Abstract
Here we present a comprehensive map of the accessible chromatin landscape of the mouse hippocampus at single-cell resolution. Substantial advances of this work include the optimization of a single-cell combinatorial indexing assay for transposase accessible chromatin (sci-ATAC-seq); a software suite, scitools, for the rapid processing and visualization of single-cell combinatorial indexing data sets; and a valuable resource of hippocampal regulatory networks at single-cell resolution. We used sci-ATAC-seq to produce 2346 high-quality single-cell chromatin accessibility maps with a mean unique read count per cell of 29,201 from both fresh and frozen hippocampi, observing little difference in accessibility patterns between the preparations. By using this data set, we identified eight distinct major clusters of cells representing both neuronal and nonneuronal cell types and characterized the driving regulatory factors and differentially accessible loci that define each cluster. Within pyramidal neurons, we identified four major clusters, including CA1 and CA3 neurons, and three additional subclusters. We then applied a recently described coaccessibility framework, Cicero, which identified 146,818 links between promoters and putative distal regulatory DNA. Identified coaccessibility networks showed cell-type specificity, shedding light on key dynamic loci that reconfigure to specify hippocampal cell lineages. Lastly, we performed an additional sci-ATAC-seq preparation from cultured hippocampal neurons (899 high-quality cells, 43,532 mean unique reads) that revealed substantial alterations in their epigenetic landscape compared with nuclei from hippocampal tissue. This data set and accompanying analysis tools provide a new resource that can guide subsequent studies of the hippocampus.
A major goal in the life sciences is to map cell types and identify the respective genomic properties of each of the cell types in complex tissues. Traditional strategies that use intact tissue are limited to averaging of the constituent cell profiles. To overcome this limitation, there has been a burst in development of unbiased single-cell genomics assays, leveraging the concept that each single cell can only occupy a single position in the landscape of cell types (Trapnell 2015). This push into the single-cell space has largely centered on the use of single-cell transcriptional profiling. Although profiling the RNA complement has produced valuable information (Zeisel et al. 2018; Saunders et al. 2018), the ability to profile chromatin status, namely, active versus inactive, has lagged behind, leaving open the question as to what extent accessible chromatin profiles are linked to cell specificity, particularly with respect to distal enhancer elements (Corces et al. 2016).
Recently, progress has been made to ascertain chromatin accessibility profiles in single cells using assay for transposase-accessible chromatin (ATAC-seq) technologies. These strategies have been applied to myogenesis (Pliner et al. 2018), hematopoietic differentiation (Buenrostro et al. 2018), fly embryonic development (Cusanovich et al. 2018b), the mouse (Preissl et al. 2018) and human cortex (Lake et al. 2018), and, most recently, an atlas of multiple tissues in the mouse, although lacking the hippocampus (Cusanovich et al. 2018a). The core concept behind the methods used in several of these studies is a combinatorial indexing schema whereby library molecules are barcoded twice, once at the transposase stage and then again at the PCR stage. This platform has also been extended to profile other properties, including transcription, genome sequencing, chromatin folding, and DNA methylation (Cao et al. 2017; Ramani et al. 2017; Vitak et al. 2017; Mulqueen et al. 2018; Yin et al. 2018). In this work, we optimized the sci-ATAC-seq assay for the analysis of fresh and frozen hippocampal tissue samples to produce single-cell chromatin accessibility profiles in high throughput, with greater information content—as measured by unique reads per cell. These improvements will also facilitate the use of this technology platform on frozen samples, enabling the assessment of banked tissue isolates.
The hippocampus is critical to the formation and retrieval of episodic and spatial memory (Scoville and Milner 1957; O'Keefe and Dostrovsky 1971; Smith and Milner 1981; Zola-Morgan et al. 1986). Historically, cell types within the hippocampus have been broadly classified by their morphology (Ramon y Cajal 1911; Lorente de No 1934) and electrophysiological properties (Kandel and Spencer 1961; Kandel et al. 1961; Spencer and Kandel 1961a,b). More recently, types have been identified by their transcriptional profiles (Lein et al. 2004; Cembrowski et al. 2016), and single-cell transcriptomics has also revealed potential subclasses within previously defined cell types (Zeisel et al. 2015; Habib et al. 2017). The defined classes of cells within the hippocampus and the existing single-cell transcriptome data allowed us to refine our sci-ATAC-seq method and provide the first single-cell epigenomics profile of the murine hippocampus.
Results
Single-cell chromatin accessibility profiles from mouse hippocampus
We used sci-ATAC-seq to profile two fresh and two frozen mouse total hippocampi to map the accessible chromatin landscape (Methods). Each sample was freshly isolated from an adult (P60) wild-type mouse (C57-Bl6) and either processed immediately or flash frozen using liquid nitrogen. Nuclei were isolated and carried through the sci-ATAC-seq protocol with several optimizations from previously described implementations (Methods; Fig. 1A; Supplemental Protocol). Briefly, nuclei were isolated by dounce homogenization of tissue in nuclei isolation buffer followed by fluorescence-assisted nuclei sorting (FANS) using DAPI as a stain to select for intact, single nuclei. One of the key improvements to our workflow was the addition of Tween 20 (Sigma-Aldrich) to the nuclei isolation buffer, which we believe increased the permeability of the nucleus and removed more of the cell membrane. We then performed sci-ATAC-seq as previously described using a 55°C tagmentation temperature (Methods; Supplemental Protocol). Sequence reads were processed, and subsequent analysis was performed using scitools (Supplemental Code).
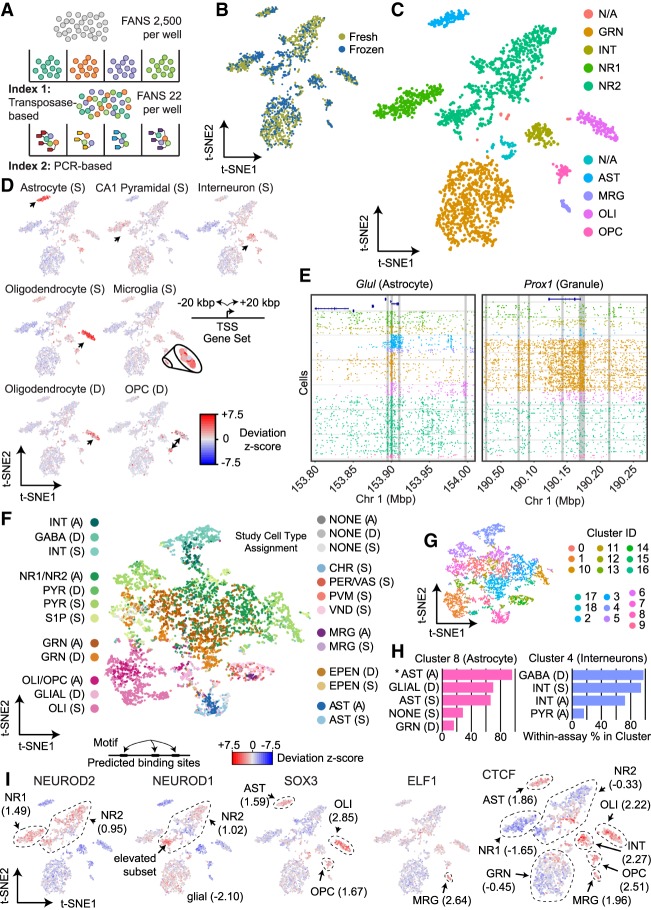
Figure 1.
sci-ATAC-seq of the murine hippocampus. (A) sci-ATAC-seq workflow. Two indexes are incorporated into library molecules for each cell, enabling single-cell discrimination. (B) LSI-t-SNE projection of single cells colored by tissue preparation method. Little variation in t-SNE space is observed between the fresh or frozen starting material. (C) LSI-t-SNE projection of cells colored by assigned cluster and cell type. (D) Enrichment of accessibility of proximal regulatory elements for marker genes as identified by single-cell RNA-seq (Smart-seq protocol [S]) (Zeisel et al. 2015) and DroNc-seq (D) (Habib et al. 2017) for each cell. The microglial population is enlarged for visibility. Black arrows indicate the cell cluster associated with the marker gene set. (E) sci-ATAC-seq read plots at Glul (astrocyte marker gene) and Prox1 (dentate granule cell marker gene). (F) Coembedding (t-SNE) of single-cell RNA-seq and DroNc-seq cells from D with our sci-ATAC-seq cells using Seurat3. Cells are colored by their study: “A” designates this study, “D” designates cells from Habib et al. (2017), and “S” designates cells from Zeisel et al. (2015). Cell type designation is from their published study (RNA) or our designations (for the sci-ATAC-seq cells). (G) PhenoGraph cluster designations on the coembedded cells. (H) Representative cluster cell compositions. The percentage of cells within each of the three assays that were assigned to the coembedding cluster using PhenoGraph is reported. For example (noted by an asterisk), in Cluster 8, 93.0% of the sci-ATAC-seq cells that were assigned to Cluster 8 were designated as astrocytes. (I) chromVAR global motif deviation Z-scores for each cell for select motifs. Dashed lines and values correspond to mean values of cell populations.
In total, we produced 2346 single cells passing quality control (1000 or more unique reads present in peaks and ≥25% of all unique reads present in peaks, alignment q ≥ 10, not aligned to chrM, unscaffolded, alternative, or random contigs) evenly represented across replicates (two frozen, two fresh). Based on existing single-cell RNA-seq studies, we assumed that our cell number should be sufficient for preliminary cell type deconvolution (Zeisel et al. 2015; Habib et al. 2017); however, we believe future innovation may enable greater numbers. Cells had a mean unique aligned read count of 29,201, which is higher than other high-throughput single-cell ATAC-seq workflows to date (Supplemental Table 1). We observed a strong correlation in ATAC signal between the aggregate profiles of the four replicates (Pearson R > 0.99), indicating high reproducibility across preparations for both fresh and frozen tissue. We did notice a statistically significant (t-test P-value = 2.2 × 10−6) increased number of unique reads per cell in the frozen samples; however, this can be attributed to greater sequencing depth (Supplemental Figs. 1, 2; Supplemental Table 1) or possibly to the freeze–thaw cycle increasing the permeability of the nucleus. Between replicates of the same preparation method, no statistically significant differences were observed. Chromatin accessibility peaks were identified by the aggregation of all cells to produce an ensemble data set containing all called peaks, resulting in a preliminary set of 93,994 high-confidence peaks, with a mean of 36.4% of reads from each cell falling within these regions. The fraction of reads in peaks for the frozen samples was greater than for the fresh samples (P-value = 1.3 × 10−4) (Supplemental Fig. 2; Supplemental Table 1).
We constructed a read count matrix of our ensemble peaks and single cells from all conditions (Supplemental Data, InVivo.counts.matrix) by tallying the number of reads for each cell at each peak. We next used scitools to perform latent semantic indexing (LSI), as previously described (Cusanovich et al. 2015, 2018b), with the exclusion of cells with reads at fewer than 1000 sites and of sites with fewer than 50 cells exhibiting signal. The LSI matrix was projected into two-dimensional (2D) space using t-distributed stochastic neighbor embedding (t-SNE) for visualization, which revealed distinct domains occupied by clusters of cells. We next used density-based clustering (Ester et al. 1996) and aggregated cells by cluster, called cluster-specific peaks, and added them to a union peak set (n = 98,043, 4% increase in peak count) for which all subsequent analysis was performed. We then identified nine major clusters (Fig. 1C), one of which likely being barcode collisions and removed from further analysis (Methods). A comparison of the proportion of cells assigned to each cluster with respect to fresh or frozen samples did not yield a significant difference (X2 = 9.85, P-value = 0.20) (Fig. 1B; Supplemental Table 2), although increased proportions of interneurons (INTs) and microglia (MRG) were observed in the frozen preparation.
To assign each of our identified clusters to a cell type, we took advantage of published single-cell RNA-seq data that produced sets of marker genes associated with cell types identified at the transcriptional level (Zeisel et al. 2015; Habib et al. 2017). For each set of cell-type–specific genes, we identified peaks 20 kbp in either direction from the transcriptional start site, which were then used to calculate the enrichment for accessible chromatin for each cell within these regions. This produced a deviation Z-score, similar to previously described methods (Buenrostro et al. 2015; Schep et al. 2017). We then visualized these scores on our t-SNE projections, which enabled us to clearly identify a number of neuronal and nonneuronal cell types, including astrocytes (ASTs), two groups of pyramidal neurons (designated neurons 1 [NR1] and 2 [NR2]), INTs, oligodendrocytes (OLIs), MRG, and OLI progenitor cells (OPCs) (Fig. 1D). To complement this strategy, we also turned to marker genes described previously in the literature that were not present in available single-cell RNA-seq data sets and assessed the chromatin accessibility at elements proximal to these genes (Fig. 1E; Supplemental Fig. 3; Lein et al. 2004; Zhang et al. 2014; Cembrowski et al. 2016). For example, the Glul gene, an established marker for ASTs (Martinez-Hernandez et al. 1977; Fages et al. 1988), showed accessibility only in the population of cells we identified as ASTs (Fig. 1E, left). Prox1, previously shown to be enriched in the dentate gyrus (Lein et al. 2004), is accessible predominantly in the dentate granule cell population (GRN) (Fig. 1E, right). Markers for particular cell types were also consistent with in situ hybridization data from the Allen Institute for Brain Science (Supplemental Fig. 3) and RNA-seq data from sorted cells (Zhang et al. 2014; Cembrowski et al. 2016). Based on our cell type assignments, the number of cells in each population reflects the proportions seen within the intact hippocampus (Supplemental Table 1; Abusaad et al. 1999). This includes the observation of 14-fold and 41-fold fewer ASTs and MRG compared with neurons, respectively, in line with previous studies (Kimoto et al. 2009).
To further confirm our cell type assignments, we used the recently released function in Seurat3 for the coembedding of single-cell ATAC-seq and single-cell RNA-seq data sets in a shared t-SNE space (Stuart et al. 2018). We first generated gene activity scores using Cicero (described below), which uses linked distal regulatory elements and promoters to approximate the putative activity of each gene (Pliner et al. 2018). These scores, along with transcript count matrices from Smart-seq and DroNc-seq publications (Zeisel et al. 2015; Habib et al. 2017), were processed using Seurat3 to identify anchors and effectively normalize them to one another to enable PCA and then visualization in a shared t-SNE space (Fig. 1F). Cells from our study were positioned proximal to cells with matching assignments in their respective publications. We next identified 18 distinct clusters (Fig. 1G) using PhenoGraph (Levine et al. 2015). Within these clusters, we quantified the percentage of cells assigned to each cell type within each of the three data sets to assess the most represented cell type that is present. This analysis further confirmed our cell type assignments with substantial concordance between the highest represented cell types across platforms (Fig. 1H). However, cross–data set assignment was far from perfect, with certain cell types performing better than others; for example, OLIs performed well versus granule cells, which did not. We suspect that the major driver of the discrepancies is because of the indirect nature of the gene activity scores for the single-cell ATAC-seq data.
Global DNA-binding motif accessibility
To assess the global activity of DNA-binding proteins, we used the recently described software tool, chromVAR (Schep et al. 2017), which aggregates the chromatin accessibility signal genome-wide at sites harboring a given motif, followed by the calculation of a deviation Z-score for each cell. This score represents the putative activity level of the DNA binding protein that corresponds to the assessed motif, which we then visualized on our t-SNE projections (Fig. 1I; heatmap in Supplemental Fig. 4). In line with expectations, our cell type clusters showed enrichment for accessibility at DNA-binding motifs concordant with the identified cell type (Fig. 1I; Supplemental Fig. 5). The analysis included the assessment of global accessibility for neuron-specific factors, such as NEUROD2, which associates with active chromatin marks (e.g., H3K27ac) in cortical tissue (Guner et al. 2017) and exhibited greater accessibility in the two pyramidal cell clusters (mean z-score (µz) = 1.49 and 0.95 for NR1 and NR2 respectively; all other cell types µz ≤ −0.74). We also observed increased accessibility of NEUROD1, also associated with active chromatin (Pataskar et al. 2016), in a portion of one of the pyramidal neuron clusters (NR2, µz = 1.02) with less accessibility across glial populations (µz ≤ −2.10). Although many studies have identified a role for SOX3 during neural differentiation, consistent with a previous expression study (Cheah and Thomas 2015), we observed increased SOX3 accessibility in AST (µz = 1.59), OLI (µz = 2.85), and OPC (µz = 1.67) populations, suggesting a glial role for this transcription factor in adulthood. ELF1, an ETS family member associated with activating interferon response in the hematopoietic lineage (Larsen et al. 2015), exhibited elevated accessibility in the microglial population (µz = 2.64), which also respond to interferon in the brain (e.g., Goldmann et al. 2015). We also observed a strong enrichment for CTCF motif accessibility in glial cell populations (AST µz = 1.86, OLI µz = 2.22, OPC µz = 2.51, MRG µz = 1.96) and INTs (µ = 2.27) compared with granule cells (µz = −0.45) or pyramidal neurons (NR1 µz = −1.65, NR2 µz = −0.33), which was reinforced by our subsequent differential accessibility analysis described below. To confirm that the observed motif accessibility increase is owing to true CTCF binding sites and not just the motif presence, we also performed a deviation analysis using peaks called from publicly available CTCF ChIP-seq data of the mouse hippocampus (Sams et al. 2016), which revealed very similar patterns of accessibility (Pearson R2 = 0.68) (Supplemental Fig. 6).
Differential accessibility by cell type
We next sought to show that accessible regions could be identified according to cell type. To provide sufficient signal, we aggregated cells within clusters in their local neighborhoods as has been described previously (Cusanovich et al. 2018b) and then performed a differential accessibility analysis for each cluster compared with the rest of the cells (Methods; Fig. 2A). Numbers of significant (Q-value ≤0.01, log2 fold-change ≥1) loci ranged from 894 (OPCs) to 7796 (granule cells), with substantial cell-type–specific signal (Fig. 2B; Supplemental Figs. 7–9; Supplemental Table 3). To characterize these loci, we performed a motif enrichment analysis to identify DNA-binding proteins that may bind within the differentially accessible regions (Fig. 2B, right). In contrast to the prior global accessibility analysis, in which all accessible loci were used to detect increased signal at sites harboring a given motif in each cell, here we are detecting enrichment of motifs in the specific subsets of loci that were determined to be differentially accessible. This strategy revealed enrichment for binding by the SOX10 transcription factor in OLIs (Claus Stolt et al. 2002) and by NEUROG2 in the dentate granule cells (Roybon et al. 2009). Within the INT population, the motif with the highest enrichment was CTCF. This is consistent with our prior global analysis of accessibility at motifs (Fig. 1I); however, this reciprocal approach suggests that a set of sites very specific to INTs harbor CTCF as opposed to sites that may be shared across numerous cell types with varying levels of accessibility. One of these regions was in an intron in the gene-encoding actin filament-associated protein 1 (Afap1) (Supplemental Fig. 10). The ChIP data revealed CTCF binding within the same intron flanking the accessible region. Previous work has suggested that CTCF may have a particular importance in this cell type (Kim et al. 2018). CTCF binding motifs were enriched in the accessible chromatin of affinity-purified parvalbumin-positive cortical INTs but not in VIP-positive INTs or excitatory neurons (Mo et al. 2015) and mice expressing one CTCF allele only in inhibitory neurons exhibit memory impairment (Kim et al. 2018). Recent data have also suggested that CTCF plays a role in the generation of cortical INTs by regulating the expression of the LIM homeodomain factor LHX6 (Elbert et al. 2019). The potential selective importance of CTCF in INTs warrants further study.
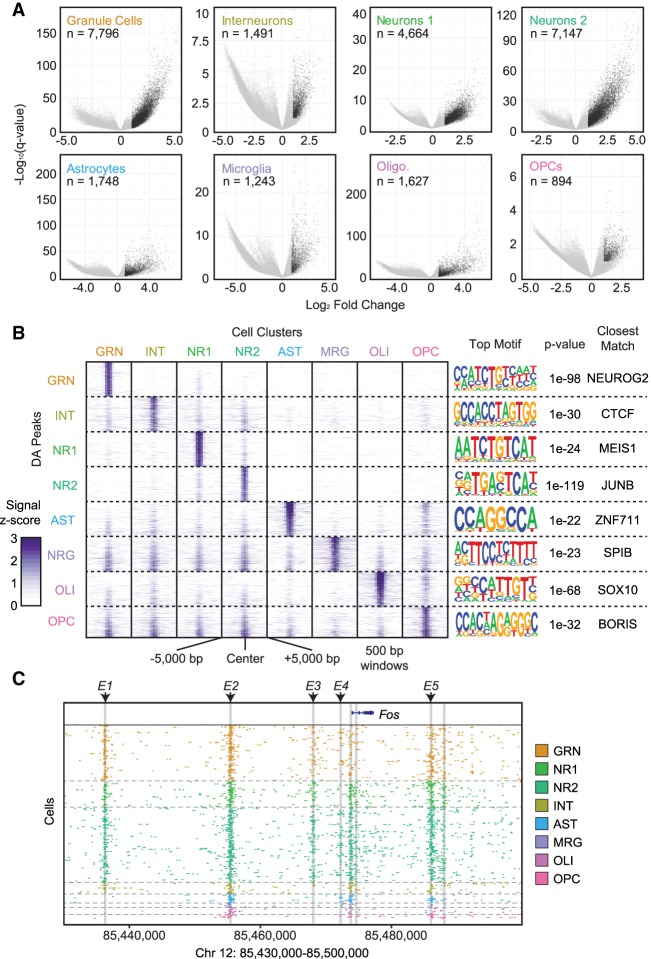
Figure 2.
Differential accessibility analysis between cell types. (A) Volcano plots −log10(Q-value) (y-axis) versus log2 accessibility signal fold-change (x-axis) showing all peaks. Each comparison is for the indicated cell population versus all other cell types. Significant peaks (number indicated, Q-value ≤0.01, log2 fold-change ≥1) are in black. (B) ATAC-seq signal plots for the top differential accessible peaks for each cell type. The most significantly enriched motif for each set is shown on the right along with the corresponding P-value and closest-matching known motif. (C) Fos locus with enhancers E1 through E5 highlighted to show cell-type–specific utilization.
To further determine the utility of our method in assigning regulatory elements to cell types, we tested whether we could parse enhancers that had been identified in the literature as inducers of target genes in response to neuronal activity. We focused on the Fos gene that has been studied previously as a general reporter of neuronal activity throughout the brain (Bullitt 1990). Specifically, five enhancers (E1–E5) have been characterized (Kim et al. 2010) for both regulation during neuronal activity and type of stimulation (Joo et al. 2015). When we examined ATAC-seq signal at the five enhancers across cell types in hippocampus, we identified cell-type–specific patterns of accessibility. Enhancers E1 and E3 were accessible only in neurons, whereas E2 and E5 were accessible in all cell types (Fig. 2C). Further, enhancer E4 was accessible in group 2 but not group 1 pyramidal neurons and was also accessible in a small portion of dentate granule cells. Our findings suggest cell-type specificity in stimuli responsiveness within the hippocampus, even between pyramidal cell subpopulations, opening the door to new studies of the basis of these signaling differences and demonstrating the utility of single-cell epigenomics over traditional bulk tissue assays.
More generally, our differential accessibility analysis was able to identify new enhancers by comparison with chromatin marks known to be associated with enhancers (Gjoneska et al. 2015). For example, during examination of the most significantly differentially accessible loci for dentate granule cells, one of the top hits was a region marked by both H3K4me1 and H3K27ac, suggesting a putative enhancer upstream of the gene Slc4a4 (Supplemental Fig. 11). Slc4a4 encodes a sodium/bicarbonate cotransporter involved in mediating both intracellular and extracellular pH (Svichar et al. 2011), and Slc4a4 expression is elevated in dentate granule neurons. Although these accessible loci were enriched only in dentate neurons, several other accessible regions were identified in dentate granule cells and in the two pyramidal neuron populations, suggesting this gene is expressed in multiple cell types and, like Fos, may exhibit variable responses in different cell types.
Pyramidal neuron subclustering
In our initial clustering, the two most prevalent pyramidal neuron populations, CA1 and CA3, were not able to be definitively resolved. We reasoned that analyzing these cells in isolation and using a recently described method for discerning themes, or “topics” of correlated signal within the data, BEDTools (Bravo González-Blas et al. 2018), may provide improved granularity. Based on a latent Dirichlet allocation framework, cisTopic identifies related sets of peaks that are classified as topics. On our NR1 and NR2 data set, the optimum number of topics was determined to be 30 (Supplemental Fig. 12), which was then used to project cells into 2D space using uniform manifold approximation and projection (UMAP) (Fig. 3A; Becht et al. 2019). Cells split into four distinct groups that were identified using PhenoGraph (Levine et al. 2015) on the topic matrix (Fig. 3B). One of the clusters was composed almost exclusively of the NR1 cells (95%), with the NR2 cells split into three groups. We did not observe any bias in cluster assignment with respect to the fresh versus frozen prepared cells (Supplemental Fig. 13). We next examined genes specifically associated with CA1 and CA3 neurons and were clearly able to assign two of the four clusters based on specific accessibility of promoters and/or cis regulatory elements at these loci (Supplemental Fig. 14). We also observed some enrichment of CA2-specific genes and genes associated with mossy cells (MCs) in two of the other clusters, suggesting that these cell types are likely present in the identified clusters; however, they may not make up the entirety of the population.
Figure 3.
Pyramidal neuron subclustering. (A) Subclustering of the NR1- and NR2-assigned cells using cisTopic and UMAP. (B) Cluster assignments identified using PhenoGraph. CA1 and CA3 neuronal populations exhibited strong signal at cell-type–specific marker genes. Asterisk indicates putative assignment based on modest enrichment at marker genes. Right panels show the NEUROD1 motif enrichment in the original t-SNE coordinates (top) that correspond to the region of cells assigned to CA3 cluster (bottom). (C) Biclustering of cisTopic topics and weights for each cell. Highlighted topics exhibit high cluster specificity. (D) Further subclustering of the Other/MC cell population produced three distinct groups, including putative mossy cells (MCs). (E) Biclustering of cisTopic topics and weights for each cell. Highlighted topics exhibit high cluster specificity. (F) Topic 5, specific to one of the subclusters, is highly enriched for AP-1–related motifs, suggesting the cells may be in a state of heightened activity.
In addition to improved sensitivity, cisTopic produces sets of peaks that are associated with one another as topics (Fig. 3C; Supplemental Fig. 15), several of which exhibited high cluster specificity. This included CA3-specific topic 13, which was enriched for NEUROD1. These cells were within the same region of the NR2 cluster that also exhibited increased NERUROD1 accessibility (Fig. 3B, right; Fig. 1I). Motif enrichment files for all topics can be found in Supplemental Data 1. We additionally performed a differential accessibility analysis between the clusters (Supplemental Fig. 16). Although none of the significant peaks were proximal to definitive marker genes, these sites may be useful to inform future functional studies.
We suspected that the fourth cluster (teal; Other/MC) might contain additional cellular subtypes based on the hierarchical clustering of topics. We therefore performed the same subclustering analysis as we did for the NR1 and NR2 groups specifically for these cells (optimum topic number = 13) (Supplemental Figs. 17, 18; Supplemental Data 2), which revealed three distinct clusters (Fig. 3D). When we assessed the topics closely associated with one of the clusters (Fig. 3E,F), we observed a very high enrichment for AP-1–associated proteins, suggesting that they may be neurons in a heightened activity state (Fig. 3F), although we did not observe enrichment of accessibility for any cell-type–specific marker genes or DNA-binding motifs, as was the case for a second cluster. We did observe increased chromatin accessibility proximal to several MC marker genes (Cembrowski et al. 2016), which was most pronounced at Pmp22 and Thbs2 (Supplemental Fig. 19).
Cis-regulatory networks in the hippocampus
Many enhancer elements reside far from the transcription start sites of the genes they regulate, making enhancer–gene associations challenging. To accomplish this, we leveraged the recently described Cicero algorithm (Pliner et al. 2018), which uses an unsupervised machine-learning framework to link distal regulatory elements to their prospective genes via patterns of coaccessibility in the single-cell regulatory landscape. We applied Cicero to our hippocampus sci-ATAC-seq data set to produce 487,156 links between ATAC-seq peaks at a coaccessibility score cutoff of 0.1 (Supplemental Data, InVivo.cicero_links.txt). Of these, 47,498 (10.5%) were links between two promoters, 146,818 (32.4%) linked a distal regulatory element to a promoter, and 259,236 (57.2%) were between two distal elements. We next compared our Cicero-linked peaks with existing chromatin conformation data that had been produced on mouse cortical tissue (Dixon et al. 2012), as no hippocampus data sets are currently available; however, a majority of topological associated domains (TADs) are conserved across cell types (Dixon et al. 2012). Consistent with expectations, we observed a 1.1- to 1.5-fold enrichment (P < 1 × 10−4 across all Cicero link thresholds out to 500 kbp) (Methods; Fig. 4A) for linked peaks that occur within the same TAD over equidistant peaks present in different TADs, suggesting that the identified links are associated with higher-order chromatin structure. We then identified cis-coaccessibility networks (CCANs) using Cicero, which uses a Louvain-based clustering algorithm, which can inform us about coregulated chromatin hubs in the genome. By using a coaccessibility score threshold of 0.15 (based on high intra-TAD enrichment) (Fig. 4A), we identified 3243 CCANs, which incorporated 102,736 sites (mean 31.7 peaks/CCAN).
Figure 4.
Cis-coaccessibility analysis using Cicero. (A) Cicero links at several coaccessibility score thresholds are heavily enriched for links that contain peaks present in the same topological associated domain (TAD) as determined by Hi-C methods (Dixon et al. 2012). The enrichment decreases at greater distances (x-axis). (B) t-SNE projection of cis-coaccessibility networks (CCANs) colored by the cell type with the greatest accessibility for the CCAN. Each point represents an individual CCAN. Networks generally group by cell type. CCAN 174, which includes the Prox1 gene shown below in D, is indicated with an arrow. (C) Accessibility Z-scores for CCANs for granule cells and microglia. (D) CCAN ID 174 including the Prox1 promoter (dentate granule marker gene). (E) CCAN 174 has the greatest accessibility signal in cells identified as dentate granule cells.
To identify the enrichment of cell-type specificity of CCANs, we aggregated ATAC-seq signal within each CCAN for each cell type and performed a Z-score normalization (Supplemental Fig. 20). We then projected the CCANs in 2D space using t-SNE and visualized them based on their enrichment to their highest matching cell type (Fig. 4B,C; Supplemental Fig. 21). This revealed distinct sets of coaccessibility networks for each cell type, with common networks falling toward the center of the projection space. CCANs with greater numbers of peaks tended to be less cell-type–specific, likely owing to the large number of genes that are encompassed by the CCAN, the majority of which are not cell-type–specific (Supplemental Fig. 22). This observation is also consistent with chromatin conformation literature (Supplemental Fig. 23; Dixon et al. 2012). We probed our cell-type–specific CCANs further by assessing networks that incorporated marker gene promoters. Prox1 (dentate granule marker) was present in a CCAN that included 89 total accessibility sites and was associated with the correct cell type (Fig. 4D,E). Although much of the CCAN did not exhibit cell-type specificity, the region centered on Prox1 (with the highest coaccessibility values) drove the assignment. To dissect out the major components of the larger CCAN, we used Cicero specifically on the dentate granule cells (Supplemental Fig. 24A). This revealed three distinct CCANs within the region, with the Prox1-containing CCAN exhibiting the greatest specificity to the dentate granule cells (Supplemental Fig. 24B). This suggests the possibility of larger chromatin networks with subsets of regulatory elements and genes joining or leaving the network based on cell type. Finally, we identified a number of CCANs that were overlapping that included mutually exclusive sets of peaks, suggesting two alternative folding patterns of chromatin within the regions dependent upon the cell type (Supplemental Fig. 25).
In vitro neurons exhibit an altered epigenetic profile
To examine how well in vitro cultured hippocampal neuronal populations match their in vivo counterparts at the epigenetic level, we isolated hippocampal neurons from P0 pups and allowed them to mature for 16–18 d in vitro (DIV). At this stage, the neurons had extended long processes and expressed markers of mature neurons such as MAP2. We performed sci-ATAC-seq as described above and produced 899 high-quality single-cell chromatin accessibility profiles passing our quality thresholds (Methods). Our mean unique read count per cell was again high, compared with currently published work, at 43,532. We then performed peak calling on the ensemble of in vitro sci-ATAC-seq profiles, resulting in 111,005 total peaks. Similar to our in vivo preparations, the ATAC-seq signal correlated well between the two replicates (Pearson R > 0.99). Subsequent filtering, LSI-t-SNE, and clustering, as described for the in vivo preparation, revealed four distinct populations (Fig. 5A). Upon examination via marker gene and DNA-binding motif accessibility enrichment, we determined one of the clusters to be the INT population (40.6% of cells), with the remainder being excitatory (59.4%).
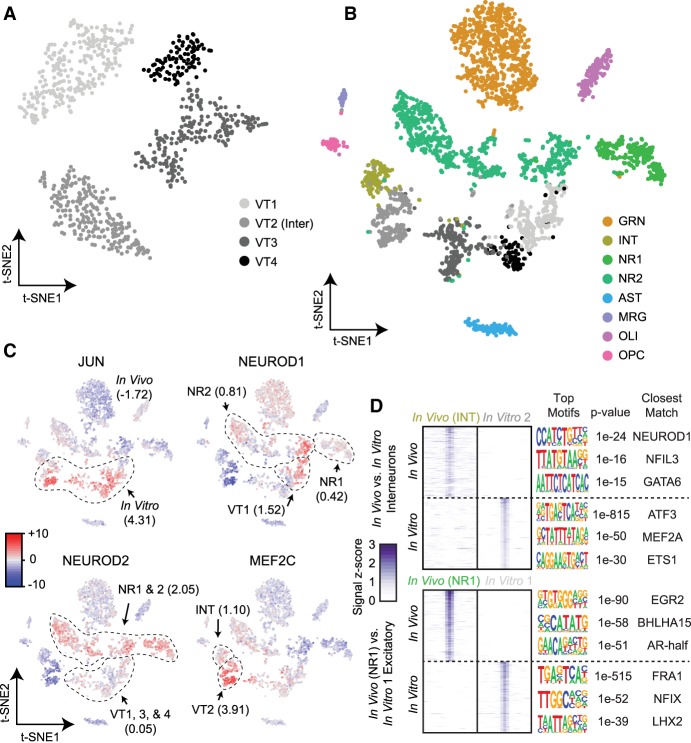
Figure 5.
Comparison of the accessible chromatin landscape of in vitro-cultured neurons with in vivo-obtained profiles. (A) LSI-t-SNE projections of in vitro-obtained cells reveals four clusters, one of which exhibits interneuron patterns (VT2) and the remaining excitatory neurons (VT1, VT3-4). (B) LSI-t-SNE projection of the combined in vivo and in vitro data sets colored by independently called clusters. Excitatory neurons in the two conditions generally cluster separately, with interneurons more closely associated. (C) chromVAR global motif deviation Z-scores for select motifs for each cell. Dashed lines and values correspond to mean values of cell populations. (D) Differential accessibility analysis between in vivo and in vitro interneurons (top; INT vs. VT2, respectively) and between two closest excitatory neuron populations between in vivo and in vitro conditions (NR1 and VT1, respectively). ATAC-seq signal is shown for the top differentially accessible loci with the top three motifs and corresponding P-values and matching motifs to the right.
We performed peak calling on the combined reads from both the in vivo and in vitro experiments and merged these peaks with those called on each set individually to produce a combined peak call set composed of 174,503 sites. It is important to note that much of the increase over the in vivo peak set was because of increased coverage at sites that may not have met the calling threshold as opposed to peaks exclusive to the in vitro cultured neurons. We then performed LSI and t-SNE on the resulting count matrix using cells produced in both experiments. Although the in vitro cultured glutamatergic neurons largely formed their own grouping independent of their in vivo counterparts, the inhibitory neurons from the in vitro preparation grouped more closely with the in vivo population (Fig. 5B).
We next examined the global DNA-binding motif accessibility of the combined population (Fig. 5C). The starkest differences between the in vivo and in vitro cell populations was in motifs associated with the AP-1 complex, that is, the FOS, JUN, ATF, and JDP families (µz = 4.32 and −1.72 for in vitro and in vivo, respectively). The AP-1 complex plays a major role in stimulus response, including cell stress (Hess 2004), which is expected for neurons grown and matured ex vivo. It has also been shown that AP-1 modulates chromatin during neuronal activation (Su et al. 2017), suggesting the possibility of an elevated activity state in neuronal cultures compared with their in vivo counterparts; however, the decoupling of the many functional roles of the AP-1 complex from one another using global accessibility is not currently possible. We also examined the motifs for several other transcription factors that are relevant to neuronal development. NEUROD1, discussed above, responsible for early differentiation (E14.5 ventricular proliferative zone) (Pataskar et al. 2016) and survival of neurons, exhibited shared accessibility enrichment in a subset of cells from both the in vivo and in vitro neuronal populations. MEF2C delineates early precursors of a subset of inhibitory INTs (Mayer et al. 2018), and we observed shared, elevated MEF2C accessibility in the INT populations, with greater accessibility in the in vitro cells (µz = 3.91) over that of the in vivo INTs (µz = 1.10). In contrast to NEUROD1 and MEF2C, NEUROD2 acts later in hippocampal development than NEUROD1 (Pleasure et al. 2000), is expressed in migrating granule neurons, and binds to a number of neuron-specific promoters. The DNA-binding motif for NEUROD2 was globally more accessible in the in vivo neurons compared with their in vitro counterpart (µz = 2.05 and µz = 0.05 for in vivo and in vitro, respectively). This finding may reflect its later developmental appearance and that the main targets of NEUROD2 are involved in layer-specific differentiation and axonal pathfinding, which are not likely to be occurring in vitro.
Differential accessibility analysis comparing in vitro and in vivo counterparts shed further light on the epigenetic differences between the two populations (Fig. 5D). A comparison of the INT populations produced 4356 and 7575 peaks significantly differentially accessible in the in vivo (INT) and in vitro (VT2) populations, respectively (Q-value ≤0.01, log2 fold-change ≥1). A motif enrichment analysis of these peak sets revealed the most significantly enriched motifs corresponded to NEUROD1 in the in vivo peaks (P = 1 × 10−24), a motif that showed low global accessibility in both INT populations (Fig. 4C). INT peaks specific to the in vitro population were significantly enriched for ATF3 (P = 1 × 10−815), which exhibits elevated accessibility of AP-1 in the in vitro cell populations and has a shared role in cell stress and interaction with the AP-1 complex (Hai and Curran 1991). We also examined differential accessibility between the most closely grouped excitatory neuronal populations, which produced 1761 and 2964 for NR1 (in vivo) and VT1 (in vitro), respectively (Q-value ≤0.01, log2 fold-change ≥1). The most significantly enriched motif in the in vivo peak set was EGR2 (P = 1 × 10−90), again a transcription factor expressed highly in migrating neural crest cells (Wilkinson et al. 1989) that may be absent in an in vitro setting where cell migration is not pertinent.
Discussion
A better understanding of the role of specific cell populations in hippocampal function is a necessary step in order to study disease processes that involve this region critical to memory and learning. Thus far, studies have used gene expression data from sorted populations (Cembrowski et al. 2016) and single cells (Zeisel et al. 2015; Habib et al. 2017) to identify subpopulations of cells and novel marker genes for the cells within the hippocampus. Here, we provide the most in-depth epigenetic analysis of the hippocampus at single-cell resolution to date. Our sci-ATAC-seq protocol (Methods) has been optimized for primary cell culture and both fresh or frozen tissue and produces unique read counts per cell in the tens of thousands, a full order-of-magnitude improvement over the initial sci-ATAC-seq publication (Cusanovich et al. 2015). The data sets released with this study can be readily analyzed using scitools (https://github.com/adeylab). This tool suite is designed to be complementary to other single-cell ATAC-seq analysis packages, such as chromVAR, cisTopic, and Cicero, and serves as an easy framework for integrating analyses and generating plots to assess data quality and facilitate biological interpretation.
We used our sci-ATAC-seq maps to identify the major cell types of the hippocampus, with sufficient depth and library complexity to profile less abundant cell types, such as MRG and OPCs. By using the recently described cisTopic analysis tool, we were able to achieve a high degree of granularity within the pyramidal neuron population, enabling the definitive identification of CA1 and CA3 neurons; a population of putative CA2 neurons; and three lower-abundance populations, likely containing MCs and two unidentified neuronal subtypes. Our analysis of global motif accessibility revealed the expected enrichment of motifs associated with specific cell populations in addition to uncovering unanticipated findings, such as increased accessibility at CTCF motifs in INT and glial populations, a finding that was also observed in our differential accessibility analysis. We used our data set to map CCANs, enabling the association of distal elements with promoters or other regulatory loci. Finally, we directly compared the accessibility profiles of neurons that were matured in vitro with their in vivo counterparts to identify altered pathways or chromatin state configurations that should be considered for future experimental design. This revealed a stark difference in the global accessibility for motifs associated with the AP-1 complex, which is involved in cell stress as well as neuronal activity. Future work to identify the cause and effect of elevated AP-1 complex activity is warranted to understand its impact on studies that use hippocampal neurons matured in vitro.
We believe that the chromatin accessibility maps we provide in this work, including the profiling of in vitro cultured neurons, and the software tools we are releasing are a valuable resource for any groups studying the hippocampus or those that wish to analyze single-cell chromatin accessibility data. Our maps complement existing single-cell transcriptional data and take the field one step closer to a comprehensive atlas of the mammalian hippocampus; however, we acknowledge that future innovation built off of the data sets we and others have produced will be required to achieve that goal.
Methods
Isolation of hippocampus tissue
All animal studies were approved by the Oregon Health and Science University Institutional Animal Care and Use Committee. Sixty-day-old C57BL/6J mice were deeply anesthetized using isoflurane. After decapitation, the brain was removed, and the total hippocampus was isolated and placed in ice-cold phosphate-buffered saline (pH 7.4).
In vitro culturing of hippocampal neurons
Pups (P0) were killed by decapitation, and the brains dissected in ice-cold Hanks basal salt solution (HBSS; pH 7.4) with 25 mM HEPES buffer. Individual hippocampi were excised without the meninges and pooled by individual animal. The tissue was treated with 2% papain and 80 ng/mL DNase I in HBSS for 10 min at 37°C. Tissue pieces were rinsed three times with hibernate A containing 2 mM GlutaMAX and 1 × B27 supplement. Neurons were dissociated carefully and filtered with a 0.4-μm mesh. Neurons were plated at a density of 1 × 106 cells per well of a six-well dish coated with 50 μg/mL Poly-l-lysine hydrobromide in boric acid buffer (50 mM boric acid, 12.5 mM sodium borate, decahydrate). The neurons were plated in neurobasal A containing 1 × B27 supplement and 2 mM GlutaMAX. After 2 h, the media was changed to remove cell debris. Media half changes occurred every 3 d with fresh neurobasal A containing 1 × B27 and 2 mM GlutaMAX. Cells were maintained at 37°C with 5% CO2 in a humidified incubator.
Sci-ATAC-seq assay and sequencing
Tissue was diced on ice using a sterile razor blade in freshly prepared nuclei isolation buffer (NIB; 500 µL 10 mM Tris-HCl at pH 0.5, 100 µL 10 mM NaCl, 150 µL MgCl2, 500 µL 0.1% Igepal, 0.1% Tween 20, 1 unit Qiagen protease inhibitor, nuclease-free water to 50 mL) followed by dounce homogenization. For cultured cells, nuclei were directly isolated by removing media and washing once with ice cold PBS, and then NIB was added to cover the dish followed by incubation on ice for 5 min, scraping using a tissue scraper, and then an additional 5-min incubation on ice. For both tissue and cultured cells, nuclei were then pelleted and resuspended in 1 mL NIB with DAPI added to a final concentration of 5 mg/mL. Nuclei were then strained in a 35-µm strainer and sorted on a Sony SH800 flow sorter and deposited into 0.2 mL PCR plates containing 5 µL of 2 × TD buffer and 5 µL of NIB, with 2500 nuclei deposited per well. Plates were placed on ice until transposition. Tagmentation was performed by the addition of 1 µL of 2.5 µM barcoded transposome (EZ-Tn5 variant) (Amini et al. 2014) and incubated for 15 min at 55°C followed by placing the plate on ice to stop the reaction. All wells were then pooled using wide-bore pipette tips, and DAPI was added to a final concentration of 5 mg/mL. Tagmented nuclei were then strained and sorted again, and 22 were deposited into each new PCR well containing 0.25 µL 20 mg/mL BSA, 0.5 µL 1% SDS, 7.75 µL nuclease-free water, 2.5 µL barcoded forward primer, and 2.5 µL reverse primer. Plates were kept on ice until all sorting was completed. After sorting, plates were incubated for 15 min at 55°C to denature the transposase followed by placing the plate on ice and adding 12 µL of PCR mix (7.5 µL NPM, 4 µL nuclease-free water, 0.5 µL 100 × SYBR Green) and then PCR amplified using the following conditions: for 5 h at 72°C, for 0:30 at 98°C, and cycles of [10 min at 98°C, 30 min at 63°C, 1 h at 72°C; plate read, 10 min at 72°C] on a Bio-Rad CFX real-time thermocycler. Reactions were pulled when mid-exponential, typically 17–22 cycles. Post-amplification, 5 µL of each reaction was pooled and cleaned up using a QIAquick PCR purification column. Libraries were quantified using a Qubit fluorimeter, diluted to ∼4 ng/µL, and assessed on an Agilent Bioanalyzer HS chip. Sequencing was performed as previously described on a NextSeq 500 (research use only) using custom primers and chemistry (Vitak et al. 2017). A detailed sci-ATAC-seq protocol is provided as a Supplemental Protocol.
For fresh replicates, nuclei were divided into two transposase plates that were processed separately. Each transposase plate was then pooled and the nuclei sorted into a full PCR plate for each preparation. The frozen hippocampi were processed using one half of a transposase plate for each biological replicate, and then all wells were pooled and sorted into a single PCR plate. We also had two biological replicates for the in vitro preparations that were processed according to the same workflow as the two frozen samples.
The scitools suite
All initial analysis was performed with scitools, a custom software package we developed to help analyze sci-ATAC-seq data and other combinatorial indexing data (sci-). The toolset is a collection of commands to perform common functions for sci- data sets, including wrappers that use existing tools, including BWA (Li and Durbin 2009), MACS2 (Zhang et al. 2008), BEDTools (Quinlan and Hall 2010), SAMtools, as well as R (R Core Team 2019) libraries: ggplot2 (Wickham 2016), chromVAR (Schep et al. 2017), chromVARmotifs, Cicero (Pliner et al. 2018), RtSNE, and DBSCAN (Ester et al. 1996). Usage of scitools for any of these functions should cite the relevant utilities. Scitools can be found at https://github.com/adeylab/scitools (an evolving tool) or as Supplemental Code for the version used at the time of this manuscript.
Sci-ATAC-seq data processing
BCL files were first converted to FASTQ files using bcl2fastq (2.19.0). We then demultiplexed our reads using scitools (fastq-dump, fastq-split) based on the two separate Tn5 tagmentation events on the P5 and P7 ends of the molecules and the following added unique PCR indexes on both sides. In order for a barcode to be considered a match, each of these four indexes constituting a barcode had to be within two Hamming edit distances away from their expected counterpart. We aligned to the mm10 genome using the scitools fastq-align function within scitools, which mapped reads using BWA-MEM. Aligned reads were filtered based on a quality score cutoff of 10 and PCR duplicates removed in a barcode-aware manner using scitools bam-rmdup. We determined whether a barcode represented a cell as opposed to it representing noise by using the mixed model approach previously presented (Vitak et al. 2017). Peaks were then called using scitools callpeak, which uses MACS2 to identify peaks, and then extended to 500 bp followed by peak merging and filtering of peaks that extend beyond chromosome boundaries.
LSI and 2D embedding
Count matrices were generated using scitools counts to produce a matrix of read counts at cells (columns) by called peaks (rows). This matrix was then filtered using scitools filter-matrix to exclude rows with fewer than 10 cells having reads (-R 10), and columns (cells) with fewer than 1000 rows with reads (-C 1000). The matrix was then carried through term-frequency inverse-document-frequency transformation using scitools tfidf followed by LSI, retaining SVD dimensions 1 through 15 using scitools lsi. The resulting LSI matrix was used in scitools t-SNE, which makes use of the RtSNE R package. All t-SNE plots were generated using scitools plot-dims using an annotation file to encode cluster ID, sample ID, or other variables, including chromVAR motif deviation Z-scores.
Coembedding of single-cell RNA-seq cells with sci-ATAC-seq cells
We used Cicero (Pliner et al. 2018) to produce gene activity scores based on the chromatin accessibility signal at the promoter and linked distal elements to each gene. These scores were loaded into Seurat3 (Stuart et al. 2018) along with the gene read count matrices from Zeisel et al. (2015) (Smart-seq), and Habib et al. (2017) (DroNc-seq). We then performed anchor identification and integration of the three data sets as decribed by Stuart et al. (2018). We then performed PCA and t-SNE on the integrated data. Clusters were identified using PhenoGraph (Levine et al. 2015) on the PCA dimensions.
Identifying transcription factor–associated changes
We applied the chromVAR (Schep et al. 2017) R package to our data to infer changes in global motif accessibility across our cell populations. This provides information on the putative binding of transcription factors and, consequently, the possible ongoing biological processes in cell populations. The mouse_pwms_v1 motif set from the chromVARmotifs R package was used in this analysis. The bias-corrected motif deviation scores were plotted on the t-SNE embedded 2D coordinates with the scitools plot-dims -M option for visualization.
Cell-type–dependent differential accessibility
To accurately identify differentially accessible peaks, we used the make_glasso_cds function from the Cicero (ver = 0.0.0.9000) package to create clusters of k = 50 cells based on their low-dimensional t-SNE coordinates. We then selected clusters with 99% cell type purity and aggregated accessibility profiles. We posited that the aggregate profiles would provide the replicates required for the DESeq2 R package (Love et al. 2014), which in turn internally corrects for technical biases such as assay efficiency. With this method, we tested (using the inherent nBinomWaldTest) for differentially accessible sites between cell types against all other cell types combined. We corrected for multiple testing at q = 0.01 and further filtered differentially accessible sites by removing peaks accessible at q = 0.2 in any of the other cell types. We also note that scitools aggregate-cells is also capable of aggregating cells in reduced dimensional space for purposes of differential accessibility analysis. We then applied HOMER (Heinz et al. 2010) (http://homer.ucsd.edu/homer/motif/) to identify potential de novo and known regulators of chromatin accessibility within the cell-type–dependent differentially accessible sites. We used all accessible peaks as background and the mm10 findMotifsGenome command.
Subclustering of pyramidal neurons
We applied cisTopic ver = 0.2.0 (Bravo González-Blas et al. 2018) to separate out subpopulations within the in vivo neuronal cell populations we found (NR1, NR2). We chose the optimal number of topics (30) (Supplemental Fig. 12) by running several models ranging from five to 50 topics and picking the model with the highest log-likelihood in the last iteration. We used the 250 burn-in iterations and 300 recording iterations for this analysis. We determined topic associated regions via topic binarization with GammaFit (included in cisTopic) on the region–topics distributions matrix (thrP = 0.975). We then projected the neuronal cells into 2D space via UMAP (Becht et al. 2019) on the topics–cell distributions matrix and observed four distinct cell groupings. We identified these clusters with the Rphenograph ver = 0.99.1 (Levine et al. 2015) package on the topics–cell distributions matrix (d = 4 clusters of k = 150). The same processing and parameters were used to perform subclustering on the cluster exhibiting high heterogeneity.
To correctly characterize these four clusters, we called potential de novo and known regulators of chromatin accessibility with HOMER (run with the mm10 genome and all sites as background using the findMotifsGenome command) on the top associated regions of topics that were enriched in individual clusters (identified via the topics–cell distributions matrix). In addition, we called differentially accessible sites unique to each of the clusters using DESeq2 (as in the preceding Methods subsection, “Cell-type–dependent differential accessibility”) and again applied HOMER for motif enrichment for these sites.
Identifying cis-regulatory networks in the hippocampus
We used the recently described Cicero package (Pliner et al. 2018) to identify CCANs according to the recommended workflow. For CCAN identification, we used a P = 0.15 threshold cutoff, which identified 2066 chromatin networks that incorporated 47,805 sites of our in vivo cell populations. Fold enrichment for links within annotated TADs (Dixon et al. 2012) was performed by calculating the proportion of distance-matched (±25 kbp of specified 50-kbp distance interval) intra-TAD links over inter-TAD links at a range of coaccessibility score cutoffs (0.05 to 0.25 at 0.05 intervals). Ten thousand permutations were then performed for each distance bin by randomly assigning two distance-matched peaks as linked and retaining the same total number of links for each coaccessibility cutoff and then calculating the fold intra-TAD enrichment as described above.
Cell-type–specific cis-regulatory networks
To assign CCANs to cell types, we first calculated the fraction of cells of each cell type that have signal at a peak and assumed that the distribution of reads per cell across cell types is close to uniform. We then Z-scored the resulting matrix across the CCANs and then visualized the separation of CCANs by cell type by biclustering and plotting the heatmap using the complexHeatmap (ver = 1.17.1) R package. We also visualized CCAN cell-type specificity by using t-SNE on the Z-scored group read fractions to embed CCANs in 2D. We assigned the cell type to each of the CCANs based on the highest Z-scored value. We next identified CCANs that contain at least one of the genes (Prox1, Dsp, Ociad2, Dkk3, Glul, Gfap, Mog, Cldn11, C1qa, Wfs1, Mobp, Pdgfra) shown to be differentially accessible in our data. We intersected ±80-kbp regions before and after transcription start sites of these genes with the CCANs using BEDTools intersect. We plotted the CCANs around genes in which the cell type assigned to the CCANs matched the cell-type specificity of the gene using the Cicero plot_connections function. We used chromVAR to further validate the relative enrichment of CCANs by using CCAN peaks as motif input files. We used scitools plot dims -M option to visualize the deviation scores for the CCANs on the t-SNE coordinates. We have to note that in order for this method to work, peaks within the CCANs had to be accessible across multiple cell types, so we decided to use only CCANs with 10 or more peaks for this analysis. We finally included a more in-depth analysis of CCAN 174 centered around Prox1. We called CCANs just within granule cells and identified three different sub CCANs, with the core of the original CCAN 174 showing even higher specify in the chromVAR deviation scores plots (Supplemental Fig. 24).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE118987 as reads as well as data tables and metadata in the form of data.gz files, which can be split into their components via “scitools split-data.” See Supplemental Note 1 and Supplemental Figure 26 for additional details on data sets provided as well as the scitools documentation, which can be found at https://github.com/adeylab/scitools (an evolving tool) or as Supplemental Code for the version used at the time of this manuscript.
Competing interest statement
F.J.S. is an employee and owner of stock of Illumina. One or more embodiments of one or more patents and patent applications filed by Illumina may encompass the methods, reagents, and the data disclosed in this manuscript.
Supplementary Material
Acknowledgments
We thank Ryan Mulqueen and Andy Fields for helpful suggestions and contributions along with other members of the Adey Laboratory. This work was supported by NIH (NIGMS R35GM124704 and NIDA R01DA047237 to A.C.A., NS099374 to G.M., U54DK107979 and DP2HD088158 to C.T.), an NSF GRFP to H.A.P. (DGE-1256082), and support from the Rett Syndrome Research Trust to G.M.
Author contributions: A.C.A., J.R.S., and G.M. designed all experiments. A.C.A., J.R.S., K.A.T., and M.W.L. wrote the manuscript. S.A.V. performed all sci-ATAC-seq preparations. A.C.A. and K.A.T. performed computational analysis and wrote software associated with this work. J.R.S. prepared all tissue samples and cultures and performed analyses and interpretation of the data. M.W.L. aided in analysis of the data and interpretation of findings. R.M.M. contributed to sci-ATAC-seq and scitools protocol development and data processing pipelines. H.A.P. and C.T. provided early access to computational tools and aided in the coaccessibility analysis and interpretation. F.J.S. provided reagents and contributed to sci-ATAC-seq method development and implementation. All authors reviewed and approved the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.243725.118.
References
- Abusaad I, MacKay D, Zhao J, Stanford P, Collier DA, Everall IP. 1999. Stereological estimation of the total number of neurons in the murine hippocampus using the optical disector. J Comp Neurol 408: 560–566. [DOI] [PubMed] [Google Scholar]
- Amini S, Pushkarev D, Christiansen L, Kostem E, Royce T, Turk C, Pignatelli N, Adey A, Kitzman JO, Vijayan K, et al. 2014. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat Genet 46: 1343–1349. 10.1038/ng.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37: 38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bravo González-Blas C, Minnoye L, Papasokrati D, Aibar S, Hulselmans G, Christiaens V, Davie K, Wouters J, Aerts S. 2018. Cis-topic modelling of single cell epigenomes. bioRxiv 10.1101/370346 [DOI] [PMC free article] [PubMed]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523: 486–490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, Majeti R, Chang HY, Greenleaf WJ. 2018. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell 173: 1535–1548.e16. 10.1016/j.cell.2018.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt E. 1990. Expression of C-fos-like protein as a marker for neuronal activity following noxious stimulation in the rat. J Comp Neurol 296: 517–530. 10.1002/cne.902960402 [DOI] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357: 661–667. 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembrowski MS, Wang L, Sugino K, Shields BC, Spruston N. 2016. Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5: e14997 10.7554/eLife.14997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah P-S, Thomas PQ. 2015. SOX3 expression in the glial system of the developing and adult mouse cerebellum. SpringerPlus 4: 400 10.1186/s40064-015-1194-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus Stolt C, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. 2002. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16: 165–170. 10.1101/gad.215802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48: 1193–1203. 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J. 2015. Epigenetics. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348: 910–914. 10.1126/science.aab1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. 2018a. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174: 1309–1324.e18. 10.1016/j.cell.2018.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Reddington JP, Garfield DA, Daza RM, Aghamirzaie D, Marco-Ferreres R, Pliner HA, Christiansen L, Qiu X, Steemers FJ, et al. 2018b. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555: 538 10.1038/nature25981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert A, Vogt D, Watson A, Levy M, Jiang Y, Brûlé E, Rowland ME, Rubenstein J, Bérubé NG. 2019. CTCF governs the identity and migration of MGE-derived cortical interneurons. J Neurosci 39: 177–192. 10.1523/JNEUROSCI.3496-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M, Kriegel HP, Sander J, Xu X. 1996. A density-based algorithm for discovering clusters in large spatial databases with noise. In KDD'96 Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, pp. 226–231. Portland, OR AAAI Press, Menlo Park, CA. [Google Scholar]
- Fages C, Khelil M, Rolland B, Bridoux A, Tardy M. 1988. Glutamine synthetase: a marker of an astroglial subpopulation in primary cultures of defined brain areas. Dev Neurosci 10: 47–56. 10.1159/000111955 [DOI] [PubMed] [Google Scholar]
- Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, Kellis M. 2015. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature 518: 365–369. 10.1038/nature14252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Zeller N, Raasch J, Kierdorf K, Frenzel K, Ketscher L, Basters A, Staszewski O, Brendecke SM, Spiess A, et al. 2015. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J 34: 1612–1629. 10.15252/embj.201490791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guner G, Guzelsoy G, Isleyen FS, Sahin GS, Akkaya C, Bayam E, Kotan EI, Kabakcioglu A, Ince-Dunn G. 2017. NEUROD2 regulates Stim1 expression and store-operated calcium entry in cortical neurons. eNeuro 4: ENEURO.0255-16.2017 10.1523/ENEURO.0255-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. 2017. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14: 955–958. 10.1038/nmeth.4407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Curran T. 1991. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci 88: 3720–3724. 10.1073/pnas.88.9.3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J. 2004. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117: 5965–5973. 10.1242/jcs.01589 [DOI] [PubMed] [Google Scholar]
- Joo JY, Schaukowitch K, Farbiak L, Kilaru G, Kim TK. 2015. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat Neurosci 19: 75–83. 10.1038/nn.4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel E, Spencer W. 1961. Electrophysiology of hippocampal neurons. II: after-potentials and repetitive firing. J Neurophysiol 24: 243–259. 10.1152/jn.1961.24.3.243 [DOI] [PubMed] [Google Scholar]
- Kandel E, Spencer W, Brinley FJ. 1961. Electrophysiology of hippocampal neurons. I: sequential invasion and synaptic organization. J Neurophysiol 24: 225–242. 10.1152/jn.1961.24.3.225 [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yu N-K, Shim K-W, Kim J-I, Kim H, Han DH, Choi JE, Lee S-W, Choi D, Kim MW, et al. 2018. Remote memory and cortical synaptic plasticity require neuronal CCCTC-binding factor (CTCF). J Neurosci 38: 5042–5052. 10.1523/JNEUROSCI.2738-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto H, Eto R, Abe M, Kato H, Araki T. 2009. Alterations of glial cells in the mouse hippocampus during postnatal development. Cell Mol Neurobiol 29: 1181–1189. 10.1007/s10571-009-9412-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake B, Cheng S, Sos B, Fan J, Yung Y, Kaeser G, Duong T, Yung YC, Gao D, Chun J, et al. 2018. Integrative single-cell analysis by transcriptional and epigenetic states in human adult brain. Nat Biotechnol 36: 70–80. 10.1038/nbt.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S, Kawamoto S, Tanuma SI, Uchiumi F. 2015. The hematopoietic regulator, ELF-1, enhances the transcriptional response to interferon-β of the OAS1 anti-viral gene. Sci Rep 5: 17497 10.1038/srep17497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Zhao X, Gage FH. 2004. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci 24: 3879–3889. 10.1523/JNEUROSCI.4710-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir ED, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. 2015. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162: 184–197. 10.1016/j.cell.2015.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente de No R. 1934. Studies on the structure of the cerebral cortex II: continuation of the study of the ammonic system. J Psychol Neurol 46: 113–177. [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hernandez A, Bell KP, Norenberg MD. 1977. Glutamine synthetase: glial localization in brain. Science 195: 1356–1358. 10.1126/science.14400 [DOI] [PubMed] [Google Scholar]
- Mayer C, Hafemeister C, Bandler RC, Machold R, Batista Brito R, Jaglin X, Allaway K, Butler A, Fishell G, Satija R. 2018. Developmental diversification of cortical inhibitory interneurons. Nature 555: 457–462. 10.1038/nature25999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. 2015. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86: 1369–1384. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulqueen RM, Pokholok D, Norberg SJ, Torkenczy KA, Fields AJ, Sun D, Sinnamon JR, Shendure J, Trapnell C, O'Roak BJ, et al. 2018. Highly scalable generation of DNA methylation profiles in single cells. Nat Biotechnol 36: 428–431. 10.1038/nbt.4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe J, Dostrovsky J. 1971. The hippocampus as a spatial map: preliminary evidence from unit activity in the freely-moving rat. Brain Res 34: 171–175. 10.1016/0006-8993(71)90358-1 [DOI] [PubMed] [Google Scholar]
- Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, Tiwari VK. 2016. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J 35: 24–45. 10.15252/embj.201591206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Collins AE, Lowenstein DH. 2000. Unique expression patterns of cell fate molecules delineate sequential stages of dentate gyrus development. J Neurosci 20: 6095–6105. 10.1523/JNEUROSCI.20-16-06095.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. 2018. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol Cell 71: 858–871.e8. 10.1016/j.molcel.2018.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. 2018. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci 21: 432–439. 10.1038/s41593-018-0079-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, Shendure J. 2017. Massively multiplex single-cell Hi-C. Nat Methods 14: 263–266. 10.1038/nmeth.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon Y, Cajal S. 1911. Histologie du Système Nerveux de L'Homme et des Vertébrés, Vol. II Maloine, Paris. [Google Scholar]
- Roybon L, Hjalt T, Stott S, Guillemot F, Li JY, Brundin P. 2009. Neurogenin2 directs granule neuroblast production and amplification while neuroD1 specifies neuronal fate during hippocampal neurogenesis. PLoS One 4: e4779 10.1371/journal.pone.0004779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams DS, Nardone S, Getselter D, Raz D, Tal M, Rayi PR, Kaphzan H, Hakim O, Elliott E. 2016. Neuronal CTCF is necessary for basal and experience-dependent gene regulation, memory formation, and genomic structure of BDNF and Arc. Cell Rep 17: 2418–2430. 10.1016/j.celrep.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. 2018. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174: 1015–1030.e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. 2017. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14: 975–978. 10.1038/nmeth.4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville W, Milner B. 1957. Loss of recent memory after bilateral hippocampal lesions. J Neurol Neurosurg Psychiatry 20: 11–21. 10.1136/jnnp.20.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Milner B. 1981. The role of the right hippocampus in the recall of spatial location. Neuropsychologia 19: 781–793. 10.1016/0028-3932(81)90090-7 [DOI] [PubMed] [Google Scholar]
- Spencer WA, Kandel ER. 1961a. Electrophysiology of hippocampal neurons, III: firing level and time constant. J Neurophysiol 24: 260–271. 10.1152/jn.1961.24.3.260 [DOI] [PubMed] [Google Scholar]
- Spencer WA, Kandel ER. 1961b. Electrophysiology of hippocampal neurons, IV: fast prepotentials. J Neurophysiol 24: 272–285. 10.1152/jn.1961.24.3.272 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Stoeckius M, Smibert P, Satija R. 2018. Comprehensive integration of single cell data. bioRxiv 10.1101/460147 [DOI] [PMC free article] [PubMed]
- Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, Kim D, Ming GL, Song H. 2017. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci 20: 476–483. 10.1038/nn.4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svichar N, Esquenazi S, Chen H-Y, Chesler M. 2011. Preemptive regulation of intracellular pH in hippocampal neurons by a dual mechanism of depolarization-induced alkalinization. J Neurosci 31: 6997–7004. 10.1523/JNEUROSCI.6088-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. 2015. Defining cell types and states with single-cell genomics. Genome Res 25: 1491–1498. 10.1101/gr.190595.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH, Carbone L, Steemers FJ, Adey A. 2017. Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14: 302–308. 10.1038/nmeth.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- Wilkinson DG, Bhatt S, Chavrier P, Bravo R, Charnay P. 1989. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature 337: 461–464. 10.1038/337461a0 [DOI] [PubMed] [Google Scholar]
- Yin Y, Jiang Y, Berletch JB, Disteche CM, Noble WS, Steemers FJ, Adey AC, Shendure JA. 2018. High-throughput mapping of meiotic crossover and chromosome mis-segregation events in interspecific hybrid mice. bioRxiv 10.1101/338053 [DOI]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347: 1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. 2018. Molecular architecture of the mouse nervous system. Cell 174: 999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34: 11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zola-Morgan S, Squire LR, Amaral DG. 1986. Human amnesia and the medial temporal region: enduring memory impairment following a bilateral lesion limited to field CA1 of the hippocampus. J Neurosci 6: 2950–2967. 10.1523/JNEUROSCI.06-10-02950.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.