Abstract
Neuraminidase inhibitors (NAIs) are first‐line agents for the treatment and prevention of influenza virus infections. As for other antivirals, the development of resistance to NAIs has become an important concern particularly in the case of A(H1N1) viruses and oseltamivir. The most frequently reported change conferring oseltamivir resistance in that viral context is the H275Y neuraminidase mutation (N1 numbering). Recent studies have shown that, in the presence of the appropriate permissive mutations, the H275Y variant can retain virulence and transmissibility in some viral backgrounds. Most oseltamivir‐resistant influenza A virus infections can be managed with the use of inhaled or intravenous zanamivir, another NAI. New NAI compounds and non‐neuraminidase agents as well as combination therapies are currently in clinical evaluation for the treatment for severe influenza infections.
Keywords: Antiviral, influenza, neuraminidase inhibitor, oseltamivir, resistance, zanamivir
Classes of antiviral agents for influenza virus infections
There are two classes of antiviral agents approved for the treatment and prevention for influenza viruses: the adamantanes and the neuraminidase inhibitors. The adamantane compounds amantadine and rimantadine act by blocking the M2 ion channel preventing viral uncoating. Influenza B viruses are intrinsically resistant to these compounds due to their lack of M2 target protein. In 1999–2000, two neuraminidase inhibitors (NAIs) were approved in many countries: the oral agent oseltamivir and the inhaled compound zanamivir. Both antiviral drugs are active against all influenza A virus subtypes as well as influenza B viruses.1 The NAIs prevent the cleavage of the terminal sialic acid residues on budding virions, a process that prevents infection of new host cells and thereby blocks virus dissemination throughout the respiratory tract. Due to high levels of resistance to the M2 blockers, the NAIs and especially oseltamivir have become the agents of choice for the treatment of individuals with severe influenza infections and for those with underlying diseases predisposing to influenza complications. As it is the case for other antivirals, the development of drug resistance is an important issue that can potentially limit the usefulness of NAIs.
Risk factors and incidence of resistance to anti‐influenza drugs
Resistance to the adamantanes has been shown to develop rapidly, that is, within 3–5 days in 30–50% of treated immunocompetent and immunocompromised individuals.2 Furthermore, resistance to this class of agents has emerged in 2004 among A(H3N2) viruses and some A(H1N1) viruses, in the absence of antiviral pressure. Also, all A(H1N1)pdm09 viruses that circulated during the 2009 influenza pandemic and up to now have been intrinsically resistant to the adamantanes. Thus, at the present time, virtually, all circulating influenza A viruses recovered from humans are resistant to these compounds.
It was previously thought that resistance to NAIs would not be an important clinical problem because the neuraminidase (NA) is a critical enzyme in the virus replicative cycle, and previous oseltamivir‐resistant viruses were found to be unfit and poorly transmissible in animal models.3 Thus, the emergence and predominance of an oseltamivir‐resistant A(H1N1) virus (A/Brisbane/59/2007, clade 2B) between 2007 and 2009 came as a surprise.4 It was subsequently shown that the good fitness of this oseltamivir‐resistant viral strain containing the H275Y (N1 numbering; H274Y in N2 numbering) NA resistance mutation was due to the presence of pre‐existing permissive NA mutations such as R222Q (N1 numbering) that increased both the activity and the surface expression of the NA.5, 6 The A/Brisbane/59/2007 strain was no longer detected after the emergence of the A(H1N1)pdm09 virus in 2009. Most circulating A(H1N1)pdm09, A(H3N2) and B viruses remain susceptible to oseltamivir with <1·5% of tested strains exhibiting phenotypic or genotypic evidence of resistance in 2011–2012.7 However, recent outbreaks of oseltamivir‐resistant A(H1N1)pdm09 viruses in Australian citizens and Dutch travelers returning from Spain have been reported in the absence of drug treatment and are reminders of the importance of continuous antiviral susceptibility monitoring.8, 9, 10 In those cases, a new set of permissive NA mutations (such as N369K and V241I in N1 numbering) may have facilitated the emergence of the H275Y resistance mutation and improved virus transmissibility. Factors associated with the selection of drug resistance at the individual level include: the use of post‐exposure prophylaxis (with the administration of lower drug dose), infection of an immunocompromised host and prolonged antiviral treatment. During the first wave of the 2009 influenza pandemic, up to 25–30% of oseltamivir‐resistant cases were reported in diverse immunocompromised individuals, and thus, immunosuppression represents the most important setting where resistance can develop.11 Of importance, during the 2011–2012 influenza season in USA, up to 74% of oseltamivir resistance cases were not associated with drug exposure,12 and this may reflect the emergence of a set of permissive NA mutations as described previously.10 A similar trend had been previously noted in 2010–2011 in United Kingdom.13 Fortunately, resistance to zanamivir has remained extremely rare in all influenza subtypes.
Assays for detecting antiviral resistance
Similar to other viruses, phenotypic and genotypic assays can be used for assessing resistance to anti‐influenza compounds. Phenotypic assays first require viral propagation and then subsequent determination of the drug 50% inhibitory concentration (IC50) value. For the adamantanes, IC50 values are assessed by the conventional plaque reduction assay. For the NAIs, enzymatic assays are preferred to plaque assays and have been shown to more reliably estimate drug susceptibilities.14 Different types of NA assays can be performed using chemiluminescent, fluorescent, or colorimetric NA substrates. The fluorometric assay allows a better discrimination between susceptible and resistant viruses, whereas the chemiluminescent assay needs less input virus for testing.15 There exists no standard definition of NAI resistance, but recent WHO guidelines have been proposed: reduced inhibition is defined by 10‐ to 100‐fold and 5‐ to 50‐fold increases in IC50 values for influenza A and B virus isolates, respectively, whereas highly reduced inhibition is defined by >100‐fold and >50‐fold increases in IC50 values.16
Because of the time needed to grow the isolates and to determine drug IC50 values, many laboratories are performing some types of genotypic assays to detect drug resistance mutations directly from the clinical samples after a RT‐PCR amplification step. Although more rapid than phenotypic assays, these tests do not determine the level of resistance and are of limited utility in case a novel NA mutation is identified in the absence of susceptibility test results. The primary approach for genotypic testing is to amplify by RT‐PCR the targeted gene, that is, M2 in the case of the adamantanes and NA for the NAIs, followed by conventional DNA Sanger sequencing. This strategy is comprehensive, potentially detecting all mutations associated with drug resistance, but it suffers from a lack of sensitivity for detection of minor variants within a viral population. Indeed, a mutant variant must be in excess of 15–20% of the total population to be identified by conventional DNA sequencing. The advent of pyrosequencing and especially next‐generation ultra‐deep sequencing has allowed the detection of minor variants in excess of 1–2%.17 Such unprecedented level of detection has improved our understanding of the evolution of drug resistance by showing the presence of drug‐resistant variants in samples of individuals before the onset of therapy and the transmission of those drug‐resistant variants along with drug‐susceptible viruses between hosts.17 This novel information is of great importance for predicting the speed at which resistance will arise and also to gain insight into the relative fitness of some drug‐resistant mutants. However, despite its undoubted potential, deep sequencing also has a number of inherent analytical difficulties including the generation of short sequence reads that could be difficult to link together as well as the problem of PCR and/or sequencing artifacts.
Mutations conferring drug resistance
Genotypic analysis of resistance to M2 blockers is relatively straightforward as only a few substitutions occurring at five codons within the M2 gene (codons 26, 27, 30, 31 and 34) have been linked to amantadine/rimantadine resistance.18 Importantly, these M2 mutants are fully virulent and transmissible between humans.19 The NA mutations conferring resistance to NAIs vary according to the viral subtype/type and the NAI20 (Table 1). In the N1 subtype, the most frequently encountered mutation is the H275Y that confers highly reduced inhibition to oseltamivir, moderate cross‐resistance to the investigational agent peramivir and susceptibility to zanamivir.21 Various amino acid changes at residue 223 (I →R/V) can also confer reduced inhibition to oseltamivir and/or to zanamivir.22, 23 In the N2 subtype, the most frequent mutations conferring highly reduced inhibition to oseltamivir are E119V and R292K, the latter being also associated with reduced inhibition to zanamivir.24, 25 Reported changes associated with NAI resistance in B viruses include mainly R150K and D197N.26, 27 Of note, some NA mutations reported at codons 136 and 151 have a questionable clinical impact because they have been almost exclusively detected after cell passages and rarely in primary clinical samples.28, 29, 30 Finally, very few zanamivir‐resistant influenza viruses have been reported in clinical samples so far which may be explained by the modest use of this inhaled antiviral and also possibly by a higher genetic barrier for resistance due to a greater structural homology to the natural substrate, sialic acid.31
Table 1.
Selected neuraminidase mutations conferring resistance to neuraminidase inhibitors (mainly reviewed in52)
| Influenza subtype | NA mutation | Virus source / NAI used for selection | Phenotype in NA inhibition assaysa | ||
|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | |||
| A(H1N1) | H275Y | Clinic / Oseltamivir | HRI | S | HRI |
| Q136K | In vitro (clinic?) / None | S | HRI | RI | |
| A(H1N1)pdm09 | N295S | Reverse Genetics | HRI | S | RI |
| H275Y | Clinic / Oseltamivir | HRI | S | RI | |
| S247N/H275Y | Clinic / None | HRI | S | RI | |
| I223V/H275Y | Clinic / Oseltamivir | HRI | S | ‐ | |
| Reverse Genetics | HRI | S | HRI | ||
| I223R/H275Y | Clinic / Oseltamivir | HRI | RI | HRI | |
| Reverse Genetics | HRI | RI | HRI | ||
| I223R | Clinic / Oseltamivir | RI | RI | ‐ | |
| Reverse Genetics | RI | RI | RI | ||
| E119G | Reverse Genetics | S | HRI | RI | |
| E119V | Reverse Genetics | RI | HRI | RI | |
| A(H5N1) | N295S | Clinic / Oseltamivir | RI | S | S |
| H275Y | Clinic / Oseltamivir | HRI | S | HRI | |
| D199G | In vitro / Zanamivir | RI | RI | S | |
| E119G | In vitro / Zanamivir | S | HRI | RI/HRI | |
| A(H3N2) | N294S | Clinic / Oseltamivir | HRI | S | ‐ |
| R292K | Clinic / Oseltamivir | HRI | ‐ | ‐ | |
| Reverse Genetics | HRI | S/RI | RI | ||
| Deletion 245–248 | Clinic / Oseltamivir | HRI | S | S | |
| D151A/D | Clinic?/ None | S | HRI | ‐ | |
| Q136K | Clinic?/ None | S | RI | ‐ | |
| E119V/I222V | Clinic / Oseltamivir | HRI | S | S | |
| E119V | Clinic / Oseltamivir | HRI | S | S | |
| B | R371K | Clinic/ None | HRI | RI | ‐ |
| N294S | Clinic/ None | HRI | ‐ | ‐ | |
| R292K | Reverse Genetics | S | RI | HRI | |
| H273Y | Clinic? / ? | RI | S | RI | |
| D197N | Clinic / Oseltamivir | RI | RI | RI | |
| R150K | Clinic / Zanamivir | HRI | RI | HRI | |
| Reverse Genetics | HRI | RI | HRI | ||
| E116A | Reverse Genetics | HRI | HRI | HRI | |
| E116D | Reverse Genetics | HRI | HRI | HRI | |
| E116G | Reverse Genetics | RI | HRI | HRI | |
| E116V | Reverse Genetics | HRI | S | HRI | |
| E105K | Clinical / None | S | RI | HRI | |
S, susceptibility or normal inhibition (<10‐fold increase in IC50 over WT for A viruses or <5‐fold increase for B viruses); RI, reduced inhibition (10‐ to 100‐fold increase in IC50 over WT for A viruses or 5‐ to 50‐fold increase for B viruses); HRI, highly reduced inhibition (>100‐fold increase in IC50 over WT for A viruses or >50‐fold increase for B viruses). ? means uncertain or unknown information.
Impact of the H275Y NA mutation in A/H1N1 viruses
Understanding the impact of the H275Y is important for several reasons: (i) it is the most frequent mutation conferring resistance to oseltamivir and (ii) it was detected and transmitted in some viral backgrounds in the absence of antiviral pressure. Indeed, close to 100% of A/Brisbane/59/2007 (H1N1)‐like viruses that circulated in 2008–2009 in Europe and North America were resistant to oseltamivir due to the H275Y NA mutation.4 Although the detection of this mutation in the more recent A(H1N1)pdm09 viral background remains limited (<1·5% of tested strains), there is a recent concern that this problem could increase due to the growing number of resistant strains detected in the absence of therapy.12 These data indicate that, in the appropriate viral background, that is, with the required permissive NA mutation(s), the H275Y mutant can retain fitness and become more transmissible.5, 32 Based on ferret experiments, many groups have shown that the oseltamivir‐resistant A(H1N1)pdm09 virus with the H275Y mutation was as virulent as its wild‐type counterpart with the exception of a reduced airborne transmission reported in some but not all studies.33, 34, 35, 36, 37
Using a mathematical model to analyze a set of in vitro experiments that allow for the full characterization of the viral replication cycle, our group showed that the primary effects of the H275Y substitution on A(H1N1)pdm09 strains were to lengthen the mean eclipse phase of infected cells (from 6·6 to 9·1 hour) and decrease (by sevenfold) the viral burst size, that is, the total number of virions produced per cell.38 However, the infectious‐unit‐to‐particle ratio of the H275Y mutant strain was 12‐fold higher than that of oseltamivir‐susceptible strain (0·19 versus 0·016 per RNA copy). A parallel analysis of the H275Y mutation in the prior seasonal A/Brisbane/59/2007 background showed similar changes in the infection kinetic parameters but, in this strain, the H275Y mutation also allows the mutant to infect cells five times more rapidly. This model estimated a basic reproductive number (Ro), which is defined by the number of secondary infections caused by a single infectious cell, that was approximately the same for the A(H1N1)pdm09 wild type and its H275Y mutant (1·7 × 103 versus 3·0 × 103, respectively), whereas it was 25 times higher for the H275Y mutant compared with the wild‐type virus in the A/Brisbane/59/2007 background (48 × 103 versus 1·7 × 103, respectively). These replication parameters help to explain why the H275Y mutant replaced the wild‐type strain in the seasonal A/Brisbane/59/2007 background, whereas it is still not the case for the 2009 pandemic virus. In addition, another group has suggested that oseltamivir‐resistant A(H1N1) strains were less susceptible to antibody inhibition than susceptible strains, which may have favored their selection and dissemination in 2008–2009.39
Management of oseltamivir‐resistant severe influenza A infections
The selection of the most appropriate antiviral therapy must take into consideration that most cases of oseltamivir resistance are due to the H275Y mutation in the N1 subtype or the E119V and R292K mutations in the N2 subtype and that the first two viral mutants remain susceptible to zanamivir. Thus, inhaled or, for more severe cases, intravenous (where available through compassionate use)40 zanamivir is the best option in the case of suspected or confirmed resistance to oseltamivir. For patients on mechanical ventilation, if intravenous zanamivir is not available, other therapeutic options include inhaled or systemic ribavirin41 or parenteral peramivir, a NAI that is currently approved in Japan and South Korea. Although the H275Y mutant exhibits highly reduced inhibition to peramivir in vitro, animal studies using the A/WSN/33 (H1N1) virus indicated that a single administration (90 mg/kg intramuscularly) or multiple daily doses (45 mg/kg × 5 days) of this compound successfully prevented mortality and significantly decreased weight loss and lung viral titers after infection with the H275Y mutant.42 Such clinical benefits are likely attributable to the high concentrations of peramivir (4000‐ to 8000‐fold higher than the IC50 value in plasma), its high binding affinity and slow off‐rate from the NA.42, 43 However, because of the emergence of the H275Y mutation in a few patients receiving peramivir during the 2009 pandemic (through an emergency access program),44 other studies are needed before recommending such therapeutic modality in that context. An algorithm for the management of oseltamivir‐resistant infections is proposed in the Figure 1.
Figure 1.
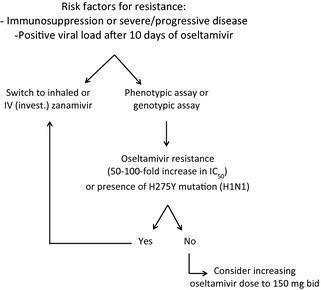
Proposed algorithm for the management of oseltamivir‐resistant influenza virus infections.
Selected investigational antivirals in clinical development for the treatment for influenza
An interim analysis of intravenous peramivir phase three clinical trials in USA has recently shown little difference with placebo on influenza outcomes, which halted the development of this drug in this country. Nevertheless, several antiviral agents are at some stages of clinical development for the treatment for influenza virus infections. In addition to intravenous zanamivir which is in phase three clinical trials in USA (ClinicalTrials.gov #NCT01231620), laninamivir octanoate is another NAI that is already approved in Japan and is in phase three clinical trial in USA (#NCT00803595). This long‐acting inhaled prodrug that lasts for 5 days is metabolized to laninamivir, which has a structure and a spectrum of activity similar to those of zanamivir.45 Favipiravir (formerly known as T‐705) is a potent inhibitor of the polymerase of influenza and several other RNA viruses administered by the oral route currently in phase two clinical trials (#NCT01068912).46 Selection of favipiravir‐resistant viruses has not been achieved so far after multiple in vitro passages. Another compound with a different mechanism of action is Fludase (formerly known as DAS181), which is an inhaled drug with activity against influenza and parainfluenza viruses.47 This drug, which is currently in phase two clinical trial (#NCT01037205) for the treatment for influenza infections, acts as a host receptor‐destroying enzyme (i.e., it has sialidase activity).
Conclusions
Along with the availability of new compounds with different viral targets, the options for the management of oseltamivir‐resistant infections should significantly expand. Consequently, combination therapies for immunocompromised patients will become a feasible strategy. Randomized trials of a triple combination therapy (amantadine, oseltamivir and ribavirin) are in progress based on synergy data for these compounds demonstrated in vitro and in vivo.48, 49 Another strategy under evaluation consists of combining antiviral agents and immunomodulators such as COX‐2 inhibitors.50 Finally, the administration of broad‐spectrum neutralizing antibodies targeting, for example, conserved epitopes of the HA protein is another promising approach.51 These strategies should increase our therapeutic options and reduce the emergence of NAI‐resistant viruses in high‐risk patients.
Conflict of interests
Dr Guy Boivin has received research grants from Roche, GlaxoSmithKline and Biota on the topic of influenza treatment.
Boivin. (2013) Detection and management of antiviral resistance for influenza viruses. Influenza and Other Respiratory Viruses 7(Suppl. 3), 18–23.
References
- 1. Mendel DB, Roberts NA. In‐vitro and in‐vivo efficacy of influenza neuraminidase inhibitors. Curr Opin Infect Dis 1998; 11:727–732. [DOI] [PubMed] [Google Scholar]
- 2. Shiraishi K, Mitamura K, Sakai‐Tagawa Y, Goto H, Sugaya N, Kawaoka Y. High frequency of resistant viruses harboring different mutations in amantadine‐treated children with influenza. J Infect Dis 2003; 188:57–61. [DOI] [PubMed] [Google Scholar]
- 3. Ives JA, Carr JA, Mendel DB et al The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res 2002; 55:307–317. [DOI] [PubMed] [Google Scholar]
- 4. Dharan NJ, Gubareva LV, Meyer JJ et al Infections with oseltamivir‐resistant influenza A(H1N1) virus in the United States. JAMA 2009; 301:1034–1041. [DOI] [PubMed] [Google Scholar]
- 5. Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 2010; 328:1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abed Y, Pizzorno A, Bouhy X, Boivin G. Role of permissive neuraminidase mutations in influenza A/Brisbane/59/2007‐like (H1N1) viruses. PLoS Pathog 2011; 7:e1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization (WHO) . Review of the 2011‐2012 winter influenza season, northern hemisphere. Wkly Epidemiol Rec 2012; 87:233–40. [PubMed] [Google Scholar]
- 8. Hurt AC, Hardie K, Wilson NJ, Deng YM, Osbourn M, Gehrig N. Community transmission of oseltamivir‐resistant A(H1N1)pdm09 influenza. N Engl J Med 2011; 365:2541–2542. [DOI] [PubMed] [Google Scholar]
- 9. Meijer A, Jonges M, van Beek P et al Oseltamivir‐resistant influenza A(H1N1)pdm09 virus in Dutch travellers returning from Spain, August 2012. Euro Surveill 2012; 17:20266. [PubMed] [Google Scholar]
- 10. Hurt AC, Hardie K, Wilson NJ et al Characteristics of a widespread community cluster of H275Y oseltamivir‐resistant A(H1N1)pdm09 influenza in Australia. J Infect Dis 2012; 206:148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hurt AC, Chotpitayasunondh T, Cox NJ et al Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 2012; 12:240–248. [DOI] [PubMed] [Google Scholar]
- 12. Storms AD, Gubareva LV, Su S et al Oseltamivir‐resistant pandemic (H1N1) 2009 virus infections, United States, 2010‐11. Emerg Infect Dis 2012; 18:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lackenby A, Moran Gilad J, Pebody R et al Continued emergence and changing epidemiology of oseltamivir‐resistant influenza A(H1N1)2009 virus, United Kingdom, winter 2010/11. Euro Surveill 2011; 16:19784. [PubMed] [Google Scholar]
- 14. Tisdale M, Daly J, Gor D. Methods for determining resistance to neuraminidase inhibitors. Int Congr Ser 2001; 1219:879–886. [Google Scholar]
- 15. Nguyen HT, Sheu TG, Mishin VP, Klimov AI, Gubareva LV. Assessment of pandemic and seasonal influenza A (H1N1) virus susceptibility to neuraminidase inhibitors in three enzyme activity inhibition assays. Antimicrob Agents Chemother 2010; 54:3671–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization (WHO) . Meetings of the WHO working group on surveillance of influenza antiviral susceptibility ‐ Geneva, November 2011 and June 2012. Wkly Epidemiol Rec 2012; 87:369–74. [PubMed] [Google Scholar]
- 17. Ghedin E, Holmes EC, Depasse JV et al Presence of oseltamivir‐resistant pandemic A/H1N1 minor variants before drug therapy with subsequent selection and transmission. J Infect Dis 2012; 206:1504–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abed Y, Goyette N, Boivin G. Generation and characterization of recombinant influenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob Agents Chemother 2005; 49:556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayden FG, Belshe RB, Clover RD, Hay AJ, Oakes MG, Soo W. Emergence and apparent transmission of rimantadine‐resistant influenza A virus in families. N Engl J Med 1989; 321:1696–1702. [DOI] [PubMed] [Google Scholar]
- 20. Abed Y, Baz M, Boivin G. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir Ther 2006; 11:971–976. [PubMed] [Google Scholar]
- 21. Pizzorno A, Bouhy X, Abed Y, Boivin G. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis 2011; 203:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pizzorno A, Abed Y, Bouhy X et al Impact of mutations at residue I223 of the neuraminidase protein on the resistance profile, replication level, and virulence of the 2009 pandemic influenza virus. Antimicrob Agents Chemother 2012; 56:1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van der Vries E, Stelma FF, Boucher CA. Emergence of a multidrug‐resistant pandemic influenza A (H1N1) virus. N Engl J Med 2010; 363:1381–1382. [DOI] [PubMed] [Google Scholar]
- 24. Whitley RJ, Hayden FG, Reisinger KS et al Oral oseltamivir treatment of influenza in children. Pediatr Infect Dis J 2001; 20:127–133. [DOI] [PubMed] [Google Scholar]
- 25. Baz M, Abed Y, McDonald J, Boivin G. Characterization of multidrug‐resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis 2006; 43:1555–1561. [DOI] [PubMed] [Google Scholar]
- 26. Mishin VP, Hayden FG, Gubareva LV. Susceptibilities of antiviral‐resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother 2005; 49:4515–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis 1998; 178:1257–1262. [DOI] [PubMed] [Google Scholar]
- 28. Hurt AC, Holien JK, Parker M, Kelso A, Barr IG. Zanamivir‐resistant influenza viruses with a novel neuraminidase mutation. J Virol 2009; 83:10366–10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dapat C, Suzuki Y, Saito R et al Rare influenza A (H3N2) variants with reduced sensitivity to antiviral drugs. Emerg Infect Dis 2010; 16:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sheu TG, Deyde VM, Okomo‐Adhiambo M et al Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother 2008; 52:3284–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McKimm‐Breschkin JL, Rootes C, Mohr PG, Barrett S, Streltsov VA. In vitro passaging of a pandemic H1N1/09 virus selects for viruses with neuraminidase mutations conferring high‐level resistance to oseltamivir and peramivir, but not to zanamivir. J Antimicrob Chemother 2012; 67:1874–1883. [DOI] [PubMed] [Google Scholar]
- 32. Bloom JD, Nayak JS, Baltimore D. A computational‐experimental approach identifies mutations that enhance surface expression of an oseltamivir‐resistant influenza neuraminidase. PLoS ONE 2011; 6:e22201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamelin ME, Baz M, Abed Y et al Oseltamivir‐resistant pandemic A/H1N1 virus is as virulent as its wild‐type counterpart in mice and ferrets. PLoS Pathog 2010; 6:e1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hamelin ME, Baz M, Bouhy X et al Reduced airborne transmission of oseltamivir‐resistant pandemic A/H1N1 virus in ferrets. Antivir Ther 2011; 16:775–779. [DOI] [PubMed] [Google Scholar]
- 35. Duan S, Boltz DA, Seiler P et al Oseltamivir‐resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog 2010; 6:e1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kiso M, Shinya K, Shimojima M et al Characterization of oseltamivir‐resistant 2009 H1N1 pandemic influenza A viruses. PLoS Pathog 2010; 6:e1001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seibert CW, Kaminski M, Philipp J et al Oseltamivir‐resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol 2010; 84:11219–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pinilla LT, Holder BP, Abed Y, Boivin G, Beauchemin CA. The H275Y neuraminidase mutation of the pandemic A/H1N1 influenza virus lengthens the eclipse phase and reduces viral output of infected cells, potentially compromising fitness in ferrets. J Virol 2012; 86:10651–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu WL, Lau SY, Chen Y et al The 2008‐2009 H1N1 influenza virus exhibits reduced susceptibility to antibody inhibition: implications for the prevalence of oseltamivir resistant variant viruses. Antiviral Res 2012; 93:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Härter G, Zimmermann O, Maier L et al Intravenous zanamivir for patients with pneumonitis due to pandemic (H1N1) 2009 influenza virus. Clin Infect Dis 2010; 50:1249–1251. [DOI] [PubMed] [Google Scholar]
- 41. Hayden FG, Sable CA, Connor JD, Lane J. Intravenous ribavirin by constant infusion for serious influenza and parainfluenza virus infection. Antivir Ther 1996; 1:51–56. [PubMed] [Google Scholar]
- 42. Abed Y, Pizzorno A, Boivin G. Therapeutic activity of intramuscular peramivir in mice infected with a recombinant influenza A/WSN/33 (H1N1) virus containing the H275Y neuraminidase mutation. Antimicrob Agents Chemother 2012; 56:4375–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shetty AK, Peek LA. Peramivir for the treatment of influenza. Expert Rev Anti Infect Ther 2012; 10:123–143. [DOI] [PubMed] [Google Scholar]
- 44. Birnkrant D, Cox E. The Emergency Use Authorization of peramivir for treatment of 2009 H1N1 influenza. N Engl J Med 2009; 361:2204–2207. [DOI] [PubMed] [Google Scholar]
- 45. Ikematsu H, Kawai N. Laninamivir octanoate: a new long‐acting neuraminidase inhibitor for the treatment of influenza. Expert Rev Anti Infect Ther 2011; 9:851–857. [DOI] [PubMed] [Google Scholar]
- 46. Furuta Y, Takahashi K, Shiraki K et al T‐705 (favipiravir) and related compounds: novel broad‐spectrum inhibitors of RNA viral infections. Antiviral Res 2009; 82:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Triana‐Baltzer GB, Gubareva LV, Nicholls JM et al Novel pandemic influenza A(H1N1) viruses are potently inhibited by DAS181, a sialidase fusion protein. PLoS ONE 2009; 4:e7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nguyen JT, Hoopes JD, Smee DF et al Triple combination of oseltamivir, amantadine, and ribavirin displays synergistic activity against multiple influenza virus strains in vitro. Antimicrob Agents Chemother 2009; 53:4115–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nguyen JT, Smee DF, Barnard DL et al Efficacy of combined therapy with amantadine, oseltamivir, and ribavirin in vivo against susceptible and amantadine‐resistant influenza A viruses. PLoS ONE 2012; 7:e31006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng BJ, Chan KW, Lin YP et al Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc Natl Acad Sci USA 2008; 105:8091–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sui J, Hwang WC, Perez S et al Structural and functional bases for broad‐spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009; 16:265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pizzorno A, Abed Y, Boivin G. Influenza drug resistance. Semin Respir Crit Care Med 2011; 32:409–422. [DOI] [PubMed] [Google Scholar]


