored overnight in 4% PFA/PBS. Brains were vibratome-sectioned coronally (80 μM), permeabilized with Triton X-100, and labeled with the following antibodies: rabbit anti-RFP (AbCAM# 62 341), rabbit anti-GFP (Life technologies #A6455) rabbit anti-NrCAM (Abcam #ab24344), mouse anti-VGluT1 (NeuroMab #75-066), mouse anti-SAP102 (NeuroMab #75-058), mouse anti-Gad67 (Millipore #MAB5406), chicken anti-GFP (Abcam #ab13970), rat anti mouse active β1-integrin (clone 9EG7 #553 715 BD). Secondary antibodies were: AlexaFluor488, AlexaFluor565, and AlexaFluor647 (1:400). Slides were mounted in Slow-fade Gold antifade mountant (Life Technologies). Digital images were obtained by confocal microscopy using Zeiss LSM700 or 710 confocal microscopes at the Microscopy Services Laboratory at UNC Chapel Hill (Pablo Ariel, Director). Objectives used were an EC Plan Neofluar 40×/1.3 oil lens or Plan Apochromat 40×/1.4 oil lens on the LSM700 and 710, respectively. Images were acquired using a pinhole size of 1 AU, filter settings optimized for each fluorophor, and with adjustments to other parameters to avoid pixel saturation. Zoom was adjusted to obtain pixel sizes of 0.13–0.14 μm.
Spines were traced and quantified on 30 μm segments of the first branch of apical or basal dendrites from confocal z-stack images as described (Demyanenko et al. 2014) using Neurolucida software (MBF Bioscience). Spines were manually scored as thin, stubby, or mushroom types on apical dendrites as in (Peters and Kaiserman-Abramof 1970). Spine neck length, head area, and volume were quantified using the quick measure tool of Neurolucida. To produce 3D reconstructions, dendritic z-stacks were deconvolved using Autoquant 3 software (Media Cybernetics) with default blind deconvolution settings and Imaris (Bitplane) software. Mean spine densities/10 μM ± SEM (standard error of the mean) and other parameters of spine morphology were compared by the t-test (2-tailed, P < 0.05). For direct integrin activation, neurons were treated with 4 mM MnCl2.4H2O (Sigma #203 734) for 1h before treatment with Sema3F-Fc or Fc.
Site-Directed Mutagenesis and Subcloning
Mouse NrCAM splice variant 1 (GenBank accession number AJ543321) and mouse Npn2 (accession number NM_001 077 404) cDNAs were used. NrCAM mutations in the Npn2 and PDZ binding sites, and Npn2 (a1 domain) mutations in the NrCAM binding site were generated by the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA), using the following mutagenic primers:
- TARΔNER: F 5′-CCAGTGCACTGCAAGGGGAGCTGCCGTCTCC-3′
- R 5′-GGAGACGGCAGCTCCCCTTGCAGTGCACTGG-3′
- ΔSFV: F 5′-CCTGTCAACGCAATGAACTAAGTCTTTAAGC-3′
- R 5′-GCTTAAAGACTTAGTTCATTGCGTTGACAGG-3′
- R126E: F 5′-GTGCACTGCAGAAAACGAGCGTG-3′
- R 5′-TGGTAGACTCCTTCATAG-3′
R126A: F 5′- GTGCACTGCAGCAAACGAGCGTG-3′
R 5′-TGGTAGACTCCTTCATAG-3′.
E56R: F 5′- CCAGAACTGTCGGTGGATTGTCTACG-3′
R 5′-TGGGAGGGATAGTCCTGG-3′
In Utero Electroporation
In utero electroporation (IUE) was performed as described (Yokota et al. 2007) to introduce vector (pCAGG-IRES-mEGFP), WT NrCAM or NrCAM ΔSFV plasmids into E14.5 embryos of pregnant females. Briefly, 1–2 μl of plasmid DNA (1.5 μg/μl) was injected into the embryonic lateral ventricles and electroporated with 5 pulses at 30 V for 50 ms at 950 ms intervals through the uterine wall using a BTX ElectroSquarePorator (ECM 830). Brains were vibratome-sectioned and immunostained for GFP for quantification of spine density.
Cortical Neuron Cultures
Neuronal cultures were prepared from brain of WT or NrCAM null embryos (E15.5), transfected at DIV11 with pCAGG-IRES-mEGFP or additional plasmids where indicated, and assayed at DIV14 for spine density as described (Demyanenko et al. 2014). Essentially, cells were cultured on laminin, poly-d-lysine coated chamber slides, fixed in 4% PFA, permeablized with Triton X-100, blocked in 10% serum, and labeled with anti-GFP to enhance visualization of spines. Spine densities were measured from confocal images on at least 10 EGFP-labeled neurons of pyramidal morphology in each of 4 replicate cultures. Mean spine densities ± SEM were compared by the t-test (2-tailed, P < 0.05). For SAP102 immunostaining, neuronal cultures transfected with pCAGG-IRES-mEGFP were fixed at DIV14 in 4% PFA, permeablized with Triton X-100, blocked in 10% horse or donkey serum, and labeled with anti-GFP, anti-NrCAM, and mouse monoclonal SAP102 (NeuroMAB). Secondary antibodies AlexaFluor488, AlexaFluor565, and AlexaFluor647 (1:500) were added for 1 h before mounting and confocal imaging.
Structural Modeling
The horseshoe shape of Ig1–Ig4 domains of NrCAM was modeled using MODELLER (Webb and Sali 2014) based upon the crystallographic structure of human Neurofascin (PDB ID 3P3Y) (Liu et al. 2011). Human Neurofascin and NrCAM are closely related homologs that share 49% overall sequence identity with 63% identity in the Ig1 domain. The model for Npn2 a1–b2 in an extended conformation was generated based on the experimental structure of Npn1 a1–b2 (PDB 4GZ9) (Janssen et al. 2012) again using MODELLER. The ClusPro protein–protein docking server was used for protein–protein interaction prediction between the NrCAM Ig1 domain and the Npn2 a1 domain (Comeau et al. 2004; Kozakov et al. 2006). Docking poses were screened for high-ranking docking solutions as defined in the Results section.
Sema3F Receptor Clustering
Cortical neuron cultures from WT or NrCAM null mice were transfected with pCAGG-IRES-mEGFP and empty vector pCMV6, WT NrCAM, or NrCAM mutants, and after 48 h were treated with Sema3F-Fc or Fc control protein (3 nM) for 5–30 min. Cultures were fixed with 4% PFA and blocked in 10% horse serum/PBS. Slides were incubated in primary antibodies (anti-PlexA3, anti-Npn2) overnight at 4 °C. Secondary antibodies (Alexa 555, Alexa 647) were added for 1 h and mounted. Confocal images were obtained with Zeiss LSM700 and LSM710 microscopes using a Plan-Apochromat 63 × 1.4 numerical aperture objective with ×2 optical zoom using Zeiss Zen software. Only apical dendrites of GFP positive cells were imaged for analysis. Colocalization of endogenous PlexA3 and Npn2 was analyzed using ImageJ plugins: Colocalization Test and Colocalization Threshold (Wright Cell Imaging Facility, Toronto Western Research Institute) (www.uhnresearch.ca/facilities/wcif/imagej). Colocalization was assessed using Pearson correlation coefficients (R-Total), which indicate the degree of correlation of the 2 fluorescent signals, and can vary between −1 (no correlation) and 1 (absolute correlation). For each time point, an average from measurements of at least 20 images, in which individual GFP positive cells were selected as regions of interest (ROIs), was reported. Heat maps of colocalization were generated from merged images of colocalized pixels as previously described (Sullivan et al. 2016).
Immunostaining of Activated β1-Integrins
Neuronal cultures transfected with pCAGG-IRES-mEGFP were fixed at DIV14 in 4% PFA, permeablized with Triton X-100, blocked in 10% horse or donkey serum, and labeled with anti-GFP (Abcam), and anti-CD29 (clone 9EG7, BD Pharmingen) directed against activated β1-integrins. Secondary antibodies AlexaFluor488, and AlexaFluor555 (1:500) were added for 1 h before mounting. Dendritic z-stacks were obtained on a confocal LSM700 microscope using constant settings. Images were deconvolved using Autoquant 3 software (Media Cybernetics) with default blind deconvolution settings. In Imaris (Bitplane) software, all images were subjected to same threshold settings. A mask was created for the green labeling, and the red pixels outside the green were excluded from the analysis. The mean intensity of red pixels (active β1 integrin) was calculated for those within the green EGFP-labeled area. Overall, 10 neurons from 2 different experiments were analyzed. Mean intensity ± SEM was compared by the t-test (2-tailed, P < 0.05).
Rap1 GTPase Assay
Rap1 activation was assayed as described (Garcia-Mata et al. 2006). After treatment with Sema3F-Fc or control Fc (3 nM) for 30 min, cells were washed with PBS and lysed with RIPA buffer. Lysates were cleared by centrifugation, and active Rap1 was precipitated with glutathione-Sepharose beads precoupled to beads containing a GST fusion protein of the Ras association domain of Ral guanine nucleotide dissociation stimulator (GST-Ral-GDS). Precipitates were washed with lysis buffer and solubilized in SDS sample buffer. A portion of the cell lysate was reserved for analysis of total Rap content. Rap1 was detected following Western blotting with anti-Rap1 antibody specific for Rap1 (Santa Cruz Biotechnology, Dallas, TX).
Transient Cell Transfection
HEK293T or COS7 cells were grown in Dulbecco’s modified Eagle’s medium-H supplemented with gentamicin/kanamycin (Life Technologies, Grand Island, NY) and 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) in a humidified incubator with 5% CO2. On the day prior to transfection, cells were seeded at 2 × 106 cells/100 mm dish. Plasmids expressing WT or mutant NrCAM, Npn2, and SAP102-GFP were transfected into cells with Lipofectamine 2000 (Life Technologies). Media was changed on the day following transfection, and cells harvested 24 h later. Protein concentrations were determined using the BCA assay.
Immunoprecipitation
Homogenates of mouse forebrain (P21) and cell lysates were prepared in lysis buffer (1% Brij98, 10 mM Tris–Cl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA), 200 μM Na3VO4, 10 mM NaF, 1× protease inhibitors (SigmaAldrich #P8340). Lysates (0.5–1 mg) were precleared for 30 min at 4 °C using Protein A/G Sepharose. Precleared lysates were incubated with NrCAM, Npn-2, GFP or normal IgG (nIg) antibodies for 2 h at 4 °C. Protein A/G Sepharose beads were added for 30 min at 4 °C prior to washing, and bound proteins were eluted by boiling in SDS-PAGE sample buffer.
Synaptoneurosome Preparation
Synaptoneurosomes were isolated as in Villasana et al. (2006). Briefly, mouse cortices (P28, n = 4) were homogenized in 8 ml of synaptoneurosome buffer (10 mM HEPES, 1 mM EDTA, 2 mM EGTA), 200 μM sodium orthovanadate, 10 mM NaF and 1× protease inhibitor cocktail (P8340 Sigma). After sonication sample was filtered through a 100 μm filter followed by a 5 μm cell strainer. The final filtrate was centrifuged at 1000 × g for 10 min at 4 °C. The pellet was resuspended in 500 μl of RIPA lysis buffer (20 mM Tris pH 7.0, 0.15 M NaCl, 5 mM EDTA, 1 mM EGTA, 1% NP-40, 1% deoxycholate, 0.1% SDS, 200 mM Na3VO4, 10 mM NaF, 1× protease inhibitors), agitated for 40 min on a rocker, and centrifuged at 16 000 × g for 10 min. The supernatant was collected as the synaptoneurosome fraction.
Western Blotting
Lysates (50 μg), protein fractions, or immunoprecipitations were subjected to SDS-PAGE and transferred to nitrocellulose. Blots were blocked in TBS-Tween containing 5% nonfat milk, and incubated overnight with antibodies directed against: NrCAM (1:1000), Npn-2 (1:1000), NR2B (1:500), PlexA3 (1:500), PSD95 (1:2000), SAP102 (1:500) or GAPDH (1:1000). Blots were washed and incubated in secondary antibodies (1:10 000) for 1 h, before developing using enhanced chemiluminescence (ThermoFisher Scientific) and exposed to film.
Results
NexCre-Driven Deletion of NrCAM From Cortical Pyramidal Neurons
NrCAM localizes to spines on apical dendrites of approximately 30% of pyramidal neurons in V1 (Demyanenko et al. 2014), but it is also expressed in some glia (Zhang et al. 2014) and in embryonic thalamic neurons, where it regulates axon targeting to sensory cortical areas (Demyanenko et al. 2011). To evaluate the consequences of NrCAM deficiency specifically in pyramidal neurons, and to ascertain whether it functions during adolescence, we developed a conditional inducible line by first generating NrCAM floxed mice from targeted murine embryonic (ES) stem cells. PCR-based genomic DNA analysis of wild type (WT), heterozygous, and homozygous NrCAM floxed mice showed the expected DNA fragments for WT (532 kb) and floxed (728 kb) alleles (Fig. 1A). NrCAM floxed mice were intercrossed with NexCre-ERT2 knock-in mice, in which a modified Cre recombinase fused to the human estrogen receptor ligand binding domain is expressed under control of endogenous regulatory sequences in the Nex1 (Neurod6) gene (Agarwal et al. 2012). Temporal regulation of recombination is achieved by tamoxifen-induced nuclear import of the modified Cre recombinase. Postnatal tamoxifen induction in Nex-CreERT2 mice has been shown to elicit cell-specific targeting of postmitotic cortical and hippocampal pyramidal neurons with no detectable targeting of interneurons, oligodendroglia, astrocytes, or non-neural cells (Agarwal et al. 2012). To permanently label pyramidal neurons in which recombination occurred, mice were also crossed to the Ai9 reporter line (Rosa-CAG-LSL-tdTomato-WPRE) (Goebbels et al. 2006; Madisen et al. 2010; Agarwal et al. 2012).
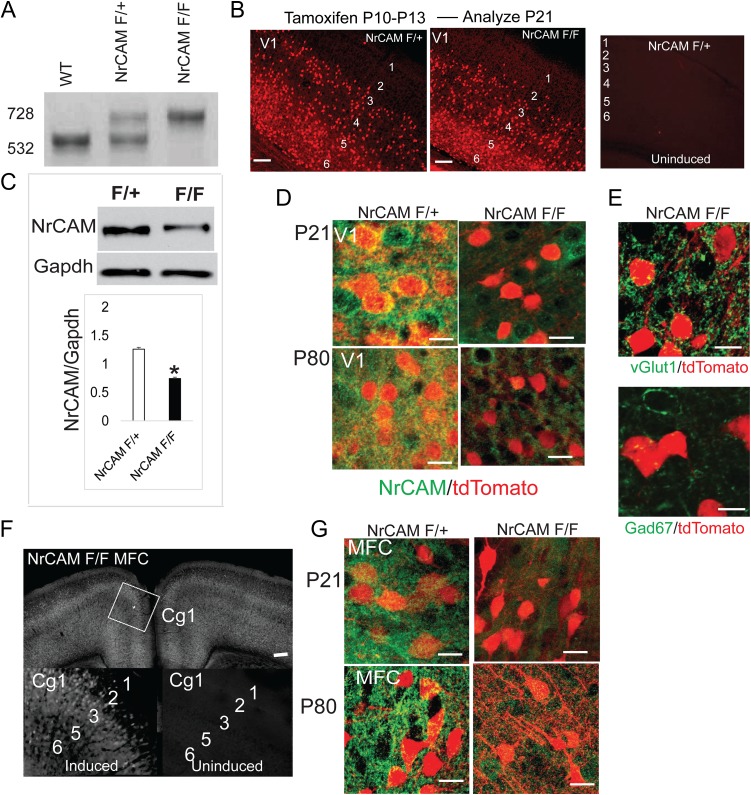
Figure 1.
Generation of Nex-CreERT2:NrCAM; tdT conditional mutant mice. (A) PCR genotyping showing diagnostic fragments for WT (532 bp), NrCAM F/+ (532 and 728 bp), and NrCAM F/F (728 bp) mice. (B) tdTomato labeling of V1 neurons in NrCAM F/+ and NrCAM F/F mice (P21) induced at P10–13, and lack of labeling in uninduced mice (bar=100 μm, layers 1–6). (C) NrCAM immunoblot from cortical lysates of NrCAM F/+ or F/F mice (P21). Histogram shows the relative pixel densities of the blot (mean ± SEM, n = 3, *P < 0.05, t test). (D) NrCAM in pyramidal neurons marked by tdTomato in NrCAM F/+ but not in NrCAM F/F mice (V1, layer 4, P21 and P80 (bar = 50 μm). (E) tdTomato+ neurons in NrCAM F/+ mice express vGlut1 but not GAD67 (bar = 50 μm). (F) tdTomato labeling of MFC neurons (Cg1) in NrCAM F/F mice (P21) after early postnatal induction at P10-13, and lack of labeling in uninduced F/F mice (bar=100 μm, layers 1–6). (G) NrCAM in pyramidal neurons of MFC colocalizes with tdTomato in NrCAM F/+ but not in NrCAM F/F at P21 and P80 (bar = 50 μm).
In the mouse, peak stages of spinogenesis (~P0–P15), spine pruning (~P15–P30), and synaptic stabilization (~P40) occur in postnatal life (Holtmaat et al. 2005). To achieve recombination in early adolescence (P21-24) (Laviola et al. 2003), NexCre-ERT2: NrCAM F/+; tdT (referred to as NrCAM F/+) and NexCre-ERT2: NrCAM F/F; tdT mice (referred to as NrCAM F/F) were given daily tamoxifen injections from P10 to P13 (Agarwal et al. 2012), then analyzed at P21. Induction of tdTomato in V1 was prominent in NrCAM F/+ and NrCAM F/F mice, and negligible in noninduced cortex (Fig. 1B), in accord with high cytoplasmic retention of Cre-ERT2 (Agarwal et al. 2012). Cortical lysates from NrCAM F/F mice showed a significant reduction in NrCAM protein compared with NrCAM F/+ mice (Fig. 1C). Residual NrCAM protein was likely due to uninduced pyramidal neurons or other cell types. Nissl staining showed no alteration in general anatomy, size, or architecture of brain structures in NrCAM F/F mice (Supplementary Fig. S1A). At P21 and P80 (adult) stages NrCAM protein localized to tdTomato+ pyramidal neurons in V1 following induction (P10–P13) in NrCAM F/+ mice, and substantially decreased in NrCAM F/F mice (Fig. 1D). The specificity of NexCre-ERT2 recombination was verified by coexpression of tdTomato and pyramidal cell marker VGlut1, but not with GABAergic interneuron marker GAD67 (Fig. 1E).
Tamoxifen induction of NrCAM F/F mice (P10–P13) resulted in tdTomato labeling of MFC pyramidal neurons at P21, as shown in layer 2/3 neurons of primary cingulate cortex (Cg1), with little labeling in uninduced Cg1 (Fig. 1F). NrCAM protein localized to pyramidal neurons in MFC at both P21 and P80 in NrCAM F/+ mice, and was efficiently lost upon induction in NrCAM F/F mice (Fig. 1G).
Temporal Regulation of Dendritic Spines Mediated by NrCAM
In NrCAM global null mice, increased density of spines and excitatory synapses on apical dendrites occurs in star pyramidal neurons, the principal recipients of thalamocortical inputs in V1, layer 4 (Demyanenko et al. 2011, 2014). Apical dendrites differ from basal dendrites in having higher levels of thalamocortical (Turner et al. 2015) and intracortical inputs (Brooks-Kayal 2010), as well as distinct temporal plasticity properties (Gordon et al. 2006). To determine if NrCAM functioned within pyramidal neurons to constrain spine density on apical dendrites in early postnatal life, NrCAM was deleted by tamoxifen treatment a (P10–P13) and spines analyzed at P21. NrCAM loss resulted in increased spine density on apical but not basal dendrites of star pyramidal neurons of V1, layer 4 at P21 (Fig. 2A,B). Apical dendrites of NrCAM F/F star pyramidal neurons displayed normal proportions of filopodial, stubby, and mushroom spines, as well as normal arborization and overall morphology (Supplementary Fig. S1B,C). The spine phenotype was stable upon maturation, as increased spine density was also observed on apical dendrites of star pyramidal neurons of NrCAM F/F mice in adulthood (P80) following deletion of NrCAM at P10–P13 (Fig. 2C,D). To determine if deletion of NrCAM in pyramidal neurons during adulthood also affected spine density, recombination was induced by tamoxifen treatment in young adults (P55–P65), and spine density analyzed in mature adults (P80). Deletion of NrCAM from pyramidal neurons in early adulthood had no effect on spine density of apical dendrites in mature adults as shown in V1 (Fig. 2E,F).
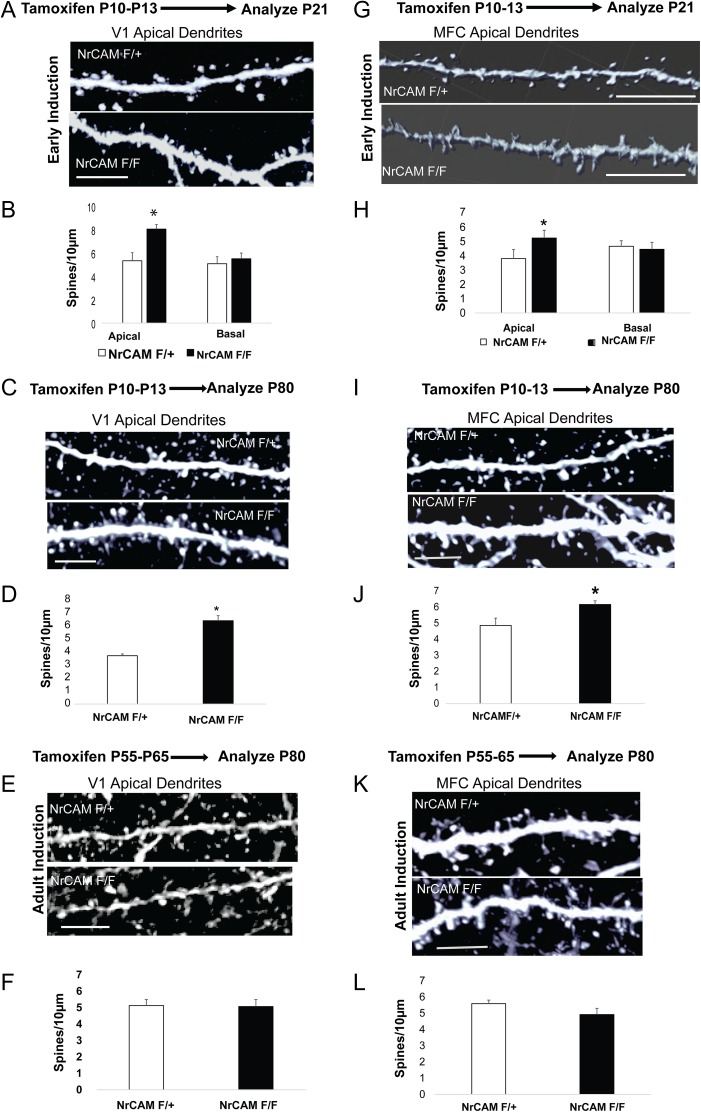
Figure 2.
NrCAM deletion in pyramidal neurons in early postnatal development increases spine density on apical dendrites in V1 and MFC. (A) Early postnatal deletion of NrCAM increased spine density on apical but not basal dendrites of V1 star pyramidal neurons at P21 in NrCAM F/F compared with F/+ mice (bar =10 μm). (B) Histogram shows mean spine density ± SEM from apical and basal dendrites of V1 cortex at P21 (n = 20, neurons from 3 brains in each condition, *P < 0.05, t test). (C) Apical dendritic spines from the adult (P80) mice, layer 4 pyramidal neurons of V1 cortex after early postnatal induction (bar=10 μm). (D) Histogram shows mean spine density from apical dendrites at P80 (n = 20, neurons from 3 brains in each condition, *P < 0.05, t test). (E) Early adult deletion of NrCAM has no effect on apical spine density in V1 star pyramidal neurons at P80 (bar = 10 μm). (F) Quantification of spine density ± SEM from apical dendrites of layer 4 pyramidal neurons in V1 at P80 after early adult induction at P55. (n = 20, neurons from 3 brains in each condition, P < 0.05, t test) (G) Early postnatal deletion of NrCAM increased spine density on apical but not basal dendrites of Cg1 layer 2/3 neurons in NrCAM F/F compared with F/+ mice at P21 (bar = 10 μm). (H) Quantification of spine density ± SEM from apical and basal dendrites of layer 2/3 pyramidal neurons in MFC cortex at P21 (n = 20, neurons from 3 brains in each condition, *P < 0.05, t test). (I) Apical dendritic spines from the adult (P80) mice layer 2, 3 pyramidal neurons of Cg1 cortex after early postnatal induction (bar=10 μm). (J) Quantification of spine density ± SEM from apical dendrites of Cg1 pyramidal neurons at P80 (n = 20, neurons from 3 brains in each condition, *P < 0.05, t test). (K) No difference in spine density on apical dendrites of pyramidal neurons in Cg1, layer 2/3 of adult NrCAM F/F mice (P80) following induction in young adults (P55) (bar = 10 μm). (L) Quantification of spine density ± SEM from apical dendrites of layer 2/3 pyramidal neurons in Cg1 at P80 after early adult induction at P55. (n = 20, neurons from 3 brains in each condition, P < 0.05, t test).
Tamoxifen-induced deletion of NrCAM at early postnatal stages (P10–P13) also resulted in increased spine density on apical but not basal dendrites in Cg1 (layers 2 and 3) of NrCAM F/F mice in early adolescence (P21) (Fig. 2G,H) and adulthood (P80) (Fig. 2I,J). Tamoxifen-induced deletion of NrCAM in the MFC of young adult mice (P55–65) did not significantly alter spine density on apical dendrites of pyramidal neurons in Cg1 (layer 2/3) in mature animals (P80) (Fig. 2K,L). Spine morphology and dendritic branching were normal in Cg1 pyramidal neurons following conditional deletion of NrCAM (not shown). We also observed a significant increase in apical spine densities in L5 and L6 pyramidal neurons in the MFC of NrCAM F/F mice compared with NrCAM F/+ at both P21 and P80 (Supplementary Fig. S1E,F).
Taken together, these results demonstrated that NrCAM functions in pyramidal neurons during adolescent stages to limit the density of spines on apical dendrites in both V1 and MFC, and that NrCAM deletion in early adulthood does not affect spine density in mature adults.
NrCAM Acts Cell Autonomously in Pyramidal Neurons to Regulate Spine Density and Requires the SAP102-Interacting PDZ Motif
To determine if NrCAM regulates spine density in cortical pyramidal neurons in a cell autonomous manner, NrCAM was re-expressed in pyramidal cell precursors of NrCAM global null embryos using IUE. Either WT NrCAM in pCAGG-IRES-mEGFP or empty vector was electroporated in utero into the lateral ventricles of NrCAM null embryos (E14.5) to achieve sparse labeling of cortical pyramidal neurons (Seiradake et al. 2013). EGFP-labeled star pyramidal neurons in V1 (layer 4) were analyzed for spine density at P21. Re-expression of NrCAM significantly decreased spine density on apical dendrites of sparsely labeled pyramidal neurons in the NrCAM null cortex compared with empty vector, consistent with a cell autonomous, postsynaptic function (Fig. 3A,B). The NrCAM cytoplasmic domain contains a carboxyl-terminal PDZ binding site (SFV) with the potential to engage scaffold proteins at the spine postsynaptic density (PSD) (Davey et al. 2005; Dirks et al. 2006; Yamagata and Sanes 2010). To determine if this motif was required for limiting spine density in vivo, a deletion mutant of NrCAM lacking the SFV motif (ΔSFV) was expressed in neural precursors following IUE of NrCAM null embryos (E14.5). Unlike WT NrCAM, the NrCAM ΔSFV mutant was unable to rescue spine density on apical dendrites of star pyramidal neurons, indicating that interactions at the PDZ binding motif were important for regulating spine density (Fig. 3A,B).
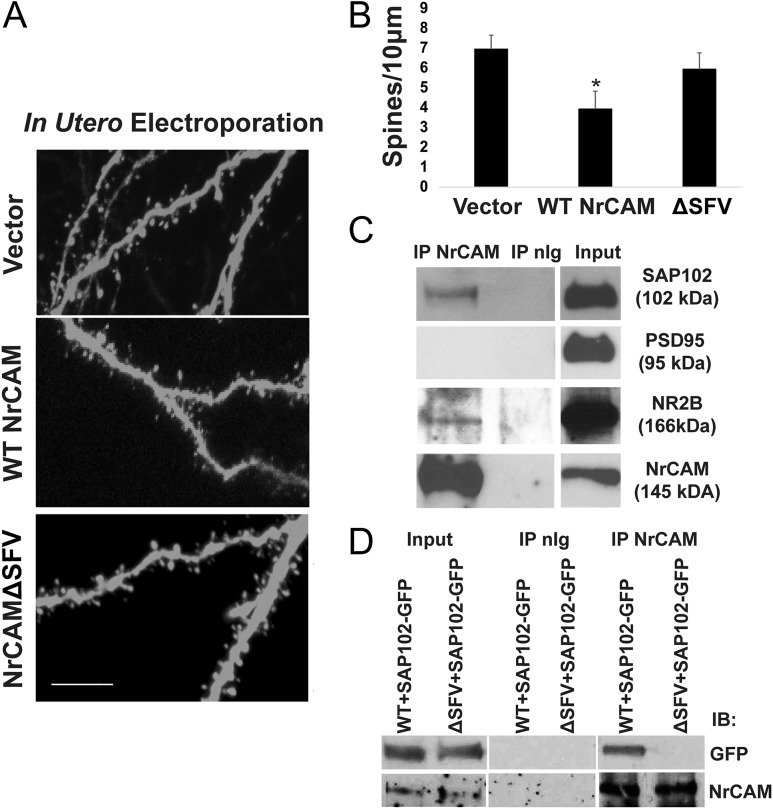
Figure 3.
NrCAM rescues spine density in null mutant pyramidal neurons, and requires the carboxyl-terminal PDZ binding motif SFV, which recruits SAP102. (A) IUE was performed in NrCAM null embryos (E14.5) with empty vector, pCAGG-IRES-WT NrCAM-mEFGP, or pCAGG-IRES-ΔSFV NrCAM-mEGFP. Apical spines were imaged in V1, layer 4 at P21 after EGFP immunostaining (scale bar 10 μm). (B) Mean spine densities on apical dendrites (n > 500 spines from 3 brains in each condition, *P < 0.05, t test). (C) Coimmunoprecipitation of NrCAM from synaptoneurosomes with SAP102 and NR2B but not PSD95. (D) SAP102 association requires the PDZ binding motif SFV in NrCAM, as shown by coimmunoprecipitation in NrCAM and SAP102-GFP transfected COS7 cells.
The PDZ-containing scaffold protein Synapse-Associated Protein (SAP102) localizes to the PSD of immature spines in fetal and neonatal brain, and is replaced by PSD95 (SAP90) at mature synapses (Murata and Constantine-Paton 2013). SAP102 loss-of-function mutations in humans are associated with nonsyndromic mental retardation (Tarpey et al. 2004; Zanni et al. 2010), while null mice have learning defects and do not thrive (Cuthbert et al. 2007). NrCAM coimmunoprecipitated with SAP102 from a synaptoneurosome fraction isolated from P28 mouse brain, but not with PSD95 even though each protein was present in the fraction (Fig. 3C). Through an amino terminal sequence distinct from its 3 PDZ domains SAP102 binds NR2B, an early developmental subunit of the NMDA receptor important for spine morphogenesis (Wei et al. 2015). NrCAM coimmunoprecipitated from synaptoneurosomes with NR2B, likely due to indirect binding of NR2B with SAP102 (Fig. 3C). Immunofluorescence staining of cortical neurons expressing EGFP in culture showed colocalization of NrCAM and SAP102 on dendritic spines (Supplementary Fig. S1D). NrCAM appeared to interact directly with SAP102 as shown coimmunoprecipitation from COS7 cells coexpressing WT NrCAM and GFP-tagged SAP102 (Fig. 3D). The association of NrCAM with SAP102 required the NrCAM SFV motif, as shown by SAP102-GFP coimmunoprecipitation with WT NrCAM but not with NrCAM ΔSFV (Fig. 3D). In other experiments (not shown) NrCAM did not coimmunoprecipitate from P21 brain lysates with the following cell adhesion, scaffold, and signaling proteins: SynCAM1, SAP97, Shank3, SrGAP2, FARP1.
NrCAM Association With Npn2 and PDZ Scaffolds Mediates Sema3F-Induced Spine Elimination
The mechanism of action of class 3 Semaphorins requires binding to transmembrane receptors that comprise heteromeric complexes of Neuropilins, Plexins, and CAMs. We have shown earlier that NrCAM forms a molecular complex with Npn2 and PlexinA3 (Demyanenko et al. 2014). To understand how NrCAM engages the Sema3F holoreceptor complex, we developed a structural model for predicting the nature of the interface between NrCAM and Npn2. We reported that the last 3 residues (NER122) of a sequence in the NrCAM Ig1 domain (TAR120 NER123) is involved in Npn2 binding (Demyanenko et al. 2014) (Fig. 4A). A conserved sequence (FASNR/KL) in the corresponding location in the Ig1 domain of L1 and CHL1 binds Npn1 (Castellani et al. 2002; Wright et al. 2007). In none of these cases is the location of the binding site on Npn1/2 identified. We modeled the structure of NrCAM Ig 1–4 based on the crystal structure of Neurofascin (PDB 3P3Y) (Liu et al. 2011), and the structure of Npn2 a1–a2 domains based on the Npn1 a1/a2/b1 structure (PDB 4GZ9) (Janssen et al. 2010). Npn1 forms a multidomain globular unit in which the a1 domain is linked to a2–b1–b2 domains by a flexible linker of ~5 residues, while the a2 domain interfaces with both the b1 and b2 domains. We used ClusPro 2.0 (https://cluspro.bu.edu/) (Chen et al. 1998; Kozakov et al. 2006) to predict candidate interfaces, which we then tested through mutagenesis. We hypothesized that the interface occurred between NrCAM TARNER motif and the Npn2 a1 domain. The NrCAM Ig1 domain was docked onto the Npn2 a1 domain using ClusPro 2.0, and high-ranking docking solutions were evaluated by (1) inclusion of the TARNER motif in the interface with Npn2, (2) lack of steric hindrance by NrCAM Ig2–Ig4 domains, and (3) compatibility of the NrCAM/Npn2 interaction within a Sema3F/Npn2/PlexA3 holoreceptor based on the Sema3A/Npn1/PlexA2 complex (PDB 4GZ9) (Janssen et al. 2010). One docking solution fit all 3 criteria. We also tested whether the Npn2 a2 domain contained an interface for NrCAM, but charge reversal mutations of residues in the Npn2 a2 domain (E270R, R287E, D431R) with the potential to bind the NrCAM TARNER motif did not perturb coimmunoprecipitation of NrCAM with Npn2 from HEK293T cells (data not shown).
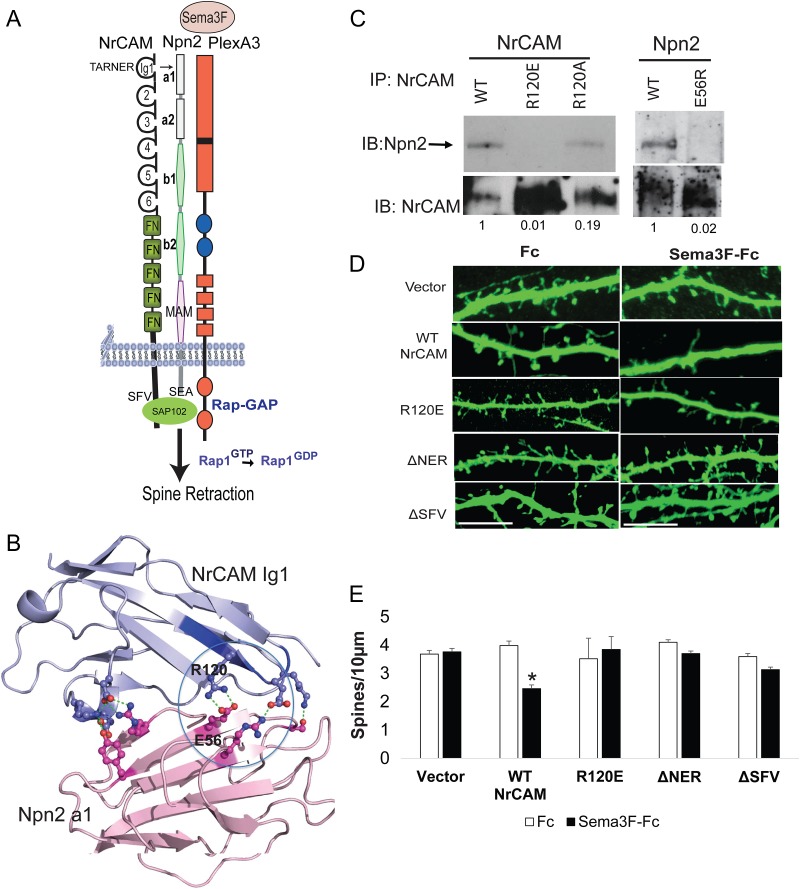
Figure 4.
NrCAM Ig1 associates with the Npn2 a1 domain through a charged interface, and requires the PDZ interaction motif for Sema3F-induced spine retraction. (A) Schematic of the Sema3F holoreceptor complex comprised of NrCAM, Npn2, and PlexA3 with intrinsic Rap-GAP activity. TARNER interface and PDZ interactions motifs SFV (NrCAM) and SEA (Npn2) are shown. (B) Structural model for NrCAM Ig1 binding to Npn2 a1 domains, showing predicted electrostatic interaction between R120 in the TAR120 NER motif of NrCAM, and E56 in Npn2. Note that modeling utilized human sequences, whereas mutagenesis tested the candidate interfaces in mouse proteins. Therefore, the residue numbering in the figure is for the mouse sequence. For NrCAM, the mouse and human sequences differ by only 2 residues out of 101 and neither substitution is near the TARNER sequence. For the Npn2 a1 domain, there are 2 substitutions between mouse and human sequences out of 116 residues, with one conservative substitution (Arg for Lys) near the predicted interface. (C) Coimmunoprecipitation of WT, R120E, and R120A NrCAM with Npn2; and NrCAM with WT Npn2 and Npn2 E56R from transfected HEK293 cells. (D) NrCAM null neurons were transfected with vector, WT NrCAM, R120E, ΔNER, or ΔSFV mutants in pCAGG-IRES-mEGFP, treated with Fc or Sema3F-Fc for 30 min at DIV14, immunostained for EGFP, and apical dendrites imaged confocally. (E) Quantification of mean spine density ± SEM on apical dendrites (n > 500 spines for each condition, t test, *P < 0.05).
In our structural prediction, NrCAM Ig1 engaged the Npn2 a1 domain through an interface where Arg120 in the TAR120NER motif interacted electrostatically with Glu56 in the Npn2 a1 domain (Fig. 4B). To test this, Arg120 was mutated to create either a charge reversal (R120E) or neutral (R120A) substitution, and binding to Npn2 assessed by coimmunoprecipitation from transfected HEK293T cells. The NrCAM mutation R120E effectively perturbed binding between NrCAM and Npn2 compared with WT NrCAM, while the neutral substitution R120A partially inhibited binding (Fig. 4C). To evaluate the corresponding role of Glu56 in the Npn2 a1 domain, a charge reversal mutation (E56R) was tested for binding to NrCAM by coimmunoprecipitation from HEK293T cells. The Npn2 E56R mutant showed little binding to NrCAM (Fig. 4C). Surface biotinylation of transfected HEK293T cells confirmed that equivalent levels of WT and mutant proteins were expressed on the cell surface (data not shown).
The NrCAM mutants were then tested in a functional assay for Sema3F-induced dendritic spine collapse in primary cortical neurons (Tran et al. 2009; Demyanenko et al. 2014). Primary cortical neurons were cultured from NrCAM null embryos (E14.5) and transfected at 11 days in vitro (DIV) with vector (pCAGGS-IRES-mEGFP) or plasmids containing WT, R120E, ΔNER, and ΔSFV NrCAM. Cells were treated at DIV14 with Sema3F-Fc or control Fc (3 nM) for 30 min, stained for EGFP, and spine density quantified on apical dendrites from confocal z-stacks. Neurons transfected with WT NrCAM exhibited decreased spine density after Sema3F-Fc treatment compared with Fc, whereas NrCAM null neurons transfected with empty vector showed no change (Fig. 4D,E), as reported (Demyanenko et al. 2014). In contrast, neurons expressing the NrCAM mutants R120E, ΔNER, or ΔSFV did not respond to Sema3F-Fc (Fig. 4D,E). These results demonstrated that the interface between the NrCAM TARNER motif and Npn2, as well as interactions involving the PDZ binding motif were crucial for Sema3F-mediated dendritic spine retraction.
NrCAM Induces Sema3F Receptor Clustering and Activates PlexA3 Rap-GAP Signaling
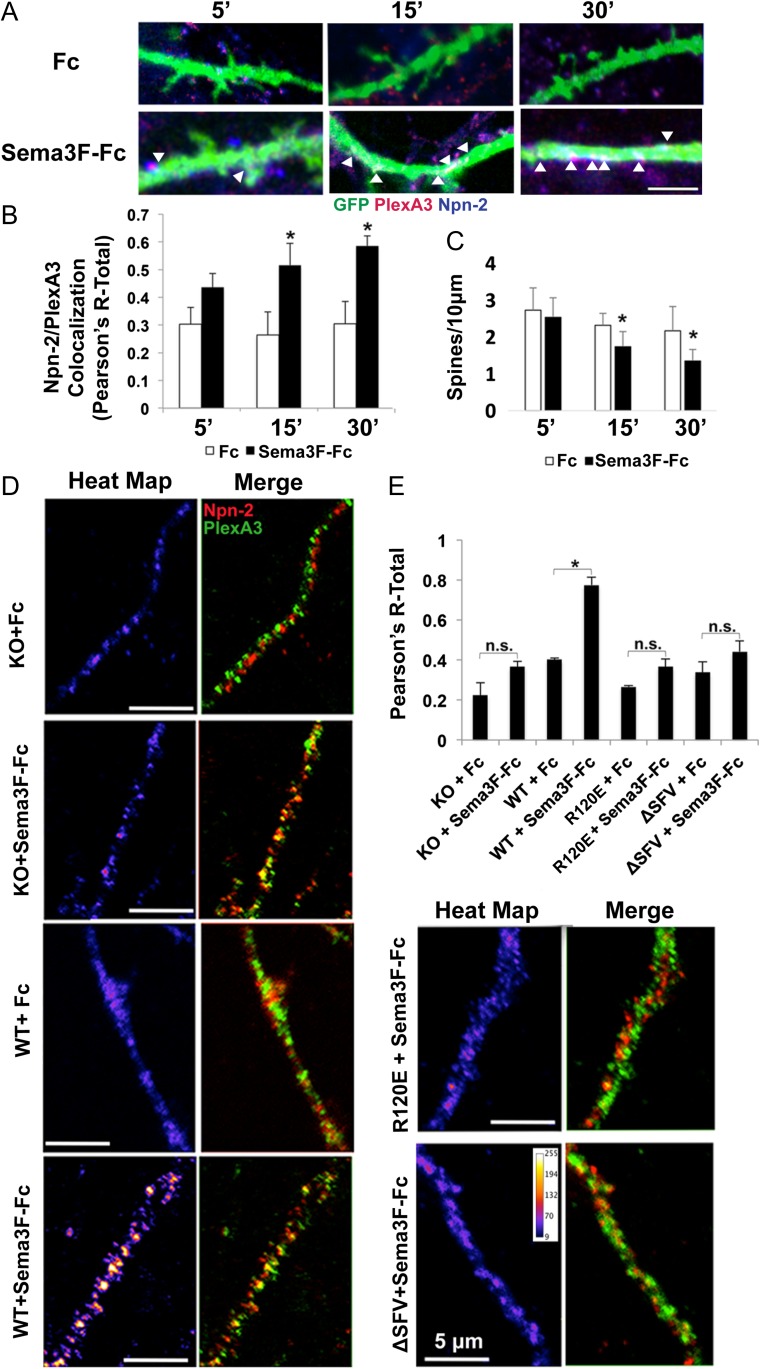
The class 3 Semaphorin-3F (Sema3F) acting through the Neuropilin-2/Plexin-A3 (Npn2/PlexA3) holoreceptor complex signals to restrain apical dendritic spine morphogenesis of cortical pyramidal neurons (Tran et al. 2009; Shiflett et al. 2015). Plexins form homodimers (or multimers) on the cell surface, and interaction with ligand induces conformational changes necessary for signal activation. Specifically, ligand-induced clustering activates the autoinhibited Rap-GAP function of PlexinAs (He et al. 2009). We used a fluorescence-based assay to analyze coclustering of Npn2 and PlexA3, analogous to one we developed to assess NCAM association with EphA3 (Sullivan et al. 2016). To establish the assay, WT cortical neuron cultures were transfected with pCAGGS-mIRES-mEGFP and treated with Sema3F-Fc or Fc (3 nM) for 0–30 min at DIV14. Immunofluorescence staining for Npn2 and PlexA3 in EGFP-labeled apical dendrites was imaged confocally, and colocalization assessed by Pearson’s correlation coefficients. Sema3F-Fc increased coclustering of Npn2 and PlexA3 within 15–30 min compared with controls (Fig. 5A,B). In parallel, there was a significant reduction in spine density in Sema3F-Fc treated neurons (Fig. 5C).
Figure 5.
NrCAM promotes Sema3F-induced coclustering of Npn2 and PlexA3 in cortical neuron cultures. (A) Enhanced colocalization of Npn2 (blue) and PlexA3 (red) in apical dendrites of Sema3F-Fc compared with Fc treated neurons (EGFP labeled) in culture after 5, 15, and 30 min (bar = 5 μM). (B) Colocalization of Npn2 and PlexA3 represented by Pearson’s correlation coefficients (R-Total). (Means ± SEM, *P < 0.05, t-test). (C) Mean spine densities in Fc and Sema3F-Fc treated neurons after 5, 15, and 30 min. (n = 20, neurons, means ± SEM, *P < 0.05, t-test). (D) Confocal images of apical dendrites of NrCAM null (KO) neurons transfected with empty vector, WT NrCAM, NrCAM R120E or NrCAM ΔSFV and stained for PlexA3 (green) or Npn2 (red). Merged images and heat maps of colocalization are shown for cells treated with Fc or Sema3F-Fc. Scale bar is 5 μM. (E) Histogram of Pearson’s colocalization coefficients (R-Total) (n = 3 replicates; *P < 0.05).
To evaluate a possible functional role for NrCAM in receptor clustering, cortical neuron cultures from NrCAM knockout (KO) mice were transfected with WT NrCAM plasmid or empty vector. Cultures were treated with Sema3F-Fc or Fc for 30 min, and analyzed for colocalization of Npn2 and PlexA3. Sema3F-Fc induced robust coclustering of Npn2 and PlexA3 on apical dendrites in neurons expressing WT NrCAM, as shown in merged images and heat maps (Fig. 5D,E). In NrCAM KO neurons, Sema3F-Fc caused a small increase in clustering that was not statistically significant. To assess the importance of NrCAM interaction with Npn2 and PDZ scaffolds, cortical neuron cultures from NrCAM KO mice were transfected with WT NrCAM, TARNER mutant (R120E), PDZ mutant ΔSFV, or empty vector. Sema3F-Fc did not significantly increase Npn2-PlexA3 clustering in neurons expressing R120E or ΔSFV compared with WT (Fig. 5D,E), demonstrating the importance of NrCAM association with Npn2 and PDZ scaffold proteins such as SAP102. Because Npn2 has a carboxyl-terminal PDZ binding motif (CEA) (Wang et al. 2003), it might additionally interact with SAP102.
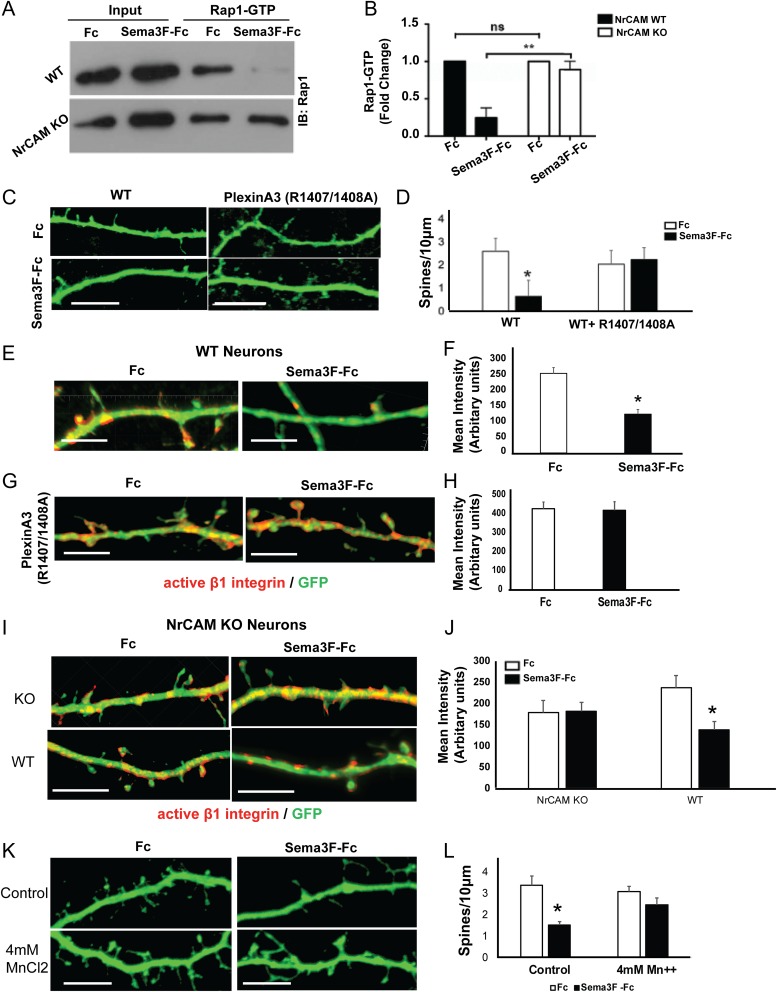
Ligand-induced dimerization of Plexin A family members relieves auto-inhibition of the cytoplasmic domain, initiating intrinsic Rap-GAP activity (He et al. 2009). In turn, Rap-GAP is able to recruit and inactivate Rap1 by hydrolyzing bound GTP to GDP. To determine if NrCAM, through its ability to promote receptor clustering, activates PlexA3 Rap-GAP signaling, we measured the activity of Rap1 GTPase in GST-pulldown assays from cortical neuronal cultures (Garcia-Mata et al. 2006). Cortical neuron cultures from WT or NrCAM null mice were treated at DIV14 with Sema3F-Fc or control Fc (3 nM) for 30 min, lysed, and assayed for active GTP-bound Rap1. Sema3F-Fc strongly decreased the levels of GTP-bound Rap1 in neurons expressing NrCAM but did not alter levels of Rap1-GTP in NrCAM null neurons (Fig. 6A,B). These results provide strong evidence that NrCAM mediates Sema3F-induced Rap1 inactivation. Although clustering was not linked directly to Rap1-GTP in these experiments, it is probably that this is due to clustering-induced activation of PlexinA3 Rap-GAP.
Figure 6.
Sema3F-induced spine remodeling in cortical neurons cultures is mediated by NrCAM-dependent Rap1 inactivation and requires active PlexA3 Rap-GAP. (A,B) Activated Rap1 pull-down assays from cortical neuron cultures show inactivation of Rap1 in Sema3F-Fc treated cultures from WT but not NrCAM null mice (n = 3, *P < 0.05). (C, D) Sema3F-induced spine retraction on apical dendrites of WT neuron cultures is inhibited by dominant negative PlexA3R1407/1408A. (Means ± SEM; n = 15 neurons, *P < 0.05, t test). (E) Immunostaining of active β1-integrin (9EG7, red) on WT cortical neurons (EGFP, green) in culture treated with Fc or Sema3F-Fc (bar=10 μm). (F) Graph depicts mean fluorescence intensity of active β1 integrin within EGFP-labeled dendrites and spines (mean ± SEM, n = 10 neurons, *P<0.05, t test). (G) Immunostaining of active β1-integrin (9EG7, red) on WT cortical neurons transfected with PlexA3R1407/1408A (EGFP) in cultures treated with Fc or Sema3F-Fc. (H) Graph depicts mean fluorescence intensity of active β1 integrins within EGFP-labeled dendrites and spines (mean ± SEM, n = 10 neurons, *P<0.05, t test). (I) Immunostaining of active β1-integrin (9EG7, red) on NrCAM KO cortical neurons transfected with vector or WT NrCAM (EGFP, green) treated with Fc or Sema3F-Fc. (J) Graph depicts mean fluorescence intensity of active β1 integrin within EGFP-labeled dendrites and spines (mean ± SEM, n = 10 neurons, P*<0.05, t test). (K,L) Sema3F-induced spine retraction on apical dendrites of WT neuron cultures is inhibited by pretreatment with 4 mM MnCl2 for 1 h, (means ± SEM; n = 10 neurons, *P < 0.05, t test). Scale bar is 10 μm.
PlexA3 binds Npn2 and coimmunoprecipitates with NrCAM in brain and transfected cells (Demyanenko et al. 2014) but its requirement for Sema3F-induced spine retraction has not been assessed. To address this, cortical neuron cultures from WT mice were transfected at DIV11 with a dominant inhibitory PlexA3 mutant (R1407/1408A), or with empty vector (pCAGGS-IRES-mEGFP). The R1407/1408A double mutation lies within the Rap-GAP catalytic site of PlexA3 and abolishes signaling (He et al. 2009). At DIV14, cultures were treated with Sema3F-Fc or control Fc (3 nM), and spine density measured on apical dendrites after 30 min. PlexA3 R1407/1408A strongly inhibited Sema3F-induced spine retraction (Fig. 6C,D), indicating that the Rap1-GAP activity of PlexA3 mediates Sema3F-induced spine retraction in cortical neuron cultures. Activated Rap1 is known to recruit talin to the β-integrin tail, triggering integrin activation (Lilja et al. 2017). We further investigated if downregulation of activated Rap1 upon Sema3F treatment inactivated β1-integrin on dendrites and spines, as a possible mechanism for de-adhesion and spine collapse. WT neurons were transfected with EGFP and treated with Sema3F-Fc or Fc (3 nM, 30 min) at DIV14. Immunostaining was carried out using monoclonal β1-integrin antibody (9EG7), which specifically recognizes the active β1-integrin conformation (Lilja et al. 2017). A significant reduction in the levels of activated β1-integrin was observed on dendrites and spines after Sema3F treatment (Fig. 6E,F). To further test the role of PlexinA3 we transfected WT neurons with dominant inhibitory PlexA3 mutant (R1407/1408A) at DIV11, and treated with Sema3F-Fc or Fc (3 nM, 30 min) at DIV14. There were no differences in active β1-integrin levels in Sema3F-Fc or Fc treated neurons expressing the PlexA3 inhibitory mutant (Fig. 6G,H). We also measured active β1-integrin levels in cultured NrCAM KO neurons transfected at DIV11 with empty vector (pCAGGS-IRES-mEGFP) or plasmid expressing WT NrCAM, then treated with Sema3F-Fc or Fc (3 nM, 30 min) at DIV14. There were no significant differences in active β1-integrin levels in NrCAM KO neurons containing empty vector upon Sema3F-Fc treatment, but a significant reduction in active β1-integrin after Sema3F-Fc treatment of neurons expressing WT NrCAM (Fig. 6I,J). We further tested if it was possible to rescue spine collapse by activating β1-integrin directly by treating neurons with Mn++, a potent activator of β1-integrins (Mould et al. 2002). EGFP-transfected WT neurons were pretreated with 4 mM MnCl2 for 1 hour prior to stimulation with Sema3F-Fc or Fc (3 nM, 30 min). Results showed that MnCl2 rescued Sema3F-mediated spine collapse, as there was no significant difference in spine density between Fc or Sema3F-Fc treated neurons that received 4 mM MnCl2 (Fig. 6K,L). Taken together, these results demonstrated that Sema3F inactivated β1-integrins leading to spine collapse, and that NrCAM and PlexinA3 are required in this process.
Discussion
Molecular mechanisms governing spine pruning during the critical juvenile to adult transition are incompletely understood. Using a novel mouse line (NexCre-ERT2: NrCAM F/F; tdT) to inducibly delete NrCAM at different developmental stages, we demonstrate that NrCAM functions cell autonomously in immature pyramidal neurons during adolescence to constrain spine number on apical dendrites in the MFC and V1. In contrast, NrCAM deletion in adulthood had no effect, and thus may not regulate the slower spine turnover associated with increasing age in mature cortical circuits (Holtmaat et al. 2005). NrCAM was found to have a novel role in promoting developmental spine pruning through its ability to induce membrane clustering/oligomerization of the Sema3F holoreceptor complex. Receptor clustering was mediated through binding between NrCAM Ig1 (TARNER) and Npn2 a1 extracellular domains, and also required the NrCAM PDZ binding motif, which engaged the immature synaptic scaffold protein SAP102. NrCAM-dependent Sema3F receptor clustering stimulated the intrinsic Rap-GAP activity of PlexA3, leading to decreased Rap1 GTPase activity, β1-integrin inactivation, and spine elimination. Our results also provide evidence that NrCAM and PlexA3 are required for Sema3F-mediated β1-integrin inactivation, which leads to spine collapse.
The molecular mechanism by which Ig recognition molecules such as NrCAM promote neuronal repellent responses has remained enigmatic. NrCAM binds Npn2 in the absence of Sema3F or PlexA3 through a motif (TARNER) in NrCAM Ig1 (Demyanenko et al. 2014), but neither the molecular determinants mediating this interaction nor the consequences on spine remodeling were known. ClusPro docking of NrCAM Ig1 with the Npn2 a1 domain predicted a binding interface involving Arg120 in the NrCAM TAR120NER motif and Glu56 in the Npn2 a1 domain. Charge reversal substitutions in NrCAM TAR120NER (R120E) or Npn2 (E56R) inhibited NrCAM-Npn2 binding, and allowed us to test the hypothesis that NrCAM promotes membrane clustering of Sema3F receptors necessary for nascent spine elimination. We found that NrCAM, through binding to Npn2, promoted clustering/oligomerization of Sema3F holoreceptors in the dendritic membrane, inducing PlexA3 Rap-GAP signaling and spine retraction. Similarly, clustering of Eph receptors transduces intracellular signaling depending on the degree of multimerization (Seiradake et al. 2013; Schaupp et al. 2014).
In WT cortical neuron cultures treated with Sema3F-Fc, Rap1 GTPase activity was inhibited as a result of PlexA3 Rap-GAP activation, and β1-integrins became conformationally inactivated in dendrites and spines. Rap1-GTPase is known to activate inside-out signaling of β1-integrins and to enhance cell adhesion through talin binding (Lilja et al. 2017). Hence, Sema3F-induced inactivation of β1-integrins in spines may promote de-adhesion as an initial step in spine retraction. Additional signaling steps on the pathway that contribute to spine loss remain to be identified, such as effectors of cytoskeletal modulation or protein degradation. In addition, NrCAM could participate in as yet unidentified Sema3F-independent events necessary for spine elimination.
Postsynaptic scaffolding of synaptic proteins can dictate spine loss versus stability (Lambert et al. 2017). The NrCAM spine phenotype was not rescued by a construct lacking the PDZ binding motif (SFV), which has been shown to bind MAGUK (membrane-associated guanylate kinase) family scaffold proteins, including SAP97, SAP102, and PSD95 (SAP90) (Davey et al. 2005; Dirks et al. 2006; Yamagata and Sanes 2010). NrCAM coimmunoprecipitated with SAP102 and NR2B from P28 synaptoneurosomes, but not with PSD95. NrCAM also colocalized with SAP102 on dendritic spines in cortical neuron cultures. SAP102 binds and regulates trafficking of NR2B at nascent spine postsynaptic densities (Zheng et al. 2010, 2011; Chen et al. 2012), whereas PSD95 preferentially traffics NR2A-NMDA receptors at mature postsynaptic densities (Elias et al. 2008). The PDZ binding motif of NrCAM was also found to be an essential determinant of both Sema3F-induced receptor clustering and spine retraction in vitro. Although the direct role of SAP102 was not demonstrated, these results implicate SAP102 with its 3 PDZ motifs, in facilitating oligomerization of Sema3F receptors at immature synapses or restricting PlexA3 signaling to sites localized in the spine or dendritic membrane.
Our results provide a new understanding of the role of NrCAM and other L1-CAMs in class 3 Semaphorin signaling. We suggest that binding of NrCAM Ig1 to the Npn2 a1 interface, together with cytoplasmic PDZ scaffold interaction, stabilizes Npn2-PlexA3 binding so that Sema3F dimers can more efficiently form an active holoreceptor signaling complex. Our study is in general accord with a low resolution (2.7 Å) crystal structure of the related Sema3A receptor, which implicated Npn1 in cross-bridging PlexA2 and Sema3A dimers to form a hexameric complex (Janssen et al. 2012). A limitation of that work was that L1 or CHL1 were not included in the model, even though their binding to Npn1 is required for Sema3A function (Wright et al. 2007; Bechara et al. 2008). Because the affinity of PlexA2 for Npn1 (Kd 66 μM) or Npn1/Sema3A (Kd 6 μM) is relatively low (Janssen et al. 2012), formation of a stable signaling complex could be limited. Based on our findings with NrCAM, L1, and/or CHL1 may similarly add stability to the Sema3A holoreceptor complex thus promoting oligomerization and downstream signaling.
Temporal restriction of NrCAM function to adolescent rather than adult development adds to accumulating evidence that neocortical mechanisms of juvenile and adult plasticity differ (Hubener and Bonhoeffer 2014). Postnatal deletion of NrCAM from developing pyramidal neurons likely enhances cortical excitability, since deletion of NrCAM in global null mice increases the number of excitatory synapses and mEPSC frequencies in pyramidal neurons, and reduces their response to visually evoked potentials (Demyanenko et al. 2011). Developmental spine pruning through NrCAM and Sema3F may cooperate with other adhesion molecules including Cadherins (Wang et al. 2014; Bian et al. 2015), SynCAM1 (Cheadle and Biederer 2012), Contactins (Gdalyahu et al. 2015), and short-range repellent ligands such as Sema5A (Duan et al. 2014) and Ephrins (Penzes et al. 2011; Um et al. 2014), which regulate diverse aspects of spine morphogenesis and dynamics.
The present study reveals a novel temporal function for NrCAM in regulating Semaphorin-mediated dendritic spine pruning in pyramidal neurons in developing neocortex. Understanding how the density of spine subpopulations is regulated by NrCAM-dependent Sema3F signaling may illuminate how appropriate cortical E/I balance is established, and provide current insight into neurodevelopmental disease.
Supplementary Material
Notes
The Knockout Mouse Project (KOMP) of UC Davis and UNC Animal Models Core (Dale Cowley, Director) are gratefully acknowledged for ES cells and generation of NrCAM floxed mice. We thank Dr Klaus Nave for providing the NexCre-ERT2 mouse line, and Drs Elizabeth Benson and Keith Burridge for reagents and assistance with GTPase assays. Plasmids were kindly provided by Drs Dirk Montag (NrCAM), Tracy Tran (Npn2), Bonnie Fierstein (SAP102), and Xuewu Zhang (PlexA R1407/1408A). Sam George, Leann Brennaman, and Shubham Upadhyay contributed to initial spine assays. Dr Pablo Ariel, Director of the Microscopy Services Laboratory (UNC Department of Pathology), provided valuable assistance with imaging. This work is dedicated to the loving memory of late Mrs Rajni Singh, mother of V.M. Conflict of Interest: None declared.
Funding
National Institute of Health grants (MH113280, MH101605, NS 090029, P30 CA016086) and pilot award (2KR361203) from North Carolina Translational and Clinical Science Institute (NC TraCS).
References
- Agarwal A, Dibaj P, Kassmann CM, Goebbels S, Nave KA, Schwab MH. 2012. In vivo imaging and noninvasive ablation of pyramidal neurons in adult NEX-CreERT2 mice. Cereb Cortex. 22:1473–1486. [DOI] [PubMed] [Google Scholar]
- Alvarez VA, Sabatini BL. 2007. Anatomical and physiological plasticity of dendritic spines. Annu Rev Neurosci. 30:79–97. [DOI] [PubMed] [Google Scholar]
- Bechara A, Nawabi H, Moret F, Yaron A, Weaver E, Bozon M, Abouzid K, Guan JL, Tessier-Lavigne M, Lemmon V, Castellani V. 2008. FAK-MAPK-dependent adhesion disassembly downstream of L1 contributes to semaphorin3A-induced collapse. EMBO J. 27:1549–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MK, Gomot M, Baron-Cohen S. 2010. Visual attention in autism families: ‘unaffected’ sibs share atypical frontal activation. J Child Psychol Psychiatry. 51:259–276. [DOI] [PubMed] [Google Scholar]
- Bian WJ, Miao WY, He SJ, Qiu Z, Yu X. 2015. Coordinated spine pruning and maturation mediated by inter-spine competition for cadherin/catenin complexes. Cell. 162:808–822. [DOI] [PubMed] [Google Scholar]
- Brooks-Kayal A. 2010. Epilepsy and autism spectrum disorders: are there common developmental mechanisms? Brain Dev. 32:731–738. [DOI] [PubMed] [Google Scholar]
- Butler MG, Rafi SK, Hossain W, Stephan DA, Manzardo AM. 2015. Whole exome sequencing in females with autism implicates novel and candidate genes. Int J Mol Sci. 16:1312–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani V, De Angelis E, Kenwrick S, Rougon G. 2002. Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J. 21:6348–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle L, Biederer T. 2012. The novel synaptogenic protein Farp1 links postsynaptic cytoskeletal dynamics and transsynaptic organization. J Cell Biol. 199:985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BS, Gray JA, Sanz-Clemente A, Wei Z, Thomas EV, Nicoll RA, Roche KW. 2012. SAP102 mediates synaptic clearance of NMDA receptors. Cell Rep. 2:1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, He Z, Bagri A, Tessier-Lavigne M. 1998. Semaphorin-neuropilin interactions underlying sympathetic axon responses to class III semaphorins. Neuron. 21:1283–1290. [DOI] [PubMed] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ. 2004. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 20:45–50. [DOI] [PubMed] [Google Scholar]
- Cuthbert PC, Stanford LE, Coba MP, Ainge JA, Fink AE, Opazo P, Delgado JY, Komiyama NH, O’Dell TJ, Grant SG. 2007. Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J Neurosci. 27:2673–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey F, Hill M, Falk J, Sans N, Gunn-Moore FJ. 2005. Synapse associated protein 102 is a novel binding partner to the cytoplasmic terminus of neurone-glial related cell adhesion molecule. J Neurochem. 94:1243–1253. [DOI] [PubMed] [Google Scholar]
- Demyanenko GP, Mohan V, Zhang X, Brennaman LH, Dharbal KE, Tran TS, Manis PB, Maness PF. 2014. Neural cell adhesion molecule NrCAM regulates Semaphorin 3F-induced dendritic spine remodeling. J Neurosci. 34:11274–11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Riday TT, Tran TS, Dalal J, Darnell EP, Brennaman LH, Sakurai T, Grumet M, Philpot BD, Maness PF. 2011. NrCAM deletion causes topographic mistargeting of thalamocortical axons to the visual cortex and disrupts visual acuity. J Neurosci. 31:1545–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks P, Thomas U, Montag D. 2006. The cytoplasmic domain of NrCAM binds to PDZ domains of synapse-associated proteins SAP90/PSD95 and SAP97. Eur J Neurosci. 24:25–31. [DOI] [PubMed] [Google Scholar]
- Douglas RJ, Martin KA. 2004. Neuronal circuits of the neocortex. Annu Rev Neurosci. 27:419–451. [DOI] [PubMed] [Google Scholar]
- Duan Y, Wang SH, Song J, Mironova Y, Ming GL, Kolodkin AL, Giger RJ. 2014. Semaphorin 5A inhibits synaptogenesis in early postnatal- and adult-born hippocampal dentate granule cells. eLife 2014;3:e04390. doi: 10.7554/eLife.04390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Gama AM, Pochareddy S, Li M, Jamuar SS, Reiff RE, Lam AT, Sestan N, Walsh CA. 2015. Targeted DNA sequencing from autism spectrum disorder brains implicates multiple genetic mechanisms. Neuron. 88:910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias GM, Elias LA, Apostolides PF, Kriegstein AR, Nicoll RA. 2008. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proc Natl Acad Sci U S A. 105:20953–20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R, Wennerberg K, Arthur WT, Noren NK, Ellerbroek SM, Burridge K. 2006. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406:425–437. [DOI] [PubMed] [Google Scholar]
- Gdalyahu A, Lazaro M, Penagarikano O, Golshani P, Trachtenberg JT, Geschwind DH. 2015. The autism related protein contactin-associated protein-like 2 (CNTNAP2) stabilizes new spines: an in vivo mouse study. PLoS One. 10:e0125633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. 2013. Dendritic spine pathology in schizophrenia. Neuroscience. 251:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA. 2006. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 44:611–621. [DOI] [PubMed] [Google Scholar]
- Gordon U, Polsky A, Schiller J. 2006. Plasticity compartments in basal dendrites of neocortical pyramidal neurons. J Neurosci. 26:12717–12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Yang T, Terman JR, Zhang X. 2009. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc Natl Acad Sci U S A. 106:15610–15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. 2009. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 10:647–658. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. 2005. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 45:279–291. [DOI] [PubMed] [Google Scholar]
- Hosseinpour M, Mashayekhi F, Bidabadi E, Salehi Z. 2017. Neuropilin-2 rs849563 gene variations and susceptibility to autism in Iranian population: a case-control study. Metab Brain Dis. 32:1471–1474. [DOI] [PubMed] [Google Scholar]
- Hubener M, Bonhoeffer T. 2014. Neuronal plasticity: beyond the critical period. Cell. 159:727–737. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. 2010. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 1309:83–94. [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR. 1979. Synaptic density in human frontal cortex—developmental changes and effects of aging. Brain Res. 163:195–205. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Malinauskas T, Weir GA, Cader MZ, Siebold C, Jones EY. 2012. Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat Struct Mol Biol. 19:1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJ, Robinson RA, Perez-Branguli F, Bell CH, Mitchell KJ, Siebold C, Jones EY. 2010. Structural basis of semaphorin-plexin signalling. Nature. 467:1118–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopaske GT, Lange N, Coyle JT, Benes FM. 2014. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 71:1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Brenke R, Comeau SR, Vajda S. 2006. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 65:392–406. [DOI] [PubMed] [Google Scholar]
- Lambert JT, Hill TC, Park DK, Culp JH, Zito K. 2017. Protracted and asynchronous accumulation of PSD95-family MAGUKs during maturation of nascent dendritic spines. Dev Neurobiol. 77:1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviola G, Macri S, Morley-Fletcher S, Adriani W. 2003. Risk-taking behavior in adolescent mice: psychobiological determinants and early epigenetic influence. Neurosci Biobehav Rev. 27:19–31. [DOI] [PubMed] [Google Scholar]
- Lilja J, Zacharchenko T, Georgiadou M, Jacquemet G, Franceschi N, Peuhu E, Hamidi H, Pouwels J, Martens V, Nia FH, Beifuss M, Boeckers T, Kreienkamp HJ, Barsukov IL, Ivaska J. 2017. SHANK proteins limit integrin activation by directly interacting with Rap1 and R-Ras. Nat Cell Biol. 19:292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Focia PJ, He X. 2011. Homophilic adhesion mechanism of neurofascin, a member of the L1 family of neural cell adhesion molecules. J Biol Chem. 286:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness PF, Schachner M. 2007. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 10:19–26. [DOI] [PubMed] [Google Scholar]
- McAllister AK. 2007. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 30:425–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin M, Carlsson B, Anckarsater H, Rastam M, Betancur C, Isaksson A, Gillberg C, Dahl N. 2006. Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology. 54:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould AP, Askari JA, Barton S, Kline AD, McEwan PA, Craig SE, Humphries MJ. 2002. Integrin activation involves a conformational change in the alpha 1 helix of the beta subunit A-domain. J Biol Chem. 277:19800–19805. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Nonneman RJ, Grossman AW, Murphy DL, D’Ercole AJ, Crawley JN, Magnuson TR, Lauder JM. 2009. Social approach in genetically engineered mouse lines relevant to autism. Genes Brain Behav. 8:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Constantine-Paton M. 2013. Postsynaptic density scaffold SAP102 regulates cortical synapse development through EphB and PAK signaling pathway. J Neurosci. 33:5040–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Gan WB. 2008. Two-photon imaging of dendritic spine development in the mouse cortex. Dev Neurobiol. 68:771–778. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. 2011. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 14:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HB, Rakic P, Kostovic I. 2011. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A. 108:13281–13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Kaiserman-Abramof IR. 1970. The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am J Anat. 127:321–355. [DOI] [PubMed] [Google Scholar]
- Phillips M, Pozzo-Miller L. 2015. Dendritic spine dysgenesis in autism related disorders. Neurosci Lett. 601:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 466:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T. 2012. The role of NrCAM in neural development and disorders—beyond a simple glue in the brain. Mol Cell Neurosci. 49:351–363. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Lustig M, Babiarz J, Furley AJ, Tait S, Brophy PJ, Brown SA, Brown LY, Mason CA, Grumet M. 2001. Overlapping functions of the cell adhesion molecules Nr-CAM and L1 in cerebellar granule cell development. J Cell Biol. 154:1259–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaupp A, Sabet O, Dudanova I, Ponserre M, Bastiaens P, Klein R. 2014. The composition of EphB2 clusters determines the strength in the cellular repulsion response. J Cell Biol. 204:409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E, Schaupp A, del Toro Ruiz D, Kaufmann R, Mitakidis N, Harlos K, Aricescu AR, Klein R, Jones EY. 2013. Structurally encoded intraclass differences in EphA clusters drive distinct cell responses. Nat Struct Mol Biol. 20:958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Cowan CW. 2010. Guidance molecules in synapse formation and plasticity. Cold Spring Harb Perspect Biol. 2:a001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett MW, Gavin M, Tran TS. 2015. Altered hippocampal-dependent memory and motor function in neuropilin 2-deficient mice. Transl Psychiatry. 5:e521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu N, Ueta Y, Mohamed AA, Hatada S, Fukuda T, Kubota Y, Kawaguchi Y. 2016. Selective thalamic innervation of rat frontal cortical neurons. Cereb Cortex. 26:2689–2704. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Kumper M, Temple BS, Maness PF. 2016. The Neural Cell Adhesion Molecule (NCAM) promotes clustering and activation of EphA3 receptors in GABAergic interneurons to induce Ras homolog gene family, Member A (RhoA)/Rho-associated protein kinase (ROCK)-mediated growth cone collapse. J Biol Chem. 291:26262–26272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J, Sulzer D. 2014. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 83:1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey P, Parnau J, Blow M, Woffendin H, Bignell G, Cox C, Cox J, Davies H, Edkins S, Holden S, et al. 2004. Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 75:318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. 2004. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 38:151–158. [DOI] [PubMed] [Google Scholar]
- Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, Kolodkin AL. 2009. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 462:1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TN, Sharma K, Oh EC, Liu YP, Collins RL, Sosa MX, Auer DR, Brand H, Sanders SJ, Moreno-De-Luca D, et al. 2015. Loss of delta-catenin function in severe autism. Nature. 520:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um K, Niu S, Duman JG, Cheng JX, Tu YK, Schwechter B, Liu F, Hiles L, Narayanan AS, Ash RT, Mulherkar S, Alpadi K, Smirnakis SM, Tolias KF. 2014. Dynamic control of excitatory synapse development by a Rac1 GEF/GAP regulatory complex. Dev Cell. 29:701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villasana LE, Klann E, Tejada-Simon MV. 2006. Rapid isolation of synaptoneurosomes and postsynaptic densities from adult mouse hippocampus. J Neurosci Methods. 158:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. 2011. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Celic I, Choi SY, Riccomagno M, Wang Q, Sun LO, Mitchell SP, Vasioukhin V, Huganir RL, Kolodkin AL. 2014. Dlg5 regulates dendritic spine formation and synaptogenesis by controlling subcellular N-cadherin localization. J Neurosci. 34:12745–12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zeng H, Wang P, Soker S, Mukhopadhyay D. 2003. Neuropilin-1-mediated vascular permeability factor/vascular endothelial growth factor-dependent endothelial cell migration. J Biol Chem. 278:48848–48860. [DOI] [PubMed] [Google Scholar]
- Webb B, Sali A. 2014. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 47:5 6 1–32. [DOI] [PubMed] [Google Scholar]
- Wei Z, Behrman B, Wu WH, Chen BS. 2015. Subunit-specific regulation of N-methyl-D-aspartate (NMDA) receptor trafficking by SAP102 protein splice variants. J Biol Chem. 290:5105–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA. 2009. Autism genetics: emerging data from genome-wide copy-number and single nucleotide polymorphism scans. Expert Rev Mol Diagn. 9:795–803. [DOI] [PubMed] [Google Scholar]
- Wright AG, Demyanenko GP, Powell A, Schachner M, Enriquez-Barreto L, Tran TS, Polleux F, Maness PF. 2007. Close homolog of L1 and neuropilin 1 mediate guidance of thalamocortical axons at the ventral telencephalon. J Neurosci. 27:13667–13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Yue W, Jia M, Ruan Y, Lu T, Gong X, Shuang M, Liu J, Yang X, Zhang D. 2007. Association of the neuropilin-2 (NRP2) gene polymorphisms with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 144B:492–495. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. 2010. Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J Neurosci. 30:3579–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Ring C, Cheung R, Pevny L, Anton ES. 2007. Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron. 54:429–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanni G, van Esch H, Bensalem A, Saillour Y, Poirier K, Castelnau L, Ropers HH, de Brouwer AP, Laumonnier F, Fryns JP, Chelly J. 2010. A novel mutation in the DLG3 gene encoding the synapse-associated protein 102 (SAP102) causes non-syndromic mental retardation. Neurogenetics. 11:251–255. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Petralia RS, Wang YX, Kachar B, Wenthold RJ. 2010. SAP102 is a highly mobile MAGUK in spines. J Neurosci. 30:4757–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CY, Wang YX, Kachar B, Petralia RS. 2011. Differential localization of SAP102 and PSD-95 is revealed in hippocampal spines using super-resolution light microscopy. Commun Integr Biol. 4:104–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.