Abstract
Little is known about the mechanism by which IFNs inhibit human cytomegalovirus (HCMV) replication. Indeed, infection of fibroblasts with HCMV initiates the expression of a subset of type I IFN-inducible genes whose role in the infectious process is unclear. We describe here the identification of a cytoplasmic antiviral protein that is induced by IFNs, by HCMV infection, and by the HCMV envelope protein, glycoprotein B (gB). Stable expression of the protein in fibroblasts inhibits productive HCMV infection, down-regulating several HCMV structural proteins (gB, pp28, and pp65) known to be indispensable for viral assembly and maturation. We have named the protein viperin (for virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible). HCMV infection causes the redistribution of the induced viperin from its normal endoplasmic reticulum association, first to the Golgi apparatus and then to cytoplasmic vacuoles containing gB and pp28. Expression before HCMV infection reduces viperin redistribution from the endoplasmic reticulum to the Golgi apparatus and prevents vacuolar localization, perhaps reflecting the mechanism used by HCMV to evade the antiviral function.
Human cytomegalovirus (HCMV) is a β-herpesvirus that commonly and persistently infects humans (1, 2). HCMV infection poses very little threat to immunocompetent individuals but causes life-threatening complications in individuals with suppressed immune systems, such as patients with AIDS, patients with cancer undergoing chemotherapy, and organ transplant recipients treated with immunosuppressants (3–5). An intact IFN response is indispensable for limiting and clearing in vivo infections by the mouse homologue of HCMV (MCMV), and in vitro incubation of cells with IFN before MCMV or HCMV infection inhibits virus production (6–9). However, the mechanism of IFN protection against CMV at the molecular level is not known. The best studied IFN-inducible antiviral proteins, such as double-stranded RNA-dependent protein kinase, Mx, and ribonuclease L, seem not to be involved (10).
Despite the inhibitory effects of IFNs on productive viral infection, both live and UV-inactivated HCMV have been shown, by differential display or microarray analysis, to induce a number of type I IFN-inducible genes (11, 12). Compton and coworkers (13) have also shown that pretreatment of human fibroblasts with glycoprotein B (gB), an HCMV envelope protein, induces at least one of these genes, ISG54K. While analyzing the IFN-γ response of human macrophages we identified a unique IFN-γ-inducible gene, fragments of which were identified as the cDNAs cig5 and cig33 cloned from HCMV-infected fibroblasts (11, 14). Although IFN-γ caused expression of the gene in primary macrophages, induction was poor in a variety of cell lines. IFN-α and -β, however, were effective in inducing expression in the majority of cell types examined. Further investigation revealed that, even though HCMV does indeed induce expression of the gene, its product inhibits HCMV replication.
Materials and Methods
cDNA Cloning and Expression.
Human primary monocytes were isolated from peripheral blood mononuclear cells by negative selection as described (15). They were plated onto 100-mm tissue-culture plates in Iscove's DMEM containing 10% human AB serum for 10 days to allow differentiation into macrophages. The cells were left untreated or treated with recombinant human IFN-γ (1,000 units/ml) for 24 h, and total RNA and mRNA were isolated by using a Qiagen RNeasy and mRNA kit. cDNA was made and amplified by using a Smart PCR cDNA synthesis kit (CLONTECH), and an IFN-γ-induced human primary macrophage subtraction library was generated by using the CLONTECH PCR-Select cDNA subtration kit. Approximately 800 clones were isolated and sequenced. The authenticity of the clones was confirmed by reverse transcription–PCR. Full-length viperin cDNA was cloned by 5′ and 3′ rapid amplification of cDNA ends (RACE) of an IFN-γ-induced human primary macrophage cDNA library made by using a Marathon cDNA Amplification kit (CLONTECH). The cDNA was cloned into the mammalian expression vector pcDNA3.1 (Invitrogen). The ORF was PCR-amplified with the addition of an N-terminal myc tag and cloned into the IRESNEO vector and also into the retroviral vector, BMNNEO. The latter was transfected into the retroviral packaging line Phoenix-Ampho, and supernatants were used to infect human fibroblasts. Full-length mouse viperin cDNA was cloned as described for human viperin from a RACE library made from IFN-γ-induced RAW264.7 macrophage cells.
HeLa M cells were transfected with IRESNEO.viperin.MycN by using lipofectamine 2000 (GIBCO/BRL). Selection with 500 μg/ml of G418 began 48 h after transfection, and the cells were cloned by limiting dilution. Telomerase-immortalized fibroblasts (a gift from T. Shenk, Princeton University, Princeton) were infected with a retrovirus (BMN.viperin.MycN) carrying the gene for 36 h. The medium was then removed and replaced with selection medium containing 500 μg/ml of G418. Polyclonal cells appeared 2 weeks after selection and were used in all experiments without further cloning.
Ab Generation and Immunoblotting.
The rabbit antiserum R.viperin.C was prepared by immunization with the COOH-terminal peptide (346 RGGKYIWSKADLKLDW 361) coupled to keyhole limpet hemocyanin. The rabbit antiserum was affinity-purified by using a peptide affinity column. For protein immunoblotting, fibroblasts were seeded at a density of 4 × 105 per well into 6-well plates. The next day, the cells were infected with the HCMV strain AD169 at a multiplicity of infection of 2 for 2 h. Total lysates were made at intervals by lysing the cells in 2X SDS sample-loading buffer containing β-mercaptoethanol. Samples containing equivalent numbers of cells were separated by 12% SDS/PAGE and electrophoretically transferred onto poly(vinylidene difluoride) membranes. Membranes were blocked with 5% nonfat dry milk in 1X PBS containing 0.2% Tween 20 overnight at 4°C. The membranes were incubated with primary Ab for 1 h, washed, and then incubated with horseradish peroxidase-conjugated secondary Ab for 1 h. The bands were visualized by developing with enhanced chemiluminescence (Pierce). We used the following mAbs: MAB810 recognizing IE1 and IE2 (Chemicon); anti-pp28 (RGI no. 1207, Rumbaugh-Goodwin Institute, Plantation, FL); 1-M-12 against gB (Research Diagnostics, Flanders, NJ); and anti-pp65 (clone 1-I-11, Research Diagnostics).
HCMV Infection and Immunofluorescence.
Primary foreskin fibroblasts or telomerase-immortalized fibroblasts (1 × 105) were seeded onto glass coverslips. The next day, the cells were infected with HCMV AD169 at a multiplicity of infection of 1.5–2 for 2 h. At the indicated times, the cells were then fixed in 4% paraformaldehyde in PBS for 30 min at room temperature, washed 3 times with PBS, and the paraformaldehyde was quenched with excess glycine. Cells were then permeabilized with 0.1% saponin in PBS containing 10% FBS for 30 min. Viperin was detected by using R.viperin.C at 1:150 dilution. The mAb to the Golgi marker GM130 (Transduction Laboratories, Lexington, KY) and the mAb to the HCMV envelope glycoprotein, gB, were used at a dilution of 1:200. Secondary Abs used, goat anti-rabbit IgG conjugated with Alexa Fluor 594 and goat anti-mouse IgG conjugated with Alexa Fluor 488, were obtained from Molecular Probes. Cells were examined by using a Zeiss Axiophot 2 microscope, and images were processed by using Adobe Systems (Mountain View, CA) photoshop.
Results and Discussion
Identification of Viperin as an IFN-Inducible Protein.
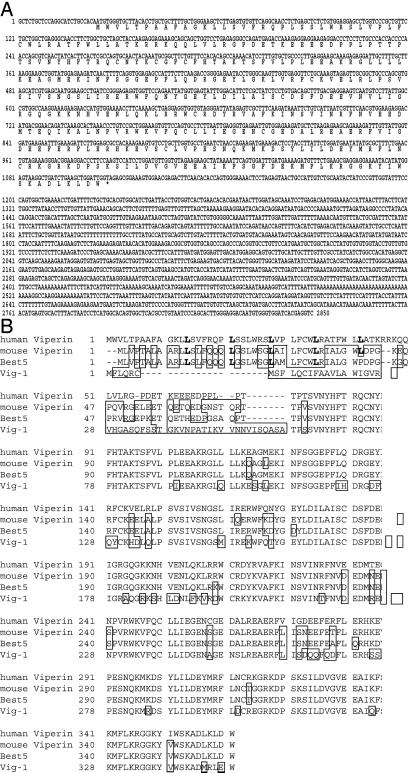
Bt using the subtractive technique of representational differential analysis, we cloned a cDNA from primary human macrophages treated with IFN-γ. The cDNA was 2858 bp in length and contained an ORF of 1,083 nucleotides, encoding a protein with 361 amino acids (Fig. 1A). However, when the human sequence was subjected to a homology search at the nucleotide level, we found it aligned with two cDNA fragments, named cig5 and cig33, which were isolated by differential display analysis of primary foreskin fibroblasts productively infected with HCMV. Cig33 was also stimulated by UV-inactivated HCMV (11). The two fragments cig5 and cig33 together yield a full-length cDNA very similar to the one we isolated. The only difference between cig (5 + 33) and the macrophage-derived clone is that cig5 contains an extra 530 nucleotides at the 5′ end of the 5′ untranslated region. This extra region seems to be a cloning artifact in that it is absent from the human genomic sequence.
Figure 1.
Sequences of human viperin and comparison with homologues from other species. (A) The human cDNA sequence encodes an ORF with 361 amino acids. (B) Comparison of viperin amino acid sequence with mouse, rat (Best 5; GenBank accession no. YO7704), and trout (Vig 1; GenBank accession no. AF076620) sequences. The leucine residues in bold type indicate the leucine zipper motif, and boxed residues are those differing from the human sequence. cDNA sequences for human and mouse viperin are available through GenBank accession nos. AF442151 and AF442152, respectively.
The amino acid sequence of viperin was homologous to BEST5, expressed during rat osteoblast differentiation (16), and to Vig-1, induced in rainbow trout infected with a fish rhabdovirus (17) (Fig. 1B). We also cloned a cDNA encoding a mouse homologue that is highly similar to the human sequence and even more similar to the rat sequence (Fig. 1B). A mouse cDNA was independently isolated as a homologue of Vig-1 by Boudinot et al. (14) who named the gene encoding it mvig. Domain searches indicated that the N-terminal 70 amino acids of human viperin, which display minimal homology to the putative rat, mouse, and fish proteins, contain a leucine zipper motif, which is commonly involved in protein–protein interactions (Fig. 1B, bold residues). This motif is also apparent in the rat and mouse sequences. In the remainder of the molecule almost every amino acid is conserved. As discussed by Boudinot et al., a stretch of amino acids from residues 71 to 182 has significant homology to the so-called MoaA/PQQIII signature in the MoaA, NIRJ, and PQQIII protein families (18). This motif has been shown to be important for iron-sulfur cluster coordination. Conservation of this region and the remaining C-terminal sequence argues for its functional importance.
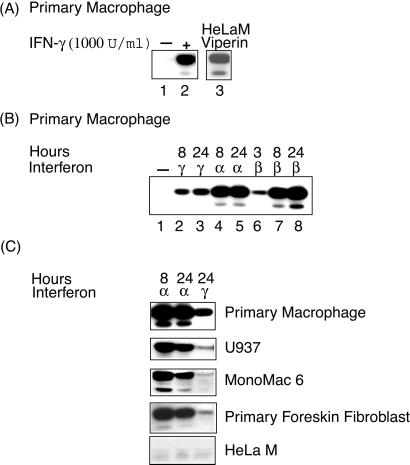
We raised a rabbit antiserum against the C-terminal peptide of viperin and used it for Western blotting of primary human macrophages treated with IFN-γ. Two bands were detected by analysis of SDS/PAGE, a major species of 43 kDa, and a minor species of 35 kDa (Fig. 2A, lane 2). The predicted Mr of viperin is 42.1 kDa. No bands were detected in untreated macrophages (Fig. 2A, lane 1). Western blotting detected the same two bands in HeLa cells transfected with viperin cDNA (Fig. 2A, lane 3), indicating that the 35-kDa band is probably a proteolysis product.
Figure 2.
Cell-type-specific induction of viperin expression by type I and II IFNs. (A) Viperin is induced in macrophages by IFN-γ. Extracts of untreated and IFN-γ-treated primary human macrophages were subjected to SDS/PAGE and viperin was detected by immunoblotting with a specific rabbit antiserum. (B) Induction of viperin in macrophages by type I and II IFNs. (C) Viperin expression is stimulated preferentially by type I IFNs in a variety of other cell types.
The expression of many genes is induced by both type I and type II IFNs, whereas others are regulated only by one type. To examine this question for viperin we treated primary human macrophages with IFN-α or -β at 500 units/ml. We observed a strong induction by both IFN-α and -β after either 8 or 24 h. Induction occurred as early as 3 h after IFN-β treatment (Fig. 2B). Viperin was less well induced by IFN-γ than by IFN-α or -β (Fig. 2B, compare lane 2 with lanes 4 and 7). We also examined the IFN-induction of viperin in the human monocytic cell lines U937 and MonoMac 6, in the epithelial cell line HeLa M, and in primary human foreskin fibroblasts. No expression was detectable in the untreated cells and little or no induction was seen in the cell lines or fibroblasts treated with IFN-γ (Fig. 2C). This finding suggests that the gene in the monocytic cell lines has lost the ability to respond effectively to IFN-γ. However, other genes, such as those encoding MHC class II molecules, were found to retain responsiveness (data not shown). The gene encoding viperin may use a different signaling pathway or regulatory elements in its promoter may be inaccessible in the cell lines. IFN-α-induced genes can be divided into two groups (19). One group is induced early and disappears about 12 h after induction. The other group is detectable at 12 h and is sustained through 24 h. All of the cells examined except HeLa M were induced to express viperin by IFN-α at both 8 and 24 h (Fig. 2C).
Viperin and HCMV Infection.
The finding that viperin is induced by IFN-α and -β suggested that it might have an antiviral function, although Zhu et al. (11) identified it as a gene induced by HCMV. Stimulation of viperin mRNA expression by HCMV does not necessarily mean that the protein does not have an antiviral function. Many viruses have evolved mechanisms to circumvent a variety of host response mechanisms. HCMV itself has a number of ways of avoiding detection by the adaptive immune system, including interfering with T cell recognition by suppressing both MHC class I and II functions (20–24).
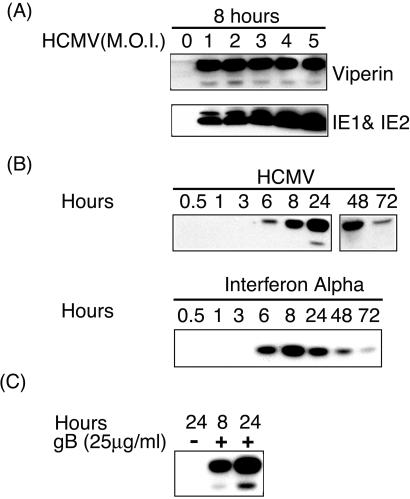
To determine whether viperin was induced by HCMV at the protein level, fibroblasts were infected at different multiplicities of infection. Expression of HCMV immediate-early protein after 8 h confirmed that the infection was successful (Fig. 3A Lower). No viperin expression was seen in uninfected cells but it was clearly expressed in cells infected with HCMV (Fig. 3A Upper). Two bands were observed, similar to those induced by both classes of IFN, indicating that the expression of viperin protein was not inhibited by HCMV infection. To examine the kinetics of induction we infected fibroblasts and harvested them at different times. In parallel, fibroblasts were incubated with IFN-α. In both cases, viperin expression was detectable after 6 h (Fig. 3B). In the IFN-α-treated cells, expression was maximal at 8 h, whereas in infected cells it was maximal at 24 h. There was no evidence for a reduction in viperin stability in HCMV-infected cells compared with cells treated with IFN-α.
Figure 3.
Induction of viperin expression by HCMV. (A) HCMV infection of primary human fibroblasts induces viperin expression detected by immunoblotting. (B) Kinetics of viperin induction by IFN-α and by HCMV infection. (C) Direct induction of viperin by gB, an HCMV envelope glycoprotein. Purified gB was added to cells, and viperin expression was assayed by immunoblotting at 8 and 24 h.
Shenk and coworkers (11) showed that induction by HCMV of cig33 message, which we now know corresponds to the 3′ region of viperin mRNA, was independent of IFN-α and could be mediated by UV-inactivated virus. This may result from interaction of the viral envelope glycoprotein, gB, with a specific receptor, because it has also been shown that incubation of fibroblasts with gB induces the MAP-kinase signaling pathway and the subsequent expression of the IFN-α-inducible gene ISG54K (13, 25). To examine this question, fibroblasts were incubated with purified soluble recombinant gB (a gift from T. Compton, University of Wisconsin, Madison) (13). Viperin was induced by gB after 8 h and was even higher after 24 h (Fig. 3C). The combined data show that HCMV can induce viperin expression in fibroblasts, and that the induction is mediated by surface binding of the gB component of the viral envelope to an unknown cell surface receptor. Induction is independent of other virion-associated molecules or virally encoded transcripts.
Viperin Inhibits HCMV Infection.
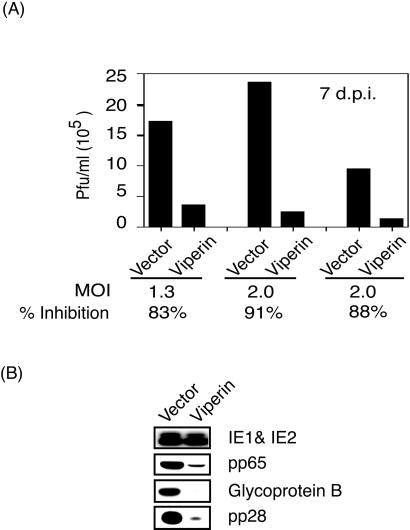
To examine viperin for antiviral effects, fibroblasts, expressing the telomerase gene to prolong their in vitro survival (26), were transduced with a retrovirus encoding viperin or with a control virus. The cells were infected with HCMV for 2 h, washed, and cultured in fresh medium. Supernatants were harvested 7 days after infection, and plaque assays were performed by using primary foreskin fibroblasts (27). Fibroblasts expressing viperin showed a reduction of virus production of ≈90% (Fig. 4A), indicating that viperin has antiviral activity for HCMV. To investigate the molecular basis of the antiviral effect we examined the expression of a number of viral proteins produced at different stages of HCMV replication. Fibroblasts were infected with HCMV for 3 days, lysed, and equal numbers of cells were examined by Western blotting. We saw no difference in expression of the products of immediate early genes (IE1 and IE2) between viperin-expressing and control fibroblasts. However, synthesis of early-late (pp65), late (gB), or true late (pp28) viral proteins was significantly reduced in the cells expressing viperin (Fig. 4B). The expression of these proteins is indispensable for productive HCMV infection (28). A possible explanation for these findings is that viperin may exert its antiviral effect by inhibiting the synthesis or function of an HCMV-encoded component critical for virus maturation and/or assembly.
Figure 4.
Viperin inhibits HCMV replication. (A) Human fibroblasts, transduced with either a retrovirus encoding viperin or with a control retrovirus, were infected with HCMV, and supernatants were harvested 7 days after infection and analyzed by a standard plaque assay. (B) Viperin expression inhibits the synthesis of late viral proteins. Fibroblasts expressing viperin or control fibroblasts were infected with HCMV and after 3 days extracts were subjected to SDS/PAGE and immunoblotted with Abs to immediate-early protein, pp65, gB, and pp28 of HCMV.
HCMV Effects on Intracellular Distribution of Viperin.
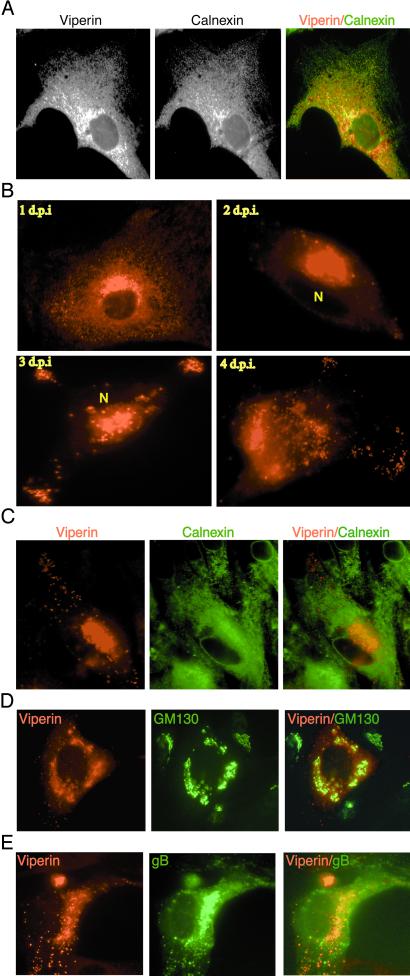
Viperin has no obvious signal sequence nor does it seem to possess a nuclear localization signal, a transmembrane region, or a defined motif that could serve as a lipid modification signal. We therefore predicted that it would be a soluble cytoplasmic protein. However, examination by indirect immunofluorescence of fixed and permeabilized IFN-α-treated fibroblasts (Fig. 5A), retrovirally transduced fibroblasts, or fibroblasts treated with recombinant gB (data not shown) showed that viperin colocalized with the endoplasmic reticulum (ER) resident protein, calnexin. The most likely explanation is that viperin associates with the cytosolic face of the ER, either directly or by an interaction with another protein, perhaps by the N-terminal leucine zipper domain.
Figure 5.
HCMV infection alters the normal cellular distribution of viperin. (A) Viperin localizes to the ER. Fibroblasts treated with IFN-α for 24 h were fixed, permeabilized, and analyzed by intracellular immunofluorescence, using a rabbit anti-viperin antiserum and a mAb to calnexin, an ER marker. (B) HCMV infection induces viperin redistribution. Fibroblasts infected with HCMV were harvested and stained with the rabbit anti-viperin serum on days 1–4 after infection. “N” indicates the location of the nucleus. (C) Viperin redistribution does not affect resident ER proteins. Fibroblasts were costained 4 days after infection for viperin and calnexin. (D) Viperin moves initially to the Golgi apparatus during HCMV infection. Fibroblasts were harvested at 3 days after infection and costained for viperin and the Golgi marker GM130. (E) Viperin redistributes after infection to the Golgi apparatus and undefined cytoplasmic vesicles expressing the HCMV envelope glycoprotein, gB.
HCMV induces the synthesis of viperin in fibroblasts even though prior expression renders them resistant to infection (Fig. 4A). Many pathogens have evolved mechanisms that alter the normal subcellular localization of host factors as a means of evading otherwise deleterious effects. Such a shift may either alter the cell to ensure survival of the pathogen or impair the functional activity of the factor. For example, in macrophages, Mycobacterium tuberculosis recruits a host factor, TACO, from its normal cytoskeletal localization to phagosomes, preventing phagosome/lysosome fusion and avoiding degradation (29). Adenovirus infection has been shown to elevate the expression of Hsp70 and to induce its transfer from the cytoplasm to the nucleus (30). This relocalization is critical for viral replication. HCMV also has been shown to induce the movement of Hsp70 from the cytoplasm into the nucleus (31), although a role for this in viral infection has not been demonstrated. Such considerations led us to ask whether HCMV might alter the distribution of viperin, perhaps in this way evading the antiviral effect.
Fibroblasts were infected with HCMV and examined by indirect immunofluorescence microscopy at different times after infection (Fig. 5B). After 24 h, viperin was again localized primarily in the ER. After 2 days we found significant redistribution of a large fraction of viperin to a perinuclear structure. A smaller fraction was also found in punctate cytoplasmic structures, probably vacuoles, and this pattern became more prominent 3 and 4 days after infection. The distribution of a classical ER marker, calnexin, was unaffected by HCMV infection (Fig. 5C). To identify the cellular structures to which viperin moves, fibroblasts were stained 3 days after infection with Abs to a cis-Golgi marker, GM130, and viperin. The perinuclear viperin staining matched that of the Golgi marker (Fig. 5D). The viperin-positive punctate structures did not stain with the anti-Golgi reagent nor did they stain with Abs to components of lysosomes, endosomes, or the ER-Golgi intermediate compartment (data not shown).
HCMV tegument proteins, such as pp150 and pp28, and the viral envelope glycoprotein gB have been detected in the Golgi region and in cytoplasmic vacuoles that did not overlap with any defined cellular compartments, including endosomes and lysosomes (32). To determine whether the viperin-positive structures corresponded to those containing HCMV proteins, infected fibroblasts were stained with Abs to viperin, to gB, and to pp28. gB (Fig. 5E) and pp28 (data not shown) were found both in the viperin-positive Golgi-related perinuclear structure as well as in the punctate viperin-positive structures. These results show that HCMV, during the course of infection, induces redistribution of viperin from its normal ER association to the Golgi, the site of viral glycoprotein maturation, and also to vacuoles presumably containing assembled, or assembling, viral particles.
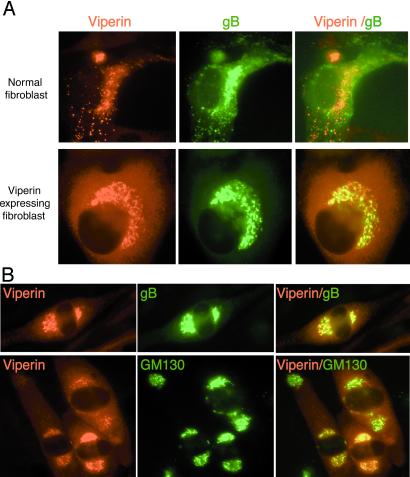
If relocalization of viperin to organelles containing HCMV components is required for a productive viral infection, one would predict that constitutively expressed viperin, which inhibits virus production (Fig. 4A), would not be similarly redistributed after infection. To test this prediction, viperin-expressing fibroblasts were examined by indirect immunofluorescence 3 days after infection by HCMV. Fig. 6A Lower shows that, in contrast to control fibroblasts (Upper), most of the viperin remains in the ER in the viperin-expressing fibroblasts. Although both viperin and gB are detectable in the Golgi area, no redistribution to the cytoplasmic vacuolar structures can be seen.
Figure 6.
Prior expression of viperin inhibits the HCMV-induced redistribution of viperin and gB. (A) Fibroblasts expressing viperin (Lower) or control fibroblasts (Upper) were infected with HCMV, harvested 3 days after infection, fixed, permeabilized, and the subcellular distribution of viperin and gB was examined by immunofluorescence. (B) Viral gene(s) sensitive to ganciclovir are required for viperin redistribution to cytoplasmic vesicles. Fibroblasts were infected with HCMV for 3 days in ganciclovir (50 μM), fixed, permeabilized, and costained with Abs to viperin and either gB or GM130.
A possible explanation for the antiviral effect is that viperin prevents the expression of virally encoded genes required for virus production. HCMV could circumvent the action of viperin by expressing a gene (or genes) that counteracts its effects sufficiently early in the infectious process to allow it to proceed to completion. The mechanism may involve inducing the intracellular redistribution that we observe. Identification of such a gene would be required to definitively prove this hypothesis, but as a preliminary step we sought to determine whether HCMV-encoded genes that depend on viral replication might be involved. To examine this question, we treated HCMV-infected cells with ganciclovir, which blocks replication by inhibiting the viral DNA polymerase. Three days after infection, the induced viperin was found predominantly in the ER with some redistribution to the Golgi area. Viperin-containing punctate structures were not apparent (Fig. 6B). Consistent with the results shown in Fig. 4B, reduced amounts of gB, also restricted to the ER and Golgi, were observed. These results indicate that viral replication is not absolutely required for movement of viperin or gB from the ER to the Golgi, but is critical for the localization of both to the cytoplasmic vacuoles that are the putative site of viral assembly.
Concluding Remarks
Viperin is a unique component of the antiviral machinery induced by IFNs. Its expression in fibroblasts before HCMV infection inhibits virus production (Fig. 4A) although viperin synthesis is induced by HCMV. In fact, its expression, along with an additional subset of IFN-inducible genes, is induced after binding of the virus, or the envelope glycoprotein, gB, to an as yet undefined membrane receptor. Boudinot et al. (14) showed that the mouse homologue of viperin is similarly directly induced by vesicular stomatitis virus, although in this case the induction seems to be restricted to dendritic cells. These observations raise three obvious, and probably interrelated, questions: What is the mechanism by which viperin inhibits viral production? How does HCMV evade the effect of virally induced viperin? Why does HCMV directly induce a protein that would normally inhibit the infectious process? Viperin seems to reside at the cytosolic face of the ER and this could be required for the antiviral effect. For example, viperin could interfere with transport of critical viral components, e.g., transmembrane glycoproteins, from the ER to the Golgi. The HCMV-induced redistribution of viperin from the ER to the Golgi, and ultimately to the observed cytoplasmic vacuoles (Fig. 5 D and E), could therefore be involved in the viral evasion mechanism. A specific HCMV gene product might antagonize the viperin effect by initiating its redistribution, perhaps binding viperin directly. Ganciclovir treatment at least partially inhibits the redistribution (Fig. 6B), consistent with the hypothesis that a replication-dependent viral component may be involved. However, one can still ask why HCMV should induce viperin expression at all when it then requires an extra function to avoid its effects. One possible explanation is that the virus actually needs viperin for a productive infection and has therefore been forced to develop the means to protect itself from it. The answer to this question clearly awaits further studies.
Acknowledgments
We thank Drs. Thomas Shenk, Theresa Compton, Jay Nelson, and David Johnson for advice and for providing valuable reagents. We also thank Dr. Kurt Cannon, Dr. Pamela Wearsch, and Suk-Jo Kang for critically reading the manuscript; and Nancy Dometios for help in its preparation. The research was funded by the Howard Hughes Medical Institute.
Abbreviations
- HCMV
human cytomegalovirus
- gB
glycoprotein B
- ER
endoplasmic reticulum
Footnotes
References
- 1.Britt W J, Alford C A. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. [Google Scholar]
- 2.Ho M. Cytomegalovirus: Biology and Infection. New York: Plenum; 1991. [Google Scholar]
- 3.Klatt E C, Shibata D. Arch Pathol Lab Med. 1988;112:540–544. [PubMed] [Google Scholar]
- 4.Rubin R H. Rev Infect Dis. 1990;12, Suppl. 7:S754–S766. doi: 10.1093/clinids/12.supplement_7.s754. [DOI] [PubMed] [Google Scholar]
- 5.Winston D J, Ho W G, Champlin R E. Rev Infect Dis. 1990;12, Suppl. 7:S776–S792. doi: 10.1093/clinids/12.supplement_7.s776. [DOI] [PubMed] [Google Scholar]
- 6.Delannoy A S, Hober D, Bouzidi A, Wattre P. Microbiol Immunol. 1999;43:1087–1096. doi: 10.1111/j.1348-0421.1999.tb03365.x. [DOI] [PubMed] [Google Scholar]
- 7.Schut R L, Gekker G, Hu S, Chao C C, Pomeroy C, Jordan M C, Peterson P K. J Infect Dis. 1994;169:1092–1096. doi: 10.1093/infdis/169.5.1092. [DOI] [PubMed] [Google Scholar]
- 8.Stinski M F, Thomsen D R, Rodriguez J E. J Gen Virol. 1982;60:261–270. doi: 10.1099/0022-1317-60-2-261. [DOI] [PubMed] [Google Scholar]
- 9.Yeow W S, Lawson C M, Beilharz M W. J Immunol. 1998;160:2932–2939. [PubMed] [Google Scholar]
- 10.Presti R M, Popkin D L, Connick M, Paetzold S, Virgin H W I. J Exp Med. 2001;193:483–496. doi: 10.1084/jem.193.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H, Cong J P, Shenk T. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, Cong J P, Mamtora G, Gingeras T, Shenk T. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle K A, Pietropaolo R L, Compton T. Mol Cell Biol. 1999;19:3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boudinot P, Riffault S, Salhi S, Carrat C, Sedlik C, Mahmoudi N, Charley B, Benmansour A. J Gen Virol. 2000;81:2675–2682. doi: 10.1099/0022-1317-81-11-2675. [DOI] [PubMed] [Google Scholar]
- 15.Koppelman B, Neefjes J J, de Vries J E, de Waal Malefyt R. Immunity. 1997;7:861–871. doi: 10.1016/s1074-7613(00)80404-5. [DOI] [PubMed] [Google Scholar]
- 16.Grewal T S, Genever P G, Brabbs A C, Birch M, Skerry T M. FASEB J. 2000;14:523–531. doi: 10.1096/fasebj.14.3.523. [DOI] [PubMed] [Google Scholar]
- 17.Boudinot P, Massin P, Blanco M, Riffault S, Benmansour A. J Virol. 1999;73:1846–1852. doi: 10.1128/jvi.73.3.1846-1852.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menendez C, Siebert D, Brandsch R. FEBS Lett. 1996;391:101–103. doi: 10.1016/0014-5793(96)00712-0. [DOI] [PubMed] [Google Scholar]
- 19.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 20.Tortorella D, Gewurz B, Schust D, Furman M, Ploegh H. Immunol Invest. 2000;29:97–100. doi: 10.3109/08820130009062289. [DOI] [PubMed] [Google Scholar]
- 21.Wiertz E, Hill A, Tortorella D, Ploegh H. Immunol Lett. 1997;57:213–216. doi: 10.1016/s0165-2478(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 22.Miller D M, Cebulla C M, Rahill B M, Sedmak D D. Semin Immunol. 2001;13:11–18. doi: 10.1006/smim.2001.0291. [DOI] [PubMed] [Google Scholar]
- 23.Tomazin R, Boname J, Hegde N R, Lewinsohn D M, Altschuler Y, Jones T R, Cresswell P, Nelson J A, Riddell S R, Johnson D C. Nat Med. 1999;5:1039–1043. doi: 10.1038/12478. [DOI] [PubMed] [Google Scholar]
- 24.Lehner P J, Karttunen J T, Wilkinson G W G, Cresswell P. Proc Natl Acad Sci USA. 1997;94:6904–6909. doi: 10.1073/pnas.94.13.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yurochko A D, Hwang E S, Rasmussen L, Keay S, Pereira L, Huang E S. J Virol. 1997;71:5051–5059. doi: 10.1128/jvi.71.7.5051-5059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bresnahan W A, Hultman G E, Shenk T. J Virol. 2000;74:10816–10818. doi: 10.1128/jvi.74.22.10816-10818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mar E C, Cheng Y C, Huang E S. Antimicrob Agents Chemother. 1983;24:518–521. doi: 10.1128/aac.24.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski U H. J Virol. 2000;74:7720–7729. doi: 10.1128/jvi.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrari G, Langen H, Naito M, Pieters J. Cell. 1999;97:435–447. doi: 10.1016/s0092-8674(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 30.Glotzer J B, Saltik M, Chiocca S, Michou A I, Moseley P, Cotten M. Nature (London) 2000;407:207–211. doi: 10.1038/35025102. [DOI] [PubMed] [Google Scholar]
- 31.Ohgitani E, Kobayashi K, Takeshita K, Imanishi J. J Gen Virol. 1999;80:63–68. doi: 10.1099/0022-1317-80-1-63. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez V, Greis K D, Sztul E, Britt W J. J Virol. 2000;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]