Abstract
Metagenomic next-generation sequencing (mNGS) for pan-pathogen detection has been successfully tested in proof-of-concept case studies in patients with acute illness of unknown etiology but to date has been largely confined to research settings. Here, we developed and validated a clinical mNGS assay for diagnosis of infectious causes of meningitis and encephalitis from cerebrospinal fluid (CSF) in a licensed microbiology laboratory. A customized bioinformatics pipeline, SURPI+, was developed to rapidly analyze mNGS data, generate an automated summary of detected pathogens, and provide a graphical user interface for evaluating and interpreting results. We established quality metrics, threshold values, and limits of detection of 0.2–313 genomic copies or colony forming units per milliliter for each representative organism type. Gross hemolysis and excess host nucleic acid reduced assay sensitivity; however, spiked phages used as internal controls were reliable indicators of sensitivity loss. Diagnostic test accuracy was evaluated by blinded mNGS testing of 95 patient samples, revealing 73% sensitivity and 99% specificity compared to original clinical test results, and 81% positive percent agreement and 99% negative percent agreement after discrepancy analysis. Subsequent mNGS challenge testing of 20 positive CSF samples prospectively collected from a cohort of pediatric patients hospitalized with meningitis, encephalitis, and/or myelitis showed 92% sensitivity and 96% specificity relative to conventional microbiological testing of CSF in identifying the causative pathogen. These results demonstrate the analytic performance of a laboratory-validated mNGS assay for pan-pathogen detection, to be used clinically for diagnosis of neurological infections from CSF.
Metagenomic next-generation sequencing (mNGS) provides a comprehensive method by which nearly all potential pathogens—viruses, bacteria, fungi, and parasites—can be accurately identified in a single assay (Chiu 2013; Chiu and Miller 2016; Gu et al. 2018; Simner et al. 2018). This approach is attractive for diagnosis of infectious diseases, as pathogens that cause an infectious syndrome commonly have nonspecific, overlapping clinical presentations (Washington 1996). Recent advances in sequencing technology and the development of rapid bioinformatics pipelines have enabled mNGS testing to be performed within a clinically actionable time frame (Cazanave et al. 2013; Naccache et al. 2014, 2015; Wilson et al. 2014; Frémond et al. 2015; Greninger et al. 2015; Quinn et al. 2016; Salzberg et al. 2016; Mongkolrattanothai et al. 2017; Parize et al. 2017; Schlaberg et al. 2017b). However, numerous challenges remain with migrating mNGS testing into the clinical microbiology laboratory. These include (1) lack of an established blueprint for mNGS clinical validation, (2) difficulty in discriminating pathogens from colonizers or contaminants, (3) paucity of bioinformatics software tailored for clinical diagnostic use, (4) concern over quality and comprehensiveness of available reference databases, and (5) requirement for regulatory compliance inherent to patient diagnostic testing in a Clinical Laboratory Improvement Amendments (CLIA) environment.
Acute neurological illnesses such as meningitis and encephalitis are devastating syndromes, remaining undiagnosed in a majority of cases (Glaser et al. 2003, 2006; Granerod et al. 2010). The diagnostic workup for many patients requires extensive, and often negative, serial testing that utilizes a combination of culture, antigen, serologic, and molecular methods, resulting in delayed or missed diagnoses and increased costs. Cerebrospinal fluid (CSF) sample volume is often limiting, so that only a fraction of desired tests are able to be performed. Given the high burden of encephalitis-associated hospitalizations in the United States (Khetsuriani et al. 2002), there is a large unmet clinical need for better and more timely diagnostics for this syndrome, both to identify and to exclude infectious etiologies.
Here, we present the development and validation of an mNGS assay for comprehensive diagnosis of infectious causes of meningitis and encephalitis from CSF, expanding on summary data presented in a previously published review (Schlaberg et al. 2017a). The analytic performance of the mNGS assay was compared to results from conventional clinical microbiological testing performed in hospital or commercial diagnostic laboratories. We also tested the assay by blinded retrospective analysis of a challenge set of 20 CSF samples collected from patients with diagnosed neurological infections at a single pediatric tertiary care hospital.
Results
Sample processing and bioinformatics analysis
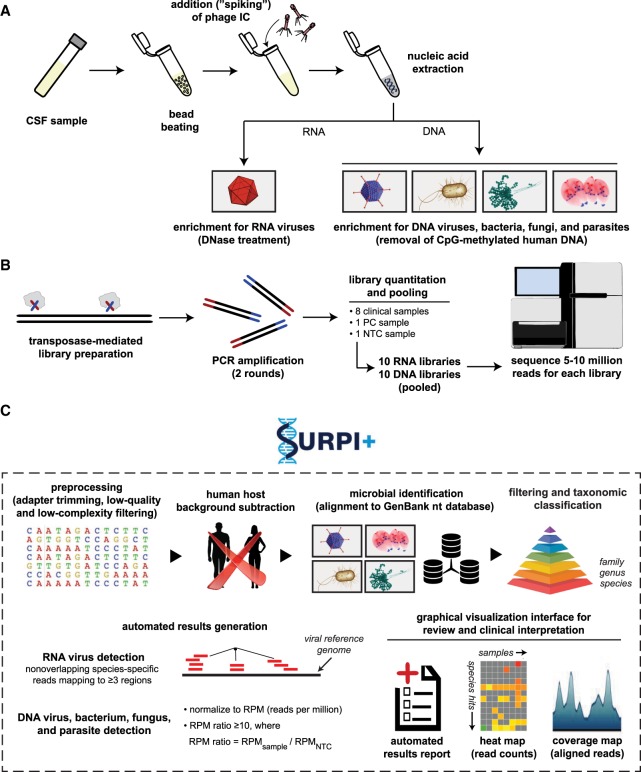
We developed an mNGS assay for pathogen identification from CSF consisting of library preparation, sequencing, and bioinformatics analysis for pathogen detection (Fig. 1) and validated the performance of the assay in a CLIA-certified laboratory. Standardized operating procedures for the “wet lab” protocols and sequencing runs for this analytic validation study were performed by state-licensed clinical laboratory scientists. For each sequencing run, NTC (“no template” control), PC (positive control), and both RNA and DNA libraries from up to eight patient CSF samples were processed in parallel. Steps included (1) microbial enrichment, (2) nucleic acid extraction, (3) Nextera library construction with two rounds of PCR, (4) library pooling in equimolar concentrations, and (5) rapid-run sequencing on an Illumina instrument, targeting five to 20 million sequences per library (Fig. 1A,B). Raw mNGS sequence data were analyzed using SURPI+, a bioinformatics analysis pipeline for pathogen identification (Naccache et al. 2014) that was modified for clinical use. Specifically, the modified pipeline incorporated filtering algorithms for confirmation of pathogen hits and taxonomic classification for accurate species-level identification.
Figure 1.
Schematic of the mNGS assay workflow. (A) CSF is extracted after lysis by bead-beating and internal control addition to allow viral, bacterial, fungal, and parasite nucleic acid retrieval. Total nucleic acid extracts are enriched for pathogen DNA by removal of methylated DNA (DNA libraries) and treatment with DNase (RNA libraries). (B) Libraries are generated using the Nextera XT protocol and amplified using two rounds of PCR. Libraries are quantified, pooled, and loaded onto the sequencer. (C) Sequences are processed using SURPI+ software for alignment and classification. Reads are preprocessed by trimming of adapters and removal of low-quality/low-complexity sequences, followed by computational subtraction of human reads and taxonomic classification of remaining microbial reads to family, genus, or species. For viruses, reads are mapped to the closest matched genome to identify nonoverlapping regions; for bacteria, fungi, and parasites, a read per million (RPM) ratio (RPM-r) metric is calculated, defined as RPM-r = RPMsample/NTC. To aid in analysis, automated result summaries, heat maps of raw/normalized read counts, and coverage/pairwise identity plots are generated for use in review and clinical interpretation.
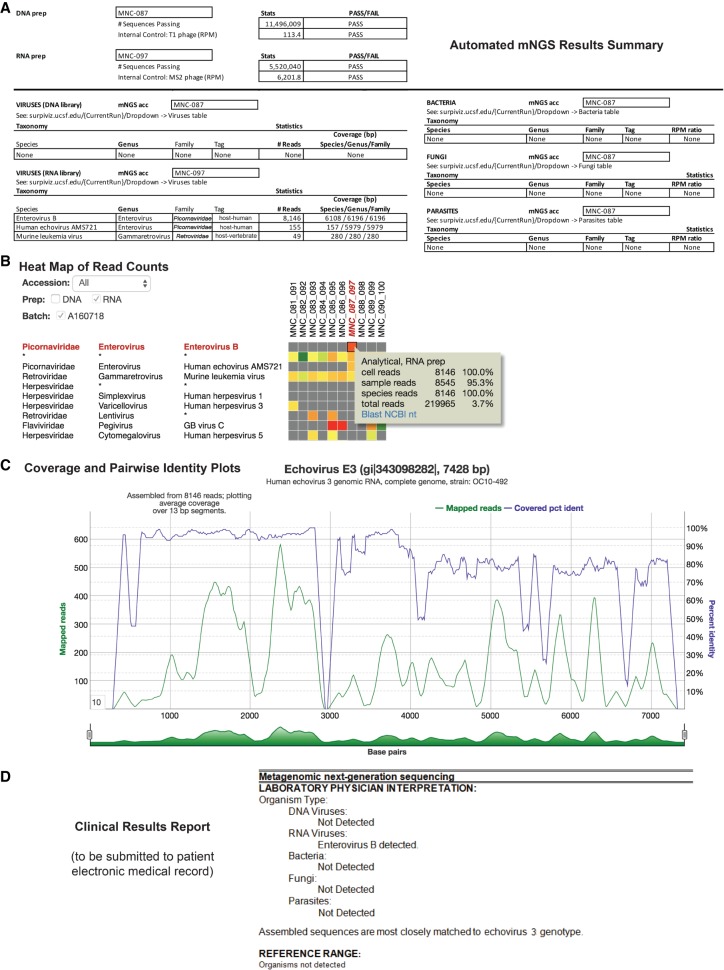
Results from SURPI+ were tabulated and used to populate a web-based graphical user interface (SURPIviz) designed to facilitate laboratory physician (e.g., microbiologist or pathologist) review and reporting of mNGS findings (Figs. 1C, 2). Results accessible to the laboratory physician included: (1) an automated mNGS results summary in Excel spreadsheet format providing QC run metrics and an overall clinical interpretation (Fig. 2A); (2) heat maps of raw and normalized read counts in each sample (Fig. 2B); (3) individual cells could be interrogated using BLASTn (Altschul et al. 1990) or downloaded in FASTA format for manual downstream analyses; and (4) coverage maps of aligned hits against the most closely matched microbial reference genome or sequence in the NCBI GenBank nucleotide (nt) database (Fig. 2C). During review of mNGS assay results, the laboratory physician assesses the displayed organism hits for clinical significance, taxonomic specificity, and potential contamination. The SURPIviz graphical tools are used to assist laboratory physicians in preparing a finalized clinical results report that is submitted to the patient electronic medical record (EMR) and available for viewing by treating clinicians (Fig. 2D). The clinical report lists detected organisms by type (virus, bacteria, fungus, or parasite), often with an interpretation and citation of relevant literature.
Figure 2.
Identification and reporting of enterovirus infection in a patient with meningoencephalitis using a clinical CSF mNGS assay. SURPI+ provides tools to aid clinical interpretation and graphical visualization (the SURPIviz package), including (A) an automated mNGS results summary, (B) a heat map of aligned reads corresponding to detected pathogens, and (C) a coverage plot (green line) of reads corresponding to a detected pathogen that are mapped to the most closely matched genome or gene in the reference database, along with a corresponding pairwise identity plot (purple line, sliding window = 10 nt). Viral hits corresponding to one CSF patient sample (MNC_087_097, column highlighted in red in B) are taxonomically identified as enterovirus (enterovirus B and echovirus AMS721 species) and murine leukemia virus, a known reagent contaminant (Zheng et al. 2011). After an interpretive review, a laboratory physician prepares a clinical results report (D) that is submitted to the patient electronic medical record (EMR).
Establishing thresholds for reporting detected pathogens
To minimize false-positive results from low-level microbial contamination, threshold criteria were established for organism detection (Fig. 1C). For viruses, we developed threshold criteria based on the detection of nonoverlapping reads from ≥3 distinct genomic regions. A viral species or genus meeting this threshold was reported as “detected”; otherwise, the virus type (DNA or RNA) was reported as “not detected.” Viruses comprising known body flora, such as anelloviruses (Maggi and Bendinelli 2010; Moustafa et al. 2017) and papillomaviruses (Foulongne et al. 2012), or known laboratory reagent contaminants (Zheng et al. 2011; Salter et al. 2014; Strong et al. 2014; Wilson et al. 2016) were not reported, nor were viruses detected in the NTC and attributed to cross-contamination.
For identification of bacteria, fungi, and parasites, we developed a reads per million (RPM) ratio metric, or RPM-r, defined as RPM-r = RPMsample/RPMNTC, with the minimum RPMNTC set to 1. This metric accounted for low-level microbial contamination by normalizing detected pathogen reads with respect to the NTC. To determine the optimal threshold value for RPM-r, we plotted receiver operating characteristic (ROC) curves at varying ratios corresponding to mNGS analysis of 95 clinical CSF samples used for accuracy evaluation (Supplemental Fig. S1), showing that an RPM-r of 10 maximized accuracy for organism detection. Thus, a minimum threshold of 10 RPM-r was designated for reporting the detection of a bacterium, fungus, or parasite as “detected” (RPM-r ≥ 10) or for reporting the pathogen type as “not detected” (RPM-r < 10).
Limits of detection
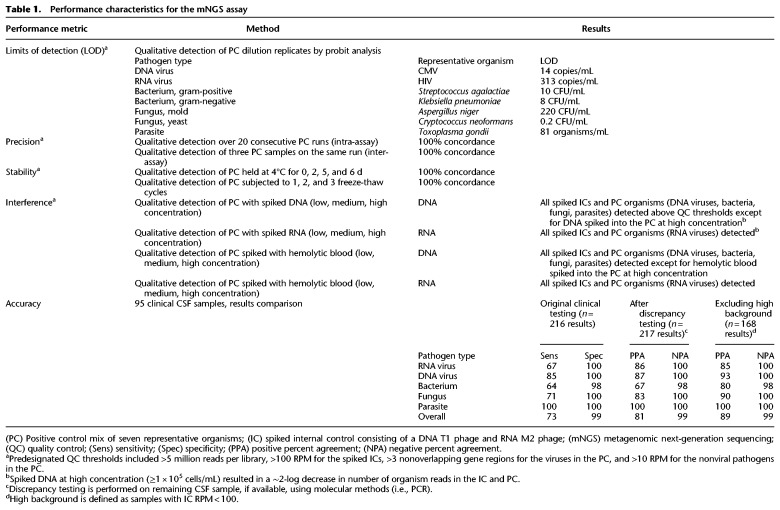
To calculate the 95% limits of detection (LOD), defined as the lowest concentration at which 95% of positive samples are detected, we evaluated the PC at concentrations ranging from 0.005 to 5000 genome equivalents/mL across a minimum 4-log dilution range per organism, testing three to 11 replicates at each concentration. Using probit analysis, a 95% limit of detection was determined for each of the seven representative organisms in the PC (Table 1). The final working PC consisted of the seven organisms spiked at concentrations in a range of 0.5- to 2-log above the 95% LOD.
Table 1.
Performance characteristics for the mNGS assay
Precision
We demonstrated inter-assay reproducibility by mNGS testing of the NTC and PC across 20 consecutive sequencing runs and intra-assay reproducibility by testing of three independently generated sets of NTC and PC on the same run. Internal spiked phage controls passed QC for every run, and only one PC RNA library (out of 46 DNA and RNA libraries) had fewer than the minimum cutoff of 5 million reads. All seven organisms were detected using pre-established threshold criteria for the intra-assay run and each replicate inter-assay run (Table 1).
Accuracy
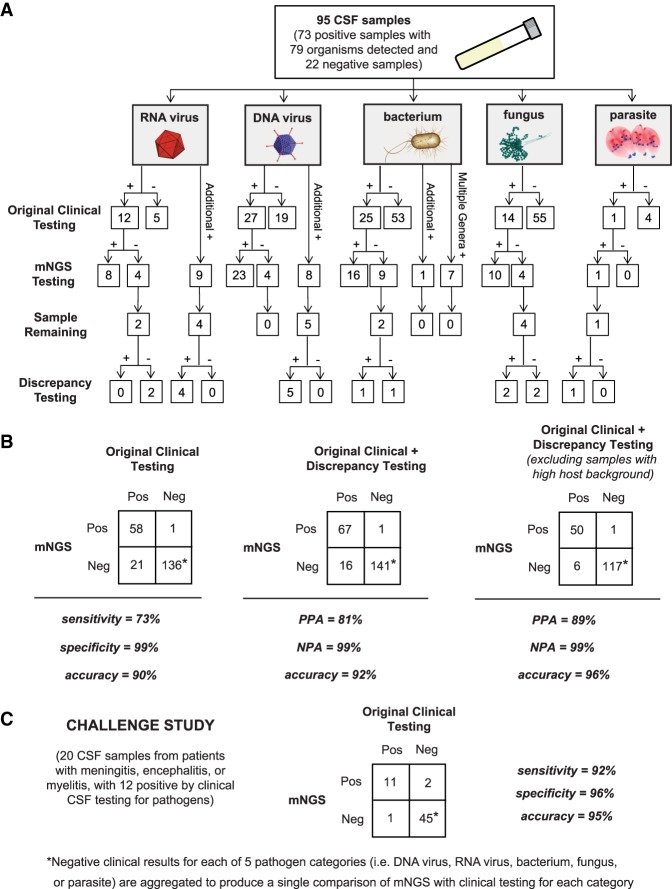
For evaluation of accuracy, a total of 95 CSF patient samples (73 positive with 79 organisms detected and 22 negative for any pathogen by conventional clinical testing) were tested using the mNGS assay. A board-certified infectious diseases physician (C.Y.C.) and pathologist (S.M.) independently performed in-depth, retrospective patient chart review to extract the results of conventional clinical testing and final microbiological diagnosis. mNGS results were compared to (1) original clinical test results, (2) results after discrepancy testing, and (3) results after discrepancy testing and exclusion of samples with high host background (Table 1; Fig. 3A,B; Supplemental Table S2).
Figure 3.
Accuracy of mNGS relative to clinical testing of CSF. (A) Flow chart of results from samples evaluated in the accuracy study. Results are separated by organism category (RNA virus, DNA virus, bacterium, fungus, and parasite). Shown are the number of samples positive or negative by clinical testing (first row), agreement between mNGS results and positive clinical results and additional positive detections by mNGS (second row), and, in samples with sufficient remaining volume (third row), the results of orthogonal confirmatory testing (fourth row). (B) 2 × 2 contingency tables comparing the performance of mNGS relative to clinical testing of CSF. The composite reference standards used are original clinical testing (left), combined original clinical and discrepancy testing (middle), and combined original clinical and discrepancy testing after excluding high host background samples (right). (PPA) Positive predictive agreement, (NPA) negative predictive agreement. (C) 2 × 2 contingency table showing the results of challenge study. Twenty CSF samples were analyzed by mNGS and compared with the results of conventional clinical testing.
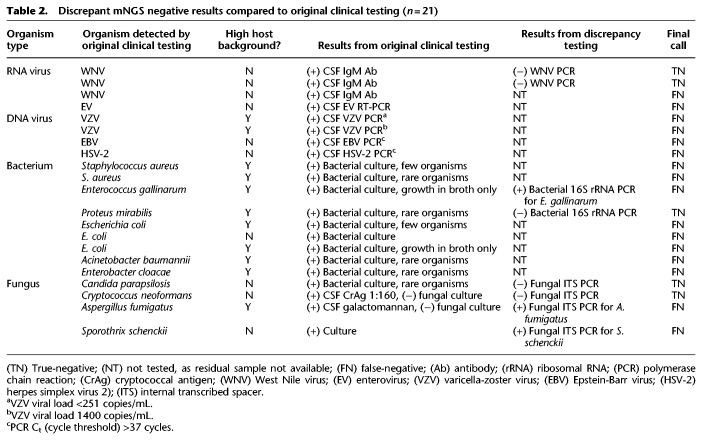
Overall, the mNGS assay showed 73% sensitivity and 99% specificity compared to original clinical test results (Table 1; Fig. 3B, left). Twenty-one cases were initially classified as mNGS false-negatives (Table 2; Supplemental Table S2); eight of 21 had sufficient residual CSF volume available for discrepancy testing. Among these eight mNGS-negative cases, five (two WNV, Proteus mirabilis, Candida parapsilosis, and Cryptococcus neoformans) were negative by follow-up clinical PCR testing and hence reclassified as true-negative cases after discrepancy testing (Table 2; Fig. 3B, middle). Three of eight mNGS-negative cases were positive by follow-up PCR testing and hence considered bona fide mNGS false-negative results. False-negative cases of Enterococcus gallinarum and Aspergillus fumigatus were probably missed by mNGS testing because they were high-background samples and weakly positive by original clinical testing (the E. gallinarum grew from broth only, and the A. fumigatus was galactomannan-positive but fungal culture-negative). A case of Sporothrix schenckii was negative by mNGS testing because the full ∼32-megabase (Mb) genome of S. schenckii, while publicly available (Cuomo et al. 2014), had not yet been deposited in the GenBank nt reference database (March 2015 build) used by SURPI+.
Table 2.
Discrepant mNGS negative results compared to original clinical testing (n = 21)
For the remaining 13 mNGS false-negative cases out of 21 that had insufficient volume for follow-up discrepancy testing, we examined the clinical data for potential explanations (Table 2). Among six viral cases, one (WNV) had been diagnosed via serology only, and four (two VZV, one EBV, one HSV-2) had low viral loads with high PCR cycle threshold (Ct) values (>37 cycles); two of four additionally had high background. For six of seven missed bacterial cases, cultures recovered few (n = 2), rare (n = 3), or broth only growth (n = 1), with all six being high-background samples.
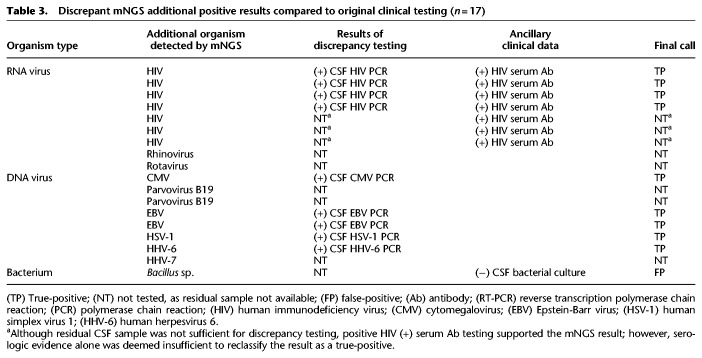
In 18 cases, additional organisms were detected by mNGS that had not been tested for clinically (Table 3). Nine cases (four HIV, one CMV, two EBV, one HSV-1, one HHV-6) had sufficient CSF sample available for discrepancy testing, and follow-up PCR testing confirmed the positive mNGS results in all nine of these cases, which were reclassified as true-positive cases. Given insufficient CSF volume, the additional organisms detected by mNGS in the nine remaining cases (three HIV, one rotavirus, one rhinovirus, two parvovirus B19, one HHV7, Bacillus sp.) could not be independently confirmed by follow-up discrepancy testing. Presumptive evidence of infection from these organisms also could not be documented from patient chart review (serum antibody [Ab] positivity alone in three cases of HIV was not deemed strong enough evidence for CSF viremia). However, mNGS detection of one case of Bacillus sp. was classified as false-positive, as it was from a culture-negative CSF sample. Thus, eight of these nine cases overall were excluded from the comparisons, as it could not be determined whether a given additional detection was a true- or false-positive. After discrepancy testing, the mNGS assay overall yielded 81% positive percent agreement and 99% negative percent agreement relative to the combined original and discrepancy testing results (Table 1; Fig. 3B, middle).
Table 3.
Discrepant mNGS additional positive results compared to original clinical testing (n = 17)
A third comparison was performed after exclusion of results from CSF samples with an IC RPM of <100, indicating potential decreased mNGS assay sensitivity due to high background. A total of 26 samples had high background (one RNA virus, three DNA virus, 19 bacteria, two fungi, one negative), and exclusion of these yielded 89% positive percent agreement and 99% negative percent agreement for the mNGS assay overall. Notably, the 19 high-background bacterial samples comprised 70.4% of the total number of culture-positive bacterial cases (n = 27), consistent with the relatively high leukocyte levels associated with bacterial meningitis.
Seven of the 95 CSF samples in the accuracy study yielded mNGS results with multiple bacterial genera detected, all seven of which were negative by original clinical testing. Detected genera corresponded to low-virulence environmental and/or skin flora organisms not typically associated with cases of meningitis and/or encephalitis (Supplemental Table S2). After clinical chart review, none of these cases were consistent with culture-negative bacterial meningitis, and residual sample was not available for discrepancy testing. Thus, results were attributed to sample contamination, reported as “multiple bacterial genera detected” (with an interpretive comment indicating likely contamination), and considered as negative for pathogen detection by mNGS.
We also evaluated contrived samples consisting of cultures from uncommon pathogenic organisms spiked into negative CSF matrix. All five organisms (Neisseria meningitidis, Streptococcus agalactiae, Candida albicans, Mycobacterium fortuitum, Mycobacterium abscessus) were correctly detected by mNGS testing.
Interference
We evaluated the effects of interference from human DNA and RNA, red blood cell hemolysis, and mixtures of related species in the same genus (Staphylococcus aureus and Staphylococcus epidermidis) on mNGS assay performance (Table 1). Addition of exogenous DNA at a level ≥1 × 105 cells/mL resulted in an approximately 2-log reduction in the number of IC and PC reads (Supplemental Fig. S2), impairing detection of DNA organisms in the PC. Addition of exogenous DNA at lower levels or RNA did not impact qualitative detection. Based on the interference results, a minimum RPM threshold of 100 was chosen for the IC phage reads, with RPM values below this level indicating that the sample library had high host background, along with an interpretive comment in the mNGS clinical report regarding decreased assay sensitivity for detection of RNA viruses (from RNA libraries) or DNA viruses, bacteria, fungi, and parasites (from DNA libraries).
Available data from 55 CSF samples in the accuracy study were used to evaluate the effect of WBC count, related to the level of host background, on recovery of IC phage sequences. Among 26 samples with IC DNA phage counts of <100 RPM, indicating high background, the average WBC was 5896 cells/mm3, while 29 samples with IC counts of >100 RPM had an average WBC count of 27 cells/mm3 (P = 0.0498 by two-tailed t-test).
Gross hemolysis (dark red CSF) resulted in decreased sensitivity for RNA virus detection (HIV-1 in the PC) by mNGS but did not affect detection sensitivity for DNA pathogens. Moderate to low levels of hemolysis (pink to light red CSF) did not affect detection sensitivity for any of the PC organisms. Analysis of spiked samples containing S. aureus and S. epidermidis with equivalent RPM-r values at baseline demonstrated accurate discrimination of species within the same genus when mixed at 1:1, 4:1, and 1:4 ratios, as both species were correctly identified and calculated RPM-r values were within 7% of that expected on the basis of the spiked proportions.
Stability
Analysis of replicates of the PC held at 4°C for 0, 2, 5, and 6 d and subjected to three freeze-thaw cycles demonstrated detection of all organisms (Table 1).
Challenge study
Blinded evaluation of the mNGS assay was performed using a set of 20 CSF samples collected prospectively from pediatric patients hospitalized at Children's Hospital Colorado (CHCO) with meningitis, encephalitis, and/or myelitis (Supplemental Table S3). The overall sensitivity and specificity of mNGS relative to conventional clinical microbiology testing (culture, serology, and/or PCR) were 92% and 96%, respectively. The causative pathogen was correctly identified in 11 of 12 previously positive cases, including cases of enterovirus (n = 8), HSV-1 (n = 1), HIV-1 (n = 1), and WNV (n = 1). The mNGS assay failed to detect WNV in a second patient with positive CSF IgM serology. Three additional organisms (Enterobacter sp., Corynebacterium sp., and EBV) were detected by mNGS testing, each from a different sample. The detection of Enterobacter sp. and Corynebacterium sp. were classified as false-positives, since the two samples had previously tested negative by CSF culture. The patient with positive CSF mNGS testing for EBV was also positive for EBV IgG antibodies in blood; however, this finding was excluded from the comparison due to the lack of residual CSF sample volume for confirmation. In addition, mNGS failed to detect organisms in four cases presumptively diagnosed by testing at sites other than CSF, including one case of Borrelia burgdorferi (blood serology), two cases of Mycoplasma encephalitis (PCR-positive respiratory but PCR-negative CSF samples), and one case of enterovius 71 infection (positive rectal swab culture but negative CSF PCR). These four cases were also excluded from the comparison, as the diagnosis had not been made directly from CSF. Negative mNGS results were concordant with negative clinical testing in four undiagnosed cases, including one case of culture-negative bacterial meningitis and three cases of idiopathic encephalitis.
Discussion
We developed and analytically validated a clinical CSF mNGS assay intended to aid in the diagnosis of infectious etiologies of meningitis, encephalitis, and/or myelitis in hospitalized patients. The mNGS assay has been subsequently evaluated for clinical utility in a 1-yr prospective diagnostic trial in hospitalized patients presenting acutely with suspected neurological infection (Wilson et al. 2019). As CSF is considered a normally sterile site, we postulated that interpretation of CSF mNGS data would be more straightforward than data from nonsterile sites such as respiratory secretions and stool. However, numerous challenges had to be overcome for successful implementation of mNGS for broad-spectrum pathogen detection in the clinical laboratory. First, a universal sequencing library preparation protocol employing two rounds of PCR was developed, robust across the wide range of human host background seen in patient CSF (0–108 leukocytes/mm3). Second, QC materials were incorporated, including external PC and NTC samples that are run in parallel with CSF samples, as well as internally spiked RNA and DNA controls. Third, reproducible threshold metrics were established and evaluated using ROC curve analysis to enable correct identification of pathogens from mNGS data above background noise and minimize false-positive results. The final clinical mNGS protocol incorporated (1) a bead-beating step for complete lysis of microbial cell walls, (2) separate construction of RNA and DNA libraries from nucleic acid extracts for detection of RNA viruses and DNA-based microorganisms, respectively, and (3) SURPI+ bioinformatics analysis using the entirety of the NCBI GenBank nt database as a comprehensive reference database.
Our clinical mNGS library preparation protocol uses a transposon-based approach (Nextera). A key advantage of this approach is the ease of use and rapid turnaround time for the protocol, which is amenable to routine clinical laboratory workflows. This method has been shown to exhibit a GC bias with respect to sequenced reads (Lan et al. 2015). However, here we tested representative organisms with GC content ranging from 35.4% to 57.4%, core genomes tend to have a lower GC bias (Bohlin et al. 2018), and detection by mNGS remains possible from less-biased genomic regions. Of note, we have not observed an appreciable difference in mNGS assay sensitivity using adapter ligation-based kits (Naccache et al. 2014; Luk et al. 2015). Nevertheless, further studies will be needed to establish whether mNGS is able to detect organisms at the extremes of GC content.
Based on the PC mix of seven representative organisms, assay limits of detection ranged from from 0.2 CFU/mL for C. neoformans to 313 copies/mL for HIV. Metagenomic sensitivity for detection of a given organism is dependent on a number of factors, including extraction efficiency, size of the genome, library preparation bias, and availability of matching reference genomes in the database. We believe that C. neoformans was detected at lower levels due to a number of factors. The relative large eukaryotic genome size (∼19 Mb) increases the amount of pathogen relative to human DNA, even for low numbers of organisms. C. neoformans may also be more susceptible to lysis than organisms with more rigid cell walls such as Aspergillus niger, so that more DNA may be released after bead-beating and hence available for sequencing. In addition, the complete sequences of all 14 C. neoformans chromosomes were available in the NCBI GenBank nt database (March 2015 build) used by SURPI+, unlike for Toxoplasma gondii, with only representation of a limited subset of genes or gene regions.
Given the untargeted nature of mNGS, a key limitation for infectious disease diagnostics is background interference, generally from human host DNA. In addition to controlling for nucleic acid extraction efficiency, the use of a spiked phage IC was found to be useful for assessing whether high host background was present, indicating decreased sensitivity of pathogen detection by mNGS. Overall, 27.4% of DNA libraries and 6.3% of RNA libraries in the accuracy study had fewer than 100 RPM IC phage reads recovered, making background interference a fairly common limitation. Thus, in high-background samples, negative mNGS findings may be less useful for excluding infection, and other diagnostic tests that are less sensitive to background should be considered, such as 16S rRNA bacterial PCR (Salipante et al. 2013) and ITS fungal PCR (Pryce et al. 2006). This is especially relevant in cases of bacterial meningitis with high leukocyte counts in CSF. Despite this limitation, mNGS was still able to detect bacterial pathogens in 12 of 19 culture-positive samples in the accuracy study with high host background.
The overall accuracy of the mNGS assay for pathogen detection relative to conventional clinical testing was 90%, with 73% sensitivity and 99% specificity. The calculated 73% sensitivity refers to clinical sensitivity—sensitivity in diagnosis of infection, and not analytical sensitivity—sensitivity in detection of pathogen nucleic acid. Factors impacting the clinical sensitivity of mNGS include (1) cases diagnosed only by serology (e.g., WNV), (2) use of clinical testing as an imperfect “gold standard,” with some samples possibly representing false-positive detections from contamination (e.g., Enterococcus faecalis with growth from broth only), (3) analysis of remnant biobanked clinical samples for mNGS accuracy testing, with degradation from prior freeze-thaw steps likely decreasing sensitivity, (4) use of robust pre-established thresholds to minimize false-positive detections, and (5) role of human host background (e.g., high CSF pleocytosis) in limiting sensitivity. Among the eight of 21 mNGS false-negative cases with sufficient remaining CSF volume, follow-up discrepancy PCR testing was negative for five of eight (62.5%), suggesting that sample degradation may have occurred over time or that the original clinical result was incorrect. Indeed, positive percent agreement rose to 81% after discrepancy testing of samples with sufficient volume, and exclusion of samples with high host background increased this further to 89%.
Only a fraction of all possible diagnostic tests for pathogens are performed in clinical microbiology laboratories given cost, limited CSF sample volume, and long turnaround times for reference (send out) laboratory testing. We decided to exclude additional organism detections by mNGS (n = 18) in the initial assessment of specificity, as no clinical reference result was available. However, in nine cases out of 18 with sufficient CSF volume for discrepancy testing, all nine were found to be analytical true-positives. Furthermore, an additional three cases had peripheral blood serology results consistent with neurological infection by the organism detected by mNGS (Table 3). Thus, at least 12 of 18 (66.7%) additional organisms identified by mNGS are likely true-positive detections, with only one of 18 (5.6%), a culture-negative case positive by mNGS for Bacillus sp., classified as a false-positive.
As with any diagnostic assay, mNGS testing is prone to contamination. Often, the identity of the species detected can provide clues as to the contamination source, such as skin flora (e.g., S. epidermidis, papillomaviruses), laboratory reagents (murine gammaretroviruses, E. coli, insect viruses), body flora (e.g., anelloviruses), or environmental flora (e.g., Thermus sp., Bacillus sp.). Cross-contamination in particular is a major concern given that the mNGS protocol involves PCR amplification. Strict processing controls to minimize contamination are essential and include unidirectional workflow, positive pressure ventilation in pre-amplification areas, and workspace separation for different assay steps. To monitor for contamination, we also developed standardized protocols for QC testing of new reagents and periodic swipe testing of instruments and laboratory surfaces (Supplemental Methods). Continual tracking of contaminants seen in the NTC or PC is also done, and conservative threshold criteria are used to minimize the reporting of false-positive results. The development and use of ultraclean reagents with no or extremely low levels of DNA contamination may also help in minimizing assay contamination (Motley et al. 2014).
Approximately 7% of the clinical samples in the accuracy study had multiple bacterial genera detected above pre-established thresholds, generally consisting of environmental or skin flora. The challenge of determining the clinical significance of detecting organisms that may be contaminants is a classical problem in clinical microbiology and often requires clinical context for interpretation. As CSF is a normally sterile site, rarely are bacterial or fungal co-infections causative for cases of meningitis or encephalitis, with the possible exception of foreign body infections or polymicrobial brain abscesses communicating with CSF (Martin et al. 2018). Thus, detection of multiple bacterial and/or fungal genera are noted in the mNGS results report as probable sample contamination and were considered negative in our evaluation of assay performance.
The challenge study evaluated CSF mNGS testing as a first-line diagnostic assay for neurological infections and demonstrated that mNGS detected the same organism identified via conventional microbiological testing of CSF in 11 of 12 (91.7%) cases. The missed case of WNV was diagnosed by serologic IgM testing of CSF. This case and a presumptive Lyme disease case diagnosed by serologic testing of peripheral blood underscore the critical dependence of mNGS detection on the presence of nucleic acid from the organism at the time of sample collection. As a direct detection method, mNGS can miss infections that are often only successfully diagnosed using serology (e.g., WNV, Lyme neuroborreliosis, and neurosyphilis), given that the causative pathogen may be absent or only transiently present in CSF. For these cases, direct detection testing approaches such as PCR and mNGS lack sensitivity, and there should be a low threshold for indirect serologic testing or laboratory testing from other body sites to establish the diagnosis (DeBiasi and Tyler 2004).
While mNGS testing can provide broad-spectrum pathogen identification, assessment of the clinical significance of the reported findings may require interpretation. Direct discussions or teleconferences can be set up with treating clinicians to clarify and review mNGS results in clinical context. These forums can also be used to communicate results of secondary analyses of mNGS data, including (1) genome assembly for characterization of predicted antibiotic or antiviral resistance mutations, (2) phylogenetic analysis for genotyping and strain-level identification, and (3) disclosure of reads from potential pathogens below formal reporting thresholds. Thus, the clinical relevance of mNGS findings can be efficiently communicated to physicians, potentially informing the next steps in management and treatment of the patient, and may also prove informative for public health surveillance and outbreak investigation (Chiu et al. 2017).
Methods
mNGS assay
We developed standard operating procedures (SOPs) in the clinical laboratory for processing and analyzing CSF samples by mNGS. Each of the “wet lab” and bioinformatics processing steps was optimized to ensure sensitive and accurate organism detection (Schlaberg et al. 2017a). The mNGS assay workflow was performed as follows (Fig. 1), with a more detailed description provided in the Supplemental Methods. Briefly, each CSF sample was first subjected to bead-beating to lyse organisms (Fig. 1A), followed by addition (“spiking”) of T1 (DNA) and MS2 (RNA) bacteriophages as an internal control (IC). Total nucleic acid was then extracted and split into two aliquots for construction of separate DNA and RNA libraries. Microbial sequences were enriched by antibody-based removal of methylated host DNA (for DNA libraries) or DNase treatment (for RNA libraries), followed by transposon-based library construction (Fig. 1B). Each sequencing run on an Illumina HiSeq instrument included up to eight samples, along with a negative “no template” control consisting of elution buffer, intended to allow for sensitive detection of contamination, and a positive control consisting of a mixture of seven representative pathogenic organisms (RNA virus, DNA virus, Gram-positive bacterium, Gram-negative bacterium, fungus, mold, and parasite).
Sequence analysis was performed using the SURPI+ computational pipeline (Fig. 1C; Supplemental Methods), an automated clinical version of the previously published SURPI (“sequence-based ultrarapid pathogen identification”) research pipeline (Naccache et al. 2014). Receiver-operator curve analyses were performed as part of the accuracy study to determine optimal threshold values for organism detection (Supplemental Methods), using 95 clinical CSF samples with established microbiological results. These pre-established thresholds were then finalized and used for all subsequent clinical mNGS runs. Each mNGS run was analyzed by experienced laboratory physicians (S.M. and C.Y.C.), and results were generated for five categories per sample (RNA virus, DNA virus, bacteria, fungi, and parasite). Run quality control (QC) metrics included a minimum of 5 million reads per library, ≥100 reads per million for the IC T1 and MS2 phages in the DNA and RNA libraries, respectively, and positive qualitative detection of each of the seven organisms in the PC.
Evaluation of mNGS analytical performance characteristics
A detailed description of the methods used to evaluate mNGS analytical performance characteristics is provided in the Supplemental Methods. Briefly, limits of detection were determined for each of the seven representative organisms in the PC by probit analysis using a series of dilutions across a minimum 4-log range. Precision was determined using repeat analysis of the PC and NTC over 20 consecutive sequencing runs (inter-assay reproducibility) and three sets of separate PCs and NTCs processed in parallel on the same run (intra-assay reproducibility). Test stability was determined using control samples held at various temperatures and subject to multiple freeze/thaw cycles. Interference was determined using PC spiked with known amounts of human DNA or RNA material. Results were assessed for qualitative detection of organisms in the PC.
Accuracy was determined using 95 clinical CSF samples comprising 73 positive samples containing 79 detected organisms in total and 22 negative samples (Fig. 3A). Samples were obtained from patients at the University of California, San Francisco (UCSF) (n = 59), Children's National Medical Center (CNMC) (n = 19), Children's Hospital Colorado (CHCO) (n = 1), and Quest Diagnostics (n = 16). Three composite reference standards were generated for comparisons of mNGS assay performance with “gold standard” clinical microbiological testing (Fig. 3B): (1) original clinical testing results; (2) combined results from original clinical testing and additional discrepancy testing of initial false-negative or false-positive samples for which sufficient residual CSF volume was available; and (3) original and discrepancy testing results after exclusion of samples with high background corresponding to the human host (see “Interference,” above). The second and third comparisons are reported as positive percent agreement (PPA) and negative percent agreement (NPA), as selective discrepancy testing can bias estimates of test sensitivity and specificity (U.S. Food and Drug Administration 2007). To evaluate mNGS detection performance for additional organism types not readily available from clinical CSF samples, the accuracy study also included mNGS testing of contrived samples of five known organisms (N. meningitidis, S. agalactiae, C. albicans, M. fortuitum, M. abscessus) spiked into negative CSF at defined concentrations.
Challenge study
The Aseptic Meningitis and Encephalitis Study (AMES) is a prospective cohort study enrolling children presenting to CHCO with culture-negative meningitis and encephalitis since 2012. A subset of CSF samples (n = 20) with sufficient residual volume of 600 µL from subjects with known and unknown etiologies was coded for mNGS testing as a challenge set. Samples were processed in a blinded fashion at UCSF and results discussed in clinical context with site investigators at CHCO over web-based teleconferencing.
Software availability
The SURPI+ computational pipeline software used by the UCSF clinical CSF mNGS assay consists of publicly available source code and proprietary binaries and is available for download on GitHub (https://github.com/chiulab/SURPI-plus-dist). Shell scripts to reconstruct the essential NCBI GenBank nt reference databases using the alignment programs (SNAP [Naccache et al. 2014], Bowtie 2 [Langmead and Salzberg 2012], and BLASTn [Altschul et al. 1990]) are also provided in this distribution. Note that the databases used are from the March 2015 distribution of GenBank nt and use GI instead of accession numbers. The source code includes the following external open-source tools: BLAST v2.7.1 (Altschul et al. 1990), Bowtie 2 v2.3.2 (Langmead and Salzberg 2012), cutadapt v1.2.1 (Martin 2011), PRINSEQ-lite v0.20.3 (Schmieder and Edwards 2011), and SNAP v0.15.4 (Naccache et al. 2014).
Data access
Metagenomic reads from patient CSF samples from this study were depleted of human host sequences and have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA516289. Sequences corresponding to the HIV-1 and CMV controls in the PC and the MS2 (RNA) phage and T1 (DNA) phage spiked IC samples from this study have been submitted to NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MK214316, MK213797, MK213795, and MK213796, respectively.
Competing interest statement
C.Y.C. is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center (VDDC) and receives research support from Abbott Laboratories, Inc. C.Y.C., S.A., D.S., S.F., and S.M. are inventors on a patent application on algorithms related to SURPI+ software titled “Pathogen Detection using Next-Generation Sequencing” (PCT/US/16/52912).
Supplementary Material
Acknowledgments
We thank the staff of the UCSF Clinical Immunology lab for their help in discrepancy testing and Gail Cunningham for her expert help in obtaining AFB organisms. We also thank Brittany Goldberg at CNMC for providing clinical samples for use in the validation. This work is supported by National Institutes of Health (NIH) grants R01 HL105704 and R21/R33 AI120977 (C.Y.C.), a UC Center for Accelerated Innovation grant funded by NIH grant U54 HL119893 and NIH NCATS UCSF-CTSI grant UL1 TR000004 (C.Y.C.), the California Initiative to Advance Precision Medicine (C.Y.C. and S.M.), research support from Abbott Laboratories, Inc. (C.Y.C.), and supplemental funding from the UCSF Medical Center (C.Y.C. and S.M.).
Author contributions: C.Y.C., S.M., E.S., and S.N.N. developed the project. E.S., E.P., S.A., B.F., and W.L. generated libraries. S.N.N., S.M., and C.Y.C. analyzed data for clinical validation. S.F., D.S., and C.Y.C. developed SURPI+ software and a graphical user interface for clinical use. W.J.B. modified the SNAP algorithm to facilitate taxonomic classification by SURPI+, and S.M.P. banked CSF samples. J.A.L., S.D., K.M., S.M., and C.Y.C. provided clinical specimens. K.M., S.D., J.A.L., S.N.N., S.M.P., C.Y.C., and S.M. conducted chart review. D.I., B.F., and S.A. conducted discrepancy testing. S.M., S.N.N., and C.Y.C. wrote the manuscript with contributions from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.238170.118.
Freely available online through the Genome Research Open Access option.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bohlin J, Eldholm V, Brynildsrud O, Petterson JH, Alfsnes K. 2018. Modeling of the GC content of the substituted bases in bacterial core genomes. BMC Genomics 19: 589 10.1186/s12864-018-4984-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazanave C, Greenwood-Quaintance KE, Hanssen AD, Karau MJ, Schmidt SM, Gomez Urena EO, Mandrekar JN, Osmon DR, Lough LE, Pritt BS, et al. 2013. Rapid molecular microbiologic diagnosis of prosthetic joint infection. J Clin Microbiol 51: 2280–2287. 10.1128/JCM.00335-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CY. 2013. Viral pathogen discovery. Curr Opin Microbiol 16: 468–478. 10.1016/j.mib.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C, Miller S. 2016. Next-generation sequencing. In Molecular microbiology: diagnostic princples and practice, 3rd ed. (ed. Persing DH, et al. ), pp. 68–79. ASM Press, Washington, DC. [Google Scholar]
- Chiu CY, Coffey LL, Murkey J, Symmes K, Sample HA, Wilson MR, Naccache SN, Arevalo S, Somasekar S, Federman S, et al. 2017. Diagnosis of fatal human case of St. Louis encephalitis virus infection by metagenomic sequencing, California, 2016. Emerg Infect Dis 23: 1964–1968. 10.3201/eid2310.161986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Rodriguez-Del Valle N, Perez-Sanchez L, Abouelleil A, Goldberg J, Young S, Zeng Q, Birren BW. 2014. Genome sequence of the pathogenic fungus Sporothrix schenckii (ATCC 58251). Genome Announc 2: e00446-14 10.1128/genomeA.00446-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBiasi RL, Tyler KL. 2004. Molecular methods for diagnosis of viral encephalitis. Clin Microbiol Rev 17: 903–925. 10.1128/CMR.17.4.903-925.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguière A, Manuguerra JC, et al. 2012. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7: e38499 10.1371/journal.pone.0038499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frémond ML, Pérot P, Muth E, Cros G, Dumarest M, Mahlaoui N, Seilhean D, Desguerre I, Hébert C, Corre-Catelin N, et al. 2015. Next-generation sequencing for diagnosis and tailored therapy: a case report of astrovirus-associated progressive encephalitis. J Pediatric Infect Dis Soc 4: e53–e57. 10.1093/jpids/piv040 [DOI] [PubMed] [Google Scholar]
- Glaser CA, Gilliam S, Schnurr D, Forghani B, Honarmand S, Khetsuriani N, Fischer M, Cossen CK, Anderson LJ, California Encephalitis Project 1998–2000. 2003. In search of encephalitis etiologies: diagnostic challenges in the California Encephalitis Project, 1998–2000. Clin Infect Dis 36: 731–742. 10.1086/367841 [DOI] [PubMed] [Google Scholar]
- Glaser CA, Honarmand S, Anderson LJ, Schnurr DP, Forghani B, Cossen CK, Schuster FL, Christie LJ, Tureen JH. 2006. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis 43: 1565–1577. 10.1086/509330 [DOI] [PubMed] [Google Scholar]
- Granerod J, Ambrose HE, Davies NW, Clewley JP, Walsh AL, Morgan D, Cunningham R, Zuckerman M, Mutton KJ, Solomon T, et al. 2010. Causes of encephalitis and differences in their clinical presentations in England: a multicentre, population-based prospective study. Lancet Infect Dis 10: 835–844. 10.1016/S1473-3099(10)70222-X [DOI] [PubMed] [Google Scholar]
- Greninger AL, Messacar K, Dunnebacke T, Naccache SN, Federman S, Bouquet J, Mirsky D, Nomura Y, Yagi S, Glaser C, et al. 2015. Clinical metagenomic identification of Balamuthia mandrillaris encephalitis and assembly of the draft genome: the continuing case for reference genome sequencing. Genome Med 7: 113 10.1186/s13073-015-0235-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Miller S, Chiu CY. 2018. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol 14: 319–338. 10.1146/annurev-pathmechdis-012418-012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetsuriani N, Holman RC, Anderson LJ. 2002. Burden of encephalitis-associated hospitalizations in the United States, 1988–1997. Clin Infect Dis 35: 175–182. 10.1086/341301 [DOI] [PubMed] [Google Scholar]
- Lan JH, Yin Y, Reed EF, Moua K, Thomas K, Zhang Q. 2015. Impact of three Illumina library construction methods on GC bias and HLA genotype calling. Hum Immunol 76: 166–175. 10.1016/j.humimm.2014.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Berg MG, Naccache SN, Kabre B, Federman S, Mbanya D, Kaptué L, Chiu CY, Brennan CA, Hackett J Jr. 2015. Utility of metagenomic next-generation sequencing for characterization of HIV and human pegivirus diversity. PLoS One 10: e0141723 10.1371/journal.pone.0141723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi F, Bendinelli M. 2010. Human anelloviruses and the central nervous system. Rev Med Virol 20: 392–407. 10.1002/rmv.668 [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martin RM, Zimmermann LL, Huynh M, Polage CR. 2018. Diagnostic approach to health care- and device-associated central nervous system infections. J Clin Microbiol 56: e00861-18 10.1128/JCM.00861-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolrattanothai K, Naccache SN, Bender JM, Samayoa E, Pham E, Yu G, Dien Bard J, Miller S, Aldrovandi G, Chiu CY. 2017. Neurobrucellosis: unexpected answer from metagenomic next-generation sequencing. J Pediatric Infect Dis Soc 6: 393–398. 10.1093/jpids/piw066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley ST, Picuri JM, Crowder CD, Minich JJ, Hofstadler SA, Eshoo MW. 2014. Improved multiple displacement amplification (iMDA) and ultraclean reagents. BMC Genomics 15: 443 10.1186/1471-2164-15-443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, Bloom K, Delwart E, Nelson KE, Venter JC, et al. 2017. The blood DNA virome in 8,000 humans. PLoS Pathog 13: e1006292 10.1371/journal.ppat.1006292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, Bouquet J, Greninger AL, Luk KC, Enge B, et al. 2014. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res 24: 1180–1192. 10.1101/gr.171934.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, Grant P, Samayoa E, Federman S, Miller S, Lunn MP, et al. 2015. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 60: 919–923. 10.1093/cid/ciu912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parize P, Muth E, Richaud C, Gratigny M, Pilmis B, Lamamy A, Mainardi JL, Cheval J, de Visser L, Jagorel F, et al. 2017. Untargeted next-generation sequencing-based first-line diagnosis of infection in immunocompromised adults: a multicentre, blinded, prospective study. Clini Microbiol Infect 23: 574 10.1016/j.cmi.2017.02.006 [DOI] [PubMed] [Google Scholar]
- Pryce TM, Palladino S, Price DM, Gardam DJ, Campbell PB, Christiansen KJ, Murray RJ. 2006. Rapid identification of fungal pathogens in BacT/ALERT, BACTEC, and BBL MGIT media using polymerase chain reaction and DNA sequencing of the internal transcribed spacer regions. Diagn Microbiol Infect Dis 54: 289–297. 10.1016/j.diagmicrobio.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Quinn RA, Navas-Molina JA, Hyde ER, Song SJ, Vazquez-Baeza Y, Humphrey G, Gaffney J, Minich JJ, Melnik AV, Herschend J, et al. 2016. From sample to multi-omics conclusions in under 48 hours. mSystems 1: e00038-16 10.1128/mSystems.00038-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salipante SJ, Sengupta DJ, Rosenthal C, Costa G, Spangler J, Sims EH, Jacobs MA, Miller SI, Hoogestraat DR, Cookson BT, et al. 2013. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One 8: e65226 10.1371/journal.pone.0065226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12: 87 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, Breitwieser FP, Kumar A, Hao H, Burger P, Rodriguez FJ, Lim M, Quiñones-Hinojosa A, Gallia GL, Tornheim JA, et al. 2016. Next-generation sequencing in neuropathologic diagnosis of infections of the nervous system. Neurol Neuroimmunol Neuroinflamm 3: e251 10.1212/NXI.0000000000000251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists. 2017a. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 141: 776–786. 10.5858/arpa.2016-0539-RA [DOI] [PubMed] [Google Scholar]
- Schlaberg R, Queen K, Simmon K, Tardif K, Stockmann C, Flygare S, Kennedy B, Voelkerding K, Bramley A, Zhang J, et al. 2017b. Viral pathogen detection by metagenomics and pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J Infect Dis 215: 1407–1415. 10.1093/infdis/jix148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simner PJ, Miller S, Carroll KC. 2018. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis 66: 778–788. 10.1093/cid/cix881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ, Xu G, Morici L, Splinter Bon-Durant S, Baddoo M, Lin Z, Fewell C, Taylor CM, Flemington EK. 2014. Microbial contamination in next generation sequencing: implications for sequence-based analysis of clinical samples. PLoS Pathog 10: e1004437 10.1371/journal.ppat.1004437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration. 2007. Guidance for industry and FDA staff: statistical guidance on reporting results from studies evaluating diagnostic tests. U.S. Food and Drug Administration, Silver Spring, MD: https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm071287.pdf. [Google Scholar]
- Washington JA. 1996. Principles of diagnosis. In Medical microbiology, 4th ed. (ed. Baron S). University of Texas Medical Branch at Galveston, Galveston, TX. [PubMed] [Google Scholar]
- Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, et al. 2014. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 370: 2408–2417. 10.1056/NEJMoa1401268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Fedewa G, Stenglein MD, Olejnik J, Rennick LJ, Nambulli S, Feldmann F, Duprex WP, Connor JH, Mühlberger E, et al. 2016. Multiplexed metagenomic deep sequencing to analyze the composition of high-priority pathogen reagents. mSystems 1: e00058-16 10.1128/mSystems.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Sample HA, Zorn KC, Arevalo S, Yu G, Neuhaus J, Federman S, Stryke D, Briggs B, Langelier C, et al. 2019. Clinical metagenomic next-generation sequencing for diagnosis of infectious meningitis and encephalitis. N Engl J Med (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Jia H, Shankar A, Heneine W, Switzer WM. 2011. Detection of murine leukemia virus or mouse DNA in commercial RT-PCR reagents and human DNAs. PLoS One 6: e29050 10.1371/journal.pone.0029050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.