Abstract
In this issue of Cancer Cell, Jiang et al. report a genomic and transcriptional analysis of triple negative breast cancers (TNBCs) from an East Asian population. Their study shows minor differences with published studies of European and North American populations and suggests a therapeutic decision tree for treatment of TNBC.
Breast cancer treatment strategies typically are determined by underlying molecular characteristics of the cancers. Historically, ductal breast cancers have been managed based on a limited number of markers as: triple negative (TN) (HER2−, ER−, PR−), hormone receptor (HR) or luminal (HER2−, ER+, PR+/−), or HER2+, which is frequently divided based on hormone receptor status into HR+/HER2+ (HER2+, ER+, PR+/−) and HR−/HER2+ (HER2+, ER−, PR−). Management of ductal breast cancer based on these subtypes has both prognostic and therapeutic relevance contributing substantively to improved outcomes for breast cancer patients. Large-scale molecular profiling beginning with gene expression measurements enabled further refinement of ductal breast cancers into molecularly distinct subtypes with different regulatory pathway usage that dictates response to pathway-targeted therapies. Initial work defined five molecular subtypes designated luminal A, luminal B, HER2, basal-like, and normal-like based on measurements of gene expression profiling (Perou et al., 2000) (Figure 1). Ongoing classification efforts based on measurements of additional molecular endpoints (genome, epigenome, microRNA, protein, etc.) are further refining the classification into different subtypes (Cancer Genome Atlas, 2012; Curtis etal., 2012).
Figure 1. Schematic Illustration of a Developing Therapeutic Decision Tree for Ductal Breast Cancer with Emphasis on the TNBC Subtype.
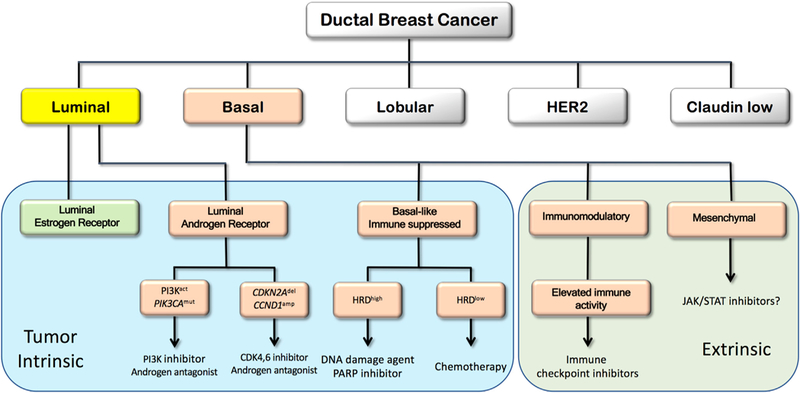
Subtypes defined in early transcriptional profiling efforts (Perou et al., 2000) are illustrated in the upper tier. TNBC subtypes (Jiang et al., 2019) are illustrated in the salmon-colored boxes. Subtypes now defined as tumor intrinsic are in the blue area, and subtypes defined as due to invading immune or other stromal cells are in the green extrinsic area. The therapeutic actions proposed in the boxes are suggested by Jiang et al. and/or by the published literature. The question mark after the JAK/STAT suggestion reflects uncertainty about the utility of drugging a pathway that may be generated by invading stromal cells.
The TN subgroup of breast cancer (TNBC) is of special interest due to their aggressive nature and poor outcome. Importantly, the TNBC subgroup is not a single disease but a set of diverse diseases (Cancer Genome Atlas, 2012; Curtis et al., 2012) that display epigenomic heterogeneity, phenotypic plasticity, and often harbor DNA repair defects (homologous recombination deficiency [HRD], as well as mutations of TP53, BRCA1, and BRCA2 [Cancer Genome Atlas, 2012; Curtis et al., 2012]). Their diversity and epigenomic plasticity have hindered the development of effective therapeutic approaches, although their high prevalence of BRCA1 and BRCA2 mutations and HRD has led to implementation of PARP inhibitors and platin-based therapy. The heterogeneity of TNBC has spawned efforts to further subset it into groups that can be targeted with greater precision and effectiveness. The first effort (Abramson et al., 2015) established six subtypes using transcriptional profiling: basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal stem cell-like (MSL), and luminal androgen receptor (LAR) that are therapeutically relevant. This has since been reduced to four subtypes (BL1, BL2, M, and LAR) based on the recognition that the IM and MSL subtypes were contributed from infiltrating lymphocytes and tumor-associated stromal cells, respectively (Lehmann et al., 2016). Importantly, the LAR subtype has many characteristics that suggest these tumors are more closely related to HR luminal ductal breast cancers, and consistent with this, they localize to the luminal and HR/HER2 enriched PAM50 subtypes (Abramson et al., 2015). The refined subtyping of TNBC has further improved our understanding of patient prognosis and therapeutic opportunities. Indeed, a number of clinical trials aimed at capitalizing on characteristics of different TNBC subsets are currently underway (NCT02457910, NCT02456857) including from Fudan University (NCT03805399).
Targeting specific TNBC molecular subtypes is highly attractive. However, genetic background and environmental exposure can have a large impact on molecular subtype. Thus, it is not clear that TNBCs that arise in geographically and ethnically distinctive populations will have the same molecular characteristics. While the majority of studies of TNBC are enriched for European ancestry, several studies have characterized African Americans (Huo et al., 2017), but studies of other ethnic groups have been more limited (Ding et al., 2019). In addition, the effects of geographically distinct environmental exposure have not been systematically evaluated, although genomic studies show the existence of carcinogen-specific genomic fingerprints (Alexandrov et al., 2013).
Jiang et al. (2019) present analyses of 465 TNBCs treated at the Fudan University Shanghai Cancer Center between 2007 and 2014 that provide some insights into these issues. Their cohort is comprised of patients with TNBC and high-quality follow-up information allowing an unbiased assessment of the frequency and prognosis of various TNBC subtypes. Samples were analyzed for DNA copy number using a microarray platform (401 samples), DNA sequence using whole-exome sequencing (279 samples), and gene expression using RNA-seq (360 samples). They present several useful comparisons between tumors from their Chinese cohort and Caucasian and African American samples. However, the comparisons need to be viewed with some caution given the possibility of subtype bias—particularly the increased frequency of LAR in this population. That said, several interesting features emerge.
The transcriptionally defined subtypes defined by Jiang et al. are strongly concordant with those defined by Abramson et al. in a mixed Caucasian and African American cohort (Abramson et al., 2015). The LAR and IM subtypes are the same in both studies. The Abramson et al. M, BL1, and BL2 subtypes map to the basal-like immune-suppressed (BLIS) subtype but do not appear as distinct clusters in the Jiang et al. study, raising a question about the robustness of these subtypes. The mesenchymal (MES) subtype defined by Jiang et al. maps to the same MSL subtype defined by Abramson et al. that was later dropped as a tumor intrinsic subtype and rather interpreted as an indicator of stromal invasion, which may not be targetable (Lehmann et al., 2016). Overall, tumors with the LAR subtype were more frequently observed in the Chinese cohort. The robustness of the Jiang et al. results, particularly the increased frequency of the LAR subtype, is supported by several smaller studies (Ding et al., 2019).
The recurrent genomic aberrations observed by Jiang et al. (Jiang et al., 2019) overall were similar to those identified in other studies, but there are some differences in the frequencies of key aberrations. For example, the Chinese TNBC cohort demonstrated higher frequencies of PIK3CA mutations than in TCGA, which was driven primarily by the differences between African American TNBCs and Chinese TNBCs; this is likely due to an increased frequency of the LAR subtype that has more frequent PI3K pathway mutations than other TNBC subtypes (Lehmann et al., 2014). Jiang et al. (2019) also identified specific mutational signatures based on the Catalog of Somatic Mutations on Cancer that inform on aspects of cancer etiology, including mutational signatures related to APOBEC deaminases, homologous recombination deficiency, and clock-like aging.
Importantly, Jiang et al. (2019) suggest subtype-specific therapeutic options for Chinese patients, which they are proposing to explore (NCT03805399), and for patients with TNBC in general. These are illustrated in Figure 1, where an attemptismadetoreconciletheirsubtypes with those defined by other groups. For example, LAR subtype patients may respond to combination therapy with PI3K pathway and androgen inhibitors (NCT02457910). Alternatively, CDKN2A loss and CCND1 amplification may render LAR tumors responsive to treatment with CDK4/6 and AR inhibitors (NCT03805399).
Patients with IM tumors showed a significantly better prognosis after adjusting for lymph node status and tumor size, which is consistent with increased activity of the adaptive immune system and IFN-γ-related pathways. Indeed, multiple trials of immune modulators are underway in this population (NCT03487666, NCT01042379, NCT03801369). The BLIS subtype was associated with reduced survival duration and lack of immune activation. A subset of BLIS tumors showed high HRD scores suggesting treatment with platinum-based therapy or PARP inhibitors. The enrichment of JAK/STAT pathway activity in MES subtype tumors was used to suggest treatment with JAK/STAT inhibitors; however, because this subtype may reflect high stromal cell content, this interpretation must be taken with caution (Lehmann et al., 2016).
Overall, Jiang et al. (2019) provide information showing that Asian TNBCs presenting at Fudan University display similar characteristics to studies from Europe and North America. However, there are some molecular features specific to the Fudan population that may guide therapeutic decisions for these patients. The study also adds a large ethnically and geographically distinct cohort to the growing international compendium of molecular information about breast cancers.
ACKNOWLEDGMENTS
L.M.H., G.B.M., and J.W.G. are supported by the National Institutes of Health grant U54 HG008100, the National Cancer Institute grants U2C CA233280 and U54 CA209988, and the Prospect Creek Foundation. L.M.H. and J.W.G. are supported by National Cancer Institute grant U54 CA209988. J.W.G. and G.B.M. are supported by the Susan G. Komen Foundation. L.M.H. and G.B.M. are supported by the Breast Cancer Research Foundation. L.M.H. is supported by The Jayne KoskinasTed Giovanis Foundation.
J.W.G. receives research support from Danaher Corporation, PDX Pharmaceuticals, Zeiss, Thermo Fisher Scientific, and Micron Technology and royalties for licensed technology from Abbott Diagnostics and the Danaher Corporation, and he has ownership positions in PDX Pharmaceuticals and Convergent Genomics. G.B.M. receives research support from AstraZeneca, Ionis, and Pfizer; is a SAB member or Consultant for AstraZeneca, Chrysallis Biotechnology, ImmunoMET, Ionis, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, and Tarveda; has stock options or other financial Interests with Catena Pharmaceuticals, ImmunoMet, SignalChem, Spindle Top Ventures, and Tarveda; and has licensed a HRD assay to Myriad Genetics and a DSP patent to Nanostring.
Footnotes
DECLARATION OF INTERESTS
REFERENCES
- Abramson VG, Lehmann BD, Ballinger TJ, and Pietenpol JA (2015). Subtyping of triplenegative breast cancer: implications for therapy. Cancer 121, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, B0rresen-Dale AL, et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N; Cancer Genome Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490,61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. ; METABRIC Group (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YC, Steele L, Warden C, Wilczynski S, Mortimer J, Yuan Y, and Neuhausen SL (2019). Molecular subtypes of triple-negative breast cancer in women of different race and ethnicity. Oncotarget 10, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, Yoshimatsu TF, Pitt JJ, Hoadley KA, Troester M, et al. (2017). Comparison of breast cancer molecular features and survival by African and European ancestry in the cancer genome atlas. JAMA Oncol. 3, 1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y-Z, Ma D, Suo C, Shi J, Xue M, Hu X, Xiao Y, Yu K-D, Liu Y-R, Yu Y, et al. (2019). Genomic and transcriptional landscape of triple negative breast cancers: subtypes and treatment strategies. Cancer Cell 35, this issue, 428–440. [DOI] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Schafer JM, Pendleton CS, Tang L, Johnson KC, Chen X, Balko JM, Gomez H, Arteaga CL, et al. (2014). PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 16, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, and Pietenpol JA (2016). Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoSONE 11, e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, S0rlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. (2000). Molecular portraits of human breast tumours. Nature 406, 747–752. [DOI] [PubMed] [Google Scholar]


