Abstract
The human genome is now investigated through high-throughput functional assays, and through the generation of population genomic data. These advances support the identification of functional genetic variants and the prediction of traits (e.g. deleterious variants and disease). This review summarizes lessons learned from the large-scale analyses of genome and exome data sets, modeling of population data and machine-learning strategies to solve complex genomic sequence regions. The review also portrays the rapid adoption of artificial intelligence/deep neural networks in genomics; in particular, deep learning approaches are well suited to model the complex dependencies in the regulatory landscape of the genome, and to provide predictors for genetic variant calling and interpretation.
Introduction
The rapidly falling cost and ease of collecting genomics data has increasingly highlighted our relatively limited abilities to interpret what this data says about traits, particularly in a medical context. In the classic words of Eric Lander, ‘Genomics: bought the book; hard to read’. Despite significant progress in understanding coding variation (changes in gene sequences, which nevertheless remain deeply challenging), making strong statements about the consequences of variation in non-coding regions, which constitute 98% of the human genome remains a key challenge in the field. These regions contribute to the regulatory networks that for example, direct development, tissue specificity, gene expression and disease perturbation. Challenges in the interpretation of the non-coding genome include a relative lack of strong landmarks (as opposed to, say the exon-intron boundaries, codon alphabet), the reliance on bioinformatic prediction of elements and functions, the dynamic nature of the regulatory apparatus and the long-range regulatory relationships.
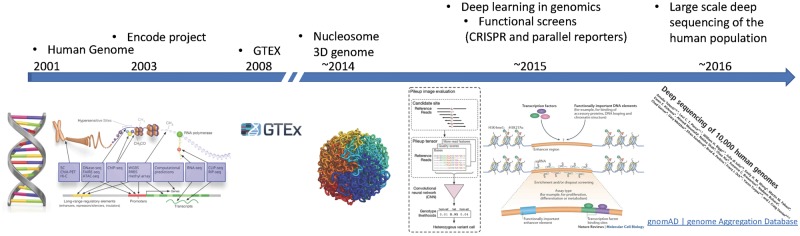
Novel technologies to study functional elements, regulatory organization of the genome and genetic variation are poised to help overcome some of the barriers limiting the study of non-coding variation and their impact on human traits and diseases (Fig. 1). Machine-learning methods allow us to learn functional relationships from data in the form of predictive models, largely free of strong assumptions about the underlying biological mechanisms (see reviews by Angermueller etal. (1) and Ching et al. (2).
Figure 1.
Convergence of data-rich technologies fueling the field of machine learning for genomics.
The canonical machine-learning workflow involves four steps: data cleaning and pre-processing, feature engineering, model fitting and evaluation. Deriving the most informative features is essential for performance, but effective feature engineering (the process of using domain knowledge of the data to create features) is labor intensive (1). Deep learning, one of the most active fields of machine learning, is so exciting in large part because it reduces, and in many cases, eliminates the need for feature engineering.
This review highlights successive steps that exploit increasingly large and high-dimensional genomic data sets. Importantly, the steps aim at decreasing the expert input in favor of an increasingly automated process. The text reports on progress in descriptive analyses, modeling, classic pipelines involving machine learning and deep learning approaches to large-scale genomics data. This review will not discuss other important areas of development such as the use of deep learning for the prediction of protein structure from sequence data (3,4). Specific studies are showcased in each section.
Descriptive Approaches
The estimated size of the human genome is 3.2 × 109 bp. As of 2017, large community and corporate efforts had identified single-nucleotide variants (SNV) across the genome: 150 million SNVs in the public database dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_summary.cgi; date last accessed April 4, 2018), 10 million coding variants in ExAC (http://exac.broadinstitute.org; date last accessed April 4, 2018) (5) and 150 million SNVs in 10 545 deeply sequenced whole genomes (http://hli-opensearch.com; date last accessed April 4, 2018) (6). The union of these resources was 242 million unique SNVs, suggesting that by the time of this assessment, at least 1 out of every 13 nucleotides in the genome has an observed variant in the population.
Telenti et al. (6) report on 10 545 whole genomes sequenced at a depth of 30× coverage to show that 84% of the genome could be sequenced reliably. The mean single nucleotide density was 57 per 1 kb of sequence. However, there were differences across chromosomes, and importantly, different rates of variation depending on the sequence context. Two papers ‘deconstructed’ the genome into k-mers [specifically, heptamers) to characterize the genome-wide rates of variation accounting their genomic context (7,8)].
There are 16 384 unique heptamers present in the human genome. Heptamers vary greatly in abundance, ranging between 1941 (TACGCGA) and 6 332 326 (AAAAAAA) counts per genome, and are not evenly distributed genomewide (8). The various genomic elements differ in the heptamer composition and each heptamer is characterized by unique rates of variation. The k-mer context was shown to explain more than 81% of variability in substitution probabilities in the human genome (7). To capture this property, di Iulio et al. (8) computed the rate and frequency of variation at the central (fourth) nucleotide of each heptamer. The metric varies 95-fold across heptamers (between 0.0046 and 0.438). It defines the expectation of variation for each nucleotide in the genome.
There are two aspects of these works that make them meaningful in the current review. First, large data sets of deep sequenced human genomes are providing increasingly precise estimates of genetic variation and accurate models of nucleotide substitution (7). Second, the conserved or hypervariable nature of any sequence in the genome can be based on observed versus expectation in a sequence-specific (k-mer) context. Conserved non-coding genomic elements regulate the most essential genes (8).
In summary, we can now describe the saturation of the human genome with variation. This basic exercise is highly informative of the constraints imposed on the genome, and thus of the underlying functional requirements. These new perspectives on genetic diversity can be explored in the frame of current knowledge on functional elements accumulated through ENCODE work (9), conservation of elements across species, and other descriptive approaches. The data sets are increasingly rich, and optimally suited for modeling and for machine learning.
Modeling and Scoring Algorithms
Analysis of protein coding regions
Predicting the functional, evolutionary or medical implications of a given genetic variant is a long-studied problem (10). The most widely used tool today is combined annotation-dependent depletion (CADD), a method (support vector machine) for integrating many diverse annotations into a single measure (C score) for each variant (11). This review highlights novel initiatives (12) that leverage three-dimensional protein structure information to impute functionality of protein domains and the deleteriousness of variants in conserved domains.
Recently, Hicks et al. (13) used data on over 140 000 human genome and exomes to understand variation in the three-dimensional structural proteome. This work first identified 26 593 structures associated with 4390 representative uniprot entries. Then, 139 535 uniprot features were mapped to the structures, to extract a three-dimensional context by defining a 5 Å radius space for each feature. The population data provided 481 708 missense variants for these proteins from the analysis of 146 426 individuals’ exomes. These data were modeled to describe functional constraints in three-dimensional protein structures. Structural intolerance data correlated with experimental functional read-outs in vitro and revealed characteristic features of ligand-binding pockets, orthosteric and allosteric sites.
Non-coding genome pathogenicity scores
While there are multiple functional predictive scores that are used to prioritize variants in the protein coding, less is known about the functional consequences of genetic variation in the non-protein coding genome. In this section, we highlight seven recently reported tools: Eigen (14), CADD (11), FunSeq2 (15), LINSIGHT (16), FATHMM (17), ReMM (18) and Orion (19) (Table 1). These scores share to certain extent a common source of labeled data (limited number of well-studied pathogenic variants in the non-coding genome), but apply a great diversity of modeling approaches. In addition, the context-dependent tolerance score (CDTS) (8) is based on the analysis of genome heptamer properties (see previous section), can also be used for the ranking of pathogenic variants (Table 1); however, it differs from other scores by being independent of existing labeled data. DeepSEA (20) will be presented below in the section on deep learning.
Table 1.
Non-coding genome pathogenicity scores
| Score | Data sources | Approach | Reference |
|---|---|---|---|
| Eigen |
|
|
(14) |
| FunSeq2 |
|
|
(15) |
| LINSIGHT |
|
|
(16) |
| CADD |
|
|
(11) |
| FATHMM |
|
|
(17) |
| ReMM |
|
|
(18) |
| Orion |
|
|
(19) |
| CDTS |
|
|
(8) |
Prediction of polygenic scores
The various scores described above generally serve to predict the functionality and deleteriousness of single variants. However, many complex traits and disorders (e.g. metabolic syndrome) are also defined by the contributions of many variants that can be represented in a single score. Typically, those variants, identified through genome-wide association studies, are included in polygenic risk scores. These scores are usually constructed as a weighted sum of allele counts, the weights being given by log odds ratios or linear regression coefficients from univariate regression tests from the originating population genotyping studies (21). We chose here to showcase progress in the field by discussing recent work by Pare et al. (22). This paper leverages the large number of SNVs and the available summary-level statistics from genome-wide association studies to calibrate the weights of SNPs contributing to the polygenic risk score, adjusting for linkage disequilibrium (instead of pruning). A limitation of the method (like in other publications) is that it is based on the premise that SNVs contribute additively to genetic variance. As indicated by Pare et al. (22), incremental improvements are to be expected with increased sample size, the inclusion of additional predictors and the availability of more precise summary association statistics.
In summary, many features are used to train models that predict the consequences of genetic variation in coding and non-coding regions of the genome. The output is expressed in scores that are used to rank and prioritize candidate variants for further investigation, or polygenic scores that summarize effects. Although continuously evolving, most approaches tend to train on limited sets of known pathogenic variants, and to use the same data resources.
Determining Sequence Variation
Traditionally, calling genetic variants from sequencing instrument data are done using complex and expert-engineered pipelines that involve multiple steps, such as alignment, generation of relevant features and parameter tuning using statistics and machine learning. Generalizing these pipelines to new sequencing technologies have proven difficult because the individual processes require manual parameter re-tuning or, with more effort, extensions to their statistical models. This constitutes a major problem in an area with such rapid technological progress (23). To illustrate the complexity of these pipelines, we present a number of case studies from our own and others’ work.
The most routinely called type of variations are SNVs and minor structural variation which are reliably returned by standard calling pipelines with error rates < 1% for germline samples. For example, the widely used GATK uses logistic regression to model base errors, Hidden Markov models to compute read likelihoods, and naive Bayes classification to identify variants, which are then filtered to remove likely false positives using a Gaussian mixture model with hand-crafted features capturing common error modes (24). These techniques allow the GATK to achieve high but still imperfect accuracy on the Illumina sequencing platform (25,26).
While SNV can largely be called, several complex and hard-to-call regions such as the major histocompatibility complex (MHC) cannot be accurately called with standard pipelines. The MHC is located on chromosome 6 of the genome and codes for genes that are involved in the adaptive immune system. The MHC region is highly variable, meaning that there are many different but also many similar alleles, making allele calling from short-read sequencing data an extremely hard problem. As DNA sequences in these genes are so variable on the DNA level, Xie et al. (27) determine the type of MHC by aligning sequencing reads to the database of known MHC alleles based on amino acid identity to retrieve a set of candidate alleles. Out of these candidates, an optimal allele set is determined based on solving a discrete optimization problem using integer linear programming, achieving close to perfect accuracy in four-digit typing, which is equivalent to amino-acid identity.
Short-tandem repeats (STRs) are repeats of hyper-mutable sequences in the human genome. While STR typing using PCR amplification is routinely used in forensics, their repetitive nature makes STRs hard to call from next-generation sequencing (NGS) data. STRs have been associated with several diseases, such as Huntington’s, and complex traits, making them important for clinical applications. As strand slippage leads to errors in DNA replication of STRs, they are also highly polymorphic. Tang et al. (28) extract four distinct sources of information from an alignment around an STR locus to estimate the length from sequencing data. Reads that completely span an STR allow accurate calling of alleles that are shorter than the read length (29). Partial reads, repeat only reads and mate pairs (a set of two reads from the same sequence fragment) spanning an STR provide individually uncertain information that Tang et al. (28) combine into a posterior distribution to infer STR lengths.
The telomeres are long stretches of highly repetitive and thus hard to align sequences that make up the tips of our chromosomes. Not only do they show variation between different individuals, but they also undergo somatic changes during our lifetime. Especially, shortening of telomeres has been associated with several aging-related disorders, including cancer. While aligning and calling of telomeric sequence variation is still an unsolved problem, methods have been developed to get a statistical estimate of the length of the telomeres. For example, Lippert et al. (30) derive an estimate of telomere length by determining the fraction of sequencing reads that are likely telomeric. Telomeres are associated with long stretches of repeats made up of a number of six-nucleotide patterns, most prominently of CCCTAA, making repeated occurrence of this pattern a distinctive feature for classification. Lippert et al. (30) tune a classification threshold based on DNA reference samples that have been repeatedly sequenced using identical technology.
Another type of important somatic variation that is not included in standard calling pipelines is sex chromosome mosaicism. During our lifetime, a fraction of our cells loses one copy of the sex chromosomes, keeping only a single X chromosome. That is, females lose one X chromosome copy and males lose their Y chromosome. Loss of Y chromosomes has been associated to an increased risk for Alzheimer’s disease. As the multiplicity of a given sequence influences that amount of sequencing reads proportionally, Lippert et al. (30) estimate the sex chromosome copy number (proportional to the degree of mosaicism) from the read depth obtained in the sex chromosomes in contrast to the read depth of the autosomes, as observed in a sequence alignment. The algorithm also requires careful modeling of confounding factors that affect the estimate. For example, sequencing depth is strongly influenced by the GC content, which is treated using non-parametric estimates of read depth of each region given its GC content.
The reliable identification of structural variation through short-read sequencing remains a challenge (31). Many algorithms detecting small and large deletion and insertions have recently been developed (https://omictools.com/structural-variant-detection-category; date last accessed April 4, 2018). Such callers mainly exploit split-read mapping or paired-end read mapping; however, there is still no single caller that can be considered a community standard, and increasingly the various callers are combined in integrated pipelines (32). Here, we discuss SV2 (33) to showcase recent work in this field. SV2 is a machine-learning algorithm for genotyping deletions and tandem duplications from paired-end whole genome sequencing data. It serves to integrate variant calls from multiple structural variant discovery algorithms into a unified callset with low rates of false discoveries and Mendelian errors with accurate de novo detection. One advantage of SV2 to comparable structural variant detection solutions is the ability to genotype breakpoints overlapping repetitive elements using read depth.
Overall, this section underscores the multiplicity of solutions that are currently available to solve complex regions of the genome using various modeling and machine-learning approaches. It shows, however, that the complexity of the data may need more advanced analytical solutions that are less reliant on expert knowledge.
Deep Learning
Deep learning is evolving from machine-learning systems, in particular from artificial neural network algorithms. Its interest for high‐throughput biology is clear: it allows to better exploit the availability of increasingly large and high‐dimensional data sets by training complex networks with multiple layers that capture their internal structure (1).
One main application of deep learning in genomics has been in functional genomics: predicting the sequence specificity of DNA and RNA-binding proteins and of enhancer and cis-regulatory regions, methylation status and control of splicing. More recently, there have been applications for applied genomics in particular for base calling, and for population genetics. An overview of the exploding field of applications and resources in presented in Box 1. The box highlights approaches to studying germline DNA. There are also important developments in proteomics, RNA and single-cell analytics that are not covered in this review.
Box 1. Deep-learning applications in genomics (DNA)
Adapted and expanded from https://github.com/hussius/deeplearning-biology; date last accessed April 4, 2018 and Jones et al. (43)
CNNs for DNA-binding prediction from sequence
DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Uses convolution layers to capture regulatory motifs, and a recurrent layer to discover a ‘grammar’ for how these single motifs work together. Based on Keras/Theano.
Basset—learning the regulatory code of the accessible genome with deep convolutional neural networks. CNN to discover regulatory sequence motifs to predict the accessibility of chromatin. Accounts for cell-type specificity using multi-task learning.
DeepBind and DeeperBind—predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Based on ChIP-seq, ChIP-chip, RIP-seq, protein-binding microarrays and others. Deeperbind adds a recurrent sequence learning module (LSTM) after the convolutional layer(s).
DeepMotif—visualizing genomic sequence classifications. Predicting binding specificities of proteins to DNA motifs. Makes use of a convolutional layers with more layers than the DeepBind network.
Convolutional neural network architectures for predicting DNA–protein binding. Systematic exploration of CNN architectures for predicting DNA sequence binding using a large compendium of transcription factor data sets.
Predicting enhancers, 3d interactions and cis-regulatory regions
PEDLA: predicting enhancers with a deep-learning-based algorithmic framework. Predicting enhancers based on heterogeneous features from (e.g.) the ENCODE project using a deep learning, HMM hybrid model.
DEEP: a general computational framework for predicting enhancers. Predicting enhancers based on data from the ENCODE project.
Genome-wide prediction of cis-regulatory regions using supervised deep-learning methods. toolkit based on the Theano) for applying different deep-learning architectures to cis-regulatory elements.
FIDDLE: an integrative deep-learning framework for functional genomic data inference. Prediction of transcription start site and regulatory regions. FIDDLE stands for Flexible Integration of Data with Deep Learning that models several genomic signals using convolutional networks (DNase-seq, ATAC-seq, ChIP-seq, TSS-seq, RNA-seq signals).
DNA methylation
DeepCpG—predicting DNA methylation in single cells. Neural network for predicting DNA methylation in multiple cells.
Predicting DNA methylation state of CpG dinucleotide using genome topological features and deep networks. Uses a stacked autoencoder with a supervised layer on top of it to predict whether CpG islands are methylated.
Variant callers, pathogenicity scores and identification of genomic elements
DeepVariant—a variant caller in germline genomes. Uses a deep neural network architecture (Inception-v3) to identify SNP and small indel variants from next-generation DNA sequencing data.
DeepLNC, a long non-coding RNA prediction tool using deep neural network. Identification of lncRNA-based on k-mer profiles.
evoNet—deep learning for population genetic inference [code][paper]. Jointly inferring natural selection and demographic history
DANN. Uses the same feature set and training data as CADD to train a deep neural network
DeepSEA—predicting effects of non-coding variants with deep-learning-based sequence model. Models chromatin accessibility as well as the binding of transcription factors, and histone marks associated with changes in accessibility.
In contrast to the methods discussed so far, where heterogeneous pipelines are built from several models that are each engineered and optimized on its own, the aim of deep learning is to build a single model by hierarchically connecting multiple building blocks together. This way, any involved model parameters can be ‘learned’ from data in an end-to-end fashion. Here we review some of the standard building blocks used in deep learning tools and some of the applications of deep learning in regulatory genomics, variant calling and prediction of pathogenicity of sequence variants. Box 2 includes a glossary of terms proper to this field.
Box 2. Glossary
Artificial intelligence (AI) is a subfield of computer science that aims at enabling computers to solve tasks commonly associated with intelligence, such as reasoning, planning, learning, natural language processing or perception. While AI has been implemented in the form of expert-defined rules, recent successes have been achieved by automatically inferring such rules from training data using machine learning.
Backpropagation is an algorithm to learn the parameters in a DNN from training data. Backpropagation consists of a forward pass, where predictions for the training data are computed based on the current parameter estimates and a backward pass, where the prediction errors are propagated back through the network to compute an update to the parameters that improves the predictions and reduce the error.
Convolutional neural networks (CNNs) are a class of DNNs that are most suitable for the analysis of spatial data such as images (2D) and sequences (1D) and aim at extracting local patterns such as edges or short sequence patters in the input by sliding a set of filters over the input sample. Instead of requiring the user to design task-specific filters, the filters in CNNs can be learned from training data using backpropagation. As the output of any convolutional layer, representing the occurrence of a feature at any location in the input, can again be interpreted as an image or sequence, CNNs can hierarchically utilize convolutions to extract complex patterns from the input. When applied to DNA sequences, a convolutional filter can be interpreted as position-specific weight matrix, a commonly used representation of motifs in functionally related sequences.
Deep learning or deep neural networks (DNNs) refer to ML methods based on connected layers of artificial neurons, inspired by neurons in the brain, that process an input signal using parameterized functions that are transmited from one neuron to another. By connecting multiple layers, DNNs can compute complex non-linear functions of the input.
Hidden Markov models (HMM) are probabilistic models for sequences, where the sequence elements are modeled in mixture models with hidden states that are dependent between neighboring positions.
An integer linear program is a mathematical optimization problem in which some of the variables are restricted to be integers, which makes determining the solution NP complete.
The posterior distribution is the probability distribution that models an uncertain quantity, conditioned on any relevant empirical data.
Long short-term memory (LSTM) units are building blocks in RNNs too. A common LSTM unit is composed of a cell, an input gate, an output gate and a forget gate. The cell is responsible for ‘memorizing’ values over arbitrary sequence intervals.
Machine learning (ML) refers to a set of methods that give omputers the ability to ‘learn’” the solutions to a task by progressively improving performance on training data.
Logistic regression is an ML model for classification that is for a sample to predict the most likely out of a finite number of classes given its input features. Based on a linear function of the input features, which is learned from the training data, the logistic model estimates the probabilities of each output class.
(Multinomial) Mixture models are hierarchical probabilistic models to infer sub-groups in a data set, without a training data set with sub-group identity information.
Multi-task learning refers to ML models in which multiple related learning tasks are solved simultaneously, while exploiting commonalities between tasks, for example by sharing convolutional filters. Doing so can improve accuracy, especially for tasks where training data are limited for individual tasks, but where large data sets exist for closely related tasks.
Naive Bayes refers to a simple probabilistic classifier based on applying Bayes’ theorem while making independent assumptions between the features.
Recurrent neural networks (RNNs) are a class of DNNs where neighboring neurons are connected to form a directed graph that processes a signal along a sequence. To enable relevant signals to be transmitted over longer distances in the graph, gating mechanisms such as LSTM have been developed to regulate the flow of information.
Support vector machines (SVMs) are ML models for classification. SVMs can perform a non-linear classification by implicitly mapping the input features into higher-dimensional feature spaces where they may be separated using a linear function.
Deep learning for DNA sequences
Instead of manually specifying sequence features prior to learning, convolutional neural networks (CNNs) extract informative sequence patterns known as position-weight matrices (PWMs) that summarize nucleotide frequencies at each position during model training. Repeatedly occurring sequence patterns boost the weight of any PWM that increases prediction accuracy. These are applied to every position on the sequence in a sliding window. By hierarchically stacking multiple convolutional layers, using the PWM scores as input to the next layer, CNNs learn dependencies between neighboring PWMs and combine them into higher-level sequence patterns to improve predictive power. A common alternative to the use of convolutional filters is to apply sequential models, in particular recurrent neural networks (RNNs), which originally had been developed for time series. Here, each position in the sequence corresponds to a single time step. While PWMs only capture short sequence patterns, recurrent layers can also capture dependencies between sequence elements that are further apart using a mechanism called memory and implemented by gating functions, such as in the long short-term memory (LSTM) unit (34). As there is no temporal direction in DNA sequences, recurrence typically is applied in both directions. It is possible to combine CNNs and RNNs. For example, it is possible to apply a recurrent layer to the output of a convolutional layer to capture higher-order sequential dependencies between PWM patterns.
Accounting for context using vector representations
In deep learning, everything is a vector. First developed to encode topics related to words in classification of text, vector representations encode the properties (topics) of a set of relevant contexts (words) that provide important context information for each learning instance (text). For example, in a model that predicts transcription factor binding to a DNA sequence (instance) in different tissues (words), we may want to account for tissue specificity of binding using vector representations. To predict the probability that a transcription factor binds in the context of a given tissue, we would condition the model on the tissue by using the corresponding word vector as an additional input to the prediction. Thus, word vectors enable us to train a single model using observations from different contexts, thus re-using the same sequence patterns for multiple prediction tasks.
Combining multiple related learning tasks
Genomic sequences often exert different behavior under different contexts. For example, the same sequence pattern may attract transcription factors differently in different tissues. Consequently, it is desirable to account for tissue-specific behavior by treating different tissues as separate learning tasks. Yet, it is desirable to re-use learned sequence patterns between the different tasks. These can be multi-task architectures that share parts of the network, including sequence features extracted from convolutional filters, between tasks, feeding these into separate, task specific, layers. During training, examples from all tasks are shown to the model, which updates any shared parameters and the corresponding task-specific parameters.
Visualizing and understanding deep models
As deep models use features that have been detected in large data sets instead of expert-derived ones and apply the features in a highly non-linear way, they are often seen as black boxes. Therefore, it is an interesting question to interpret the features that are being detected. For example, given a sequence pattern, we can ask the question: ‘how the prediction derived from the network is influenced by each individual letter in the sequence?’ For example, to compute how much the prediction would change if we replaced a particular letter in the input sequence, we may compute the gradient of the network with respect to the corresponding letter. If we performed this with each letter, we get a so-called saliency map (35). The general problem of identifying the key input data used to make a prediction, known as the attribution problem, is a very active area of machine-learning research (e.g. see http://www.unofficialgoogledatascience.com/2017/03/attributing-deep-networks-prediction-to.html; date last accessed April 4, 2018 and (36)). DeepLIFT (Deep Learning Important FeaTures) (37), is a method for decomposing the output prediction of a neural network on a specific input by backpropagating the contributions of all neurons in the network to every feature of the input. DeepLIFT has been applied to simulated genomic data, and show significant advantages over gradient-based methods.
Selected Deep-Learning Algorithms
Deepbind
Deepbind predicts transcription factor binding affinity based on sequence motifs learned in the form of convolutional filters. The input to the convolutional layer is the sequence and the output is a prediction of whether the prediction factor binds or not. Training this model thus jointly determines suitable sequence motifs as well as a classifier for transcription factor binding. Alipanahi et al. (38) used publically available sequence data, spanning thousands of public protein-binding microarray (PBM) data, RNAcompete, ChIP-seq and HT-SELEX experiments to train a separate model for each of 538 distinct transcription factors and 194 distinct RNA binding proteins. At the time, Alipanahi et al. outcompeted all competitive models in several comparisons, including the DREAM5 challenge. Hassanzadeh and Wang (39) further improved performance by feeding convolutional filters into a recurrent layer. Qin and Feng (40) jointly modeled binding for different transcription factors specific to different cell lines by learning dictionaries for each of transcription factors and cell lines, respectively.
DeepVariant
Variant calling is a process to identify differences between an individual’s genome from a reference genome based on NGS data. Variant calling is challenging because there are many signals from the NGS data that are involved in making the decision, which makes the process of writing rules difficult for humans. Worse, many exogenous factors, such as the variety of sequencing technologies and library preparation protocols, affect the underlying distribution of signals obtained. DeepVariant (23) approaches this decision-making problem by providing these signals to a deep-learning architecture (Inception-v3) and using standard deep-learning training regimes to let it learn directly from data. This bypasses the need to manually inject knowledge or craft rules, and makes it easier to generalize to much more data with much more variety. DeepVariant encodes information from the sequencing instrument data in the form of multi-channel tensors. In the first version that won the performance award in the 2016 PrecisionFDA Truth challenge, the data representation used only three channels, and so could be encoded in a standard (RGB) image. In the latest release on GitHub [link], DeepVariant extends the representation to many more channels, encoding the information about the bases, the quality of the bases and the mapping, whether a read supports alt (alternate base) or whether it matches ref (reference base), among others in each channel. This newer representation, along with a more robust training scheme, allows DeepVariant to be even more accurate. We observe a lower number of FN (False Negative) and FP (False Positive) calls by comparing DeepVariant 2016 version and the latest release version: FN for Indel: 4175 to 2384; FP for Indel: 2839 to 1811; FN for SNV: 1689 to 735; FP for SNV: 832 to 363. It also brings the latest DeepVariant model to the highest SNV and Indel performance compared with all submissions in the 2016 PrecisionFDA Truth challenge. DeepVariant is a powerful testimonial for deep learning: variant calling is one of the most well-studied areas in bioinformatics. Despite substantial efforts, in recent years the improvement on accuracy has only been incremental. Many researchers believe that we might have reached the best accuracy we can do on this problem, but DeepVariant has demonstrated that it is still possible to achieve even higher accuracy with deep-learning techniques.
Base calling for other technologies, such as those based on nanopore sequencing, also make use of deep RNNs. DeepNano is an open-source base caller for the MinION nanopore sequencing platform (41). The program Chiron couples a CNN with an RNN and a Connectionist Temporal Classification (CTC) decoder to directly translate raw nanopore signal to DNA sequence (42).
DeepSEA
Accurate sequence-based prediction of chromatin features requires a flexible quantitative model capable of modeling complex dependencies. The development of DeepSEA (20) is based on three convolution layers. DeepSEA differentiates from previous approaches to functional effects of non-coding variants (presented in Table 1), by predicting, with single-nucleotide sensitivity, the effects of non-coding variants on transcription factor binding, DNA accessibility and histone marks of sequences. Among the key features of the approach are the use of a wider sequence context, as sequences surrounding the variant position may determine the regulatory properties of the variant—whereas previous studies for transcription factor binding prediction have focused on small sequence windows directly associated with the binding sites, DeepSEA uses a context sequence size of up to 1 kb to improve the performance of the model. A second feature is the deployment of an ‘in silico saturated mutagenesis’ approach that analyzes the effects of each base substitution on chromatin feature predictions, thereby identifying which sequence features are most informative for a specific chromatin effect prediction. Finally, the authors trained boosted logistic regression classifiers for predicting Human Gene Mutation Database-annotated non-coding regulatory mutations, non-coding eQTLs and non-coding trait-associated SNVs identified in GWAS studies on the basis of predicted chromatin effects and evolutionary features.
Conclusions
Genomics, like many fields in biomedical and computational biology, is enjoying exponential growth of data generation. Its analysis is transitioning from descriptive statistics and data modeling through machine learning, and increasingly, through deep learning. This evolution is all about automating pipeline generation: the underlying principle is the switch from expert-based processes toward more and more automated data-driven and learned approaches.
There are many settings in which studies of DNA sequence variation will benefit from deep learning and related approaches beyond the inherited germline DNA that we emphasized in this review. For example, mtDNA analysis, mosaicism beyond the sex chromosomes, phasing genomes and de novo assembly, heterogeneous single-cell settings, and especially tumor genomes and somatically acquired DNA sequence mutations/variations—all of which pose unique challenges.
However, there are limits to deep learning that should be taken into consideration given the broad excitement for these new approaches: it requires large amounts of data that may not be available when working with experimental biological systems, it is limited in the capacity to discern mechanistic components, and can reflect biases and inaccuracies inherent in the data fed to them.
Acknowledgements
We thank Alex Wells for compiling information in Table 1, Mikael Huss for the excellent resource in Github on implementation of deep-learning methods in genomics and Ali Torkamani, Pejman Mohammadi and Eric Topol for comments and discussion.
Conflict of Interest statement. P.-C.C. and M.DeP. are employees of Google Inc. A.T. and C.L. declare no conflict of interest.
Funding
Work of A.T. is supported by the Qualcomm Foundation and the NIH Center for Translational Science Award (CTSA, grant number 5UL1 TR001114).
References
- 1. Angermueller C., Parnamaa T., Parts L., Stegle O. (2016) Deep learning for computational biology. Mol. Syst. Biol., 12, 878.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ching T., Himmelstein D.S., Beaulieu-Jones B.K., Kalinin A.A., Do B.T., Way G.P., Ferrero E., Agapow P.-M., Xie W., Rosen G.L.. et al. (2017) Opportunities and obstacles for deep learning in biology and medicine. bioRxiv, 142760, doi: 10.1101/142760. [DOI] [PMC free article] [PubMed]
- 3. Paliwal K., Lyons J., Heffernan R. (2015) A short review of deep learning neural networks in protein structure prediction problems. Adv. Tech. Biol. Med., 3, 139. [Google Scholar]
- 4. Wang S., Sun S., Li Z., Zhang R., Xu J. (2017) Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol., 13, e1005324.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B.. et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Telenti A., Pierce L.C., Biggs W.H., di Iulio J., Wong E.H., Fabani M.M., Kirkness E.F., Moustafa A., Shah N., Xie C.. et al. (2016) Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. U. S. A., 113, 11901–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aggarwala V., Voight B.F. (2016) An expanded sequence context model broadly explains variability in polymorphism levels across the human genome. Nat. Genet., 48, 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. di Iulio J., Bartha I., Wong E.H.M., Yu H.-C., Hicks M.A., Shah N., Lavrenko V., Kirkness E.F., Fabani M.M., Yang D.. et al. (2018) The human non-coding genome defined by genetic diversity. Nat. Genet., 50, 333–337. [DOI] [PubMed] [Google Scholar]
- 9. Consortium E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cassa C.A., Weghorn D., Balick D.J., Jordan D.M., Nusinow D., Samocha K.E., O'Donnell-Luria A., MacArthur D.G., Daly M.J., Beier D.R.. et al. (2017) Estimating the selective effects of heterozygous protein-truncating variants from human exome data. Nat. Genet., 49, 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glusman G., Rose P.W., Prlic A., Dougherty J., Duarte J.M., Hoffman A.S., Barton G.J., Bendixen E., Bergquist T., Bock C.. et al. (2017) Mapping genetic variations to three-dimensional protein structures to enhance variant interpretation: a proposed framework. Genome Med., 9, 113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hicks M., Bartha I., di Iulio J., Abagyan R., Venter J.C., Telenti A. (2017) Functional characterization of 3D-protein structures informed by human genetic diversity. bioRxiv, 182287; doi: 10.1101/182287. [DOI] [PMC free article] [PubMed]
- 14. Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. (2016) A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet., 48, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fu Y., Liu Z., Lou S., Bedford J., Mu X.J., Yip K.Y., Khurana E., Gerstein M. (2014) FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol., 15, 480.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang Y.F., Gulko B., Siepel A. (2017) Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet., 49, 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shihab H.A., Rogers M.F., Gough J., Mort M., Cooper D.N., Day I.N., Gaunt T.R., Campbell C. (2015) An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics, 31, 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smedley D., Schubach M., Jacobsen J.O.B., Kohler S., Zemojtel T., Spielmann M., Jager M., Hochheiser H., Washington N.L., McMurry J.A.. et al. (2016) A whole-genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. Am. J. Hum. Genet., 99, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gussow A.B., Copeland B.R., Dhindsa R.S., Wang Q., Petrovski S., Majoros W.H., Allen A.S., Goldstein D.B. (2017) Orion: detecting regions of the human non-coding genome that are intolerant to variation using population genetics. PLoS One, 12, e0181604.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou J., Troyanskaya O.G. (2015) Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods, 12, 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. So H.C., Sham P.C. (2017) Improving polygenic risk prediction from summary statistics by an empirical Bayes approach. Sci. Rep., 7, 41262.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pare G., Mao S., Deng W.Q. (2017) A machine-learning heuristic to improve gene score prediction of polygenic traits. Sci. Rep., 7, 12665.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poplin R., Newburger D., Dijamco J., Nguyen N., Loy D., Gross S.S., McLean C.Y., DePristo M.A. (2017) Creating a universal SNP and small indel variant caller with deep neural networks. bioRxiv, 092890, doi: 10.1101/092890. [DOI] [PubMed]
- 24. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M.. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H. (2014) Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics, 30, 2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldfeder R.L., Priest J.R., Zook J.M., Grove M.E., Waggott D., Wheeler M.T., Salit M., Ashley E.A. (2016) Medical implications of technical accuracy in genome sequencing. Genome Med., 8, 24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie C., Yeo Z.X., Wong M., Piper J., Long T., Kirkness E.F., Biggs W.H., Bloom K., Spellman S., Vierra-Green C.. et al. (2017) Fast and accurate HLA typing from short-read next-generation sequence data with xHLA. Proc. Natl. Acad. Sci. U. S. A., 114, 8059–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang H., Kirkness E.F., Lippert C., Biggs W.H., Fabani M., Guzman E., Ramakrishnan S., Lavrenko V., Kakaradov B., Hou C.. et al. (2017) Profiling of short-tandem-repeat disease alleles in 12,632 human whole genomes. Am. J. Hum. Genet., 101, 700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gymrek M., Golan D., Rosset S., Erlich Y. (2012) lobSTR: a short tandem repeat profiler for personal genomes. Genome Res., 22, 1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lippert C., Sabatini R., Maher M.C., Kang E.Y., Lee S., Arikan O., Harley A., Bernal A., Garst P., Lavrenko V.. et al. (2017) Identification of individuals by trait prediction using whole-genome sequencing data. Proc. Natl. Acad. Sci. U. S. A., 114, 10166–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. English A.C., Salerno W.J., Hampton O.A., Gonzaga-Jauregui C., Ambreth S., Ritter D.I., Beck C.R., Davis C.F., Dahdouli M., Ma S.. et al. (2015) Assessing structural variation in a personal genome—towards a human reference diploid genome. BMC Genomics, 16, 286.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin K., Smit S., Bonnema G., Sanchez-Perez G., de Ridder D. (2015) Making the difference: integrating structural variation detection tools. Brief Bioinform., 16, 852–864. [DOI] [PubMed] [Google Scholar]
- 33. Antaki D., Brandler W.M., Sebat J. (2017) SV2: Accurate Structural Variation Genotyping and De Novo Mutation Detection. bioRxiv, 113498, doi: 10.1101/113498. [DOI] [PMC free article] [PubMed]
- 34. Hochreiter S., Schmidhuber J. (1997) Long short-term memory. Neural Comput., 9, 1735–1780. [DOI] [PubMed] [Google Scholar]
- 35. Simonyan K., Vedaldi A., Zisserman A. (2013) Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv, 1312.6034v2.
- 36. Sundararajan M., Taly A., Yan Q. (2017) Axiomatic attribution for deep networks. arXiv, 1703.01365v2.
- 37. Shrikumar A., Greenside P., Kundaje A. (2017) Learning important features through propagating activation differences. arXiv, arXiv: 1704.02685v1.
- 38. Alipanahi B., Delong A., Weirauch M.T., Frey B.J. (2015) Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol., 33, 831–838. [DOI] [PubMed] [Google Scholar]
- 39. Hassanzadeh H.R., Wang M.D. (2016) DeeperBind: enhancing prediction of sequence specificities of DNA. arXiv, 1611.05777v1. [DOI] [PMC free article] [PubMed]
- 40. Qin Q., Feng J. (2017) Imputation for transcription factor binding predictions based on deep learning. PLoS Comput. Biol., 13, e1005403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boza V., Brejova B., Vinar T. (2017) DeepNano: deep recurrent neural networks for base calling in MinION nanopore reads. PLoS One, 12, e0178751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teng H., Hall M.B., Duarte T., Cao M.D., Coin L. (2017) Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. bioRxiv, 179531, doi: 10.1101/179531. [DOI] [PMC free article] [PubMed]
- 43. Jones W., Alasoo K., Fishman D., Parts L. (2017) Computational biology: deep learning. Emerg. Top. Life Sci., 1, 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]