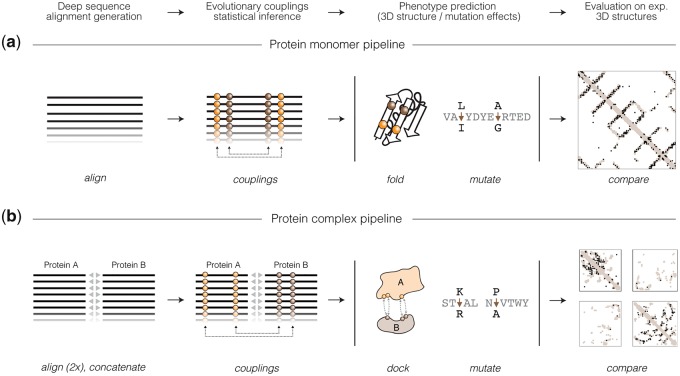
Fig. 1.
The EVcouplings Python framework. (a) The protein monomer EVcouplings pipeline entails multiple sequence alignment generation (align stage), EC inference (couplings stage), de novo folding (fold stage), mutation effect prediction (mutate stage) and comparison to experimental structure (compare stage). (b) The protein complex pipeline extends the monomer pipeline to protein interactions by pairing putatively interacting homologs (concatenate stage) and providing restraints for molecular docking (dock stage)