Abstract
Oligonucleotide DNA microarrays were used for a genome-wide analysis of immune-challenged Drosophila infected with Gram-positive or Gram-negative bacteria, or with fungi. Aside from the expression of an established set of immune defense genes, a significant number of previously unseen immune-induced genes were found. Genes of particular interest include corin- and Stubble-like genes, both of which have a type II transmembrane domain; easter- and snake-like genes, which may fulfil the roles of easter and snake in the Toll pathway; and a masquerade-like gene, potentially involved in enzyme regulation. The microarray data has also helped to greatly reduce the number of target genes in large gene groups, such as the proteases, helping to direct the choices for future mutant studies. Many of the up-regulated genes fit into the current conceptual framework of host defense, whereas others, including the substantial number of genes with unknown functions, offer new avenues for research.
Innate immunity is the first-line defense of multicellular organisms that operates to limit infection after exposure to microbes. Invertebrates and vertebrates share a common ancestry for this defense system, illustrated by the striking conservation of the intracellular signaling pathways that regulate the rapid transcriptional response to infection in the fruit fly Drosophila and in mammals (1, 2).
Because of its flexible genetics, Drosophila has emerged as a powerful model system for the study of innate immunity. Prominent among the innate immunity reactions is the phagocytosis or encapsulation of the invading organism by the hemocytes (3) and the massive synthesis of antimicrobial peptides by the fat body (4, 5), a functional equivalent of the liver. Transcriptional induction of antimicrobial peptide genes is known to be controlled by at least two distinct pathways, Toll and Imd (6).
Although much has been learned about Drosophila immunity through genetic screens and biochemical analyses, many questions remain. For example, what gene products are responsible for recognition of invading pathogens and how do they activate the Toll or Imd pathways? What genes other than the antimicrobial peptide genes are induced after immune challenge and what roles do these genes play in the innate immune response? To complement the genetic approaches currently underway, transcriptional profiling experiments were carried out to survey the majority of Drosophila genes for their response to bacterial and fungal infection, using Affymetrix (Santa Clara, CA) GeneChips. The induction of the various Drosophila antimicrobial peptides correlated well with many earlier studies based on Northern blotting experiments (7, 8), confirming the accuracy of the microarray methodology used. In addition, a large number of genes previously unknown to be induced by infection were identified. The potential role of these genes in recognition, signaling, and effector mechanisms of the Drosophila immune response can now be assessed by using reverse genetic tools available in Drosophila.
Materials and Methods
Drosophila Stocks.
Cinnabar brown flies (cn bw) were reared on standard cornmeal medium in vials held in humid culture rooms, at either 18 or 25°C. Adult male flies were removed from the colonies at 1-day-old and kept at 18°C until 3 days old. At this age, flies were either inoculated or designated as controls. Control and infected flies were snap-frozen in liquid nitrogen and stored at −80°C before extraction of total RNA.
Microbial Challenge of Flies.
Inoculation with bacteria.
The bacteria Escherichia coli and Micrococcus luteus were precultured in LB medium. Pellets taken when the cultures were in the log phase of growth were resuspended in a small amount of culture medium, and sharpened needles dipped into these suspensions were used to inoculate the flies. Flies were harvested at 6, 12, and 48 h after inoculation.
Natural infection with fungi.
Flies anaesthetized with CO2 were shaken for a few minutes in a Petri dish containing a sporulating culture of Beauveria bassiana. Flies covered with spores were placed in fresh tubes of Drosophila medium and kept at 25°C. Flies were collected 3 days after infection.
Sample Preparation and Analysis.
For each time point and infection type, three samples were analyzed by using the microarrays. Five control samples were also analyzed. Total RNA was prepared by using a mixer mill and RNeasy midipreps, according to the manufacturer's instructions (Qiagen, Hilden, Germany). The RNA was then processed, labeled, and hybridized to Affymetrix GeneChips according to the manufacturer's instructions with slight modifications. Data analysis was carried out by using genespring software (Silicon Genetics, Redwood City, CA). Full details of these methods are presented in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org, along with the Multiplex Quantitative real-time reverse transcription (RT)-PCR protocols, primer, and probe sequences.
Results and Discussion
Male flies were subjected to three types of immune challenge: inoculation with the Gram-negative E. coli or the Gram-positive M. luteus, or natural fungal infection with B. bassiana. RNA, extracted 6, 12, or 48 h after bacterial challenge or 3 days after fungal infection, was hybridized to Affymetrix Drosophila GeneChips. Expression values were derived from the average of at least 3 microarrays. For the individual genes discussed in the body of this article, the CG identifier and the highest level of expression for each treatment are given in Table 1.
Table 1.
Absolute and relative expression values for genes discussed in text
| Function*/ gene identity† | Identity‡/homology§ | Highest
absolute value‖ (SD)**/relative
value‡‡
|
|||
|---|---|---|---|---|---|
| Naive | E. coli | M. luteus | B. bassiana | ||
| Caspase | |||||
| CG7486 (Dredd) | Death related ced-3/Nedd2-like protein | 0.42 (0.17) | 0.72/1.7 | 0.74/1.8 | 0.46 (0.08)/1.1 |
| CG7788 (Ice) | Interleukin-1 beta-converting enzyme | 0.44 (0.27) | 1.07 (0.52)/2.5 | 0.92 (0.49)/2.1 | 1.35 (0.25)/3.1 |
| CG14902 (Decay) | Death executioner caspase related to Apopain | 0.65 (0.19) | 0.59 (0.16)/0.9 | 0.66 (0.02)/1.0 | 1.22 (0.44)/1.9 |
| CG18188 (Daydream) | Death Associated Molecule related to Mch2 | 0.32 (0.08) | 0.42 (0.15)/1.3 | 0.79 (0.25)/2.5 | 0.61 (0.07)/1.9 |
| Defense or immunity protein | |||||
| CG11709 (PGRP-SA) | Peptidoglycan recognition protein-SA | 1.09 (0.40) | 3.9 (1.4)/3.6 | 3.9 (1.7)/3.6 | 2.82 (0.85)/2.6 |
| CG9681 (PGRP-SB1) | Peptidoglycan recognition protein-SB1 | 6.2 (2.6) | 28.6 (9.0)/4.6 | 20.3 (7.7)/3.2 | 5.2 (3.0)/0.8 |
| CG14745 (PGRP-SC2) | Peptidoglycan recognition protein-SC2 | 3.5 (1.7) | 22.0 (7.1)/6.3 | 16.4 (6.2)/4.7 | 7.7 (2.0)/2.2 |
| CG7496 (PGRP-SD) | Peptidoglycan recognition protein-SD | 0.49 (0.21) | 3.0 (1.3)/6.1 | 2.38 (0.94)/4.9 | 0.97 (0.25)/2.0 |
| CG14704 (PGRP-LB) | Peptidoglycan recognition protein-LB | 1.58 (0.55) | 4.2 (1.5)/2.7 | 4.1 (1.8)/2.6 | 1.47 (0.40)/0.9 |
| CG10146 (AttA) | Attacin-A | 5.7 (2.1) | 61 (14)/10.8 | 66 (10)/11.7 | 16.8 (4.1)/2.9 |
| CG18372 (AttB) | Attacin-B | 13.9 (5.8) | 79 (21)/5.7 | 82 (17)/5.9 | 23.5 (5.3)/1.7 |
| CG4740 (AttC) | Attacin-C | 7.3 (3.3) | 52 (16)/7.1 | 47 (15)/6.4 | 8.6 (2.5)/1.2 |
| CG7629 (AttD) | Attacin-D | 0.32 (0.16) | 9.5 (4.6)/29.5 | 5.3 (3.5)/16.4 | 0.41 (0.08)/1.3 |
| CG1365 (CecA1) | Cecropin A1 | 10.7 (4.7) | 73 (27)/6.8 | 77 (23)/7.2 | 13.6 (3.0)/1.3 |
| CG1367 (CecA2) | Cecropin A2 | 4.8 (2.2) | 63 (15)/13.1 | 69.5 (12.9)/14.4 | 8.5 (2.2)/1.8 |
| CG1878 (CecB) | Cecropin B | A¶ | 1.7 (0.9)/ | 3.2 (1.6)/ | ††/ |
| CG1373 (CecC) | Cecropin C | 0.45 (0.16) | 9.9 (3.0)/22.0 | 11.1 (5.2)/24.7 | 0.61 (0.10)/1.4 |
| CG12763 (Dpt) | Diptericin A | 19.8 (7.7) | 70 (37)/3.5 | 53 (12)/2.7 | 15.3 (3.7)/0.8 |
| CG10794 (DptB) | Diptericin B | 12.7 (5.6) | 63 (32)/5.0 | 56 (13)/4.4 | 13.1 (3.4)/1.0 |
| CG1385 (Def) | Defensin | 4.3 (2.2) | 26.5 (7.9)/6.2 | 18.2 (8.0)/4.3 | 3.76 (0.87)/0.9 |
| CG10816 (Dro) | Drosocin | 14.1 (6.6) | 53.5 (14.7)/3.8 | 47 (12)/3.3 | 20.1 (3.9)/1.4 |
| CG10810 (Drs) | Drosomycin | 18.5 (7.4) | 49 (16)/2.6 | 58 (21)/3.1 | 51 (23)/2.8 |
| CG8175 (Mtk) | Metchnikowin | 30 (12) | 85 (33)/2.9 | 76 (32)/2.5 | 54 (17)/1.8 |
| CG12780 | Gram-negative binding protein | 1.93 (0.93) | 3.1 (1.6)/1.6 | 3.6 (1.3)/1.9 | 4.3 (1.6)/2.2 |
| CG13422 | Gram-negative binding protein | 2.4 (1.2) | 25.5 (7.0)/10.6 | 30.5 (5.4)/12.7 | 23.5 (3.9)/9.8 |
| CG7052 (TepII) | Thiolesther containing protein II | 0.31 (0.017) | 2.19 (0.90)/7.1 | 2.3 (1.7)/7.6 | 2.51 (0.70)/8.2 |
| CG1361 (Anp) | Andropin | 48 (14) | 50.0 (8.1)/1.0 | 58 (10)/1.2 | 40 (12)/0.8 |
| Endopeptidase | |||||
| CG10882 | Kallikrein | 0.35 | 0.72 (0.24)/2.0 | 0.75 (0.17)/2.1 | 0.46 (0.10)/1.3 |
| CG16821 | Kallikrein | 0.18 | 0.29/1.6 | 0.39 (0.27)/2.2 | 0.19 (0.03)/1.1 |
| CG4920 (ea) | Easter | 0.52 (0.10) | 0.63/1.2 | 0.60 (0.07)/1.2 | 0.65/1.2 |
| CG2045 | Easter serine protease | 2.17 (0.76) | 2.72 (0.74)/1.2 | 6.1 (1.8)/2.8 | 6.8 (1.7)/3.1 |
| CG3505 | Easter serine protease | 1.24 (0.44) | 2.6 (1.1)/2.0 | 2.63 (0.84)/2.1 | 2.92 (0.66)/2.4 |
| CG16705 | Easter serine protease | 4.7 (1.7) | 5.4 (1.9)/1.2 | 12.1 (6.6)/2.6 | 18.6 (3.4)/4.0 |
| CG7996 (snk) | Snake | 0.59 (0.14) | 0.70 (0.01)/1.2 | 0.78 (0.13)/1.3 | 0.38 (0.14)/0.6 |
| CG11841 | Snake—coagulation factor XI | 4.6 (1.6) | 3.28 (0.98)/0.7 | 5.9 (2.8)/1.3 | 10.0 (3.2)/2.2 |
| CG11842 | Snake—trypsin-like serine protease | 0.94 (0.24) | 1.53 (0.71)/1.6 | 2.7 (1.1)/2.9 | 3.06 (0.80)/3.3 |
| CG11843 | Snake—trypsin-like serine protease | 0.26 | 0.42 (0.11)/1.6 | 0.49 (0.44)/1.8 | 0.68 (0.20)/2.6 |
| CG9645 | Stubble stubbloide—transmembrane domain | 2.06 (0.91) | 2.9 (1.1)/1.4 | 6.9 (2.3)/3.4 | 4.9 (1.1)/2.4 |
| CG9372 | Stubble—hemocyte protease-I | 1.50 (0.51) | 1.32 (0.38)/0.9 | 2.35 (0.85)/1.6 | 3.09 (0.99)/2.1 |
| CG2105 | Corin “transmembrane mosaic serine protease” | 0.21 (0.59) | 0.51 (0.02)/2.5 | 0.74/3.6 | 0.59 (0.11)/2.9 |
| CG11459 | Cathepsin L | 0.74 (0.12) | 1.42 (0.54)/1.9 | 1.86 (0.79)/2.5 | 3.6 (1.8)/4.9 |
| CG11836 | Hepsin—coagulation factor X precursor | 0.18 | 0.37 (0.09)/2.1 | 0.37/2.1 | 0.15/0.9 |
| CG6639 | Masquerade | 0.29 | 0.50 (0.03)/1.7 | 10.7 (8.0)/36.4 | 17.8 (4.4)/60.5 |
| Growth factor | |||||
| CG9224 (sog) | Short gastrulation | 0.3 | 0.45 (0.13)/1.5 | 0.62 (0.16)/2.1 | 0.29 (0.13)/1.0 |
| CG4559 (Idgf3) | Imaginal Disc Growth Factor 3 | 4.8 (1.7) | 10.3 (4.5)/2.2 | 10.5 (4.0)/2.2 | 7.6 (2.4)/1.6 |
| CG9885 (dpp) | Decapentaplegic | 0.13 | 0.32/2.4 | / | |
| Motor protein | |||||
| CG16910 (key) | Kenny | 0.61 (0.20) | 0.80 (0.39)/1.3 | 0.78 (0.33)/1.3 | 0.74 (0.15)/1.2 |
| Serpins | |||||
| CG6687 | Necrotic-like—squamous cell carcinoma antigen 1 | 3.0 (1.3) | 8.1 (3.1)/2.7 | 14.9 (9.0)/5.0 | 22.2 (4.1)/7.4 |
| CG1857 (nec) | Necrotic | 3.3 (1.4) | 7.2 (2.9)/2.2 | 15.4 (5.8)/4.6 | 16.2 (4.9)/4.9 |
| CG9453 (sp1) | Serpin 1 | 1.65 (0.63) | 1.95 (0.74)/1.2 | 1.76 (0.65)/1.1 | 2.14 (0.55)/1.3 |
| CG8137 (sp2) | Serpin 2 | 1.15 (0.50) | 1.76 (0.57)/1.5 | 2.0 (1.1)/1.8 | 0.95 (0.30)/0.8 |
| CG9334 (sp3) | Serpin 3 | 4.0 (1.9) | 5.5 (1.5)/1.4 | 5.4 (2.5)/1.4 | 3.3 (1.1)/0.8 |
| CG9456 (sp4) | Serpin 4 | 2.21 (0.97) | 2.8 (1.0)/1.3 | 4.2 (1.4)/1.9 | 5.4 (1.3)/2.4 |
| CG18525 (sp5) | Serpin 5 | 0.96 (0.33) | 2.27 (0.78)/2.4 | 3.0 (1.2)/3.1 | 2.04 (0.39)/2.1 |
| CG10913 (sp6) | Serpin 6 (not on Affymetrix microarray) | ||||
| Signal transduction protein | |||||
| CG5576 (Imd—BG5) | Death Domain | 0.23 | 0.35/1.5 | 0.38/1.6 | / |
| Specific RNA polymerase II transcriptional factor | |||||
| CG6667 (dl) | Dorsal | 0.11 | 0.75 (0.33)/6.6 | 1.00 (0.36)/8.8 | 0.30 (0.19)/2.6 |
| CG11992 (Rel) | Relish | 0.86 (0.42) | 2.8 (1.4)/3.3 | 3.0 (1.3)/3.5 | 2.79 (0.71)/3.3 |
| CG6794 (Dif) | Dorsal-related immunity factor | 0.36 (0.02) | 0.37/1.0 | 0.25/0.7 | 0.26 (0.004)/0.7 |
| Transcription factor associated | |||||
| CG5848 (cact) | Cactus | 1.48 (0.69) | 2.2 (1.1)/1.5 | 4.5 (1.4)/3.1 | 4.0 (1.2)/2.7 |
Absolute and relative levels of expression for Drosophila genes discussed within the text, following challenge the Gram-negative E. coli, the Gram-positive M. luteus, or the fungus B. bassiana, where function or gene identity was given in Flybase on or before June 1, 2001.
Functional categories of genes imposed by FlyBase.
Gene identifier or CG number applied to each gene following from complete sequencing of the Drosophila genome by Celera.
Fully identified genes are indicated in bold type and the associated gene acronyms are indicated in brackets after the CG number.
Genes with homology and/or proposed function, but no clear identity, are indicated in ordinary type.
Absolute level of gene expression in naive flies was less than 0.01; therefore, no relative values were calculated.
The absolute values are the maximum levels of induction observed, regardless of when this occurred during the time course.
SD for highest absolute value.
The relative values given are the maximum absolute level of induction as a ratio of the absolute expression level seen in naive animals. Where relative gene induction exceeds the 2-fold or more threshold, the relative values are indicated in bold.
No data for a particular gene/treatment combination.
In all of the experiments, Andropin, a challenge-independent antimicrobial peptide gene that is constitutively produced in the male genital tract (9), was expressed at constant levels. This constancy of expression after a variety of immunological challenges indicated the utility of Andropin as a reference gene for baseline expression in the present context.
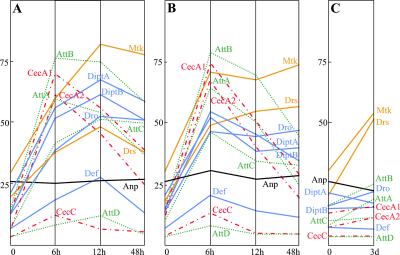
With the exception of four of the seven Drosomycin genes identified (5), all of the antimicrobial peptide genes were represented on the array. The expression of some or all of these antimicrobial peptide genes was induced by the three types of immune challenge (Fig. 1). Bacterial infection induced the expression of all antimicrobial peptide genes, although, because of the lack of signal in naive flies, the expression of CecropinB cannot be confirmed with the current data. High absolute levels of expression were observed in the case of Gram-negative infection for Metchnikowin, Attacins (A, B, and C), Diptericins (A and B), Cecropins (A1 and A2), and Drosocin, whereas Drosomycin and Defensin showed moderate levels of expression. CecropinC and AttacinD genes were expressed at very low levels, even after microbial challenge. In these experiments, the Cecropin and Attacin genes seemed to react more rapidly than the other antimicrobial peptide genes, with expression levels peaking as early as 6 h after challenge (as compared with 12 h for the other antimicrobial peptide genes). Gram-positive infection gave results similar to those with Gram-negative infection, with two exceptions. (i) All antimicrobial peptide genes showed maximum expression at 6 h, and (ii) Metchnikowin and Drosomycin expression persisted at elevated levels until at least 48 h after infection. In the case of natural fungal infection, only Metchnikowin and Drosomycin were found to be strongly up-regulated after 3 days.
Figure 1.
Absolute expression levels of antimicrobial peptide genes after microbial challenge. Expression of antimicrobial peptide genes in cn bw flies was assessed at 6, 12, and 48 h after inoculation with (A) E. coli and (B) M. luteus, and 3 days after natural infection with B. bassiana (C). Mtk and Drs (antifungal peptide genes), orange lines; Att gene family, green dotted lines; Cec gene family, red dash-dotted lines; Anp, black line; Dpt, Dro, and Def genes, blue lines.
The results obtained were globally similar to those found in earlier studies based on Northern blot analysis (7, 8). In particular, they show the differential induction of antimicrobial peptide genes by bacterial challenge vs. fungal infection. However, as no other method has been used to simultaneously resolve all of the variants of the Attacin, Cecropin, and Diptericin gene families, a more detailed comparison of any two approaches is not possible. Concurrently with microarray analysis, multiplex real-time quantitative PCR was used to assess the induction levels for three of the antimicrobial peptide genes, Diptericin, Drosomycin, and Attacin A, after the various immune challenges. The data were in agreement with the results obtained with the microarray technique (data not shown). It was therefore concluded that the array methodology, as used under the conditions presented here, gave a valid assessment of transcript levels in the context of the Drosophila immune response.
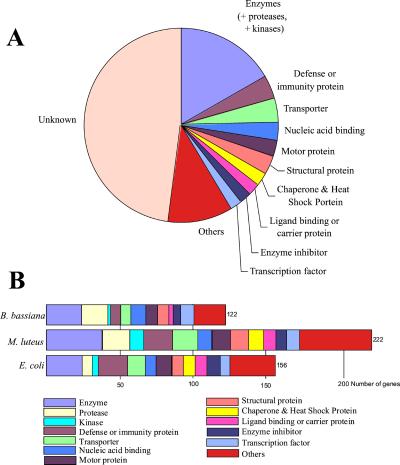
Global analysis of the expression data revealed that 8,459 of the 13,600 genes represented on the array were expressed in naïve flies. Bacterial or fungal immune challenge induced expression levels for 543 genes by at least 2-fold. Our confidence limit for detecting meaningful changes in expression was ≈1.8-fold, and this provided the primary criteria for selecting an induction threshold of 2-fold. For each type of infection, between 219 and 351 genes were up-regulated by 2-fold or more, with many up-regulated by more than one pathogen. Approximately half of these immune-responsive genes have been fully described or have been ascribed a putative function through their similarity to known genes from other organisms (10).
Genes from many different functional classes were up-regulated by 2-fold or more by immune challenge. These categories included actin-associated, calcium binding, cell adhesion, chaperones, heat-shock proteins, enzyme inhibitors, growth factors, carrier proteins, motor proteins, nucleic acid-binding factors, structural proteins (cytoskeleton, cuticle, and muscle), transcription factors, and others (classification as per FlyBase on June 1, 2001; http://flybase.bio.indiana.edu) (Fig. 3). The complete data set of genes induced by at least a factor of 2 is available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. The consequent analysis was focused on addressing the following questions: (i) How are the genes of known immune intracellular signaling pathways affected by infection? (ii) What are the most strongly induced genes? (iii) Do the profiles provide novel insights into the Drosophila host defense system?
Figure 3.
Drosophila genes up-regulated 2-fold or more during microbial challenge according to gene functional group. (A) Genes up-regulated 2-fold or more regardless of microbial challenge, including genes with unknown function. Genes were assigned to functional categories according to the protocol described in Table 2 (which is published as supporting information on the PNAS web site) with the additional fusion of enzymes, proteases, and kinases into one category and the regroupment of all categories containing six genes or less into “other.” (B) Number of genes whose expression was up-regulated 2-fold or more according to microbial challenge, excluding genes of unknown function. Genes were assigned to functional categories according to the protocol described in Table 2 with the regroupment of all categories containing six genes or less into “other.”
(i) How Are the Genes of Known Immune Intracellular Signaling Pathways Affected by Infection?
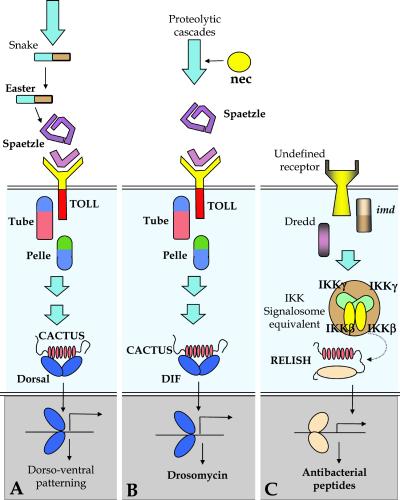
Genetic and biochemical analyses to date have implicated two major intracellular signaling pathways in the control of expression of immune-responsive genes (Fig. 3). The Toll pathway directs expression of Drosomycin, Metchnikowin, and a large number of defense-related polypeptides (Drosophila immune-induced molecules, proteases, serpins, etc.; ref. 11). The Imd pathway regulates expression of the antibacterial peptide genes (12). More than 20 genes involved in the Toll and Imd signaling pathways have now been identified. It was not anticipated that these genes themselves would necessarily be strongly induced by immune challenge, in contrast to the circulating recognition and effector polypeptides described below. Nevertheless, practically all known genes of the Toll signaling cascade were at least modestly up-regulated. More surprisingly, the dorsal gene was strongly induced (×8) by bacterial challenge. As antimicrobial peptide gene expression is unperturbed in dorsal mutants (13), induction of this Rel protein gene suggests that it may play an as yet undescribed role in the immune response. The cactus gene was also noticeably up-regulated (×3) by Gram-positive bacterial and fungal challenge, in keeping with the immune role of the Cactus protein of retaining the Rel proteins Dorsal and Dif in the cytoplasm (11). Up-regulation of cactus expression by immune challenge ultimately contributes to shutting-off expression of the immune-responsive genes, which is observed for a large proportion of genes 12–48 h after their initial induction by challenge. In contrast to dorsal, no marked induction of the Dif gene was observed. This finding was unexpected given the published evidence of Dif involvement in the Toll pathway (14). One explication may be that Dif expression peaked in between the time points used in these experiments. Alternatively, the lack of a Dif signal from these experiments could indicate the inability of the arrays used here to detect this gene.
With regard to the genes of the Imd signaling pathway, there was a noticeable up-regulation of Relish (×3), which was similar for all three immune challenges. Interestingly, the level of Relish expression in naive adults was severalfold that of dorsal. As these genes are at roughly the same point in their appropriate signaling pathways (Fig. 2), there are two possible explanations for the difference in background levels. The first is that the level of gene expression in naive flies may indicate the degree of “readiness” of each system to be set in motion, implying that the Imd pathway would be able to respond more rapidly or at an immediately higher level than that of Toll. A second possibility is that Relish fulfills a separate, as yet unknown, metabolic role in the unchallenged adult fly. Finally, the Imd pathway genes encoding the Death Domain protein Imd (15), the caspase-8 homologue Dredd (16), and the IKKγ/NEMO homologue Kenny (17), were modestly up-regulated (less than 2-fold).
Figure 2.
Distal components of the dorso-ventral, Toll, and Imd signaling pathways. (A) The distal part of the embryonic dorso-ventral patterning pathway, showing the end of the proteolytic cascade that leads to the cleavage of Spaetzle and intracellular signaling. (B) Established elements of the distal part of the Toll pathway, where a serpin, nec, regulated a proteolytic cascade leading to the cleavage of Spaetzle. This pathway reuses many of the components seen in embryonic dorso-ventral patterning, and it is induced by infection with Gram-positive bacteria and fungi. (C) Established elements of the Imd pathway, which is induced by Gram-negative bacteria. At present, none of components in the extracellular cascades are known for this pathway.
(ii) What Are the Most Strongly Induced Genes?
With a few exceptions, the genes most strongly induced by immune challenge in this study encode polypeptides known or presumed to be secreted into the hemolymph. These include the antimicrobial peptides discussed above, putative microbial pattern-recognition proteins, proteases, and their inhibitors. The recognition proteins belong to two categories initially identified in other insect species: the Gram-negative binding proteins (GNBPs; refs. 18–20) and peptidoglycan recognition proteins (PGRPs; ref. 21). The Drosophila genome contains five genes predicted to encode GNBPs. Only one of these five genes (CG13422) is very strongly up-regulated, and this up-regulation is observed for all three types of infection. Of the 13 predicted PGRP genes, 5 are markedly immune-responsive (to all types of challenges) and 4 of these encode putatively secreted proteins (PGRP-SA, PGRP-SB1, PGRP-SC, and PGRP-SD). It is interesting to note that the GNBP and PGRP genes that are strongly induced are also expressed at relatively high levels in naive adults. The flies used in the experiments were not raised in aseptic conditions and, as both gene families are possibly involved in the recognition of non-self, the base-line levels of the immune-inducible family members may indicate the presence of minor microbial infections or stress within the naive population.
A large number of proteases, along with a few protease-like polypeptides and protease inhibitors, were induced by infection. Many of these are zymogens, inactive protease precursors containing N-terminal disulfide-knotted motifs (CLIP domains) that are assumed to play a role in regulating the processing of the proenzyme form to the active enzyme (22). It is interesting to put these data in perspective with the activation of the Toll signaling pathway, which directs expression of a large variety of immune-responsive genes. Toll is activated through a proteolytic amplification cascade resulting in the cleavage of the cytokine Spaetzle. The proteases of this cascade have not been identified and it is assumed, on the basis of genetic evidence, that they are distinct from the Snake and Easter proteases, which are involved in Spaetzle cleavage in early embryos during dorsal-ventral patterning (Fig. 2). No obvious up-regulation of snake and easter was observed, but several genes encoding Easter- and Snake-like serine proteases were induced, particularly in response to Gram-positive bacteria and fungi. These genes are good candidates for fulfilling the roles of easter and snake in activating Toll during the immune response. Several other genes encoding secreted proteases (e.g., cathepsin-, Stubble-, and hepsin-like) were also up-regulated and, as for the easter- and snake-like genes, their precise roles in the host defense remain to be established.
The large spectrum of protease genes reacting to immune challenge is paralleled by induced expression of numerous protease-inhibitor genes, most of which are predicted to be secreted. Three of the genes belong to the Kunitz family of inhibitors and several others are serine-protease inhibitors (serpins). A strongly induced member of this family is the necrotic gene, previously shown to be involved in the regulation of the hemolymph protease cascade that leads to the cleavage of Spaetzle and activation of Toll (23). The data point to similar up-regulation of a necrotic-like gene, which is a plausible candidate for regulation of further proteolytic cascades in the hemolymph of challenged flies.
The majority of the other induced serpin genes encode proteins with similarity to the anti-thrombins. Several members of a newly described serpin family, referred to as sp1-sp6 (24), seem to be specifically up-regulated in response to either fungal infection (sp1) or bacterial infection (sp2, sp5).
The most strongly induced of all of the genes analyzed in the study was a masquerade-like gene, minimally up-regulated by Gram-negative challenge, but responding dramatically to Gram-positive infection (36-fold) and fungal infection (60-fold), both of which trigger the Toll pathway. Among the features of the predicted protein for the gene is trypsin-like serine proteinase domain at the C terminus, probably not active due to a serine to glycine replacement in the active site. The Masquerade-like proteins discovered in other invertebrates are suggested to act as antagonists of serine proteases, sequestering the protease targets and thus regulating enzymatic activity (25–27).
Finally, it is noteworthy to mention in this section that the gene encoding the thiolester-containing protein dTEPII, a member of the complement factor C3/α2 macroglobulin superfamily, is also strongly up-regulated by all three types of challenge. dTEPII is a secreted protein that has been proposed to participate in the host defense (28).
The observed up-regulation of putative circulating recognition proteins and proteases agrees with the current view that the systemic antimicrobial response is triggered when microorganisms interact with hemolymph proteins and activate proteolytic amplification cascades. Although there are 421 known proteases in Drosophila (http://flybase.bio.indiana.edu), only 26 were up-regulated by a factor of 2 or more, and only 8 by more than 3-fold in this study, illustrating how the microarray approach can narrow down a large number of potential candidate genes to a more tractable number.
(iii) Do the Profiles Provide Novel Insights into the Drosophila Host Defense System?
In addition to genes whose increased expression could be reasonably expected, there were also a large number of induced genes that had not been linked previously to the immune response in Drosophila. Presented here are some examples for which future experiments in the context of host defense seem warranted.
Among the functions of the kallikrein-kininogen-kinin system in mammals are responses to septic shock and the initiation of inflammation, involving tissue-specific and plasma factors (29). Two kallikrein-like genes were induced by bacterial challenge, one encoding a secreted protein (CG10882) and the other a transmembrane protein (CG16821). Both have an active protease site, and the combination of circulating and membrane-bound proteases raises the possibility that Drosophila relies on an equivalent of a kallikrein-kininogen-kinin system, which could have, as in vertebrates, a proinflammatory function. In keeping with this idea, some of the recently identified small-sized Drosophila immune-induced molecules found in immune-induced hemolymph (30) are good candidates for a kinin-like role in this system.
Two particularly interesting genes encoding type II transmembrane proteins were up-regulated. The first was up-regulated (×3) by all three immune challenges and encoded a homologue of mammalian corin (CG2105), a mosaic protein with two frizzled-like cysteine-rich motifs, a macrophage scavenger receptor-like domain, and a trypsin-like protease domain in the extracellular region. The second gene was one of the Stubble-like genes up-regulated during fungal infection (CG9645), encoding a transmembrane serine protease with structural similarities to mammalian hepsin. Both hepsin and corin are involved in blood circulation and coagulation in mammals (31, 32). Because almost nothing is known about the process of coagulation in Drosophila, the genes induced are of interest as they may play similar roles to their mammalian counterparts.
Caspases are known to play a role in apoptosis; however, the function of a number of caspases is still unclear (33). Several caspases were induced: drICE (up-regulated in all three types of immune challenges), Decay (induced by fungal infection), Daydream (induced by fungal and Gram-positive bacterial infection), and Dredd (implicated in the activation of the Imd pathway and up-regulated by bacteria). As nonapoptotic roles have been found for a number of caspases (33), the exact function of those up-regulated during the immune response remains to be clarified.
Finally, the genes encoding the growth factors Short Gastrulation, Imagical Disk Growth Factor 3, and Decapentaplegic are up-regulated by bacterial challenge. These genes function downstream of Toll, or later, during dorsal-ventral patterning in the embryo. Several other factors involved in dorsal-ventral patterning are known to be involved in the Toll immune response pathway, and it is interesting to consider whether these genes may also play a role during the immune response.
The data at hand did not point to a clear-cut signature for each of the three types of infection beyond what was already known. Rather, the same categories of genes were induced by all infections. Bacterial challenge induced a larger number of known defense proteins, reflecting the induction of the numerous antimicrobial peptide genes following septic injury. B. bassiana and M. luteus seemed to induce more protease genes than E. coli, possibly as a result of their preferential induction of the Toll pathway, which is regulated by a protease cascade. Clearly more experiments with distinct bacterial and fungal species are required to furnish a more complete picture of such pathogen-specific responses. It is important, however, to reiterate that in this study 47% of the genes induced by 2-fold or more after immune challenge are for the time being labeled as unknowns, lacking apparent similarities to known genes in the databases. Further, the groups of genes up-regulated by each of the three challenges differ significantly more among the cohorts of unknown genes than among genes with known or proposed functions. Future analysis of these unknown genes will undoubtedly increase the understanding of pathogen-specific immune responses.
In conclusion, this genome-wide analysis of immune-challenged Drosophila with DNA microarrays provided results that agree with previous findings as well as identifying a significant number of novel immune-induced genes. Some of the previously unseen immune-induced genes fit into the current conceptual framework of the host defense, whereas others offer new avenues for research. Singling-out these genes from the whole genome sequence will help to establish priorities for precise functional studies based on mutant analysis. Surprisingly, the data has also shown that nearly half of the immune-induced genes in Drosophila have not yet been ascribed a putative function and do not show obvious similarities to any known genes. This finding is a dramatic illustration of the distance that still separates us from a reasonable understanding of the prototypical innate immune response of this model organism.
Supplementary Material
Acknowledgments
Thanks go to Martine Schneider for expert technical assistance and to Sophie Rutschmann for PCR primers and probes. P.I. was financed by European Community Research and Training Network Contract HPRN-CT-2000-00080. The work carried out in Strasbourg and South San Francisco was supported jointly by the Centre National de la Recherche Scientifique and Exelixis.
References
- 1.Hoffmann J A, Kafatos F C, Janeway C A, Jr, Ezekowitz R A B. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 2.Borregaard N, Elsbach P, Ganz T, Garred P, Svejgaard A. Immunology. 2000;21:68–70. doi: 10.1016/s0167-5699(99)01570-4. [DOI] [PubMed] [Google Scholar]
- 3.Carton Y, Nappi A J. Immunogenetics. 2001;52:157–164. doi: 10.1007/s002510000272. [DOI] [PubMed] [Google Scholar]
- 4.Meister M, Hetru C, Hoffmann J A. Curr Top Microbiol Immunol. 2000;248:17–36. doi: 10.1007/978-3-642-59674-2_2. [DOI] [PubMed] [Google Scholar]
- 5.Khush R S, Lemaitre B. Trends Genet. 2000;16:442–449. doi: 10.1016/s0168-9525(00)02095-3. [DOI] [PubMed] [Google Scholar]
- 6.Imler J L, Hoffmann J A. Curr Opin Microbiol. 2000;3:16–22. doi: 10.1016/s1369-5274(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 7.Lemaitre B, Reichhart J-M, Hoffmann J A. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedengren M, Borge K, Hultmark D. Biochem Biophys Res Commun. 2000;279:574–581. doi: 10.1006/bbrc.2000.3988. [DOI] [PubMed] [Google Scholar]
- 9.Samakovlis C, Kylsten P, Kimbrell D A, Engstrom A, Hultmark D. EMBO J. 1991;10:163–169. doi: 10.1002/j.1460-2075.1991.tb07932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers E W, Sutton G G, Delcher A L, Dew I M, Fasulo D P, Flanigan M J, Kravitz S A, Mobarry C M, Reinert K H, Remington K A, et al. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 11.Imler J L, Hoffmann J A. Trends Cell Biol. 2001;11:304–310. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- 12.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross I, Georgel P, Kappler C, Reichhart J M, Hoffmann J A. Nucleic Acids Res. 1996;24:1238–1245. doi: 10.1093/nar/24.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutschmann S, Jung A C, Hetru C, Reichhart J M, Hoffmann J A, Ferrandon D. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 15.Georgel, P., Naitza, S., Kappler, C., Ferrandon, D., Zachary, D., Swimmer, C., Kopczynski, C., Duyk, J., Reichhart, J.-M. & Hoffmann, J. A. (2001) Dev. Cell, in press. [DOI] [PubMed]
- 16.Leulier F, Rodriguez A, Khush R S, Abrams J M, Lemaitre B. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutschmann S, Jung A C, Zhou R, Silverman N, Hoffmann J A, Ferrandon D. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 18.Lee W J, Lee J D, Kravchenko V V, Ulevitch R J, Brey P T. Proc Natl Acad Sci USA. 1996;93:7888–7893. doi: 10.1073/pnas.93.15.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin S W, Sang W S, Mi G K, Soon S K, Lee W-J, Brey P T, Park H-Y. Insect Biochem Mol Biol. 1998;28:827–837. doi: 10.1016/s0965-1748(98)00077-0. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y-S, Ryu J-H, Han S-J, Choi K-H, Nam K-B, Jang I-H, Lemaitre B, Brey P T, Lee W-J. J Biol Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- 21.Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. Proc Natl Acad Sci USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H, Kanost M R. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- 23.Green C, Levashina E, McKimmie C, Dafforn T, Reichhart J M, Gubb D. Genetics. 2000;156:1117–1127. doi: 10.1093/genetics/156.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J-H, Zhang H-Y, Min G-S, Kemler D, Hashimoto C. FEBS Lett. 2000;468:194–198. doi: 10.1016/s0014-5793(00)01224-2. [DOI] [PubMed] [Google Scholar]
- 25.Kwon T H, Kim M S, Choi H W, Joo C H, Cho M Y, Lee B L. Eur J Biochem. 2000;267:6188–6196. doi: 10.1046/j.1432-1327.2000.01695.x. [DOI] [PubMed] [Google Scholar]
- 26.Huang T S, Wang H, Lee S Y, Johansson M W, Soderhall K, Cerenius L. J Biol Chem. 2000;275:9996–10001. doi: 10.1074/jbc.275.14.9996. [DOI] [PubMed] [Google Scholar]
- 27.Dimopoulos G, Richman A, Muller H M, Kafatos F C. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagueux M, Perrodou E, Levashina E A, Capovilla M, Hoffmann J A. Proc Natl Acad Sci USA. 2000;97:11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma J N, Mohsin S S. Exp Pathol. 1990;38:73–96. doi: 10.1016/s0232-1513(11)80241-0. [DOI] [PubMed] [Google Scholar]
- 30.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann J A, Bulet P. Proc Natl Acad Sci USA. 1998;95:11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Q. Front Biosci. 2001;6:D192–D200. doi: 10.2741/a604. [DOI] [PubMed] [Google Scholar]
- 32.Yan W, Sheng N, Seto M, Morser J, Wu Q J. J Biol Chem. 1999;274:14926–14935. doi: 10.1074/jbc.274.21.14926. [DOI] [PubMed] [Google Scholar]
- 33.Vernooy S Y, Copeland J, Ghaboosi N, Griffin E E, Yoo S J, Hay B A. J Cell Biology. 2000;150:F69–F75. doi: 10.1083/jcb.150.2.f69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.