Abstract
Summary
As the number and complexity of biosimulation models grows, so do demands for tools that can help users understand models and compose more comprehensive and accurate systems from existing models. SemGen is a tool for semantics-based annotation and composition of biosimulation models designed to address this demand. A key SemGen capability is to decompose and then integrate models across existing model exchange formats including SBML and CellML. To support this capability, we use semantic annotations to explicitly capture the underlying biological and physical meanings of the entities and processes that are modeled. SemGen leverages annotations to expose a model’s biological and computational architecture and to help automate model composition.
Availability and implementation
SemGen is freely available at https://github.com/SemBioProcess/SemGen.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Biosimulation models are used throughout the life sciences to test hypotheses about biological systems and explore the effects of perturbations. Such models are written in a variety of languages and they can simulate a wide variety of biological processes across physical scales. Given the growing complexity of biosimulation models, model-archiving initiatives such as BioModels (Glont et al., 2017) and the Physiome Model Repository (PMR) (Yu et al., 2011) have been established to make models more accessible and reproducible across research groups. While reproducibility is itself an important goal, to best build from the efforts of others, researchers should not only be able to access and replicate what has been done in the past but also adapt and extend that work for their own models. One approach for enhancing the retrieval and integration tasks required for repurposing models is to leverage models’ semantic annotations. Semantic annotations are machine-readable metadata that capture the meaning of model elements. They are useful in the context of model reuse and integration because biological modelers do not use an agreed-upon set of identifiers to indicate the meaning of model elements. For example, a variable named ‘x’ could be used to represent cytosolic calcium concentration in one model, heart rate in another. By linking model elements to terms in controlled knowledge resources such as the Gene Ontology (Ashburner et al., 2000), UniProt (UniProt Consortium, 2015), etc., semantic annotations act as a common ground for disambiguating model elements across models and modeling formats. Software tools can then leverage these annotations to facilitate cross-repository model search, quantify the biological similarity between models, enhance model exploration through the use of common visualization schemes, align models during composition and establish links between models and experimental datasets (Neal et al., 2018). Such capabilities can help biological researchers more easily discover, understand, reproduce and repurpose biosimulation models.
SemGen is a software system designed to help the broader biological modeling community apply semantic annotations to models and leverage these annotations to enhance model visualization and facilitate model composition. SemGen is being developed as part of an effort to promote model reuse and repurposing among the broader community of biological researchers. Therefore, our focus has been on supporting models encoded in declarative formats designed for exchange among research groups and which support machine-readable semantic annotation, including the Systems Biology Markup Language (SBML) (Hucka et al., 2003) and CellML (Cuellar et al., 2003). SemGen can also convert models into SemSim models, which are encoded in the Web Ontology Language (OWL—https://www.w3.org/OWL/), and which we use to explore the application of automated inference tools to reason over the knowledge contained in biosimulation models (Neal et al., 2016).
2 Annotating models
When annotating a model using SemGen’s Annotator tool, users provide composite annotations (Gennari et al., 2011) that precisely describe the biological properties of entities and processes simulated by the model. For example, a chemical network model may represent the chemical concentration of calcium in the cytosol, or the cytosolic fluid volume. In these cases, composite annotations link physical entities (calcium ions, cytosol) represented in publicly available knowledge resources with their associated physical properties (chemical concentration, fluid volume) represented in the Ontology of Physics for Biology (Cook et al., 2011). SemGen leverages these semantic annotations to help modelers more accurately understand the biology represented in models and to identify potential points of coupling between models that are merged together. The SemGen development team maintains a protocol for annotating a model using composite annotations at https://github.com/SemBioProcess/SemGen/wiki/Annotation-protocol. This protocol lists the recommended knowledge resources to use when creating composite annotations and also indicates which model components should be annotated and at what level of detail.
For established model exchange formats such as SBML and CellML, SemGen stores semantic annotations as Resource Description Framework (RDF) triples (https://www.w3.org/RDF/). These statements can either be stored within the model file itself, or stored separately within a COMBINE archive (Bergmann et al., 2014) that also contains the model.
3 Visualizing models
SemGen provides node-and-edge model visualizations based on D3 technology (Bostock et al., 2011) to help users better understand the computational and biological structure of models. SemGen provides three types of visualizations: a computational dependency network representing mathematical relationships in the model, a ‘PhysioMap’ (Cook et al., 2013) of a model’s physical processes and entities, and a hierarchical ‘submodel’ view. These three views allow researchers to track the mathematics of the model, trace physiological process flows, and identify pre-existing modules available for use in composition tasks. Since SemGen supports models encoded in various formats, these visualizations give users the ability to investigate models from multiple archiving initiatives, including BioModels and the PMR, using a common visualization scheme.
4 Composing models
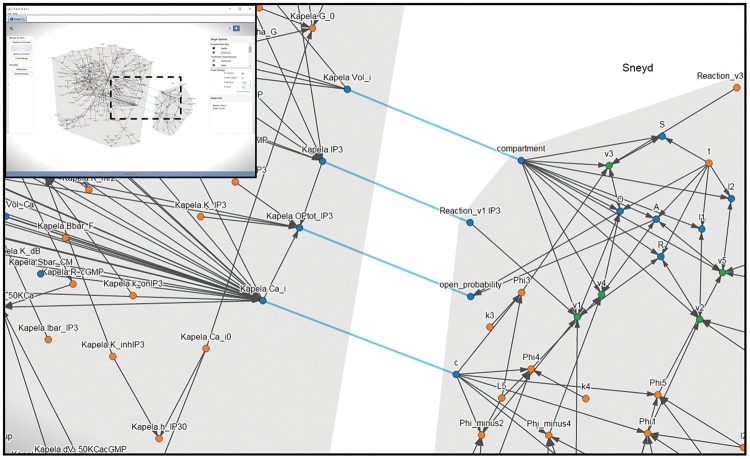
To facilitate model reuse and repurposing, SemGen provides capabilities for model extraction and merging. Extraction capabilities are important because researchers may only be interested in a reusing a subset of the processes a model simulates when attempting to repurpose it. Users can select and extract a subset of nodes as a new model by using one of the three visualizations described above. These extractions can be immediately merged into another model and can also be exported to a variety of modeling formats. Figure 1 shows an example of SemGen’s cross-format visualization and merging capabilities. In this example, a CellML model of vascular smooth muscle calcium dynamics (Kapela et al., 2008) is merged with an SBML model (BioModels ID BIOMD0000000057) simulating multistate inositol trisphosphate (IP3) control of endoplasmic reticulum calcium release (Sneyd and Dufour, 2002). The Kapela, et al. model is broader in scope and uses a simplified representation of how IP3 concentration affects calcium levels in the cytosol; therefore, a reasonable modeling task is to combine these models, creating a more comprehensive and hopefully more accurate model of calcium dynamics. The figure illustrates the points at which the computational dependency networks of the two models overlap in terms of the biological properties they simulate.
Fig. 1.
Visualizing coupling points during model merging. When performing merging tasks such as the one described in the text, SemGen can illustrate the coupling points between the computational dependency networks of the merged models. The four non-directional lines in the center of the image connect model variables that have the same biological meaning. For example, the variables ‘Kapela.Ca_i’ and ‘c’ both represent cytosolic calcium concentration. Other lines indicate computational dependencies between variables. Inset shows full SemGen interface; dashed box indicates area of detail
SemGen model merging is a multi-step process as exemplified previously (e.g. Neal et al., 2015). First, the user collects and prepares models for merging. As part of this step, portions of models that are not of interest or would be replicated in the destination model might be removed. Next, the computational links between the models are established. In contrast to other model composition approaches that rely on pre-defined interfaces to control the ways models can be coupled, SemGen applies a more flexible, semantics-based, ‘white box’ approach that, as argued previously (Neal et al., 2014), requires no prior component modularization or module interface definitions. Rather, inter-model links are determined at the time of merging based on the common biological properties shared by the models. When a merging task is initiated, SemGen automatically scans each source model to identify and list those biological properties that are represented in both models. SemGen lists these points of overlap so the user can choose which to represent in the final, merged system. To facilitate these decisions, SemGen computes and displays networks of computational dependencies to visualize the mathematical consequences of a given choice. SemGen then automatically rewrites equations for the coupled system according to the user’s choices. In our example merging task, SemGen identified four points of overlap, including the concentrations of cytosolic calcium and IP3. The Supplementary Material details the merging task, including how points of overlap were resolved. A tutorial on SemGen model merging is available on the SemGen GitHub page (https://github.com/SemBioProcess/SemGen).
Based on our experience with such merging tasks, it is unlikely that semantics-based model merging will ever be fully automated; merging choices depend on scientific intent and not simply on model code. SemGen requires user input to resolve semantic overlap between models, and code-level adjustments may be required to align models prior to merging. Thus, we do not aim to fully automate the merging process, but rather minimize the amount of code-level investigations and manipulations required for this process. SemGen helps in this regard by automatically identifying critical points of semantic overlap between source models that would otherwise require manual inspection of model code and by automatically generating equations in the merged model based on the user’s design decisions. SemGen can also detect and resolve units of measure conflicts that arise during the model alignment process, and it offers a way to include conversion factors for ensuring unit balance in the new equations resulting from the merging process. Much like software version-control systems such as Git, SemGen’s features are intended to do the ‘heavy lifting’ involved in code-level integration tasks, and some degree of code-level manipulation and assessment of integrated code will often be necessary during merging operations.
SemGen can output merged model code for simulation in a variety of declarative modeling formats, including SBML and CellML. The semantic annotations on the model are preserved for each of these formats on output. We emphasize that optimization of model parameters for simulating merged models is currently outside SemGen’s scope and best handled with existing model editors.
5 Community, evaluation and next steps
SemGen and our composite annotation approach are in use by the COmputational Modeling in BIology NEtwork (COMBINE, http://co.mbine.org/), a community of researchers developing biosimulation standards. To promote a standard approach to annotation and model-sharing, SemGen supports use of the COMBINE archive, where annotations are saved as a companion file to the original source model so that semantic and computational aspects of a model remain independent. In the Supplementary Material, we provide COMBINE archives for the models used in the example merging task described above and shown in Figure 1.
We have carried out an initial evaluation of SemGen for usability by external computational modelers in an IRB-approved study that evaluated user satisfaction as well as ease-of-use and collected formative information for improvements to the user interface. Overall, participants found SemGen useful for annotation of biosimulation models. In the future, we plan to perform formal evaluations of its compositional capabilities, and extend the software to support additional modeling languages, to provide cross-repository search capabilities, and to improve its merging capabilities through the use of semantic similarity metrics. As the biological modeling community continues to explore semantics-based approaches for improving model discovery, reproducibility and reuse, we hope that SemGen will provide useful, fundamental capabilities for semantics-based annotation and integration.
Funding
This work was partially funded by National Institutes of Health grants R01 LM011969 and P41 GM109824.
Conflict of Interest: none declared.
Supplementary Material
References
- Ashburner M. et al. (2000) Gene Ontology: tool for the unification of biology. Nat. Genet., 25, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann F.T. et al. (2014) COMBINE archive and OMEX format: one file to share all information to reproduce a modeling project. BMC Bioinformatics, 15, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock M. et al. (2011) D3 data-driven documents. IEEE Trans. Vis. Comput. Graph, 17, 2301–2309. [DOI] [PubMed] [Google Scholar]
- Cook D.L. et al. (2011) Physical properties of biological entities: an introduction to the Ontology of Physics for Biology. PLoS One, 6, e28708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D.L. et al. (2013) Representing physiological processes and their participants with PhysioMaps. J. Biomed. Semantics, 4, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar A.A. et al. (2003) An overview of CellML 1.1, a biological model description language. Simulation, 79, 740–747. [Google Scholar]
- Gennari J.H. et al. (2011) Multiple ontologies in action: composite annotations for biosimulation models. J. Biomed. Inform., 44, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glont M. et al. (2017) BioModels: expanding horizons to include more modelling approaches and formats. Nucleic Acids Res., 46, D1248–D1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M. et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics, 19, 524–531. [DOI] [PubMed] [Google Scholar]
- Kapela A. et al. (2008) A mathematical model of Ca2+ dynamics in rat mesenteric smooth muscle cell: agonist and NO stimulation. J. Theor. Biol., 253, 238–260. [DOI] [PubMed] [Google Scholar]
- Neal M.L. et al. (2014) A reappraisal of how to build modular, reusable models of biological systems. PLoS Comput. Biol., 10, e1003849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal M.L. et al. (2018) Harmonizing semantic annotations for computational models in biology. Brief. Bioinform., doi: https://doi.org/10.1101/246470. [DOI] [PMC free article] [PubMed]
- Neal M.L. et al. (2016) Qualitative causal analyses of biosimulation models. CEUR Workshop Proc., 1747. [PMC free article] [PubMed] [Google Scholar]
- Neal M.L. et al. (2015) Semantics-based composition of integrated cardiomyocyte models motivated by real-world use cases. PLoS One, 10, e0145621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneyd J., Dufour J.-F. (2002) A dynamic model of the type-2 inositol trisphosphate receptor. Proc. Natl. Acad. Sci. USA, 99, 2398–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res., 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T. et al. (2011) The physiome model repository 2. Bioinformatics, 27, 743–744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.