Abstract
Sir2p is an NAD+-dependent histone deacetylase required for chromatin-dependent silencing in yeast. In a cell-based screen for inhibitors of Sir2p, we identified a compound, splitomicin, that creates a conditional phenocopy of a sir2 deletion mutant in Saccharomyces cerevisiae. Cells grown in the presence of the drug have silencing defects at telomeres, silent mating-type loci, and the ribosomal DNA. In addition, whole genome microarray experiments show that splitomicin selectively inhibits Sir2p. In vitro, splitomicin inhibits NAD+-dependent histone deacetylase activity (HDA) of the Sir2 protein. Mutations in SIR2 that confer resistance to the drug map to the likely acetylated histone tail binding domain of the protein. By using splitomicin as a chemical genetic probe, we demonstrate that continuous HDA of Sir2p is required for maintaining a silenced state in nondividing cells.
Keywords: chemical genetics‖histone deacetylase‖silencing‖SIR2
Portions of the eukaryotic genome can be maintained in a transcriptionally inactive, or silenced, state as the result of the local chromatin structure. Silent chromatin may encompass regions ranging from a few thousand base pairs, as in the silent mating-type genes of the yeast Saccharomyces cerevisiae (1), to whole chromosomes such as the inactive X chromosome in mammals (2). The formation of silent chromatin, which is best understood at the S. cerevisiae silent mating-type loci HMR and HML, and telomeres depends on DNA elements, or silencers. The HM silencers are located in proximity to the genes that they regulate and contain a combination of binding sites for Rap1p, Abf1p, and the origin recognition complex (1). These proteins recruit the SIR (silent information regulator) protein complex (Sir2p–4p) through protein–protein interactions. Once recruited to silencers, the SIR complex is thought to spread along the chromatin through binding of Sir3p and Sir4p to the NH2-terminal tails of histone H3 and H4 (reviewed in ref. 3). Among the many requirements for silent chromatin (reviewed in ref. 4), posttranslational modification (i.e., acetylation, phosphorylation, methylation, and ubiquitination) of histones seems to be critical. For example, the NH2-terminal tails of histones H3 and H4 are hypoacetylated in silent chromatin compared with other regions of the genome (5). Of the SIR proteins, Sir2p has been shown to be an NAD+-dependent histone deacetylase and is responsible for the hypoacetylated state of histones in silent chromatin (6–9). Sir2p also acts at the rRNA-encoding DNA (rDNA) in the RENT protein complex, which does not include Sir3p or Sir4p (10).
The yeast SIR2 gene is the defining member of a broadly conserved family of NAD+-dependent deacetylases found in organisms ranging from bacteria to humans (11). In S. cerevisiae alone four additional homologues have been identified (see below). However, it shares the greatest similarity with genes found in other eukaryotes, where it is believed that these closely related homologues serve a comparable role in silencing. Interestingly, SIR2 and its homologues have been implicated in the genetic regulation of aging in both yeast and Caenorhabditis elegans (12, 13) and in metazoan development (M. I. Rosenberg and S. M. Parkhurst, personal communication), although the details of how it affects these fundamental processes are still mysterious.
To provide a new tool to dissect the functional role of Sir2p in vivo further, we undertook a phenotypic screen for small molecule inhibitors of the HDA of Sir2p. Our approach exploits the preexisting knowledge of Sir2p function in a drug screen to identify compounds that recreate the effect of a sir2 loss-of-function mutation. Here we report the identification of a compound that phenocopies the sir2 mutant in S. cerevisiae and inhibits the NAD+-dependent deacetylase activity of Sir2p in vitro. In a chemical genetic sense, we demonstrate that the inhibitor's action on wild-type Sir2p is equivalent to a loss-of-function mutation. By using this agent, we show that the deacetylase activity of Sir2p is required continuously for the maintenance of the silenced state in nondividing cells.
Materials and Methods
Yeast Media.
All strains were grown in synthetic complete medium or selective synthetic drop-out medium containing 2% glucose.
Compound Screening.
Drug screening was performed in 96-well plates. Each well was inoculated with 150 μl of yeast culture [strain UCC2210 MATα ppr1 adh4∷URA3-TEL(VII–L)] containing 1 × 105 cell per ml in uracil-deficient medium. Compounds dissolved in DMSO were applied at three different concentrations: 0.5, 5, and 50 μM. Cultures were incubated for 36–48 h, and growth in individual wells was tested by OD660 measurements and visual inspection. Splitomicin was purified from a mixture of compounds in a sample of NSC-112546.
Drug-Resistant SIR2 Mutants.
The conserved core region of SIR2 was amplified by using error-prone PCR and integrated into a SIR2-containing centromeric plasmid (pRS314-SIR2) by cotransformation into a sir2Δ strain with a URA3 telomeric marker [strain AB14053 MATα sir2 ppr1 adh4∷URA3-TEL(VII–L)]. Transformants from selective (−trp) medium were pooled, and aliquots were plated onto selective medium containing 5-fluoroorotic acid and 10 μM splitomicin. Plasmid DNA was recovered from the individual colonies and retransformed into the test strain to assure that drug resistance was conferred by SIR2-containing plasmid. All SIR2 ORFs from 20 independent plasmids conferring splitomicin resistance were sequenced. Mutations were introduced into a plasmid containing galactose-inducible SIR2 (pAR14; ref. 5) by using gap repair or site-directed mutagenesis to make GAL-SIR2-Y298N and GAL-SIR2-H286Q.
HDA Assay.
Histone H4 was acetylated chemically by using the HDAC assay kit (Upstate Biotechnology, Lake Placid, NY). The whole-cell extract was prepared as described (8) from an hst2Δ strain containing 2μ plasmid with galactose-inducible wild-type SIR2 (pAR14; ref. 5), mutant SIR2 (GAL-SIR2-Y298N or GAL-SIR2-H286Q), or empty vector. For histone deacetylase assays, 50 μg of yeast whole-cell protein extract was incubated with [3H]-acetylated histone H4 peptide (40,000 cpm) with or without 500 μM NAD+ in a 100-μl reaction. The buffer contained 150 mM NaCl, 25 mM sodium phosphate, pH 7.4, and 1 mM DTT. Reactions were incubated at 30°C for 16 h and stopped by the addition of 25 μl of 1 N HCl and 0.15 N acetic acid. Released [3H]acetate was extracted with 400 μl of ethyl acetate.
Microarrays.
Strains for the DNA array experiments were obtained from Research Genetics, Huntsville, AL (wild-type BY4741: MATa his3, leu2, met15, ura3, or isogenic sir2, hst1, hst2, hst3, and hst4 deletion mutants). Several colonies from fresh cultures were inoculated into synthetic complete medium with 2% glucose, grown overnight at 30°C, diluted to 0.5–1 × 106 cell per ml, and grown for an additional 6–9 h until reaching a density of 0.5–1 × 107 cells per ml. For experiments with splitomicin, drug or the solvent (DMSO) was added at the beginning of the final 9-h growth phase. In experiments with cycloheximide, cells were treated with 50 μg/ml of cycloheximide for 40 min before the addition of splitomicin. Total RNA was extracted by using the hot acid phenol method. Microarray construction and hybridization protocols were modified from those described elsewhere (14). Yeast microarrays were constructed by employing a set of ≈6,200 ORF-specific PCR primer pairs (Research Genetics), which were used to amplify each ORF of the yeast genome. Individual PCR products were verified as unique via gel electrophoresis and purified by using ArrayIt 96-well PCR purification kits (TeleChem International, Sunnyvale, CA). Purified PCR products were “spotted” mechanically in 3× SSC (450 mM sodium chloride and 45 mM sodium citrate, pH 7.0) onto polylysine-coated microscope slides by using an OmniGrid high-precision robotic gridder (GeneMachines, San Carlo, CA). The protocol used for cDNA labeling was a modification of a protocol described elsewhere (cmgm.stanford.edu/pbrown/protocols/aadUTPCouplingProcedure.htm).
Briefly, labeled cDNA targets were prepared by reverse transcription of 30 μg of total RNA using oligo dT(18) primer in the presence of 0.2 mM 5-(3-aminoallyl)-dUTP (Sigma–Aldrich), 0.3 mM dTTP, and 0.5 mM each of dATP, dCTP, and dGTP. After cDNA synthesis, either Cy3 or Cy5 monoreactive fluors (Amersham Pharmacia) were coupled covalently to the cDNA-incorporated aminoallyl linker in the presence of 50 mM sodium bicarbonate (pH 9.0). Two-color expression profiles were generated by using microarrays in which reference and experimental cDNA targets were labeled with different fluors. After cohybridization to the chip, a fluorescent image of the microarray was collected at both emission wavelengths by using a GenePix 4000 fluorescent scanner (Axon Instruments, Foster City, CA), and image analysis was performed by using GENEPIX PRO microarray acquisition and analysis software.
Three competitive hybridizations for each experimental group (sir2, hst1, hst2, hst3, and hst4 versus wild type, splitomicin-treated wild type versus wild type, and splitomicin plus cycloheximide versus cycloheximide alone) were performed by using three separate cultures and log2 of the expression ratio calculated for every ORF. To assess the intrinsic variation of expression level for different ORFs, nine wild type versus wild type hybridizations were performed by using nine separate cultures. The Student's t test was used to assess whether the difference between the log2 of the expression ratio for ORF in the experimental and control group (wild type versus wild type) is significant. Table 1, containing the mean log2 of the expression ratios and P values for all experiments, is published as supporting information on the PNAS web site, www.pnas.org.
Results and Discussion
Cell-Based Chemical Screen for the Sir2p Inhibitor.
To find inhibitors of the deacetylase activity of Sir2p, we screened for compounds that perturbed silencing at each of the loci at which Sir2p is known to act in S. cerevisiae: telomeres, HML, HMR, and the rDNA. The cell-based positive selection screen was designed such that inhibition of Sir2p activity permitted normal cell growth to avoid identifying cytotoxic compounds.
When the URA3 gene is in close proximity to a telomere in S. cerevisiae, it is repressed by telomeric chromatin (15). Because Ura3p is required for uracil biosynthesis, cells with the silenced telomeric URA3 gene are unable to grow in media lacking uracil. Accordingly, genetic perturbation of silencing activates URA3 expression and enables cells to grow in the absence of uracil (16). By using a strain with a telomeric URA3 gene, we screened 6,000 compounds from the National Cancer Institute repository for those that disrupted telomeric silencing. Eleven structurally unrelated compounds identified in this primary screen (data not shown) were analyzed further to determine whether silencing at the HML and HMR loci was also affected.
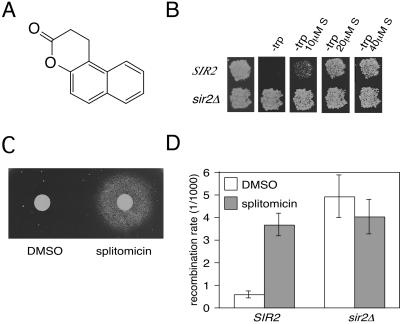
A strain with a TRP1 gene integrated at the silent HMR locus cannot grow in media lacking tryptophan (17). One of the 11 compounds enabled cells to grow in media lacking tryptophan (Fig. 1B), indicating a loss of silencing at HMR. This compound (1,2-dihydro-3H-naphtho[2,1-b]pyran-3–one, Fig. 1A), hereafter referred to as splitomicin, also disrupted silencing at HML. When haploid MATa cells are exposed to the mating pheromone α factor, they arrest in the G1 phase of the cell cycle. The loss of silencing at the HMLα locus in MATa cells results in expression of α mating-type genes (18). The coexpression of α and a genes creates a pseudodiploid state; cells are immune to α factor and unable to mate. In the presence of splitomicin, MATa cells lost responsiveness to α factor (Fig. 1C) and were defective for mating (data not shown). Thus, treatment with splitomicin disrupted silencing at HML, HMR, and telomeric loci. Because the remaining 10 compounds did not disrupt silencing at HML and HMR, they were not tested in subsequent assays.
Figure 1.
(A) Chemical structure of splitomicin. (B) Activation of a TRP1 reporter at the silent HMR mating locus by splitomicin (S). Wild-type (SIR2) or sir2Δ cells with TRP1 integrated into HMR (19). Cells were replica-plated onto complete synthetic medium or medium lacking tryptophan (−trp) without or with the indicated concentrations of splitomicin. (C) Loss of responsiveness to α factor in the presence of splitomicin. Logarithmically growing MATa cells were imbedded into agar with synthetic medium containing 2.5 μM α factor. The paper discs with 5 μl of DMSO or 5 mM splitomicin were placed onto the agar, and the plate was incubated at 30°C for 2 days. The halo of cells indicates those able to grow. (D) Splitomicin increases recombination of an ADE2 reporter integrated within the ribosomal DNA array. Logarithmic-phase cells were exposed to splitomicin (15 μM) or DMSO for 6 h and plated onto rich medium. The recombination rate was calculated directly from the frequency of loss of the ADE2 gene in the first division after plating by counting half-sectored red colonies. Three independent determinations were performed for each experimental group.
Splitomicin Increases Recombination at the rDNA.
Silencing within the rDNA locus is manifested in two ways. It can weakly repress expression of an inserted reporter gene (19), and it reduces recombination between tandem copies of the ribosomal RNA genes (20). Splitomicin disrupted silencing of a reporter gene within the rDNA locus just as it did at telomeres and the HM loci (data not shown). Recombination was analyzed by measuring the loss rate of an ADE2 gene integrated into the rDNA array (21). Treatment with splitomicin increased the recombination rate at the rDNA locus 7-fold, which is similar to rates observed in a sir2 mutant (Fig. 1D). There was no effect on rDNA recombination in sir2 cells treated with the compound, indicating that splitomicin was acting specifically through the SIR2 pathway. Taken together, these data suggest that splitomicin treatment creates a phenocopy of a sir2 mutant.
Transcriptional Profiles of Splitomicin-Treated Cells Mimic Profiles of sir2 and hst1 Mutants.
In addition to SIR2, the S. cerevisiae genome encodes four SIR2 homologues: HST1–4 (homologue of sir two; ref. 22). Hst2p is located in the cytoplasm and is responsible for virtually all the NAD+-dependent deacetylase activity detected in a cellular lysate (8). Its relevant biological substrate is unknown. Hst1p is required for the transcriptional repression of meiotic genes (23), whereas little is known about the cellular function of Hst3p or Hst4p. To determine whether the antisilencing effects of splitomicin were mediated solely by Sir2p and whether splitomicin affected any of the Hst proteins, we compared the expression profile of wild-type cells grown in the presence of splitomicin to that of sir2, hst1, hst2, hst3, or hst4 deletion mutants by whole-genome DNA microarray analysis. The transcriptional effects of splitomicin correlated most highly with those of a sir2 mutation (correlation coefficient, 0.748; Fig. 2A and Table 1). Genes adjacent to telomeres such as COS12 and the α1 and α2 genes from the HML locus were up-regulated significantly in both conditions (Fig. 2A and Table 1). The expression of MATa-specific (e.g., MFA1, STE2, STE6, and BAR1) and haploid-specific genes (e.g., FUS1 and STE5) was down-regulated in both splitomicin-treated and sir2 cells (Fig. 2 A and B). Splitomicin also up-regulated a small number of genes that were not altered in sir2 cells including meiosis-specific genes (e.g., SPS1) that seem to be regulated by HST1 (Fig. 2B). There was no overlap between splitomicin and HST2-, HST3-, or HST4-regulated genes (supporting Table 1, Fig. 2D). Thus the majority of all transcriptional changes (88%) induced by splitomicin were mediated through SIR2, and a smaller subset (9%) was mediated through HST1 (Fig. 2B). These results indicate that splitomicin inhibits Sir2p and to a lesser extent Hst1p.
Figure 2.
Splitomicin-treated wild-type (wt) cells and sir2Δ cells display similar transcriptional changes relative to untreated wild-type cells. (A) Correlation of transcriptional changes between genetic and chemical inactivation of Sir2p. Transcriptional changes were determined by competitive hybridization to DNA microarrays containing >6,000 yeast ORFs. The correlation plot shows transcriptional changes in a sir2Δ mutant relative to wild type (sir2Δ/wt) on the vertical axis and changes in wild-type cells treated with splitomicin relative to untreated wild-type cells (15 μM splitomicin/no treatment) on the horizontal axis. (B) A Venn diagram comparing genes up-regulated (Left) and down-regulated (Right) more than 2-fold relative to wild-type or untreated cells and sir2Δ, hst1Δ, or splitomicin-treated wild-type cells. (C) Correlation of transcriptional changes in wild-type cells in response to splitomicin treatment with and without cycloheximide. The correlation plot shows transcriptional changes in splitomicin- and cycloheximide-treated wild-type cells relative to cells treated with cycloheximide alone (15 μM splitomicin CYH/CYH) on the vertical axis and changes in wild-type cells treated with splitomicin relative to untreated wild-type cells (15 μM splitomicin/no treatment) on the horizontal axis. (D) Venn diagrams comparing transcriptional changes (up- or down-regulation) in hst2Δ, hst3Δ, and hst4Δ cells and splitomicin-treated cells (split).
Identifying Direct Targets of SIR2.
Sir2p is critical for silencing, yet the majority of the transcriptional changes induced by either chemical or genetic inactivation of the enzyme constituted transcriptional down-regulation (Fig. 2 A and B). A number of these changes are known to be indirect. For instance, haploid-specific genes are down-regulated by the gene products of the derepressed HMLα and HMRa loci (18). Splitomicin, combined with the protein synthesis inhibitor cycloheximide, afforded a unique opportunity to identify genes that are regulated directly by Sir2p. Such an examination has not been possible before, because conditional alleles of SIR2 are not available. The addition of cycloheximide did not affect the up-regulation of genes by splitomicin treatment. In contrast, virtually all transcriptional down-regulation was abolished in the absence of new protein synthesis, confirming that the direct effect of Sir2p is to repress transcription (Fig. 2C). With the exception of a single gene, BPH1, the only genes that were up-regulated as a result of Sir2p inactivation in the presence of cycloheximide were subtelomeric genes and silent mating-type loci (supporting Table 1), indicating that Sir2p activity does not affect transcription outside of these regions. Overall, these results are consistent with a recent study examining the location of Sir2p by genome-wide chromatin immunoprecipitation (24).
Splitomicin Inhibits the Deacetylase Activity of Sir2p.
Inhibition of the HDA of Sir2p was the most likely mechanism by which splitomicin caused its phenotypic changes. Therefore, we tested whether splitomicin inhibited the HDA of Sir2p in vitro. We used a [3H]-acetylated histone H4 peptide and measured the NAD+-dependent release of free [3H]acetate in the presence of whole yeast cell extract from an hst2 strain overexpressing yeast SIR2. [A cell extract obtained from a SIR2-overexpressing hst2 strain had robust NAD+-dependent HDA derived exclusively from Sir2p (Fig. 3 A and B)]. Splitomicin induced dose-dependent inhibition of HDA in the yeast extract, with an IC50 of 60 μM (Fig. 3A). This result established Sir2p deacetylase activity as a direct target of splitomicin.
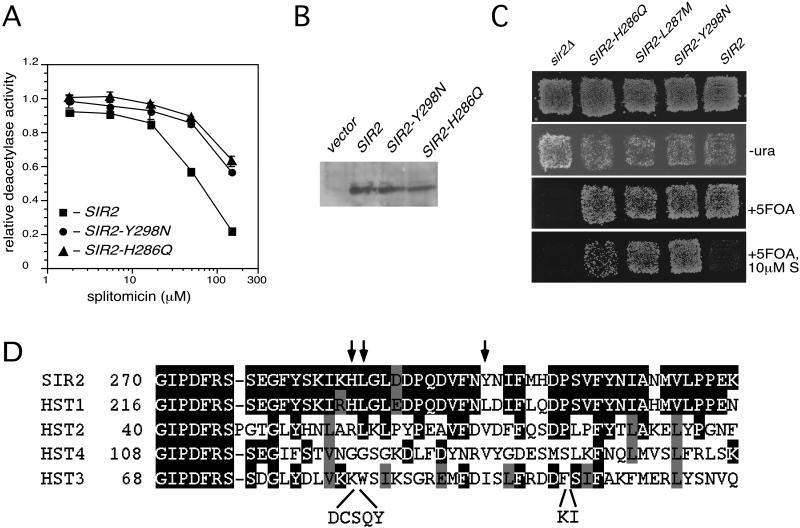
Figure 3.
(A) Inhibition of NAD-dependent HDA of Sir2p by splitomicin. The effect of splitomicin on NAD+-dependent HDA in wild-type and drug-resistant Sir2p mutants is shown. Chemically [3H]-acetylated histone H4 peptide (40,000 cpm per reaction) was incubated with whole-cell protein extracts (50 μg) prepared from hst2Δ strain containing overexpressed wild-type SIR2 or two drug-resistant SIR2 alleles (SIR2-Y298N and SIR2-H286Q), NAD+, and splitomicin at 30°C for 16 h. The assays were done in triplicate. The NAD+-dependent activity in the extract without splitomicin was 1,776 ± 25 cpm for wild-type SIR2, 1,620 ± 44 cpm for SIR2-Y298N, 1,795 ± 36 cpm for SIR2-H286Q, and 28 ± 14 cpm for cells containing the empty vector. (B) Immunoblot of Sir2p in whole-cell lysates containing overexpressed wild-type or drug-resistant mutant SIR2. The whole-cell lysates (25 μg) prepared from hst2Δ strain containing empty vector or overexpressed wild-type SIR2 and two drug-resistant SIR2 alleles (SIR2-Y298N and SIR2-H286Q) were probed with an anti-Sir2p antibody (Santa Cruz Biotechnology). (C) Telomeric silencing in SIR2, sir2Δ, and drug-resistant SIR2 mutants. Cells from a sir2Δ strain with telomeric URA3 gene containing either empty plasmid (sir2Δ), a plasmid with wild-type SIR2, or drug-resistant alleles SIR2-H286Q, SIR2-L287M, and SIR2-Y298N were replica-plated onto selective medium lacking leucine (for plasmid selection), selective medium lacking uracil (−ura), or selective medium to which 5-flouroorotic acid was added (+5-FOA) with or without 10 μM splitomicin and incubated at 30°C for 2 days. Expression of the telomeric URA3 gene kills cells, because Ura3p converts 5-fluoroorotic acid into a toxic metabolite. (D) Sequence alignment between yeast Sir2p and Hst1–4p. The region displayed in the alignment contains the putative substrate-binding site. Arrows indicate the positions of residues that, when mutated in Sir2p, confer splitomicin resistance.
Mutations Conferring Drug Resistance to Splitomicin Map to the Putative Acetyl-Peptide Binding Cleft.
To obtain insight into the molecular mechanism by which splitomicin inhibited deacetylase activity of Sir2p we generated mutant forms of Sir2p that were resistant to the compound. We identified three alleles of SIR2 (SIR2-H286Q, SIR2-L287Q, and SIR2-Y298N) that render yeast cells resistant to the antisilencing effects of splitomicin. Silencing was at normal levels in the drug-resistant mutants in the absence of drug, but disruption of silencing in the mutants required higher concentrations of splitomicin than in wild-type strains (Fig. 3C and data not shown). In vitro, when compared with equivalent amounts of wild-type Sir2p, mutant proteins exhibited similar HDA in the absence of drug, with increased resistance to the inhibitory effect of splitomicin (Fig. 3 A and B).
The three mutations lie in close proximity within a region that is highly similar to human SIRT2. Most interestingly, the crystal structure of SIRT2 defines this region to be a hydrophobic cavity that is hypothesized to be the binding site for acetylated lysine peptides (25, 26). As noted above, the expression profile of splitomicin-treated cells has no overlap with mutant hst2, hst3, or hst4 strains but does have some overlap with the hst1 profile. Intriguingly, of all the HST genes, Hst1p has the highest sequence similarity (86% identity) to Sir2p in the 50-aa region containing the splitomicin resistance mutations (Fig. 3D). Because Hst1p also acts to repress gene expression via hypoacetylation of histones (27, 28), it seems likely that this shared region defines a common binding pocket for acetylated histone tails in both proteins. Thus, splitomicin likely inhibits Sir2p deacetylase activity by altering or blocking access to the acetylated histone binding pocket.
Continuous Deacetylase Activity of Sir2p Is Required for the Maintenance of the Silent State in Nondividing Cells.
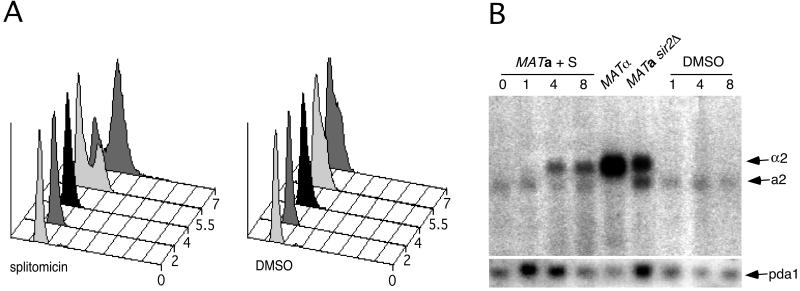
The establishment of silencing in previously active chromatin is a cell cycle-dependent event that can be accomplished only during S phase (29, 30). Once established, the silent state needs to be maintained between cell divisions: after mitosis, in G1, and into the next S phase. Studies with a temperature-sensitive allele of SIR3 demonstrated that silencing is lost quickly in G1-arrested cells after cells are shifted to the nonpermissive temperature (31). In contrast, removal of the DNA silencer elements from the HMLα locus in G1-arrested cells does not disrupt silencing (32). The study with the temperature-sensitive allele of SIR3 suggests that the presence of the entire SIR complex is required for the maintenance of a silent state. However, we were curious whether the deacetylase activity of Sir2p was required for the maintenance of a silent state in nondividing cells or whether it was dispensable once silent chromatin was formed. We took advantage of splitomicin's ability to inhibit the HDA of Sir2p to address this issue. MATa cells were arrested first in G1 by using α factor and then treated with splitomicin. Although untreated cells remained arrested in G1, those treated with splitomicin progressed through the cell cycle (Fig. 4A). This progression presumably was caused by a loss of mating competence, a consequence of expression of the α2 gene from the “silent” HML locus. To test this idea more directly, a MATa strain with a single G1 cyclin gene (CLN3), which is under control of a galactose-inducible promoter (33), was arrested in G1 by replacing galactose with glucose in the medium. Once the cells arrested in G1, they were treated with splitomicin or DMSO control. While the cells remained arrested in G1 under both conditions, α2 mRNA expression from HML was detected only in the splitomicin-treated cells (Fig. 4B). The lag period of several hours between the application of splitomicin and the appearance of α2 mRNA was similar to the delay before cell cycle progression was observed in the α factor-arrested cells treated with splitomicin (described above). These results demonstrate that the deacetylase activity of Sir2p is required continuously for maintenance of the silent state in nondividing cells.
Figure 4.
(A) Cell cycle analysis of α factor arrested MATa cells treated with splitomicin. Logarithmically growing MATa cells were treated first with α factor for 90 min. At time 0, splitomicin (20 μM) or DMSO was added to the culture. DNA content of the cells was determined by flow cytometry at several time points after the addition of splitomicin. (B) α2 mRNA expression from the silent HML locus in G1-arrested cells treated with splitomicin. A strain containing the galactose-inducible CLN3 gene in which genomic G1 cyclin genes were deleted [MATa cln1Δ, cln2Δ, cln3Δ, GAL-CLN3 (35)], was arrested in G1 by exposure to glucose for 90 min. Splitomicin (20 μM) or DMSO was added to the culture of these G1-arrested cells, and the expression of MATα from the silent HML locus was assessed at several time points. Both splitomicin- and DMSO-treated cells remained arrested in G1 as judged by flow cytometry (data not shown). The RNA from MATα and MATa sir2Δ cells is included for comparison. The weak lower molecular weight band is caused by cross hybridization to a2 mRNA. The blot was stripped and reprobed for the PDA1 mRNA as a loading control.
This finding suggests that Sir2p must remain diligent in its maintenance of the silent state, presumably counteracting the constant activity of histone acetylases. The acetylases may gain access to the chromatin in a targeted manner via transcriptional activators (34, 35) or be part of a global histone acetylation maintenance system (36). These results also support the idea that silent chromatin is not a static, rigid structure, but rather that it is in a dynamic equilibrium with silencing factor exchanging on and off the chromatin even when cells are not dividing (37).
Our study underscores the power of phenotypic screening in model systems to identify new compounds that are useful for dissecting complex biological processes such as silencing in vivo. To this end, the identification of an inhibitor of Sir2p complements the existing inhibitors of histone deacetylases [i.e., trapoxin and trichostatin (38)]. Because SIR2 homologues exist in other organisms, the identification of splitomicin serves as a paradigm for applying chemical genetic methods to study the role of Sir2p-like enzymes in silencing, development, and aging in a variety of species. In addition to histones, many other proteins are regulated by acetylation including pRb, E2F, and p53 proteins (39–41). Two recent reports (42, 43) implicate deacetylation of p53, by Sir2, in down-regulation of transcriptional and proapoptotic activities of p53 in response to DNA damage. Toxicity assays using splitomicin and a variety of DNA-damaging agents have shown that splitomicin sensitizes mammalian cells to these agents (A.B., J. R. Lamb, and J.A.S., unpublished observation), which is consistent with splitomicin abrogating Sir2p activity on p53. Thus, splitomicin may serve a useful role in the evaluation of Sir2p-like deacetylases as drug targets for treating cancer and other diseases (44, 45).
Supplementary Material
Acknowledgments
We are grateful to J. Broach, L. Guarente, and D. Shore for plasmids and yeast strains, J. Delrow for assistance with the microarray experiments, C. Kooperberg for advice on the statistical analysis of the microarray data, J. Johnson and R. Schultz of the Developmental Therapeutics Program National Cancer Institute for compounds, members of the Gottschling and Simon labs for valuable discussions, and B. Berg, S. Parkhurst, J. Roberts, T. Tsukiyama, and E. Foss for comments on the manuscript. This work was supported by National Heart, Lung, and Blood Institute Grant HL04211 (to A.B.), National Institutes of Health Grant GM43893 (to D.E.G.), and National Cancer Institute Grant CA78746 (to J.A.S.).
Abbreviations
- SIR
silent information regulator
- rDNA
rRNA-encoding DNA (rRNA gene)
- HDA
histone deacetylase activity
Note
During the preparation and review of this manuscript, a study reporting identification of α-substituted β-naphthol compounds that inhibit Sir2p and are related to splitomicin structurally was published by Grozinger et al. (46).
References
- 1.Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 2.Lyon M F. Curr Biol. 1999;9:R235–R237. doi: 10.1016/s0960-9822(99)80151-1. [DOI] [PubMed] [Google Scholar]
- 3.Gartenberg M R. Curr Opin Microbiol. 2000;3:132–137. doi: 10.1016/s1369-5274(00)00064-3. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Grunstein M. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 5.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 6.Moazed D. Curr Opin Cell Biol. 2001;13:232–238. doi: 10.1016/s0955-0674(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 7.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 8.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. . (First Published May 16, 2000; 10.1073/pnas.110148297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straight A F, Shou W, Dowd G J, Turck C W, Deshaies R J, Johnson A D, Moazed D. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 11.Frye R A. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 12.Tissenbaum H A, Guarente L. Nature (London) 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 13.Sinclair D A, Guarente L. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 14.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 15.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 16.Singer M S, Kahana A, Wolf A J, Meisinger L L, Peterson S E, Goggin C, Mahowald M, Gottschling D E. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck S W, Shore D. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 18.Marsh L, Neiman A M, Herskowitz I. Annu Rev Cell Biol. 1991;7:699–728. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- 19.Smith J S, Boeke J D. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb S, Esposito R E. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 21.Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 23.Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon A K. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieb J D, Liu X, Botstein D, Brown P O. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 25.Finnin M S, Donigian J R, Pavletich N P. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 26.Min J, Landry J, Sternglanz R, Xu R M. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 27.Rusche L N, Rine J. Genes Dev. 2001;15:955–967. doi: 10.1101/gad.873601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutton A, Heller R C, Landry J, Choy J S, Sirko A, Sternglanz R. Mol Cell Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y C, Cheng T H, Gartenberg M R. Science. 2001;291:650–653. doi: 10.1126/science.291.5504.650. [DOI] [PubMed] [Google Scholar]
- 30.Kirchmaier A L, Rine J. Science. 2001;291:646–650. doi: 10.1126/science.291.5504.646. [DOI] [PubMed] [Google Scholar]
- 31.Miller A M, Nasmyth K A. Nature (London) 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- 32.Holmes S G, Broach J R. Genes Dev. 1996;10:1021–1032. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- 33.Cross F R. Mol Cell Biol. 1990;10:6482–6490. doi: 10.1128/mcb.10.12.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aparicio O M, Gottschling D E. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 35.Sekinger E A, Gross D S. Cell. 2001;105:403–414. doi: 10.1016/s0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- 36.Vogelauer M, Wu J, Suka N, Grunstein M. Nature (London) 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 37.Cheng T H, Gartenberg M R. Genes Dev. 2000;14:452–463. [PMC free article] [PubMed] [Google Scholar]
- 38.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 39.Chan H M, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue N B. Nat Cell Biol. 2001;3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Balbas M A, Bauer U M, Nielsen S J, Brehm A, Kouzarides T. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo J, Su F, Chen D, Shiloh A, Gu W. Nature (London) 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 42.Vaziri H, Dessain S K, Eaton E N, Imai S I, Frye R A, Pandita T K, Guarente L, Weinberg R A. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 43.Luo J, Nikolaev A Y, Imai S I, Chen D, Su F, Shiloh A, Guarente L, Gu W. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 44.Wolffe A P. Oncogene. 2001;20:2988–2990. doi: 10.1038/sj.onc.1204322. [DOI] [PubMed] [Google Scholar]
- 45.Tycko B, Ashkenas J. J Clin Invest. 2000;105:245–246. doi: 10.1172/JCI9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grozinger C M, Chao E D, Blackwell H E, Moazed D, Schreiber S L. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.