Abstract
Baboons (genus Papio) are broadly studied in the wild and in captivity. They are widely used as a nonhuman primate model for biomedical studies, and the Southwest National Primate Research Center (SNPRC) at Texas Biomedical Research Institute has maintained a large captive baboon colony for more than 50 yr. Unlike other model organisms, however, the genomic resources for baboons are severely lacking. This has hindered the progress of studies using baboons as a model for basic biology or human disease. Here, we describe a data set of 100 high-coverage whole-genome sequences obtained from the mixed colony of olive (P. anubis) and yellow (P. cynocephalus) baboons housed at the SNPRC. These data provide a comprehensive catalog of common genetic variation in baboons, as well as a fine-scale genetic map. We show how the data can be used to learn about ancestry and admixture and to correct errors in the colony records. Finally, we investigated the consequences of inbreeding within the SNPRC colony and found clear evidence for increased rates of infant mortality and increased homozygosity of putatively deleterious alleles in inbred individuals.
Baboons are Old World monkeys commonly found in open woodlands and savannahs across sub-Saharan Africa and the southern portion of the Arabian Peninsula. They are closely related to humans, with an estimated divergence time of ∼30 million yr (Perelman et al. 2011; Zinner et al. 2013; Pozzi et al. 2014), and like humans, baboons are social, omnivorous, and highly adaptable. They originated in southern Africa and over the past two million yr have expanded their range and evolved into six distinct morphotypes or species: olive (P. anubis), yellow (P. cynocephalus), hamadryas (P. hamadryas), Guinea (P. papio), chacma (P. ursinus), and kinda (P. kindae) baboons (Jolly 1993; Newman et al. 2004; Zinner et al. 2009; Boissinot et al. 2014). Because they share so many similarities with humans, baboons have been studied intensively both in the wild and in captivity since the early 1960s and are considered a useful model in a wide array of research areas.
Over the past several decades, the baboon has become an important nonhuman primate model in biomedical research, second only to macaques (genus Macaca) (for review, see VandeBerg et al. 2009). Baboons have been used to study normal physiology and development as well as various diseases that commonly affect humans, including diabetes, atherosclerosis, osteoporosis, obesity, hypertension, epilepsy, and addiction (e.g., Aufdemorte et al. 1993; Comuzzie et al. 2003; Guardado-Mendoza et al. 2009; VandeBerg et al. 2009; Szabó et al. 2012; Mahaney et al. 2018). Much of this research has been facilitated by the Southwest National Primate Research Center (SNPRC), which houses the world's largest captive baboon colony, containing >1000 individuals at any given time. The colony was established in the 1960s with olive and yellow baboon founders from southern Kenya. Since then, a complete pedigree for the colony spanning seven generations has been maintained, providing relatedness and ancestry information for all captive-bred individuals, along with recorded birth and death dates. Biological samples and phenotype data have also been kept as a resource for biomedical studies.
Relative to other model organisms, however, there are few genomic resources available for baboons. These resources are essential for modern evolutionary and biomedical studies, and their absence hinders baboons’ usefulness as a model organism. The recently published baboon reference genome, Panu_3.0, is somewhat fragmented and was assembled into chromosomes based on synteny with the rhesus macaque (Macaca mulatta) genome (Rogers et al. 2019). Thus, very little is known about baboon genomic variation of single nucleotide polymorphisms (SNPs) or haplotypes, and the latest baboon genetic map is based on only 284 microsatellite markers (Cox et al. 2006).
In this study, we take a step toward developing baboon genomic resources by generating high-coverage whole-genome sequence data from 100 SNPRC baboons. We used these genomes to generate a fine-scale map of recombination rates (relative to the Panu_2.0 assembly), for use as a resource in future studies and to highlight potentially misassembled regions of the reference genome (we used this older assembly due to the unavailability of Panu_3.0 until 2019). We also estimated olive versus yellow baboon ancestry in the sequenced individuals and used this to identify errors in the pedigree file. Lastly, we examined rates of infant mortality and patterns of putatively deleterious variation to investigate the consequences of inbreeding in the colony. We expect the resources and results from this study will provide a useful foundation for future studies of baboons in captivity and in the wild.
Results
Resources to enable future baboon genomic research
We sequenced the complete genomes of 100 baboons from the SNPRC colony to high coverage (21–42×), including 33 founders, and mapped reads to the Panu_2.0 genome assembly (Supplemental Table S1). In light of the recent release of the Panu_3.0 genome assembly, we also generated liftOver (Hinrichs et al. 2006) chain files as a resource for converting the genomic coordinates in this study (using Panu_2.0) to coordinates in the new assembly (see Methods). In total, we identified 56.4 million SNPs, 5.87 million indels, and 5.52 million other complex variants. The raw density of variants was noticeably higher on unplaced scaffolds than on the chromosomes (74.6 variants/kb on scaffolds vs. 19.1 variants/kb on autosomes), implying a higher error rate in regions that have not been incorporated into chromosomal assemblies, perhaps due to repetitive sequence content. We focused solely on high-quality biallelic SNPs located on the autosomes for our analyses. After applying quality filters (Methods), our data set contained 20,352,729 variants distributed across 20 autosomes. Of these, >10.5 million variants were common (minor allele frequency >0.05) within the sequenced baboon founders, and >620,600 SNPs were highly differentiated (FST > 0.8) between genetically identified olive and yellow baboon founders (see below). Variation within individuals was comparable to what was observed previously (Wall et al. 2016), with per-individual heterozygosity values ranging from 1.16 to 3.03 heterozygous genotypes per kilobase.
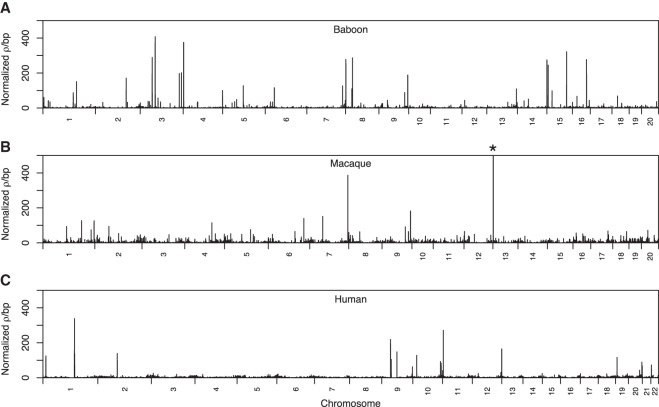
Additionally, we used the variation present within 24 olive founders to generate a fine-scale linkage-disequilibrium-based genetic map of the baboon genome with LDhelmet (Chan et al. 2012). Our estimates of recombination rates are in terms of ρ (= 4Ner), the population-scaled recombination rate (see Methods). Since LDhelmet is sensitive to a parameter called the “block penalty,” which is a smoothing parameter, we repeated the analysis with a range of values. Our results were largely consistent across block penalty values of 5, 25, and 50 (Supplemental Fig. S1). Here, we focus on results obtained with a block penalty of 5, which the manual suggests is the appropriate value for humans (vs. 50 for Drosophila). The genome-wide average ρ is ∼3.55 per kilobase. We compared our estimates of total ρ for each chromosome to the genetic map lengths inferred from microsatellite data in a previous study (Rogers et al. 2000) and observed a modest but statistically significant correlation between the results from these two methods (Supplemental Fig. S2). We then calculated ρ/bp in nonoverlapping 100-kb windows across the genome and found that estimated rates varied widely, from a low of 3.17 × 10−7 to a high of 1.45 ρ/bp within each window. We noted that there were several distinct peak regions with exceptionally high recombination rates (Fig. 1A). In total, we identified 45 different 100-kb windows with ρ/bp greater than 100-fold above the mean. Such high recombination rates are biologically implausible and are most likely due to errors in haplotype phasing, reference genome assembly structure, or in the rate estimation itself.
Figure 1.
Recombination rates across the baboon, macaque, and human reference genomes. Rates were inferred from genetic variation in 24 unadmixed olive founder baboons (A), 24 Indian rhesus macaques (B), and 24 unrelated African (Yoruban) individuals (C). Rates were calculated in nonoverlapping 100-kb windows across the genome and normalized by dividing raw rates by the mean rate inferred within each data set (mean ρ/bp: baboons, 3.55 × 10−3; macaques, 1.79 × 10−3; humans: 5.87 × 10−3). Here, a block penalty of 5 was used. Extremely high recombination rates, evident in the large number of high peaks across the genome, highlight putative errors in the Panu_2.0 genome assembly. (*) Peak height = 963.133.
To determine whether excessive peaks of recombination rate are expected when the reference genome is well-assembled, we repeated our analysis with a comparable data set of 24 unrelated Indian rhesus macaque genomes (obtained from the Macaque Genotype and Phenotype Resource at https://mgap.ohsu.edu) and a data set of 24 African (Yoruban) genomes (Fig. 1B,C; Supplemental Fig. S3; JD Wall, E Stawiski, A Ratan, et al., unpubl.). In macaques, we found 10 100-kb windows with ρ/bp greater than 100 times the mean (note that the macaque genome assembly is slightly longer than the baboon assembly and contains 3.64% more windows overall). In humans, we found 21 100-kb windows with ρ/bp greater than 100 times the mean (the human genome assembly contains 8.26% more windows overall). The number of windows with exceptionally high ρ in the baboon data set is significantly greater than in the macaque and human data sets (macaques, P < 2.2 × 10−16; humans, P = 5.49 × 10−7). We found that the windows with high ρ in the baboon genome were not enriched for gaps in either the Panu_2.0 or Panu_3.0 assemblies, or for repetitive sequence content, but did contain significantly reduced SNP densities relative to other windows (P < 2.2 × 10−16). Some regions with exceptionally high estimated recombination rates might reflect structural errors in the baboon reference genome assembly (e.g., due to chromosomal rearrangements or structural variants fixed between baboons and rhesus) rather than a true biological signal. For example, out of 20 large syntenic differences between a de novo Hi-C-based baboon assembly and Panu_2.0, 11 show an extremely strong recombination hotspot (corresponding to a complete breakdown of linkage disequilibrium) near at least one of the breakpoints (SS Batra, M Levy-Sakin, J Robinson, et al., unpubl.).
Analysis of founder origins and admixture status
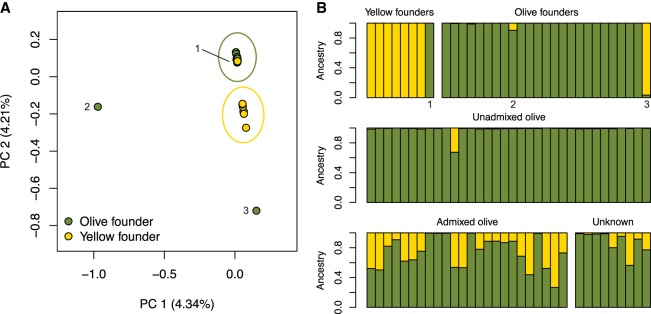
Our data allow us to determine whether species designations in the pedigree were concordant with genetic assignments and to determine whether any of the founders showed evidence of hybrid origin. The original founders of the SNPRC colony were captured near what is now known to be a large admixture zone between olive and yellow baboons in Eastern Africa (Samuels and Altmann 1986; Tung et al. 2008, Charpentier et al. 2012; Wall et al. 2016). Olive and yellow baboons are phenotypically distinct, and all 33 founders in our sample were originally labeled based on sampling location and physical appearance. Principal component analysis (PCA) with these 33 founders revealed two primary groups corresponding to putative olive and yellow baboons, as well as two distinct outlier individuals (ID: 1X0812 and 1X4384), both originally labeled as olive baboons (Fig. 2A). The PCA results are not consistent with a recent olive-yellow hybrid origin for these individuals, since hybrids would be expected to fall in between the olive and yellow baboon clusters. Additionally, one unambiguous olive founder was mislabeled as a yellow baboon (ID: 1X3576). Identity-by-state (IBS) clustering showed the same qualitative patterns (Supplemental Fig. S4) and also suggests that one of the two mystery founders (ID: 1X4384) is genetically more similar to yellow baboons. Without additional information, it is unclear whether the two outlier individuals were members of diverged olive/yellow baboon populations or were from other baboon species entirely. We also saw little variation in genetic distances between individuals within the olive and yellow clusters, suggesting our sample of founders does not include individuals that were closely related (Supplemental Fig. S4).
Figure 2.
Genetic ancestry patterns in baboons from the SNPRC. (A) PCA shows that olive and yellow baboon founders form distinct clusters, but one olive baboon was mislabeled as a yellow baboon (1: 1X3576), and two other individuals are extreme outliers (2: 1X0812, 3: 1X4384). (B) Similarly, ADMIXTURE results under a model of two ancestral populations (K = 2) show that patterns of olive and yellow baboon ancestry are not always concordant with given species labels. Individuals labeled as 1, 2, and 3 in the PCA are indicated. Individuals are grouped according to their classification from the colony records (see Supplemental Table S1).
We ran ADMIXTURE (Alexander et al. 2009) with 31 founders (two mystery individuals were excluded) under a range of K-values (K = 1–6) to provide another approach for looking at ancestry and population structure (Supplemental Fig. S5; Supplemental Table S2). The most likely number of genetic partitions within the 31 founders was K = 2, indicating a clear distinction between 24 genetically olive and seven genetically yellow baboons, consistent with the PCA and IBS cluster results. Runs with higher K values had higher cross-validation error rates and generated inconsistent partitions within the olive founders. Overall, we found no evidence of recent hybrid ancestry in the founders, and olive and yellow baboons are clearly differentiated. As a resource for future investigations of admixture between olive and yellow baboons, either in the SNPRC colony or in wild populations, we developed a list of >24,000 ancestry informative markers, which are sites where the olive and yellow baboon founders were fixed for different alleles. We calculated that the overall value of FST between genetically olive and genetically yellow baboons genome-wide is 0.366, comparable to previous estimates—FST = 0.3069 from Boissinot et al. (2014) and FST = 0.33 from Wall et al. (2016).
We next used the projection method in ADMIXTURE to assign ancestry proportions to the remaining individuals, assuming two parental populations (K = 2) (Fig. 2B; Supplemental Table S2). According to the pedigree and original species labels of the founders, our sample of captive-bred individuals contained three groups: unadmixed olive individuals, admixed individuals, and individuals with no designation (“unknown”). Of 34 putatively unadmixed captive-born olive baboons, one was a recent hybrid (15197: 32.5% yellow ancestry); of 24 “admixed” individuals, five appeared to have pure olive ancestry (>99% olive ancestry); of nine “unknown” individuals, four were clearly admixed (>5% and <95% olive ancestry). In total, nine out of 91 individuals (10%) were incorrectly labeled on the basis of species or admixture status. In contrast, the recorded sex of all 100 individuals was correct, which we confirmed by evaluating the ratio of mean coverage on the X Chromosome relative to the autosomes (Supplemental Fig. S6). Our results reveal that even in a well-documented captive population with a full pedigree, a nonnegligible proportion of individuals may be erroneously categorized. These errors can propagate through the pedigree by affecting the labels of individuals in subsequent generations.
Comparison of pedigree-based and genomic estimates of inbreeding
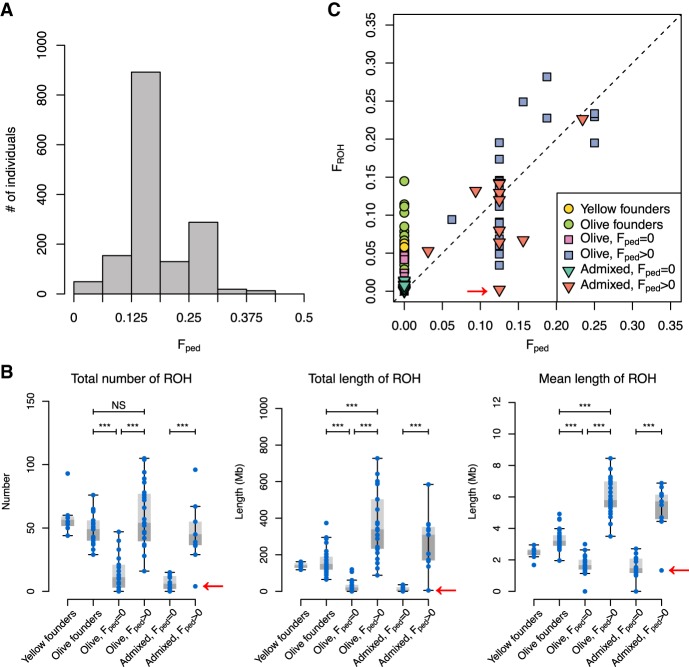
A subset of baboons within the SNPRC colony are inbred, making it an ideal system for studying the genomic impact of inbreeding and the link between inbreeding and fitness (i.e., inbreeding depression). The pedigree spans seven generations and contains 16,973 individuals born in 1966–2015. We calculated pedigree-based inbreeding coefficients (Fped) for all 16,973 individuals from the pedigree and found that 1700 had Fped > 0 (Fig. 3A). Mating within the colony is, for the most part, controlled, and deliberate inbreeding has been used to investigate medically relevant phenotypes. The most common form of inbreeding in the colony is unions between half-siblings or between uncles/aunts and nieces/nephews, resulting in offspring with Fped = 1/8 = 0.125 (n = 783). The next most common form of inbreeding is mating between parents and offspring (Fped = 1/4 = 0.25, n = 285). The maximum Fped was 0.40625 (n = 3). Thus, the degree of inbreeding within the colony is far greater than what has been observed in human populations (McQuillan et al. 2008; Bittles and Black 2010; Stevens et al. 2012).
Figure 3.
Inbreeding and ROH in the captive baboon colony. (A) Histogram showing the distribution of inbreeding coefficients in the pedigree (Fped). Individuals with Fped = 0 are not shown. (B) Inbred ancestry (Fped > 0) increases the total number, total length, and mean length of ROH in the genome, regardless of admixture status. Founder individuals also show indications of inbreeding in their ancestry. Sample sizes: yellow founders, 8; olive founders, 25; olive, Fped = 0: 18; olive, Fped > 0: 24; admixed, Fped = 0: 14; admixed, Fped > 0: 11. The putatively inbred individual that does not appear to be inbred based on lack of ROH is indicated with red arrows. (NS) Not significant below a threshold of 0.05, (***) P < 0.001. All P-values were multiplied by 4 to correct for multiple tests. (C) The proportion of the genome contained within ROH (FROH) can vary substantially from Fped. The dashed line represents the line y = x.
Pedigree-based inbreeding coefficients provide the expected values for the proportion of the genome that is autozygous, or inherited identically-by-descent. As expected, we found that inbred ancestry (Fped > 0) is associated with larger numbers of long (>1 Mb) runs of homozygosity (ROH), greater total lengths of ROH in the genome, and longer ROH lengths on average (Fig. 3B). All comparisons between inbred and noninbred groups among captive-born individuals were statistically significant (P ≤ 6.19 × 10−4). Both olive and yellow baboon founders in our data set appear to have signatures of inbreeding. Olive baboon founders have greater mean lengths, total numbers, and total lengths of ROH than noninbred olive baboons born within the colony (P ≤ 1.22 × 10−6). Long ROH within the founders may reflect nonrandom mating within wild populations, which have sex-biased dispersal (females are philopatric) and complex social hierarchies that determine access to mates, particularly among males (Alberts et al. 2003).
Next, we compared Fped and FROH, the proportion of the genome within ROH, and found that these values were positively correlated, but FROH exhibited variance around the predicted values from the pedigree (Fig. 3C). The realized proportion of the genome that is autozygous can vary from the expected value due to inherent randomness in recombination and chromosomal segregation during meiosis. Further, FROH was greater than Fped in 79% of cases, a statistically significant difference (one-tailed Wilcoxon signed-rank test, P = 6.83 × 10−7). This result is consistent with recent studies comparing genomic estimates of inbreeding to pedigree-based estimates and reflects the fact that genomic data can reveal inbreeding that is not captured in the pedigree (Kardos et al. 2015). Finally, we identified one putatively inbred individual (ID: 8465; Fped = 0.125) that does not contain autozygous genomic segments consistent with inbred ancestry (FROH = 0.00206), suggesting a possible error in the pedigree. Subsequent investigation of animal housing records confirmed that the parentage of this individual in the pedigree is incorrect. Overall, our results confirm that genomic measures provide greater resolution and accuracy for quantifying the proportion of the genome that is identical by descent, even relative to estimates from a large pedigree of more than 16,000 individuals over several generations.
Impacts of inbreeding on infant mortality and deleterious variation
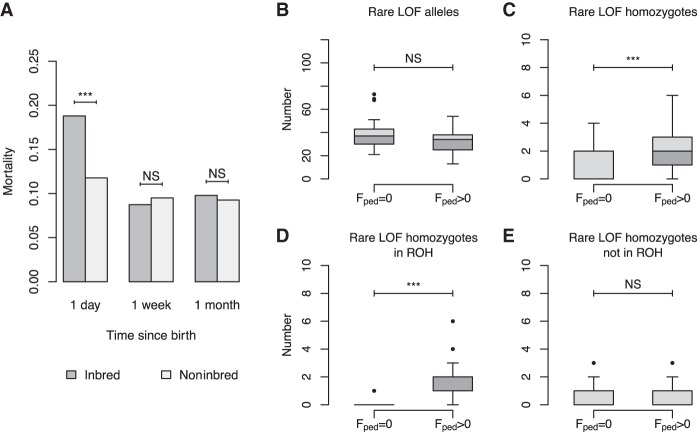
To determine whether inbred ancestry is associated with reduced fitness, we calculated the mortality rate of individuals at 1 d, 1 wk, and 1 mo after birth in 13,313 individuals, 1393 of which were inbred (Fig. 4A). Of noninbred individuals, 16.1% died on their day of birth versus 23.0% of inbred individuals. This difference was highly statistically significant (P = 5.24 × 10−11). After the day of birth, mortality rates within the first week and first month of life were similar between inbred and noninbred groups. These results imply that the reduced survival of inbred individuals is sharply reduced at birth, but this effect does not persist or is too slight to be detected with our analysis. We were unable to test whether inbred individuals have shorter lifespans or reduced reproduction, since breeding within the colony is controlled, and “exit” dates in the pedigree may not reflect natural mortality (Methods). Nonetheless, we found a dramatic difference in mortality on day of birth between inbred and noninbred groups, corresponding to an odds ratio of 1.57. Overall, we find that the offspring of related parents show evidence for reduced fitness, suggesting the SNPRC baboon colony would be a useful system for studying the genetic basis of inbreeding depression.
Figure 4.
Rates of infant mortality and the burden of LOF mutations as a function of inbreeding in captive-born baboons. (A) Inbred baboons (Fped > 0) have substantially higher rates of infant mortality on the day of birth. (B–E) Box plots showing that the homozygosity of putatively deleterious rare LOF mutations is affected by inbreeding. The total number of alleles is not changed by inbreeding (B), but the number of homozygous LOF mutations increases due to increased ROH content in the genome (C,D). Outside of ROH, the number of rare LOF homozygotes is unchanged between inbred and noninbred individuals (E). (NS) Not significant below a threshold of 0.05, (***) P < 0.001.
Higher rates of infant mortality in inbred baboons may be due to increased rates of recessive deleterious alleles within ROH. Strongly deleterious recessive alleles can persist in large outbred populations, since these alleles are not exposed to selection when they are rare and most often present in the heterozygous state. Such mutations are often exposed through inbreeding, resulting in inbreeding depression (Charlesworth and Willis 2009). We compared the total numbers of rare loss-of-function (LOF) alleles and the total numbers of homozygous LOF genotypes between inbred and noninbred individuals (Fig. 4B,C). LOF mutations are predicted to diminish or eliminate gene function and are therefore expected to be deleterious, especially when homozygous and located within genes required for normal function. The total number of LOF alleles was equivalent between inbred and noninbred groups, consistent with the fact that inbreeding alters genotype frequencies but does not impact allele frequencies in the absence of other factors (e.g., selection, drift). However, inbred individuals had significantly higher numbers of rare homozygous LOF mutations (P = 1.41 × 10−4), specifically due to an increased number of homozygous rare LOF mutations within ROH (P = 1.69 × 10−9) (Fig. 4D). Outside of ROH, rare homozygous LOF mutations were equally prevalent in inbred and noninbred genomes (Fig. 4E), demonstrating that the increased number of rare homozygous LOF mutations in inbred genomes is due to their higher proportions of ROH and suggesting a possible role in the reduced fitness of inbred individuals.
Discussion
The primary goal of our project was to help generate resources that enable future genome-wide studies in baboons, with a focus on the SNPRC baboon colony. To this end, the variants identified in this study could be used for future SNP or capture array designs, while the genetic map will be useful for future evolutionary or genotype-phenotype association studies. In addition, since the method used for estimating recombination rates required phased haplotypes, we also have a computationally phased data set of 24 olive baboon founders that can be used as a reference panel for future genotype imputation in baboons. Although only the Panu_2.0 genome assembly was available at the time of our analyses, our results are expected to be largely robust to changes in the Panu_3.0 assembly, and we further provide chain files for converting coordinates from our study to the new genome (Methods).
Preliminary analyses of our data highlighted several novel findings. First, anomalies in our genetic map identified regions of the Panu_2.0 assembly that might be problematic. Second, genetic analyses identified several errors in the existing animal records related to species identity, admixture status, and inbred ancestry. These inconsistencies may have resulted from mistakes in recordkeeping that were never corrected, unexpected matings between baboons in different enclosures, or challenges in distinguishing recently diverged species from one another. Third, we exploited the unique pedigree structure of the colony to quantify the effects of inbreeding on infant mortality and genetic load of rare, homozygous, putatively harmful mutations. Further work on the inbred baboons in the SNPRC colony will provide an ideal opportunity to study the effects of recessive, deleterious mutations in a nonhuman primate model.
Finally, we want to emphasize that the SNPRC baboon colony, as a mixture of olive and yellow baboons, is also an ideal system for studying the effects of admixture between diverged populations. The yellow and olive baboon founders in our study are highly differentiated (FST = 0.366), more so than between continental human populations (The 1000 Genomes Project Consortium 2010) or between isolated human groups (Wall et al. 2008). This differentiation may be useful for the mapping of phenotypic traits (e.g., using admixture mapping) or for evolutionary studies of selection and adaptation. Although little attention has been paid to possible differences between olive and yellow baboons with regard to medically relevant traits (however, see Jolly and Phillips-Conroy 2006), olive and yellow baboons are phenotypically distinguishable and differ in important behavioral and physical traits such as age at dispersal and time to reproductive maturity (Alberts and Altmann 2001; Charpentier et al. 2008). Olive and yellow baboons form a natural hybrid zone in the wild, and the Amboseli Baboon Research Project has continuously observed several baboon troops in this hybrid zone for over 45 yr. Research in Amboseli has revealed that hybrids between olive and yellow baboons may have higher fitness relative to unadmixed members of either parental species (Charpentier et al. 2008). We expect that future work enabled by the development of genomic resources will compare the genetic and phenotypic effects of both inbreeding and admixture in the wild and in captivity.
Methods
Sequencing and genotype calling
We extracted DNA from archived buffy coats or liver samples from 100 individuals from the SNPRC colony for high-coverage (>20×) whole-genome sequencing. These individuals included 33 founders of the colony and 67 captive-born descendants (Supplemental Table S1). Initially, ∼200 wild-caught founders were used to establish the SNPRC colony, but incomplete records prevented us from obtaining a comprehensive list of founders. To maximize the variation captured in our data set, founders with many offspring that made the greatest contribution to the colony and individuals with many offspring alive in the colony at the time of sequencing were prioritized for inclusion in our study.
DNA was extracted using the QIAamp DNA Mini kit (Qiagen) according to the manufacturer's instructions, and DNA quality was assessed using the Qubit BR Assay kit (Thermo Fisher Scientific) with a Qubit Fluorometer according to the manufacturer's instructions. DNA was quantified using the KAPA Human Genomic DNA Quantification and QC kit (KAPA Biosystems) according to the manufacturer's instructions. Sequencing libraries were prepared using the TruSeq DNA PCR-Free High Throughput Library Prep kit (Illumina) according to the manufacturer's instructions, with quality assessed using a Bioanalyzer 2100 (Agilent) and quantified using KAPA Library Quantification Kits for Illumina Platform (KAPA Biosystems).
Paired-end reads were generated with Illumina HiSeq 2500 technology and then trimmed to remove adapters and low-quality sequence with ea-utils (https://github.com/ExpressionAnalysis/ea-utils). Trimmed reads were processed with Sentieon Genomics tools (v201611) (Freed et al. 2017) to generate variant calls; briefly, reads were aligned to the olive baboon reference genome (Panu_2.0) with BWA-MEM (v0.7.12) (Li 2013), duplicate reads were removed, and genotypes were called with HaplotypeCaller (McKenna et al. 2010). The mean depth of coverage across individuals was 33.5× (see Supplemental Table S1 for mean coverage per individual). We restricted our analysis to the 21 chromosome-level scaffolds of the reference genome (20 autosomes and one X Chromosome), excluding the mitochondrial genome and all unplaced contigs. The X Chromosome was used only to calculate the ratio of read depth on the X Chromosome versus the autosomes in order to infer each individual's sex. All other analyses were conducted using only the autosomal data. Raw VCF files are available for download from https://doi.org/10.5281/zenodo.2583266.
We incorporated various filters to minimize the inclusion of erroneous genotypes. We masked repetitive regions using the “soft-masked” Panu_2.0 reference FASTA available from the UCSC Genome Browser, which annotates repeats based on RepeatMasker and Tandem Repeats Finder (with period of 12 or less) (http://www.repeatmasker.org; Benson et al. 1999). Further details are available at https://github.com/priyamoorjani/baboon. We applied recommended hard filters (QD < 2.0 , FS > 60.0, MQ < 40.0, MQRankSum < − 12.5, ReadPosRankSum < − 8.0, SOR > 3.0) and excluded variants with excess total depth (DP > 4767, the 99th percentile of total depth) or low quality (QUAL < 30). Individual genotypes were also filtered to exclude calls with low quality (GQ < 20), low coverage (individual read depth < 8), or excessive coverage (individual read depth > 99th percentile, by individual). Heterozygous genotypes with extreme allele imbalance (ratio of reads with the reference vs. alternate allele <0.2 or >0.8) were also excluded. Finally, variants that were not biallelic single nucleotide polymorphisms or that had high missingness (>20%) or excess heterozygosity (>50%) were excluded.
Creation of chain files to convert coordinates between Panu_2.0 and Panu_3.0
We generated chain files for converting coordinates from Panu_2.0 to Panu_3.0 following the protocol described at the UCSC Genome Browser (http://genomewiki.ucsc.edu/index.php/Minimal_Steps_ For_LiftOver). We split the Panu_3.0 genome into 200-kb chunks, each of which was then aligned to the Panu_2.0 genome using pblat (Wang and Kong 2019), which is the parallelized version of BLAT (Kent 2002), as described in the protocol. We used the Genome Browser utilities from http://hgdownload.cse.ucsc.edu/admin/exe/ as described in the minimal steps protocol. We then used liftOver (Hinrichs et al. 2006) to convert coordinates from Panu_2.0 to Panu_3.0 using the chain files. Chain files for all 20 autosomes are available for download from https://doi.org/10.5281/zenodo.2583292.
Fine-scale recombination rate estimation
We used LDhelmet (v1.9) (Chan et al. 2012) to infer recombination rates across the genome within 24 unadmixed olive baboon founder genomes, 24 unrelated Indian rhesus macaque genomes, and 24 unrelated African (Yoruban) human genomes (GA001430, GA001442, GA001443, GA001444, GA001445, GA001446, GA001447, GA001448, GA001449, GA001450, GA001451, GA001452, GA001453, GA001454, GA001455, GA001456, GA001457, GA001458, GA001459, GA001460, GA001461, GA001462, GA001463, GA000405). The macaque data set was downloaded from the Macaque Genotype and Phenotype Resource (https://mgap.ohsu.edu). The human data set was derived from a VCF file generated in a separate study (JD Wall, E Stawiski, A Ratan, et al., unpubl.). The human sequence read data are available under NCBI BioProject PRJNA476341.
Since LDhelmet requires phased input, we phased the genomes using Beagle (v5.0) (Browning and Browning 2007). We excluded singletons (of either the reference or alternative allele) since they are uninformative for haplotype phasing and pruned variants so that no two were within 10 bp of each other, leaving 8.48 million SNPs in the baboon data set, 7.75 million SNPs in the macaque data set, and 8.50 million SNPs in the human data set. We changed the default effective population size parameter to 40,000 for baboons, consistent with the Ne of olive baboons estimated by Boissinot et al. (2014), 50,000 for macaques (Xue et al. 2016), and 20,000 for humans. Default values were used for all other parameters. Next, we executed the LDhelmet pipeline following the steps outlined in the program manual, using default parameter values except where noted. In particular, we set the population-scaled mutation rate, θ, to 0.0016 in baboons, which is an approximation of the value of θ we calculated from the number of variants in the input data prior to pruning, to 0.002 in macaques (Xue et al. 2016), and to 0.001 in humans. We used a window size of 50 and ran the Markov chain Monte Carlo (MCMC) inference for 1 × 106 iterations, following a burn-in period of 1 × 105 iterations. The estimation was conducted under a range of block penalty values (5, 25, 50). Recombination rate estimates are given in units of ρ/bp, where ρ (= 4Ner) is the population-scaled recombination rate parameter and r is the recombination rate per nucleotide per generation. Finally, we converted the recombination rates emitted by LDhelmet, which are given as mean ρ/bp for each SNP interval, into rates in nonoverlapping 100-kb windows across the genome. The final window of each chromosome was excluded, since these windows contained fewer sites. A binomial test was used to determine whether there are significantly more windows with extreme recombination rate in the baboon data set. Here, the number of successes was defined as the number of nonoverlapping 100-kb windows in the baboon data set with an estimated recombination rate greater than 100 times the mean (45), the sample size was defined as the number of windows in the baboon genome (25,801), and the expected rate of successes was defined as the proportion of high recombination rate windows observed in the macaque (10/26,739) or human data sets (21/27,933). We used one-tailed Mann–Whitney U tests to determine whether windows with high ρ in the baboon data set were significantly enriched for assembly gaps or repetitive sequences or were significantly depleted for SNP density relative to all other windows in the genome. The phased baboon VCF files and recombination rate maps are available for download at https://doi.org/10.5281/zenodo.2583292.
Inference of genetic clusters, FST, and admixture
We used SNPRelate (v1.4.2) (Zheng et al. 2012) to perform PCA and IBS hierarchical clustering and to calculate a genome-wide value of FST between olive and yellow baboon founder individuals. Variants within the 33 founder individuals were extracted and pruned for linkage disequilibrium in SNPRelate (threshold = 0.2), leaving 127,935 variant sites for PCA and IBS clustering. The IBS clustering analysis consists of constructing a pairwise distance matrix, which is then used to construct a dendrogram to represent genetic similarity between individuals. To estimate genome-wide FST between olive and yellow baboons, we excluded two founders that were extreme outliers in the PCA (ID: 1X0812 and 1X4384), leaving 17,789,625 unpruned SNPs from the remaining 31 founders. We used VCFtools (v0.1.13) (Danecek et al. 2011) to calculate FST on a per-site basis from positions with no missing data. FST was calculated using the method of Weir and Cockerham (1984) in both SNPRelate and VCFtools. A list of >24,000 ancestry informative SNP markers can be downloaded at https://doi.org/10.5281/zenodo.2583292.
We used ADMIXTURE (v1.3.0) (Alexander et al. 2009) to investigate the possibility of admixed ancestry in the founders and to infer admixture proportions in the captive-born individuals. First, we used the 31 founders that were clearly olive or yellow baboons based on the clustering analyses described above to infer the most likely number of ancestral populations. Here, we ran ADMIXTURE unsupervised under a range of K-values (K = 1–6) and then examined the cross-validation errors from each run to determine the most likely K-value. Next, we used the projection method in ADMIXTURE to assign ancestry to the remaining 69 individuals, which is the recommended method for a data set containing related individuals. The projection analysis consisted of using the population allele frequencies learned from the founders under the most likely K-value to assign ancestry in the remaining individuals.
Analysis of inbreeding and infant mortality in the pedigree
We analyzed a complete pedigree of the baboon colony containing information for individuals born between April 3, 1966 and November 4, 2015. Typically, the sex, birth date, and identity of at least one parent were known for each individual. We used GENLIB (v1.0.6) (Gauvin et al. 2015) to calculate inbreeding coefficients of all individuals. GENLIB requires that all individuals must be labeled as either male or female, so in cases where the sex of an individual was missing, but the individual was identified as a mother or as a father elsewhere in the pedigree, we filled in the missing sex. In cases where sex was unknown, we arbitrarily changed the sex to female. These individuals have no offspring within the pedigree; therefore, their sex is irrelevant for downstream analyses. In general, we defined inbred individuals as any individuals with Fped > 0, unless otherwise noted.
To compare rates of infant mortality in inbred and noninbred individuals, we calculated the lifespan of all individuals from their recorded birth and “exit” dates. Exit dates can either represent the date an individual died or the date it left the colony for other reasons, which may not reflect natural mortality. Exit dates within one month of birth were assumed to be due to (natural) mortality. Individuals with no recorded exit date but that were known to be alive as of November 6, 2015 were given exit dates of November 6, 2015 so that they could be included in analyses of early life survival. Nonnested survival rates within 1 d, 1 wk (1–7 d), and 1 mo (8–28 d) of birth were calculated for inbred and noninbred individuals born between January 1, 1983 and October 7, 2015 (1 mo before this version of the pedigree was last updated). All individuals born earlier than 1983, which was the first year that any inbred individuals were born, were excluded from mortality analyses. These individuals were excluded since they may have experienced different environmental conditions than inbred individuals, possibly affecting survival in early life. For individuals born late in the pedigree, the 1-d, 1-wk, and 1-mo survival rates are known with certainty for all individuals born on or before October 7, 2015. χ2 tests were used to determine whether rates of infant mortality were significantly different between inbred and noninbred groups.
Identification of runs of homozygosity
To assess the effects of recent inbreeding in baboon genomes, we used PLINK (v1.9) (Chang et al. 2015) to identify large tracts of autozygosity, which indicate genomic regions inherited identically-by-descent from a recent common ancestor shared by both parents. We used the default parameters in PLINK to identify ROH in a pruned set of SNPs as recommended in the manual (--indep-pairwise 50 5 0.5). We calculated the proportion of the genome contained within long ROH (1 Mb or longer) as a measure of the realized level of inbreeding (FROH) in each of the sequenced genomes. Here, the numerator for FROH is the summed length of ROH across the autosomes, divided by the total length of the autosomal genome (2,581,196,250 nt). We used the asymptotic Wilcoxon–Mann–Whitney test from the coin library (v1.2.2) (Hothorn et al. 2008) in R (v3.3.1) (R Core Team 2018) to test for significant differences in the total number, total length, and mean length of ROH between groups. Raw P-values were multiplied by four to correct for multiple testing (four tests for each ROH statistic evaluated). A BED file with the coordinates of ROH identified in this study is available at https://doi.org/10.5281/zenodo.2583292.
Variant annotation and analysis of LOF mutations
We used SnpEff (v4.3t) (Cingolani et al. 2012) to identify the type and impact of mutations, according to the Panu_2.0 genome annotation (Panu_2.0.86). We focused specifically on loss-of-function mutations, which are predicted to severely disrupt or eliminate gene function and are therefore most likely to have a negative effect on fitness. This category includes mutations that introduce premature stop codons (“stop_gained”), eliminate start or stop codons (“start_lost”, “stop_lost”), or disrupt splice sites (“splice_acceptor_variant”, “splice_donor_variant”). Only mutations in protein-coding genes were included. Further, we concentrated on LOF mutations that were rare within the founders (<5% frequency), since common LOF mutations are unlikely to be strongly deleterious. We note that these putative LOF mutations may not actually knock out gene expression or function, and follow-up studies are needed to verify their functional impact. We used the asymptotic Wilcoxon–Mann–Whitney test from the coin library (v1.2.2) (Hothorn et al. 2008) in R (v3.3.1) (R Core Team 2018) to test for significant differences in the burden of rare LOF mutations between inbred and noninbred individuals. A table of LOF mutations identified in this study is available at https://doi.org/10.5281/zenodo.2583292.
Data access
The baboon whole-genome sequence data generated in this study have been submitted to the NCBI BioProject database (http://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA433868. A list of accession numbers for each sample is available in Supplemental Table S1.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health grants R24 OD017859 (to J.D.W. and L.A.C.), R01 GM115433 (to J.D.W.), and R24 OD021324 (to B.F.). We thank Priya Moorjani for providing the repeat-masked baboon file used in this project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.247122.118.
References
- The 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts SC, Altmann J. 2001. Immigration and hybridization patterns of yellow and anubis baboons in and around Amboseli, Kenya. Am J Primatol 53: 139–154. 10.1002/ajp.1 [DOI] [PubMed] [Google Scholar]
- Alberts SC, Watts HE, Altmann J. 2003. Queuing and queue-jumping: long-term patterns of reproductive skew in male savannah baboons, Papio cynocephalus. Anim Behav 65: 821–840. 10.1006/anbe.2003.2106 [DOI] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufdemorte TB, Fox WC, Miller D, Buffum K, Holt GR, Carey KD. 1993. A non-human primate model for the study of osteoporosis and oral bone loss. Bone 14: 581–586. 10.1016/8756-3282(93)90197-I [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Black ML. 2010. Consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci 107: 1779–1786. 10.1073/pnas.0906079106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Alvarez L, Giraldo-Ramirez J, Tollis M. 2014. Neutral nuclear variation in Baboons (genus Papio) provides insights into their evolutionary and demographic histories. Am J Phys Anthropol 155: 621–634. 10.1002/ajpa.22618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing data inference for whole genome association studies by use of localized haplotype clustering. Am J Hum Genet 81: 1084–1097. 10.1086/521987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AH, Jenkins PA, Song YS. 2012. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet 8: e1003090 10.1371/journal.pgen.1003090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat Rev Genet 10: 783–796. 10.1038/nrg2664 [DOI] [PubMed] [Google Scholar]
- Charpentier MJE, Tung J, Altmann J, Alberts SC. 2008. Age at maturity in wild baboons: genetic, environmental and demographic influences. Mol Ecol 17: 2026–2040. 10.1111/j.1365-294X.2008.03724.x [DOI] [PubMed] [Google Scholar]
- Charpentier MJ, Fontaine MC, Cherel E, Renoult JP, Jenkins T, Benoit L, Barthes N, Alberts SC, Tung J. 2012. Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Mol Ecol 21: 715–731. 10.1111/j.1365-294X.2011.05302.x [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzie AG, Cole SA, Martin L, Carey KD, Mahaney MC, Blangero J, VandeBerg JL. 2003. The baboon as a nonhuman primate model for the study of the genetics of obesity. Obes Res 11: 75–80. 10.1038/oby.2003.12 [DOI] [PubMed] [Google Scholar]
- Cox LA, Mahaney MC, VandeBerg JL, Rogers J. 2006. A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics 88: 274–281. 10.1016/j.ygeno.2006.03.020 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed DN, Aldana R, Weber JA, Edwards JS. 2017. The Sentieon Genomics Tools – a fast and accurate solution to variant calling from next-generation sequence data. bioRxiv 10.1101/115717 [DOI]
- Gauvin H, Lefebvre JF, Moreau C, Lavoie EM, Labuda D, Vézina H, Roy-Gagnon MH. 2015. GENLIB: an R package for the analysis of genealogical data. BMC Bioinformatics 16: 160 10.1186/s12859-015-0581-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardado-Mendoza R, Davalli AM, Chavez AO, Hubbard GB, Dick EJ, Majluf-Cruz A, Tene-Perez CE, Goldschmidt L, Hart J, Perego C, et al. 2009. Pancreatic islet amyloidosis, β-cell apoptosis, and α-cell proliferation are determinants of islet remodeling in type-2 diabetic baboons. Proc Natl Acad Sci 106: 0906471106 10.1073/pnas.0906471106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Hornik K, van de Wiel MA, Zeileis A. 2008. Implementing a class of permutation tests: the coin package. J Stat Softw 28: 1–23. 10.18637/jss.v028.i0827774042 [DOI] [Google Scholar]
- Jolly CJ. 1993. Species, subspecies and baboon systematics In Species, species concepts, and primate evolution (ed. Kimbel WH, Martin LB), pp. 67–107. Plenum Press, New York. [Google Scholar]
- Jolly CJ, Phillips-Conroy JE. 2006. Testicular size, developmental trajectories, and male life history strategies in four baboon taxa In Reproduction and fitness in baboons: behavioral, ecological, and life history perspectives (ed. Swedell L, Leigh SR), pp. 257–275. Springer, Boston. [Google Scholar]
- Kardos M, Luikart G, Allendorf FW. 2015. Measuring individual inbreeding in the age of genomics: Marker-based measures are better than pedigrees. Heredity 115: 63–72. 10.1038/hdy.2015.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
- Mahaney MC, Karere GM, Rainwater DL, Voruganti VS, Jr DE, Owston MA, Rice KS, Cox LA, Comuzzie AG, VandeBerg JL. 2018. Diet-induced early-stage atherosclerosis in baboons: lipoproteins, atherogenesis, and arterial compliance. J Med Primatol 47: 3–17. 10.1111/jmp.12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polasek O, Tenesa A, et al. 2008. Runs of homozygosity in European populations. Am J Hum Genet 83: 359–372. 10.1016/j.ajhg.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TK, Jolly CJ, Rogers J. 2004. Mitochondrial phylogeny and systematics of baboons (Papio). Am J Phys Anthropol 124: 17–27. 10.1002/ajpa.10340 [DOI] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. 2011. A molecular phylogeny of living primates. PLoS Genet 7: e1001342 10.1371/journal.pgen.1001342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi L, Hodgson JA, Burrell AS, Sterner KN, Raaum RL, Disotell TR. 2014. Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol 75: 165–183. 10.1016/j.ympev.2014.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, et al. 2000. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics 67: 237–247. 10.1006/geno.2000.6245 [DOI] [PubMed] [Google Scholar]
- Rogers J, Raveendran M, Harris RA, Mailund T, Leppälä K, Athanasiadis G, Schierup MH, Cheng J, Munch K, Walker JA, et al. 2019. The comparative genomics and complex population history of Papio baboons. Sci Adv 5: eaau6947 10.1126/sciadv.aau6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels A, Altmann J. 1986. Immigration of a Papio anubis male into a group of Papio cynocephalus baboons and evidence for an anubis–cynocephalus hybrid zone in Amboseli, Kenya. Int J Primatol 7: 131–138. 10.1007/BF02692314 [DOI] [Google Scholar]
- Stevens EL, Heckenberg G, Baugher JD, Roberson ED, Downey TJ, Pevsner J. 2012. Consanguinity in Centre d'Etude du Polymorphisme Humain (CEPH) pedigrees. Eur J Hum Genet 20: 657 10.1038/ejhg.2011.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó CÁ, Knape KD, Leland MM, Cwikla DJ, Williams-Blangero S, Williams JT. 2012. Epidemiology and characterization of seizures in a pedigreed baboon colony. Comp Med 62: 535–538. [PMC free article] [PubMed] [Google Scholar]
- Tung J, Charpentier MJ, Garfield DA, Altmann J, Alberts SC. 2008. Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Mol Ecol 17: 1998–2011. 10.1111/j.1365-294X.2008.03723.x [DOI] [PubMed] [Google Scholar]
- VandeBerg JL, Williams-Blangero S, Tardif SD, ed. 2009. The baboon in biomedical research. Springer, New York. [Google Scholar]
- Wall JD, Cox MP, Mendez FL, Woerner A, Severson T, Hammer MF. 2008. A novel DNA sequence database for analyzing human demographic history. Genome Res 18: 1354–1361. 10.1101/gr.075630.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Schlebusch SA, Alberts SC, Cox LA, Snyder-Mackler N, Nevonen KA, Carbone L, Tung J. 2016. Genomewide ancestry and divergence patterns from low-coverage sequencing data reveal a complex history of admixture in wild baboons. Mol Ecol 25: 3469–3483. 10.1111/mec.13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Kong L. 2019. pblat: a multithread blat algorithm speeding up aligning sequences to genomes. BMC Bioinformatics 20: 28 10.1186/s12859-019-2597-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. 10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- Xue C, Raveendran M, Harris RA, Fawcett GL, Liu X, White S, Dahdouli M, Rio Deiros D, Below JE, Salerno W, et al. 2016. The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences. Genome Res 26: 1651–1662. 10.1101/gr.204255.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28: 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinner D, Groeneveld LF, Keller C, Roos C. 2009. Mitochondrial phylogeography of baboons (Papio spp.) –indication for introgressive hybridization? BMC Evol Biol 9: 83 10.1186/1471-2148-9-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C. 2013. Baboon phylogeny as inferred from complete mitochondrial genomes. Am J Phys Anthropol 150: 133–140. 10.1002/ajpa.22185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.