A Neurospora meiotic drive element known as Spore killer-2 (Sk-2) achieves biased transmission through sexual reproduction by killing siblings that inherit a competing allele...
Keywords: Genetics of Sex, meiotic drive, meiotic silencing, RNA editing, spore killing, transmission ratio distortion
Abstract
Sk-2 is a meiotic drive element that was discovered in wild populations of Neurospora fungi over 40 years ago. While early studies quickly determined that Sk-2 transmits itself through sexual reproduction in a biased manner via spore killing, the genetic factors responsible for this phenomenon have remained mostly unknown. Here, we identify and characterize rfk-1, a gene required for Sk-2-based spore killing. The rfk-1 gene contains four exons, three introns, and two stop codons, the first of which undergoes RNA editing to a tryptophan codon during sexual development. Translation of an unedited rfk-1 transcript in vegetative tissue is expected to produce a 102-amino acid protein, whereas translation of an edited rfk-1 transcript in sexual tissue is expected to produce a protein with 130 amino acids. These findings indicate that unedited and edited rfk-1 transcripts exist and that these transcripts could have different roles with respect to the mechanism of meiotic drive by spore killing. Regardless of RNA editing, spore killing only succeeds if rfk-1 transcripts avoid silencing caused by a genome defense process called meiotic silencing by unpaired DNA (MSUD). We show that rfk-1’s MSUD avoidance mechanism is linked to the genomic landscape surrounding the rfk-1 gene, which is located near the Sk-2 border on the right arm of chromosome III. In addition to demonstrating that the location of rfk-1 is critical to spore-killing success, our results add to accumulating evidence that MSUD helps protect Neurospora genomes from complex meiotic drive elements.
IN eukaryotic organisms, alleles of nuclear genes are typically inherited in a Mendelian manner. However, some genes possess the ability to improve their own transmission rate through meiosis at the expense of a competing locus. These “selfish” genes are often referred to as meiotic drive elements (Zimmering et al. 1970). The genomic conflict caused by meiotic drive elements may impact processes ranging from gametogenesis to speciation (Lindholm et al. 2016). Meiotic drive elements are found across the eukaryote tree of life (Burt and Trivers 2008; Bravo Núñez et al. 2018), and classic examples include SD in fruit flies (Larracuente and Presgraves 2012), the t-complex in mice (Lyon 2003; Sugimoto 2014), and Ab10 in Zea mays (Rhoades 1952; Kanizay et al. 2013). In the fungal kingdom, the known meiotic drive elements achieve biased transmission through spore killing (Raju 1994) and a handful of spore killer systems have been studied in detail. While the prion-based spore-killing mechanism of het-s in Podospora anserina is well characterized (Dalstra et al. 2003; Saupe 2011), the mechanisms by which other fungal meiotic drive elements kill spores are mostly unknown [e.g., see Grognet et al. (2014), Hu et al. (2017), and Nuckolls et al. (2017)].
Two fungal meiotic drive elements have been identified in the fungus Neurospora intermedia (Turner and Perkins 1979). This species is closely related to the genetic model N. crassa (Davis 2000), and the mating processes in both fungi are essentially identical. Mating begins with the fertilization of an immature fruiting body (protoperithecium) by a mating partner of the opposite mating type. After fertilization, the protoperithecium develops into a mature fruiting body called a perithecium. The nuclei from each parent multiply within the developing perithecium, and a single nucleus from each parent is sequestered into a tube-like meiotic cell (Raju 1980). Meiosis begins with the fusion of parental nuclei and ends with the production of four recombinant daughter nuclei. Each recombinant nucleus proceeds through a single round of mitosis, resulting in a total of eight nuclei in the meiotic cell. A process known as ascosporogenesis then constructs cell walls and membranes around each nucleus to produce sexual spores called ascospores. Maturing ascospores accumulate a dark pigment and develop into the shape of a spindle; thus, at the end of ascosporogenesis, a mature meiotic cell appears to contain eight miniature black American footballs (Figure 1A). The meiotic cells also serve as ascospore sacs (asci) and a single perithecium can produce hundreds of asci, each derived from a unique meiotic event.
Figure 1.
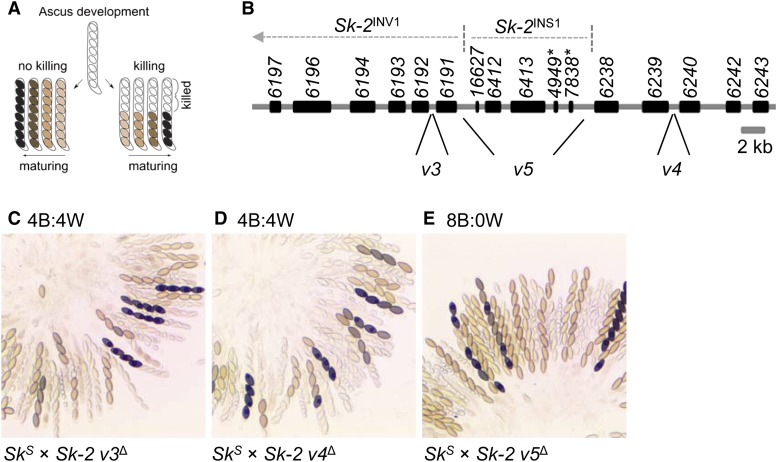
Sk-2INS1 harbors a genetic element required for spore killing. (A) The diagram illustrates phenotypic differences between spore killing and normal ascus development. Asci that have undergone spore killing contain four black and four white ascospores. Viable ascospores may appear tan, brown, or black, depending on their level of maturity. (B) Annotation of the rfk-1 region as described by Harvey et al. (2014). Genes and pseudogenes are depicted as black rectangles. Gene names (e.g., 6197) are listed above the rectangles. Pseudogene names are appended with an asterisk. Labels v3, v4, and v5 mark DNA intervals of the rfk-1 region, which were deleted and replaced with an hph selectable marker. (C–E) The images depict asci from crosses between SkS and Sk-2 strains lacking different intervals of the rfk-1 region. The predominant phenotype of the asci produced by each cross is listed above its image. Crosses are as follows: (C) F2-23 × ISU-3023, (D) F2-23 × ISU-3017, and (E) F2-23 × ISU-3029. B, black; W, white.
During an effort in the 1970s to collect and characterize Neurospora isolates from around the world, Turner and Perkins discovered pairs of compatible mating partners that did not produce asci with eight viable ascospores (Perkins 1974; Turner and Perkins 1979). This outcome was more common when crosses were performed between isolates from widely separated populations, and in some cases, the abnormal asci were attributed to heterozygosity of chromosome rearrangements between mating partners. However, for a few isolates of N. intermedia, asci with atypical phenotypes were found to be due to chromosomal factors called Spore killer-2 (Sk-2) and Spore killer-3 (Sk-3). Sk-2 and Sk-3 are not single genes; rather, they are complexes of genes that span ∼30 cM of chromosome III and are transmitted through meiosis as single units due to a recombination suppression mechanism thought to be enforced by inversions (Turner and Perkins 1979; Campbell and Turner 1987; Hammond et al. 2012; Harvey et al. 2014; Svedberg et al. 2018). Unlike standard genetic elements, which display a Mendelian transmission rate of 50% through sexual reproduction, Sk-2 and Sk-3 are transmitted at levels approaching 100% (Turner and Perkins 1979). This biased transmission occurs because Sk-2 and Sk-3 kill ascospores that do not inherit resistance to spore killing (Raju 1979; Turner and Perkins 1979). For example, in Sk-2 × Spore killer-sensitive (SkS) crosses, asci with four black ascospores and four clear (“white”) ascospores are produced (Figure 1A). This phenotype can be symbolized as 4B:4W. The four black ascospores are typically viable and nearly always of the Sk-2 genotype, while the four white ascospores are inviable and presumed to be of the SkS genotype. The same phenomenon occurs in Sk-3 × SkS crosses, except that the four black ascospores are of the Sk-3 genotype.
Although the Spore killers have not yet been detected in wild isolates of N. crassa, Sk-2 and Sk-3 have been introgressed into this species for genetic analysis. Introgression of Sk-2 and Sk-3 has allowed resistance to spore killing to be discovered in natural populations of N. crassa (Turner and Perkins 1979; Turner 2001). An Sk-2-resistant isolate from Louisiana [Fungal Genetics Stock Center (FGSC) 2222] was found to carry a resistant version of a gene whose function is best described by its name: resistant to Spore killer (rsk). Crosses of rskLA × Sk-2, where rskLA is the allele of rsk carried by FGSC 2222, produce asci with an 8B:0W phenotype because ascospores inherit either rskLA or Sk-2, and both alleles are sufficient for resistance to Sk-2-based spore killing (Hammond et al. 2012). Discovery of rskLA made identifying other rsk alleles possible, some of which do not provide resistance to the known Spore killers. For example, the Oak Ridge rsk allele (rskOR), typical of most laboratory strains of N. crassa, is resistant to neither Sk-2 nor Sk-3. Additionally, some rsk alleles confer resistance to Sk-3 but not Sk-2. An example is rskPF5123, found in an N. intermedia isolate from French Polynesia. Sk-2 and Sk-3 also carry resistant versions of rsk, referred to as rskSk-2 and rskSk-3, respectively. Crosses homozygous for Sk-2 (i.e., Sk-2 × Sk-2) or Sk-3 produce asci with an 8B:0W phenotype because each ascospore inherits a resistant rsk allele. In contrast, heterozygous crosses between different Spore killers (e.g., Sk-2 × Sk-3) produce asci with a 0B:8W phenotype (Turner and Perkins 1979) because each ascospore inherits either rskSk-2 or rskSk-3, but not both (rskSk-2 ascospores are killed by Sk-3, while rskSk-3 ascospores are killed by Sk-2).
The Killer-Neutralization (KN) model has been proposed to explain how Sk-2 and Sk-3 achieve biased transmission through sexual reproduction (Hammond et al. 2012). In this model, Sk-2 and Sk-3 each use a resistance protein and a killer protein (or nucleic acid), and both proteins are active throughout meiosis and ascosporogenesis; thus, in an SkS × Sk-2 (or Sk-3) cross, both the resistance protein and the killer protein diffuse freely throughout the meiotic cell during early stages of meiosis. This unrestricted movement of both proteins allows the resistance protein to neutralize the killer throughout the cell. However, once ascospores are separated from the cytoplasm by ascospore delimitation, the resistance protein becomes restricted to the ascospores that produce it (i.e., Sk-2 ascospores). Ascospores that do not carry a resistant version of rsk (i.e., SkS ascospores) are subsequently killed. This model requires the killer to move between ascospores after ascospore delimitation. Alternatively, the killer may have a long half-life that allows it to remain functional in sensitive ascospores after ascospore delimitation.
Evidence for the KN model is seen in the outcome of SkS × Sk-2 rskΔSk-2 crosses, where the latter strain has been deleted of its rsk allele. These crosses do not produce ascospores. Instead, SkS × Sk-2 rskΔSk-2 crosses produce asci that abort meiosis before ascospore production (Hammond et al. 2012). Meiotic cells of these crosses lack a resistant RSK, which likely causes the killing process to begin early in meiosis (at the ascus level) rather than during ascosporogenesis (at the ascospore level). The KN model is also supported by the existence of different rsk alleles. For example, previous studies have demonstrated that the sequence of RSK is the most important factor toward determining which killer it neutralizes, suggesting that RSK and the killer may interact by a “lock-and-key” mechanism (Hammond et al. 2012). However, to test this hypothesis, the killer must first be identified.
As described above, SkS × Sk-2 rskΔSk-2 crosses fail to produce viable ascospores, presumably because the killer protein is present and the resistance gene is not. We recently used this ascospore production defect to screen for mutations that disrupt spore killing (Harvey et al. 2014). Specifically, we fertilized an SkS mating partner with mutagenized Sk-2 rskΔSk-2 conidia (asexual spores that also function as fertilizing propagules), reasoning that only an Sk-2 rskΔSk-2 conidium that was mutated in a gene “required for killing” (rfk) would allow viable ascospores to be produced. With this method, we isolated six rfk mutants (ISU-3211 through ISU-3216). Complementation analysis of these mutants suggested that they are all mutated at the same locus, which was subsequently named rfk-1 and mapped to a 45-kb region within Sk-2 on chromosome III.
In this study, we identify and characterize rfk-1. Our results indicate that rfk-1 is required for spore killing and that it is located next to the right border of Sk-2. This location allows it to escape silencing caused by a genome defense process called meiotic silencing by unpaired DNA (MSUD) (Shiu et al. 2001). We propose a model of rfk-1 gene structure that includes four exons, three introns, and a coding region for a protein of at least 102 amino acids (aa). We also present evidence that the stop codon at “position 103” in the rfk-1 transcript undergoes RNA editing during sexual development. According to standard rules of mRNA translation, the edited rfk-1 transcript contains the code for a 130-aa protein. Possibilities that the shorter and/or longer RFK-1 protein variants are required for the meiotic drive mechanism are discussed.
Materials and Methods
Strains, media, and crossing conditions
The strains used in this study are listed along with genotype information in Table 1. Vogel’s minimal medium (Vogel 1956), with supplements as required, was used to grow and maintain all strains. Hygromycin B and nourseothricin sulfate were used at a working concentration of 200 and 45 μg/ml, respectively. Synthetic crossing medium (pH 6.5) with 1.5% sucrose, as described by Westergaard and Mitchell (1947), was used for crosses. Crosses were unidirectional and performed on a laboratory benchtop at room temperature under ambient lighting (Samarajeewa et al. 2014). After fertilization, crosses were allowed to mature for 12–16 days before perithecial dissection in 25 or 50% glycerol. Asci were then examined with a compound microscope (Leica DMBRE) and imaging system. Ascus phenotype designations were based on qualitative observations. More than 90% of the asci from a cross had to display the same phenotype to receive one of the following designations: 8B:0W, 4B:4W, or aborted.
Table 1. Strains used in this study.
| Name (alias) | Genotypea |
|---|---|
| F2-19 | rid; fl; Sk-2 A |
| F2-23 (RTH1005.1) | rid; fl A |
| F2-26 (RTH1005.2) | rid; fl a |
| FGSC 1766 | N. intermedia A |
| FGSC 1767 | N. intermedia a |
| FGSC 7426 | N. intermedia Sk-2 A |
| ISU-3017 (RKS2.1.2) | rid?; Sk-2 leu-1 v4Δ::hph; mus-51? a |
| ISU-3023 (RKS1.1.6) | rid?; Sk-2 leu-1 v3Δ::hph; mus-51? a |
| ISU-3029 (RKS3.2.5) | rid; Sk-2 leu-1 v5Δ::hph; mus-51Δ:bar a |
| ISU-3036 (RTH1623.1) | rid; fl; sad-2Δ::hph A |
| ISU-3037 (RTH1623.2) | rid; fl; sad-2Δ::hph a |
| ISU-3211 (RTH1158.8) | rid; Sk-2 rskΔ::hph rfk-13211; mus-51Δ::bar a |
| ISU-3212 (RTH1158.13) | rid; Sk-2 rskΔ::hph rfk-13212 a |
| ISU-3213 (RTH1158.24) | rid; Sk-2 rskΔ::hph rfk-13213 a |
| ISU-3214 (RTH1158.34) | rid; Sk-2 rskΔ::hph rfk-13214; mus-51Δ::bar A |
| ISU-3215 (RTH1158.35) | rid; Sk-2 rskΔ::hph rfk-13215 a |
| ISU-3216 (RTH1158.44) | rid; Sk-2 rskΔ::hph rfk-13216 a |
| ISU-3222 (RTH1249.14) | rid; Sk-2 rfk-13211; mus-51Δ::bar a |
| ISU-3223 (RTH1294.17) | Sk-2 leu-1; mus-51Δ::bar A |
| ISU-3224 (HAH8.1.3) | rid his-3+::AH4Sk-2-hph; A |
| ISU-3228 (HAH10.1.1) | rid his-3+::AH6Sk-2-hph; A |
| ISU-3243 (HAH16.1.1) | rid his-3+::AH14Sk-2-hph a |
| ISU-3311 (RDS1.1) | Sk-2 leu-1 v31Δ::hph; mus-51Δ∷bar A |
| ISU-3313 (RDS2.3) | Sk-2 leu-1 v32Δ::hph; mus-51Δ∷bar A |
| ISU-3315 (RDS3.9) | Sk-2 leu-1 v33Δ::hph a |
| ISU-3318 (RDS4.8) | Sk-2 leu-1 v34Δ::hph A |
| ISU-3321 (RDS5.9) | rid; Sk-2 leu-1 v35Δ::hph; mus-51Δ::bar a |
| ISU-3478 (RDS13.9.1) | rid; Sk-2 v37Δ::hph; mus-51Δ::bar A |
| ISU-3482 (RDS14.4.2) | rid; Sk-2 v38Δ::hph A |
| ISU-3483 (RDS15.1.1) | rid; Sk-2 v39Δ::hph A |
| ISU-3485 (RDS16.4.1) | rid; Sk-2 v40Δ::hph A |
| ISU-3656 (HAH42.1) | rid his-3+::AH30Sk-2-hph A |
| ISU-3658 (HAH43.1) | rid his-3+::AH31Sk-2-hph A |
| ISU-3660 (HAH44.1) | rid his-3+::AH32Sk-2-hph A |
| ISU-4269 (RAH64.1.1) | rid his-3+::AH37Sk-2-hph; mus-52Δ::bar a |
| ISU-4271 (RAH63.1.2) | rid his-3+::AH36Sk-2-hph; mus-52Δ::bar A |
| ISU-4273 (HNR12.6.1) | rid his-3+::AH36Sk-2-hph A |
| ISU-4275 (HNR10.4.2) | rid his-3+::AH363211-hph A |
| ISU-4344 (RAY1.13) | rid; Sk-2 v140Δ::hph; mus-51Δ::bar a |
| ISU-4348 (RAY6.5) | rid; fl; v150Δ::hph A |
| ISU-4551 (RNR29.1) | rid his-3+::AH36Sk-2[G27945A]-hph; mus-52? A |
| ISU-4552 (RNR28.1) | rid his-3+::AH36Sk-2[G27972A]-hph; mus-52? A |
| ISU-4553 (RNR27.1) | rid his-3+::AH36Sk-2[G28052A]-hph; mus-52? A |
| ISU-4554 (RNR26.1) | rid his-3+::AH36Sk-2[G28104A]-hph; mus-52? A |
| ISU-4555 (RNR25.1) | rid his-3+::AH36Sk-2[G28300A]-hph; mus-52? A |
| ISU-4556 (RNR30.1) | rid his-3+::AH36Sk-2[G28326A]-hph; mus-52? A |
| ISU-4557 (RNR129.1.3) | rid; Sk-2 v199Δ::hph-ccg-1(P)-ATG; mus51? A |
| ISU-4559 (RNR108.1.12) | rid; fl; ncu06238Δ::hph mus-52Δ::bar a |
| ISU-4561 (RNR109.3.2) | rid; Sk-2 ncu06238Δ::hph; mus51Δ::bar A |
| ISU-4562 (HNR92.1) | rid; Sk-2 v160Δ::nat1; mus-51Δ::bar a |
| ISU-4563 (HNR100.11.1) | rid; Sk-2 v140Δ::hph; mus-51Δ::bar a |
| ISU-4576 (HAA108.1.1) | rid; Sk-2 v214Δ::hph-ccg-1(P)-ATGT; mus-51Δ A |
| ISU-4584 (HAA117.1.1) | rid; Sk-2 v221Δ::hph-ccg-1(P)-TAA; mus-51Δ A |
| P8-42 | rid; mus-51Δ::bar a |
| P8-43 | rid; mus-52Δ::bar A |
| P15-53 (RTH1122.22) | rid; Sk-2; mus-51Δ::bar A |
The rid?, mus-51?, and mus-52? designations are used if the genotype has not been determined for the indicated allele. Genotypes of critical alleles were determined by PCR and/or lineage analysis.
Genetic modification of N. crassa, genotyping, and sequence confirmation
A technique called double-joint polymerase chain reaction (PCR) was used to construct all deletion vectors (Yu et al. 2004; Hammond et al. 2011). Transgene-insertion vectors were designed to insert transgenes along with a hygromycin resistance cassette (hph) next to his-3 on chromosome I. Construction details for deletion and insertion vectors are provided in Supplemental Material, Tables S1–S5. Transformations of N. crassa were performed by electroporation of conidia (Margolin et al. 1997). Homokaryons were derived from heterokaryotic transformants with a microconidium isolation technique (Ebbole and Sachs 1990) or by crossing the transformants to a standard laboratory strain (F2-23 or F2-26) to obtain homokaryotic ascospores. Site-directed mutagenesis was performed essentially as described for the QuikChange II Site-Directed Mutagenesis Kit (Revision E.01; Agilent Technologies). Additional details are provided in Table S5. Genotypes were confirmed by PCR assays on genomic DNA isolated from lyophilized (freeze-dried) mycelia with the IBI Scientificʼs Mini Genomic DNA Kit (Plant/Fungi). Sanger sequencing was used to confirm sequences and/or identify mutations in PCR products and plasmids.
Analysis of RNA sequencing data
Six data sets containing paired-end sequences of poly(A)-enriched RNA from perithecial cultures of N. intermedia were downloaded from the Sequence Read Archive (Leinonen et al. 2011). The accession numbers for the three SkS (FGSC 1767) × Sk-2 (FGSC 7426) data sets are SRR7700963, SRR7700964, and SRR7700965. Additionally, the accession numbers for the SkS (FGSC 1767) × SkS (FGSC 1766) data sets are SRR7700966, SRR7700967, and SRR7700968. Sequences from each data set were aligned to the AH36 interval of N. intermedia FGSC 7426 and the actin gene of N. intermedia FGSC 1766/8761 with Bowtie 2 (v2.3.4.1) (Langmead and Salzberg 2012). Alignment parameters were as follows: –local, –no-unal, –no-discordant, –no-mixed, –no-overlap, –no-contain, -X 1000. Custom Perl and Python scripts were then used to count each time a position in the reference was covered by a read. Only aligned reads with an edit distance less than four were considered in the coverage calculations. Reads from similar crosses were combined for calculations and charts were generated with Microsoft Excel (data were not normalized among replicate data sets). N. intermedia AH36 and actin DNA sequences were obtained from the genome sequences of FGSC 7426 and FGSC 1766/8761, respectively (Svedberg et al. 2018).
cDNA analysis
For vegetative tissues, total RNA was isolated from 36-hr mycelial cultures grown in liquid Vogel’s medium at 32°. For sexual tissues, total RNA was isolated from 6-day-old perithecia from a unidirectional cross between SkS (F2-26) and Sk-2 (P15-53), where SkS was used as the female. Vegetative tissues were lyophilized before RNA isolation. In contrast, fresh sexual tissues were directly ground in TRIzol reagent (Invitrogen, Carlsbad, CA) with a mortar and pestle. For both vegetative and sexual total RNA isolations, TRIzol was used at a ratio of 1.5 ml TRIzol per 100 mg of tissue. The remainder of the TRIzol-based RNA isolation procedure was performed according to the manufacturer’s recommendations. The RNA pellets from the TRIzol isolation were purified with the PureLink RNA Mini Kit (Invitrogen) by following the manufacturer’s instructions for RNA isolation with an on-column DNAse treatment. First-strand cDNA synthesis was then performed with the ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs, Beverly, MA) and the kit’s included oligo-d(T) primer. Following first-strand cDNA synthesis, the reaction mixture was diluted with sterile water to a final volume of 50 µl. cDNA fragments were then amplified with standard PCR reactions. Amplified cDNA fragments were purified by agarose gel extraction and analyzed by Sanger sequencing without cloning. Sanger sequencing data and chromatograms were visualized with BioEdit (Hall 1999).
Data availability
All strains and plasmids generated during this study are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7836692.
Results
Deletion of a DNA interval within Sk-2INS1 eliminates spore killing
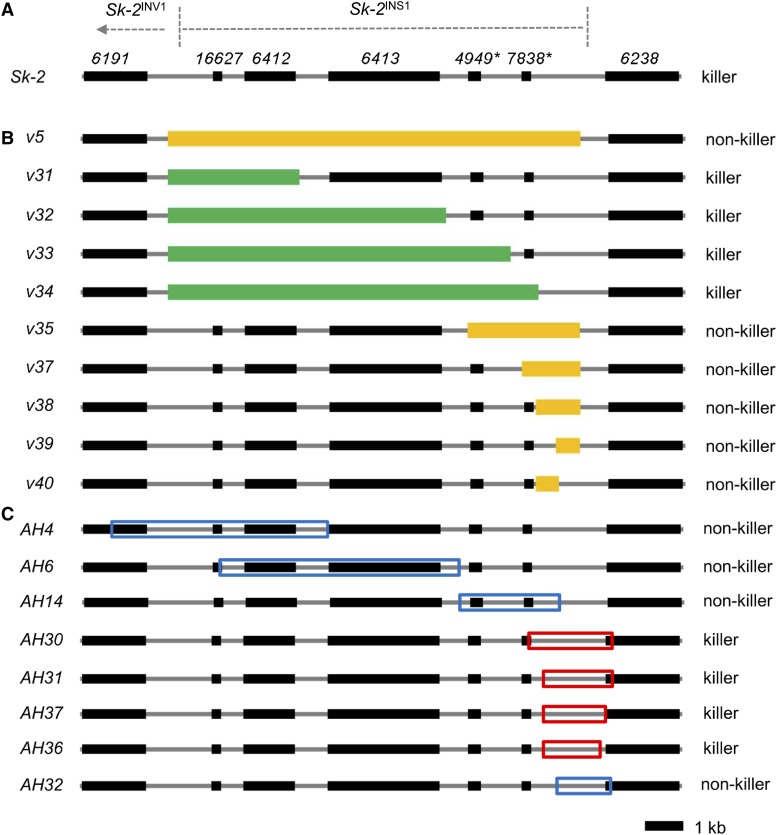
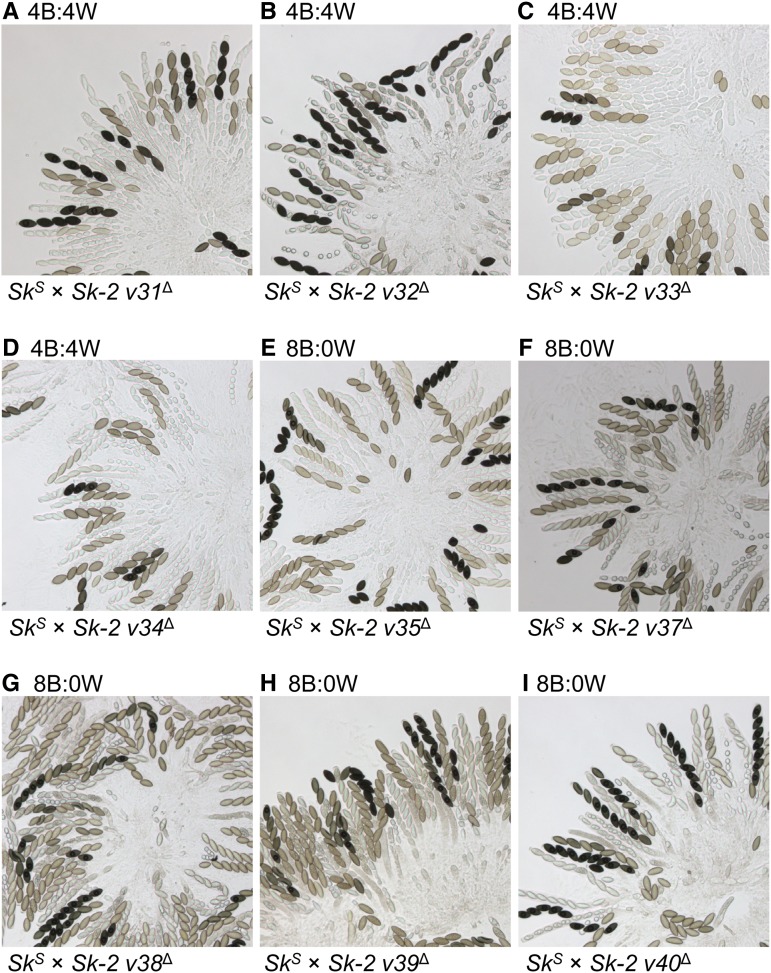
As described in the Introduction, we previously mapped rfk-1 to a 45-kb interval of Sk-2. Additionally, this interval was predicted to contain 14 protein-coding genes, two pseudogenes (denoted with an asterisk), an inverted sequence (Sk-2INV1), an inversion breakpoint, and an 11-kb insertion (Sk-2INS1) (Figure 1B; GenBank: KJ908288.1; Harvey et al. 2014). To further refine the location of rfk-1 within the 45-kb interval, we deleted subintervals v3, v4, and v5 (Figure 1B and Table 2). The resulting deletion strains were then crossed to an SkS mating partner. While spore killing was functional in both SkS × Sk-2 v3∆ and SkS × Sk-2 v4∆ crosses (asci were 4B:4W; Figure 1, C and D), it was absent in an SkS × Sk-2 v5∆ cross (asci were 8B:0W; Figure 1E). These results suggest that v5 contains rfk-1. To refine the position of rfk-1 within v5, we constructed nine deletion strains (Figure 2, A and B and Table 2) and crossed each one to an SkS mating partner. Surprisingly, while deletion of the previously annotated genes and pseudogenes within Sk-2INS1 had no effect on spore killing (Figure 3, A–D), deletion of the intergenic region between ncu07838* and ncu06238 eliminated it (Figure 3, E–I). These results suggest that rfk-1 is found within the intergenic region between ncu07838* and ncu06238.
Table 2. Interval positions.
| Name | Start | Stop |
|---|---|---|
| Miscellaneous intervals of the 45-kb rfk-1 region | ||
| Sk-2INS1 | 18,118 | 29,151 |
| rfk-1 coding | 28,264 | 28,968/29,106 |
| Repetitivea | 28,384 | 28,722 |
| Intervals deleted from the 45-kb rfk-1 region | ||
| v3 | 15,640 | 15,664 |
| v4 | 36,166 | 36,426 |
| v5 | 18,042 | 28,759 |
| v31 | 18,042 | 21,464 |
| v32 | 18,042 | 25,268 |
| v33 | 18,042 | 26,951 |
| v34 | 18,042 | 27,667 |
| v35 | 25,837 | 28,759 |
| v37 | 27,242 | 28,759 |
| v38 | 27,602 | 28,759 |
| v39 | 28,126 | 28,759 |
| v40 | 27,602 | 28,198 |
| v140 | 29,381 | 29,401 |
| v160 | 27,740 | 29,401 |
| v175 | 29,489 | 31,883 |
| v199 | 28,131 | 28,263 |
| v214 | 28,131 | 28,263 |
| v221 | 28,131 | 28,263 |
| Intervals transferred from the 45-kb rfk-1 region to SkS | ||
| AH4 | 16,579 | 22,209 |
| AH6 | 19,408 | 25,648 |
| AH14 | 25,632 | 28,324 |
| AH30 | 27,528 | 29,702 |
| AH31 | 27,900 | 29,702 |
| AH32 | 28,304 | 29,702 |
| AH36 | 27,900 | 29,380 |
| AH37 | 27,900 | 29,512 |
The coordinates of each interval correspond to the GenBank sequence KJ908288.1.
Contains repeats of a 46–48-bp sequence.
Figure 2.
Deletion and insertion maps. (A) A diagram of Sk-2INS1 and its immediate neighbors. (B) Various intervals of Sk-2INS1 were deleted and replaced with hph. For convenience, each interval was named according to its deletion vector (e.g., interval v5 is named after deletion vector v5). Mango rectangles mark intervals that disrupt spore killing upon deletion. Green rectangles mark intervals that do not disrupt spore killing when deleted. (C) Eight intervals of Sk-2INS1 were transferred to the his-3 locus of an SkS strain. Intervals were labeled according to the name of the plasmid used to clone each interval (e.g., interval AH4 is named after plasmid pAH4). Red and blue boxes mark killer (i.e., abortion-inducing) and nonkiller intervals, respectively.
Figure 3.
Deletion of a genetic element between pseudogene 7838* and the right border of Sk-2INS1 eliminates spore killing. (A–I) The images depict asci from crosses between an SkS strain and an Sk-2 mating partner. Each Sk-2 mating partner has been deleted of a different subinterval of v5. Crosses are as follows: (A) F2-26 × ISU-3311, (B) F2-26 × ISU-3313, (C) F2-23 × ISU-3315, (D) F2-26 × ISU-3318, (E) F2-23 × ISU-3321, (F) F2-26 × ISU-3478, (G) F2-26 × ISU-3482, (H) F2-26 × ISU-3483, and (I) F2-26 × ISU-3485. B, black; W, white.
An ascus-aborting element exists between ncu07838* and ncu06238
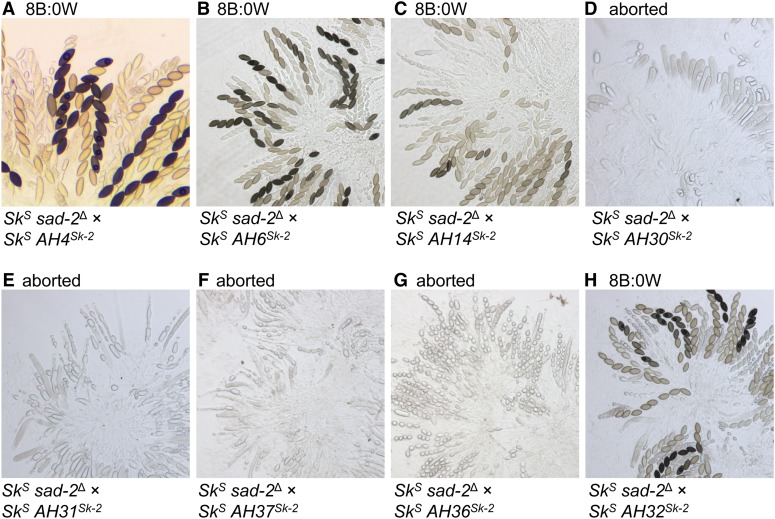
To determine if a segment of v5 could induce spore killing when transferred to an SkS genetic background, we genetically modified eight SkS strains to carry different v5 subintervals (Figure 2C and Table 2). However, none of the v5-subintervals caused spore killing when crosses were performed between subinterval-carrying strains and a standard SkS mating partner (Figure S1). These results can be explained by the existence of MSUD, a genome defense process that silences unpaired DNA during meiosis (e.g., transgenes that are not found at the same location in the genomes of both parents of a cross; Aramayo and Selker 2013; Hammond 2017). Therefore, to prevent MSUD from silencing the v5-subinterval transgenes, we crossed the v5-subinterval-carrying strains to an SkS sad-2∆ mating partner [the sad-2∆ allele suppresses MSUD when present in one parent of a cross (Shiu et al. 2006)]. In these crosses, four of the v5 subintervals had no discernable effect on ascus development (AH4, AH6, AH14, and AH32), while four others correlated with an ascus abortion phenotype (AH30, AH31, AH36, and AH37) (Figure 2C and Figure 4). It seems likely that ascus abortion occurs when a subinterval carries a functional rfk-1 allele. A traditional 4B:4W ascus phenotype is not possible in these crosses because both parents lack a resistant rsk allele. In summary, these experiments demonstrate that rfk-1 should reside within a subinterval of v5 that is shared among AH30, AH31, AH36, and AH37.
Figure 4.
A genetic element within v5 causes ascus abortion upon its transfer to SkS. (A–H) Images depict asci from crosses between SkS sad-2Δ and an SkS mating partner. Each SkS mating partner carries a different subinterval of v5. Crosses are as follows: (A) ISU-3037 × ISU-3224, (B) ISU-3037 × ISU-3228, (C) ISU-3036 × ISU-3243, (D) ISU-3037 × ISU-3656, (E) ISU-3037 × ISU-3658, (F) ISU-3036 × ISU-4269, (G) ISU-3037 × ISU-4271, and (H) ISU-3037 × ISU-3660. B, black; W, white.
A point mutation disrupts the ascus-aborting ability of AH36Sk-2
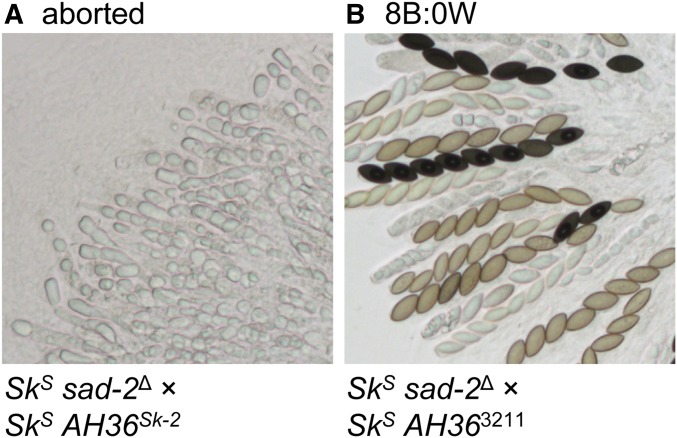
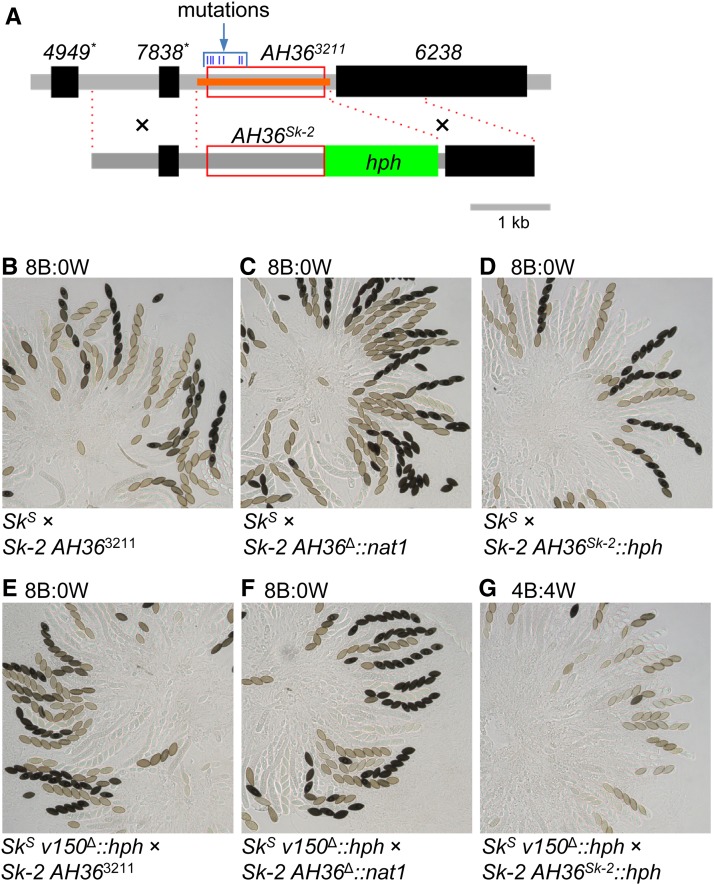
AH36 is the shortest (1481 bp) of the four abortion-inducing intervals identified above and is nested within each of them (Figure 2C and Table 2). More specifically, AH36 is found between positions 27,899 and 29,381 of the previously defined 45-kb rfk-1 region (GenBank: KJ908288.1 and Table 2). We predicted that the AH36 interval from ISU-3211 (a genetically defined rfk-1 mutant strain) would not cause ascus abortion if it were transferred to an SkS strain and analyzed in a cross with an SkS sad-2∆ mating partner. Indeed, comparison of SkS sad-2∆ × AH36Sk-2 and SkS sad-2∆ × AH363211 crosses indicates that the AH36 interval from Sk-2 (called AH36Sk-2) causes ascus abortion and that the AH36 interval from ISU-3211 (called AH363211) does not (Figure 5).
Figure 5.
The AH36 interval from an rfk-1 mutant does not cause ascus abortion. Images depict asci from crosses between an SkS sad-2Δ strain and an SkS strain, carrying either the AH36 interval from (A) F2-19 (rfk-1+) or (B) ISU-3211 (an rfk-1 mutant). Crosses are as follows: (A) ISU-3037 × ISU-4273 and (B) ISU-3037 × ISU-4275. B, black; W, white.
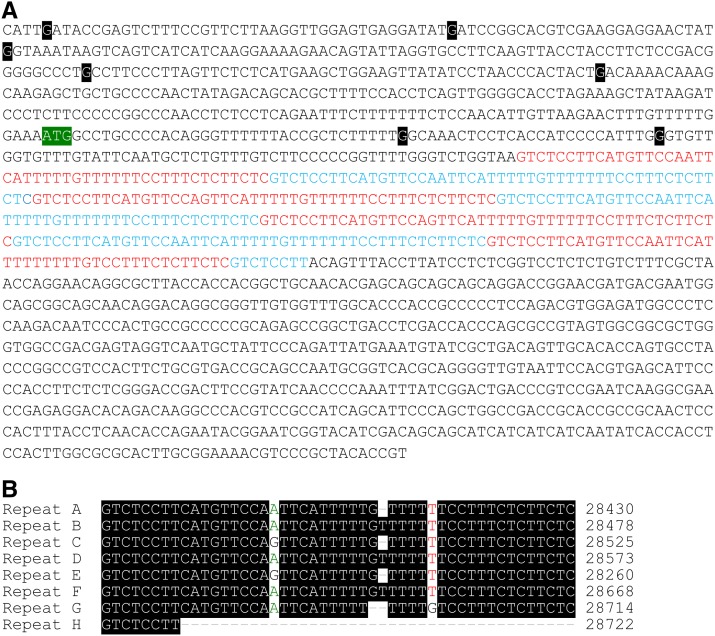
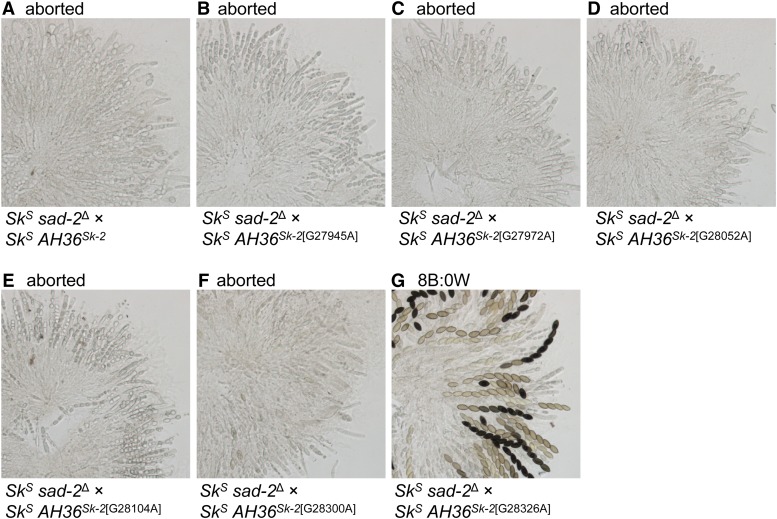
The inability of AH363211 to cause ascus abortion suggests that it differs from AH36Sk-2 at the sequence level. By sequencing AH363211, we identified seven guanine-to-adenine transition mutations relative to AH36Sk-2 (Figure 6A; G27904A, G27945A, G27972A, G28052A, G28104A, G28300A, and G28326A). We then used site-directed mutagenesis to determine which, if any, of these mutations eliminated AH363211’s ability to abort asci. For each mutation, this involved mutating the base in a clone of AH36Sk-2, placing the mutated interval (e.g., AH36Sk-2[G27945A]) in an SkS strain, and crossing the transgenic strain to an SkS sad-2Δ mating partner. Of the six mutations tested, only G28326A eliminated the ascus-aborting ability of AH36Sk-2 (Figure 7).
Figure 6.
The AH36 interval from an rfk-1 mutant contains seven point mutations. (A) The 1481-bp sequence of AH36Sk-2 is shown. A region containing 7.17 repeats of a 46–48-bp sequence is highlighted with red font and blue font. The colors alternate with each iteration of the repeated sequence. The sequence of AH363211 contains seven G-to-A transition mutations. The position of each mutation is marked by a white character on a black background with the nonmutated base shown. The mutations in AH363211 are (from left to right): G27904A, G27945A, G27972A, G28052A, G28104A, G28300A, and G28326A. The putative rfk-1 start codon is depicted with white characters on a green background. (B) Alignment of the repetitive sequences highlighted in (A).
Figure 7.
A point mutation within AH36 eliminates its ability to abort asci. Six of seven point mutations in AH363211 were examined for a potential role in eliminating ascus abortion. Each mutation was placed individually in AH36Sk-2 by site-directed mutagenesis. (A–G) Images depict asci from crosses between an SkS sad-2Δ strain and an SkS mating partner carrying AH36Sk-2 or one of its mutated derivatives. Crosses are as follows: (A) ISU-3037 × ISU-4273, (B) ISU-3037 × ISU-4551, (C) ISU-3037 × ISU-4552, (D) ISU-3037 × ISU-4553, (E) ISU-3037 × ISU-4554, (F) ISU-3037 × ISU-4555, and (G) ISU-3037 × ISU-4556. B, black; W, white.
A start codon for RFK-1
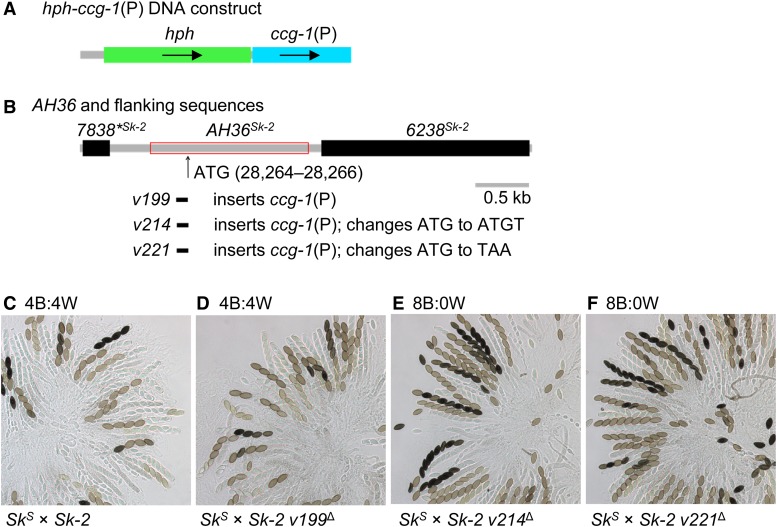
The G28326A mutation is found ∼60 bp downstream of a putative start codon at positions 28,264–28,266 (Figure 6A). To test if this “ATG” is the start codon for an RFK-1 protein, we constructed three gene replacement vectors: v199, v214, and v221. Vector v199 inserts the N. crassa ccg-1 promoter [ccg-1(P)] directly upstream of the ATG (Figure 8, A and B). Vector v214 is nearly identical to v199, except that it changes the ATG to ATGT (i.e., it inserts an extra base and induces a frameshift). Vector v221 is also nearly identical to v199; however, it changes the ATG to TAA (i.e., a stop codon). With these vectors, we found that attaching ccg-1(P) to the ATG (with v199) has no effect on spore killing (Figure 8, compare C and D) and that changing the ATG to ATGT (with v214) or TAA (with v221) eliminates spore killing (Figure 8, compare C, E, and F). Together, these results suggest that positions 28,264–28,266 code for a functional rfk-1 start codon and that rfk-1 is a protein-coding gene.
Figure 8.
A putative start codon for RFK-1 exists within AH36. (A) A DNA construct consisting of hph (a hygromycin resistance cassette) and the promoter for the N. crassa ccg-1 gene was used to make three transformation vectors: v199, v214, and v221. (B) Vector v199 replaces 133 bp of AH36 while fusing ccg-1(P) to a putative start codon at position 28,264. Vector v214 and v221 are nearly identical to v199, except that they change the start codon to ATGT and TAA, respectively. (C–F) Crosses were performed to determine the effect of each allele on spore killing. Images depict asci from the following crosses: (C) F2-26 × P15-53, (D) F2-26 × ISU-4557, (E) F2-26 × ISU-4576, and (F) F2-26 × ISU-4584. With respect to the image labels, v199∆, v214∆, and v221∆ are abbreviations of v199∆::hph-ccg-1(P)-ATG, v214∆::hph-ccg-1(P)-ATGT, and v221∆::hph-ccg-1(P)-TAA, respectively. B, black; W, white.
The arrangement of rfk-1 within Sk-2 protects it from MSUD
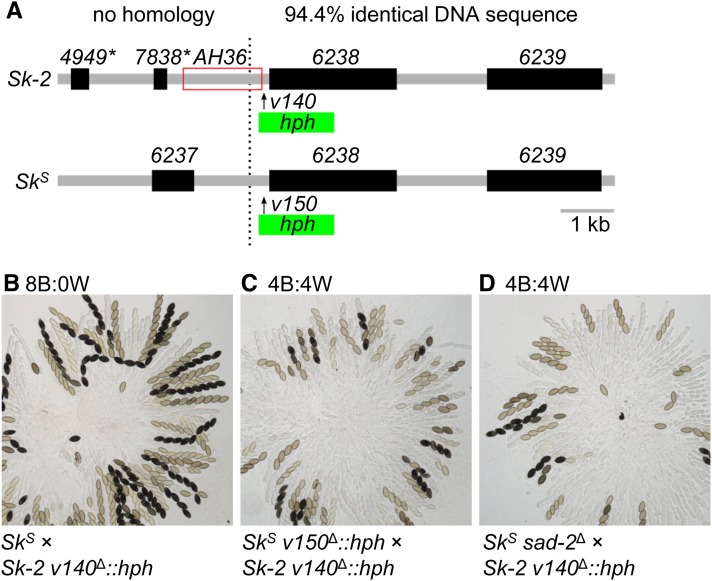
The right border of Sk-2 is thought to be located at (or very near) position 29,151 (Figure 9A, dotted line; Table 2; Harvey et al. 2014). This hypothesis is based on the similarity of DNA sequences found on the centromere-distal side (to “the right”) of this position in both Sk-2 and SkS strains. For example, a ClustalW-based alignment (Thompson et al. 1994) found that Sk-2 positions 29,152 through 35,728 are 94.4% identical to the corresponding positions within SkS (GenBank: CM002238.1, positions 2,011,073 to 2,017,662). In contrast, the sequences found on the centromere-proximal side (“to the left”) of the Sk-2 right border are completely unrelated between Sk-2 and SkS strains (Figure 9A). Because most of AH36 is found on the centromere-proximal side of the Sk-2 border (Figure 9A), most of this interval (including rfk-1) should be unpaired during meiosis in SkS × Sk-2 crosses. Assuming rfk-1 is unpaired during meiosis, how does it avoid silencing by MSUD? While the molecular details of how MSUD detects unpaired DNA are mostly unknown, we considered the possibility that the short distance of rfk-1 from a “paired” sequence allows it to avoid MSUD (e.g., note that the ncu06238 genes in Sk-2 and SkS are paired and that AH36 is located relatively close to these paired genes; Figure 9A). To test this hypothesis, we inserted a hygromycin resistance cassette (hph) between AH36 and the Sk-2 right border in a standard Sk-2 strain (Figure 9A). We refer to this particular hph transgene as v140Δ::hph. The v140Δ::hph transgene increased the distance of rfk-1 from paired sequences by a length of 1391 bp. As a control, we inserted a similar transgene called v150Δ::hph at the corresponding location in an SkS strain. In agreement with our hypothesis, an SkS × Sk-2 v140Δ::hph cross lacked signs of spore killing, while spore killing was normal in an SkS v150Δ::hph × Sk-2 v140Δ::hph cross (Figure 9, B and C). These results suggest that if rfk-1 is located too far from paired DNA, it is detected by MSUD and silenced. As an additional test of this hypothesis (rfk-1 must be located near paired DNA to avoid MSUD), we analyzed asci from an SkS sad-2Δ × Sk-2 v140Δ::hph cross and found spore killing to be normal (Figure 9D). This outcome can be explained by the presence of sad-2Δ, which suppresses MSUD and makes the distance of rfk-1 from paired DNA irrelevant to its expression during meiosis.
Figure 9.
The native arrangement of rfk-1 protects it from meiotic silencing by unpaired DNA. (A) Interval AH36 spans the right border of Sk-2 (marked by a black dotted line). An hph marker was placed immediately to the right of AH36 in an Sk-2 strain (with vector v140) to create the v140Δ::hph allele. An hph marker was also placed at the corresponding location in an SkS strain (with vector v150) to create the v150Δ::hph allele. (B–D) Crosses were performed to determine the effect of each allele on spore killing. Images depict asci from the following crosses: (B) F2-23 × ISU-4344, (C) ISU-4348 × ISU-4344, and (D) ISU-3036 × ISU-4344. B, black; hph, hygromycin resistance cassette; W, white.
The rfk-1 gene does not include ncu06238
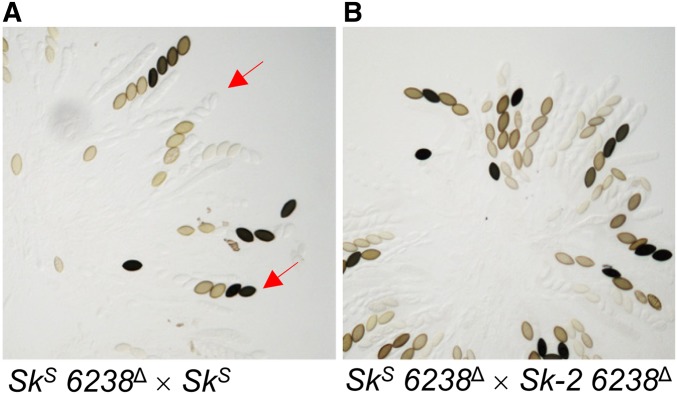
To confirm that ncu06238, the gene to the right of rfk-1 (as depicted in Figure 9A), is neither part of rfk-1 nor required for spore killing, we deleted ncu06238 from both SkS and Sk-2 and then analyzed ascus phenotypes in crosses involving the ncu06238 deletion strains. Interestingly, an SkS ncu06238Δ × SkS cross produced asci with varying numbers of fully developed ascospores (Figure 10A), suggesting that ncu06238 deletion strains have defects in ascosporogenesis that are independent of Sk-2 and spore killing. As a result, we could not use ascus phenotype to determine if spore killing is functional in an SkS ncu06238Δ × Sk-2 ncu06238Δ cross, where asci also produce varying numbers of ascospores (Figure 10B). To overcome this obstacle, we used PCR to check genotypes of progeny from an SkS ncu06238Δ × Sk-2 ncu06238Δ cross. If Sk-2-based spore killing occurred in this cross, most progeny would have an Sk-2 genotype. Indeed, we found that 34 of 35 progeny had the Sk-2 genotype (Figure S2), indicating that Sk-2-based meiotic drive functions normally in the absence of ncu06238. These results strongly suggest that the rfk-1 gene does not overlap or include positions occupied by ncu06238.
Figure 10.
The ncu06238 gene is not required for spore killing. (A) Asci from a cross between two SkS strains (ISU-4559 × P8-43), where one of the strains has had its ncu06238 coding sequence deleted. While some normal asci are detected, many asci are abnormal and some mimic the Spore killer phenotype (red arrows). (B) Asci from a cross between an SkS strain and an Sk-2 strain (ISU-4559 × ISU-4561), where both strains have had their ncu06238 coding sequence deleted. Nearly all viable progeny isolated from this cross have the Sk-2 genotype (34 out of 35; Figure S2).
Replacement of AH363211 with AH36Sk-2 restores spore killing to an rfk-1 mutant
The rfk-1 mutant strain ISU-3211 carries seven mutations within its AH36 interval (Figure 6A and Figure 11A). Although we have already shown that introduction of the G28326A mutation carried by AH363211 into AH36Sk-2 eliminates AH36Sk-2’s ability to abort asci (Figure 7G), we have not directly shown that one or more of the mutations within AH363211 causes ISU-3211’s loss-of-spore-killing phenotype. To address this shortcoming, we replaced AH363211 in a descendant of ISU-3211 (ISU-3222) with AH36Sk-2::hph (Figure 11A) and crossed it to SkS and SkS v150Δ::hph mating partners. As expected, we found that replacing AH363211 with AH36Sk-2::hph restored spore killing as long as the hph marker was “paired” during meiosis (Figure 11, B–G). These results demonstrate that mutations within the AH36 interval are solely responsible for the loss of spore killing in ISU-3211 and its rfk-1 descendants.
Figure 11.
Replacement of AH363211 with AH36Sk-2 restores spore killing to an rfk-1 mutant. (A) Strain ISU-4562 was constructed by replacing AH363211 in ISU-3222 (upper diagram, red box) with nat1 (using vector v160, with deleted positions indicated with an orange bar; ISU-3222 is a descendant of ISU-3211). Strain ISU-4563 was then constructed by replacing AH36Δ::nat1 in ISU-4562 with AH36Sk-2::hph (lower diagram, red box and green rectangle). The locations of the two recombination flanks used to replace AH36Δ::nat1 with AH36Sk-2::hph are indicated with black crosses and red dotted lines. (B–D) Asci are from crosses between F2-23 and (B) ISU-3222, (C) ISU-4562, and (D) ISU-4563. (E–G) Asci are from crosses between ISU-4348 and (E) ISU-3222, (F) ISU-4562, and (G) ISU-4563. See Figure 9 for a description of the v150Δ::hph allele. B, black; hph, hygromycin resistance cassette; W, white.
rfk-1 is transcribed
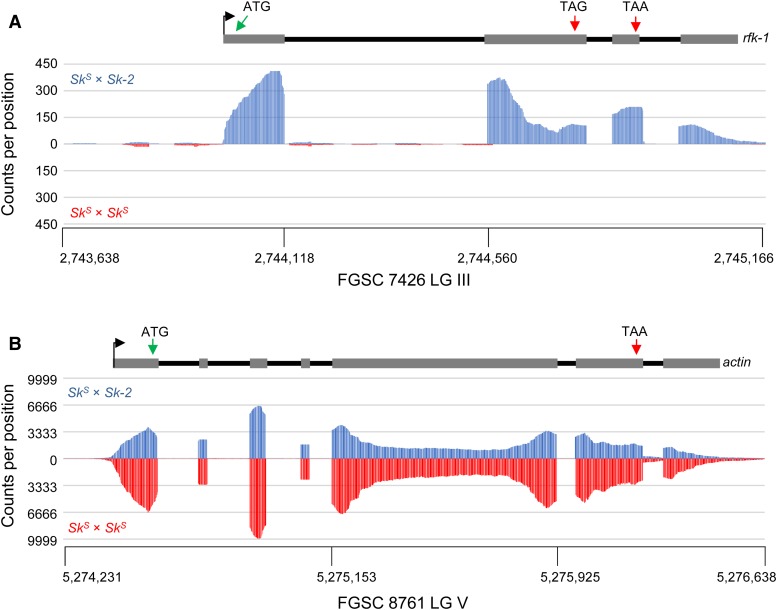
To learn more about rfk-1’s gene structure, we analyzed RNA sequencing data sets of poly(A)-enriched total RNA from perithecial cultures of N. intermedia SkS × Sk-2 and SkS × SkS crosses (Svedberg et al. 2018). The AH36 intervals in N. crassa F2-19 (referred to as AH36Sk-2) and N. intermedia FGSC 7426 (AH367426) are nearly identical, except for one single-nucleotide polymorphism and a 48-bp insertion/deletion (Figure S3). As expected, while relatively few reads from SkS × SkS perithecia align to AH367426, hundreds of reads from the SkS × Sk-2 perithecia do align to this sequence (Figure 12A). In contrast, SkS × SkS and SkS × Sk-2 perithecia contain similar levels of reads that align to the actin gene (Figure 12B). By assuming that the majority of AH367426-aligning reads from SkS × Sk-2 perithecia are derived from rfk-1 transcripts, we can predict that the rfk-1 gene contains four exons and three introns (Figure 12A and Figure S3). Furthermore, by using the ATG at positions 28,264–28,266 (Figure 8) as the putative rfk-1 translational start codon, we can detect a translational stop codon upstream of the second intron (“TAG”; Figure 12A and Figure S4). This stop codon should terminate translation after production of a 102-aa RFK-1 protein. However, if the stop codon were to undergo RNA editing (discussed in more detail below), a different translational stop codon (“TAA”; Figure 12A and Figure S4) should terminate translation after production of a 130-aa RFK-1 protein.
Figure 12.
rfk-1 is transcribed in cultures undergoing sexual reproduction. (A) Sequences of SkS × Sk-2 poly(A)-enriched RNA (top, FGSC 1767 × FGSC 7426) and SkS × SkS poly(A)-enriched RNA (bottom, FGSC 1767 × FGSC 1766) were aligned to AH367426. The y-axis values indicate the number of times each position within AH367426 is found in an aligned RNA sequence. The x-axis values indicate the position of each base within AH367426 with respect to chromosome III of N. intermedia FGSC 7426. These data suggest that rfk-1 is transcribed in SkS × Sk-2 perithecial cultures and that rfk-1 RNA is specific to SkS × Sk-2 crosses. A model of rfk-1 gene structure is shown above the chart. (B) Sequences of SkS × Sk-2 (top, FGSC 1767 × FGSC 7426) and SkS × SkS (bottom, FGSC 1767 × FGSC 1766) poly(A)-enriched RNA were aligned to actin. In contrast to the AH367426 alignment data, RNA sequences from both SkS × Sk-2 and SkS × SkS crosses similarly align to actin with respect to number and pattern. The x-axis values are for the actin locus on chromosome V of FGSC 1766.
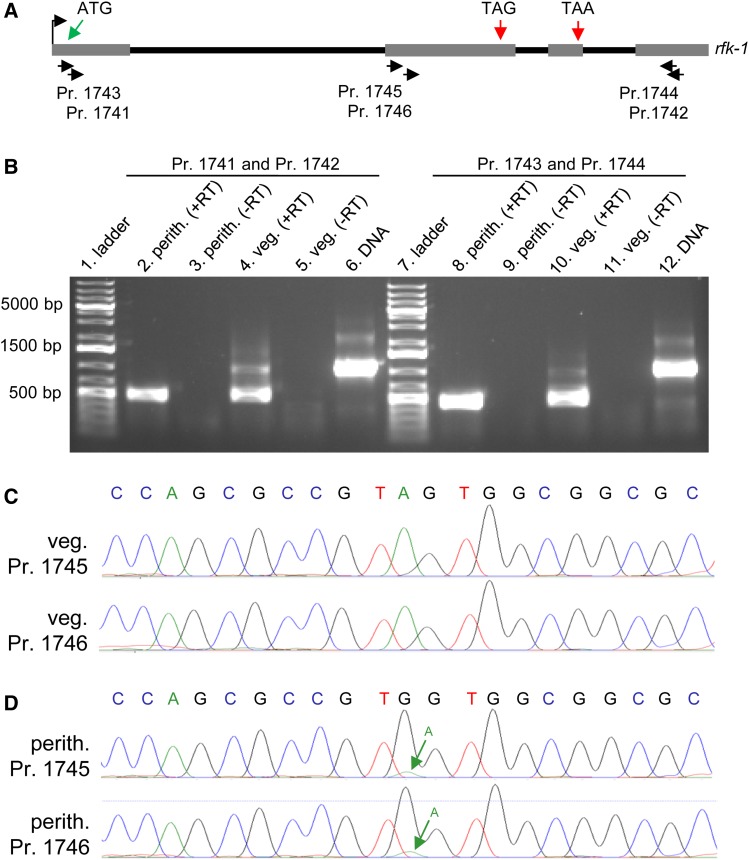
The rfk-1 transcript undergoes RNA editing during sexual development
During the aforementioned analysis of RNA sequencing reads from perithecial cultures of N. intermedia SkS × Sk-2 crosses, we noticed that the majority of reads mapping to the putative “TAG” stop codon located upstream of intron 2 (Figure 12A and Figure S4) contained evidence of RNA editing. More specifically, the codon appears to be edited from UAG to UGG (or more likely U-Inosine-G, see Discussion and Figure S5). In light of this discovery, we amplified partial rfk-1 cDNA fragments from N. crassa Sk-2 total RNA extracts and directly analyzed the cDNA fragments by Sanger sequencing (without subcloning). We found that rfk-1 cDNA fragments from vegetative tissue showed no evidence of RNA editing, while cDNA fragments from sexual tissue contained the expected UAG to UGG edit (Figure 13). These data demonstrate that the early UAG stop codon within rfk-1 transcripts undergoes RNA editing during sexual development.
Figure 13.
The rfk-1 mRNA undergoes RNA editing in perithecial tissue. (A) The transcriptional start site (bent arrow), four exons (gray rectangles), and three introns (black lines) of the rfk-1 gene model are depicted in a diagram. The approximate binding sites of six PCR primers (1741 through 1746) within rfk-1 are indicated. (B) RT-PCR analysis was performed on total RNA samples isolated from 6-day-old perithecia of an SkS (F2-26) × Sk-2 (P15-53) cross and from Sk-2 (P15-53) mycelia (36 hr liquid culture). First-strand synthesis was performed with an oligo-dT primer plus reverse transcriptase (+RT). The absence of contaminating DNA was confirmed by leaving reverse transcriptase out of some reactions (−RT). PCR was performed with primers 1741 and 1742 (left) or primers 1743 and 1744 (right), using first-strand cDNA or Sk-2 (P15-53) genomic DNA as template. RT-PCR products were then analyzed by standard agarose gel electrophoresis and visualized with ethidium bromide staining. (C and D) The sequences of the bases surrounding rfk-1 codon 103 [the “TAG” in (A)] within the ∼500-bp cDNA fragments depicted in panel B’s lane 4 and lane 8 were determined directly by Sanger sequencing (without subcloning). (C) Chromatograms from sequencing of the ∼500-bp product from panel B’s lane 4 with primer 1745 or 1746. No editing of codon 103 (TAG) is detected in the chromatograms. (D) Chromatograms from sequencing of the ∼500-bp product from panel B’s lane 8 with primer 1745 or 1746. The green arrow points to a small peak whose presence in the chromatograms suggests that a fraction of the cDNA population contains the original A, instead of the edited G, at this position. Perith., perithecia; Pr., primer; veg., vegetative.
The rfk-1 complementation group possesses codon-altering mutations
We previously identified six rfk-1 mutants, ISU-3211 through ISU-3216, and all strains were determined to be rfk-1 mutants by complementation group analysis (Harvey et al. 2014). If our rfk-1 gene model is accurate, each strain should carry a mutation that alters rfk-1’s transcriptional or coding potential in a manner that is consistent with our model. Indeed, the sequencing of rfk-1 alleles in ISU-3211 through ISU-3216 identified putative codon-altering mutations in each strain (Figure 14A). For example, ISU-3211 contains the G28326A mutation (examined by site-directed mutagenesis in Figure 7). This mutation changes the 21st rfk-1 codon to a stop codon (nonsense mutation). ISU-3212 carries a single-base insertion that causes a frameshift after the eighth rfk-1 codon. ISU-3213 possesses a G28348A mutation, which changes the amino acid specified by the 29th codon from alanine to threonine. With respect to the remaining three strains—ISU-3214, ISU-3215, and ISU-3216—they all contain rfk-1 sequences that are identical to that of ISU-3211, suggesting that all four were derived from the same mutagenized conidium (via fertilization of an SkS protoperithecium; Harvey et al. 2014). In summary, all known genetically defined rfk-1 mutants carry mutations that alter the predicted coding region of rfk-1 and support our current model of rfk-1 gene structure.
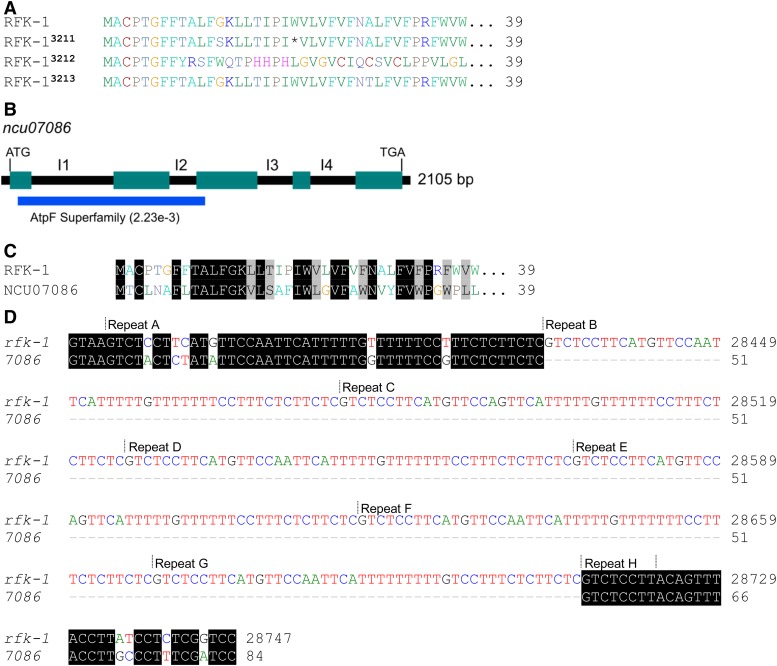
Figure 14.
RFK-1 is related to NCU07086. (A) All known rfk-1 mutations alter the predicted amino acid sequence of RFK-1. (B) A coding region of 2105 bp is predicted for gene ncu07086 (from start codon to stop codon, including introns). Predicted introns are labeled I1 through I4. An AtpF superfamily domain (National Center for Biotechnology Information conserved domain database, accession number cl28522) can be identified within the N-terminal end of the NCU07086 protein sequence. (C) The first 39 amino acids of RFK-1 and NCU07086 are similar. (D) The repetitive sequence identified within AH36 (see Figure 6) appears to have originated from within the first intron of ncu07086. To highlight the expansion of the 46–48 repeat within the first intron of rfk-1 relative to the first intron of ncu07086, the 5′ end (84 bases) of ncu07086’s intron 1 was manually aligned to the 5′ end (368 bases) of rfk-1’s intron 1.
RFK-1 is related to NCU07086
To investigate the origin of rfk-1, we downloaded a list of predicted N. crassa proteins from the National Center for Biotechnology Information (NCBI)’s Genome Database (accession number GCA_000182925.2) and performed a BLASTP search (Basic Local Alignment Search Tool - Protein; Camacho et al. 2009) with the 130-aa RFK-1 sequence as query (Figure S4). We found that the most significant match (Expect = 2e−5) to RFK-1 is a hypothetical 362-aa protein called NCU07086 (GenBank: EAA31115.1). NCU07086 is encoded by the ncu07086 gene on N. crassa chromosome VI, and it is predicted to contain four introns (Figure 14B, I1 through I4; NCBI Gene ID, 3876500). A search of NCBI’s conserved domain database (CDD v3.16; Marchler-Bauer et al. 2015) with the predicted sequence of NCU07086 identified a region with a low-scoring match to the AtpF superfamily (Expect = 2.23e−3; Figure 12B). The similarity between RFK-1 and NCU07086 appears to be reserved to the N-terminal ends of each protein. Specifically, the first 39 amino acids of RFK-1 are highly similar to those of NCU07086 (Figure 14C). Furthermore, it appears that a 47-bp sequence within ncu07086’s first intron (Figure 14D) expanded to become the 46–48-bp repeated element within rfk-1’s first intron (Figure 6, A and B, and Figure 14D). In summary, these findings suggest that rfk-1 evolved from a partial duplication of the ncu07086 gene.
Discussion
The mechanism used by the Neurospora Spore killers to achieve biased transmission is believed to involve a resistance protein and a killer protein. In a previous work, we isolated six rfk mutants (ISU-3211 through ISU-3216) and provided evidence that each is mutated at the same locus, subsequently named rfk-1 (Harvey et al. 2014). The rfk-1 locus in ISU-3211 was mapped to a 45-kb region of Sk-2. We began this study with the goal of identifying rfk-1. At first, we intended to use three-point crossing assays to further refine the position of rfk-1 within the 45-kb region. These assays were to be performed with hph markers inserted between genes ncu06192 and ncu06191 (with vector v3) and between genes ncu06239 and ncu06240 (with vector v4); therefore, deletion vectors v3 and v4 were designed to delete relatively small intervals from the rfk-1 region (Table 2) and, as such, they were not expected to influence spore killing. In agreement with our predictions, deletion of intervals v3 and v4 had no effect on spore killing (Figure 1, C and D). In contrast, v5 was designed to delete a 10,718-bp interval spanning most of the Sk-2INS1 sequence, with the goal of deleting rfk-1 should it exist within the interval (Table 2). Fortunately, deletion of v5 was fruitful and its removal from Sk-2 eliminated Sk-2’s ability to kill ascospores (Figure 1E). We were thus able to focus our efforts on deleting subintervals of v5, which allowed us to pinpoint rfk-1 to the intergenic region between ncu07238* and ncu06238 (Figure 2 and Figure 3).
We also tested various subintervals of v5 for the presence of rfk-1 by transferring them to an SkS strain and crossing them to an SkS sad-2Δ mating partner (Figure 4). For this assay to yield positive results, rfk-1 must be sufficient for spore killing. Indeed, we found this to be the case when we identified four intervals (AH30, AH31, AH36, and AH37) that triggered ascus abortion when transferred to an SkS genetic background. These four intervals all have the 1481 bp of AH36 in common, and the ascus abortion phenotype associated with each interval is likely due to the presence of rfk-1 without a compatible resistance gene. For example, the KN model holds that the resistance protein (RSK) and the killer are both active during early stages of meiosis (Hammond et al. 2012). Lack of a resistant version of RSK, along with expression of the killer, appears to cause asci to abort meiosis before ascospore delimitation. This phenomenon explains the abortion phenotypes of AH30, AH31, and AH37. However, for succinctness, we also refer to the phenotype associated with AH36 as ascus abortion, although it may be more accurate to refer to it as a “bubble” phenotype (see Figure 4G). The bubble phenotype was originally described by Raju et al. (1987), and it is thought to arise when asci and/or ascospores abort shortly after ascospore delimitation. Therefore, an explanation for the existence of the two phenotypic classes (clear ascus abortion vs. bubble phenotype) is that ascus development progresses a bit further with AH36 than it does with AH30, AH31, and AH37. In turn, one could speculate that rfk-1 expression is lower from AH36 than it is from AH30, AH31, and AH37, because AH36 is the shortest of the abortion-inducing intervals and may lack some of the regulatory sequences needed for full expression of rfk-1.
In this study, we have provided evidence that the critical rfk-1 coding region is found between a start codon beginning at position 28,264 and either a stop codon ending at position 28,968 or a stop codon ending at position 29,106. For example, (1) a putative nonsense mutation at position 28,326 disrupts the ascus-aborting ability of interval AH36 (Figure 6A, Figure 7G, and Figure 14A); (2) spore killing functions when a nonnative promoter is attached to the putative rfk-1 start codon at position 28,264 (Figure 8, B and D); (3) spore killing fails when the putative rfk-1 reading frame is shifted by one position by the insertion of a single base immediately after the putative start codon (Figure 8, B and E); and (4) spore killing fails when the putative rfk-1 start codon is changed to a stop codon (Figure 8, B and F). Additionally, all six of the known rfk-1 mutants carry codon-altering mutations that are consistent with our proposed rfk-1 gene model (Figure 14A). Furthermore, although we have not identified the transcript cleavage site or the location at which the poly(A) tail is added, we have shown that the gene immediately downstream of rfk-1 (ncu06238) is not required for rfk-1 function (Figure 10B and Figure S2) and we have detected rfk-1 cDNAs using reverse transcription reactions primed with an oligo-d(T) primer (Figure 13B). In summary, our data strongly suggest rfk-1 to be a protein-coding gene with four exons and three introns.
Adenosine-to-inosine (A-to-I) RNA editing was recently discovered in two Sordariomycete fungi: Fusarium graminearum and N. crassa (Liu et al. 2016, 2017). Interestingly, while the enzymes directing A-to-I edits of Soradriomycete RNAs are unknown, the process appears to be specific to the sexual cycle (Liu et al. 2016, 2017). While modeling rfk-1 gene structure with RNA sequencing data, we noticed that the sequences of most of the reads that aligned to the first rfk-1 stop codon (codon 103) contained mutations relative to the reference sequence and that these mutations (which appear as a “G” in sequencing data) are consistent with A-to-I editing events (Figure S5). We confirmed this finding by amplifying and sequencing rfk-1-specific cDNA fragments (Figure 13). Interestingly, and consistent with the presumed sexual phase-specific nature of A-to-I RNA editing in Neurospora (Liu et al. 2017), rfk-1 editing was detected in sexual RNA but not vegetative RNA. This raises the possibility that the RNA edit allows rfk-1 to encode protein variants with different functions, one for the vegetative phase and one for the sexual phase. Although the mechanism of A-to-I editing in fungi is unknown, edited bases are often found at the end of hairpin loops (Liu et al. 2017). Accordingly, we found that the edited rfk-1 stop codon is predicted to exist within the loop of a hairpin with a nearly perfect 18-bp stem region (Figure S6). Overall, the discovery of rfk-1 transcript editing has fascinating implications for RFK-1 function. For example, it is tempting to speculate that only the edited RNA produces a killer protein and that editing is timed to mitigate potential fitness costs associated with inopportune expression of a functional killer.
While the work presented here represents a significant step toward understanding the mechanism of Sk-2-based spore killing, many questions remain unanswered. For example, although it appears that rfk-1 evolved from ncu07086, the function of NCU07086 is unknown. NCU07086 contains a region with slight homology to the AtpF superfamily (Figure 14B). Interestingly, the atpF gene in Escherichia coli (also known as uncF; NCBI Gene ID 948247) encodes subunit b of the F-type ATP synthase complex (Walker et al. 1984; Dunn 1992; McLachlin and Dunn 1997; Revington et al. 1999). This hints that RFK-1 could mediate spore killing by targeting eukaryotic F-type ATP synthases, which are associated with mitochondrial membranes in eukaryotes (Stewart et al. 2014). However, NCU07086 in N. crassa has not been investigated, and a much more likely candidate for the b subunit of N. crassa’s F-type ATP synthase is found in NCU00502 (Kyoto Encyclopedia of Genes and Genomes oxidative phosphorylation pathway: ncr00190, release 87.0, Kanehisa and Goto 2000; Kanehisa et al. 2016). Therefore, at this point in time, a role for RFK-1 in disrupting mitochondrial function as part of the spore-killing process is purely speculative.
Although the primary goal of this work was to identify rfk-1, the identity of which has been of interest to meiotic drive researchers since the discovery of Sk-2 nearly four decades ago, we unexpectedly discovered the strongest evidence to date that genomes in some, if not all, lineages of eukaryotic organisms possess elaborate defense processes to protect themselves from complex meiotic drive elements. With respect to Neurospora genomes, this defense process appears to be MSUD. The first hint that MSUD defends Neurospora genomes from meiotic drive appeared in 2007, when it was discovered that Sk-2 and Sk-3 can suppress certain instances of MSUD (Raju et al. 2007). Next, in 2012, it was found that the position of rsk within Sk-2 allows it to pair with rsk in the SkS genome during Sk-2 × SkS crosses (Hammond et al. 2012). If rsk is not paired during these crosses (e.g., if it is deleted from the SkS mating partner), rsk is silenced by MSUD and the entire ascus is killed by the killer protein, which we now know to be RFK-1. In the work presented here, we found that the specific location of rfk-1 next to the right border of Sk-2 is critical to the success of meiotic drive because it allows rfk-1 to escape inactivation by MSUD. Unlike rsk, rfk-1 is only found in Sk-2 strains and it cannot be paired in Sk-2 × SkS crosses. It appears that the evolutionary forces that drove Sk-2 into existence found a way to circumvent this problem (i.e., the lack of an rfk-1 pairing partner in SkS strains) by positioning rfk-1 close to sequences that are paired during meiosis (i.e., close to ncu06238, see Figure 9A). While previous work has clearly demonstrated the ability of MSUD to target an unpaired transposon for silencing (and thus defend the genome from transposition events during meiosis; Wang et al. 2015), our findings add to accumulating evidence that MSUD also antagonizes the evolution of meiotic drive elements by placing significant constraints on the arrangement of critical genes within these elements. In the future, it will be interesting to learn which process (defense against transposable elements or defense against meiotic drive elements) better explains why MSUD has evolved, and why it has been maintained, in various fungal lineages.
Acknowledgments
We thank members of the Brown, Hammond, Johannesson, and Shiu laboratories for their assistance with various technical aspects of this work; and the Fungal Genetics Stock Center, whose preservation and distribution of Neurospora isolates helped make this work possible (McCluskey et al. 2010). We acknowledge use of materials generated by grant P01 GM-068087 “Functional analysis of a model filamentous fungus.” We dedicate this paper to David Perkins, Namboori Raju, Barbara Turner, and others for their pioneering work on the Neurospora Spore killers. This project was supported by a grant from the National Science Foundation (NSF) (MCB# 1615626) (T.M.H.). P.K.T.S. was supported by the NSF (MCB# 1715534) and the University of Missouri Research Board and Research Council. H.J. was supported by a European Research Council grant (under the program H2020, ERC-2014-CoG, project 648143) and the Swedish Research Council. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). The USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7836692.
Communicating editor: D. Barbash
Literature Cited
- Aramayo R., Selker E. U., 2013. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb. Perspect. Biol. 5: a017921 10.1101/cshperspect.a017921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Núñez M. A., Nuckolls N. L., Zanders S. E., 2018. Genetic villains: killer meiotic drivers. Trends Genet. 34: 424–433. 10.1016/j.tig.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A., Trivers R., 2008. Genes in Conflict: The Biology of Selfish Genetic Elements. Harvard University Press, Cambridge, MA. [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L., Turner B. C., 1987. Recombination block in the Spore killer region of Neurospora. Genome 29: 129–135. 10.1139/g87-022 [DOI] [PubMed] [Google Scholar]
- Dalstra H. J. P., Swart K., Debets A. J. M., Saupe S. J., Hoekstra R. F., 2003. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc. Natl. Acad. Sci. USA 100: 6616–6621. 10.1073/pnas.1030058100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., 2000. Neurospora: Contributions of a Model Organism. Oxford University Press, New York. [Google Scholar]
- Dunn S. D., 1992. The polar domain of the b subunit of Escherichia coli F1F0-ATPase forms an elongated dimer that interacts with the F1 sector. J. Biol. Chem. 267: 7630–7636. [PubMed] [Google Scholar]
- Ebbole D., Sachs M., 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37: 17–18. [Google Scholar]
- Grognet P., Lalucque H., Malagnac F., Silar P., 2014. Genes that bias Mendelian segregation. PLoS Genet. 10: e1004387 10.1371/journal.pgen.1004387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Hammond T. M., 2017. Sixteen years of meiotic silencing by unpaired DNA. Adv. Genet. 97: 1–42. 10.1016/bs.adgen.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Hammond T. M., Xiao H., Rehard D. G., Boone E. C., Perdue T. D., et al. , 2011. Fluorescent and bimolecular-fluorescent protein tagging of genes at their native loci in Neurospora crassa using specialized double-joint PCR plasmids. Fungal Genet. Biol. 48: 866–873. 10.1016/j.fgb.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Hammond T. M., Rehard D. G., Xiao H., Shiu P. K. T., 2012. Molecular dissection of Neurospora Spore killer meiotic drive elements. Proc. Natl. Acad. Sci. USA 109: 12093–12098. 10.1073/pnas.1203267109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey A. M., Rehard D. G., Groskreutz K. M., Kuntz D. R., Sharp K. J., et al. , 2014. A critical component of meiotic drive in Neurospora is located near a chromosome rearrangement. Genetics 197: 1165–1174. 10.1534/genetics.114.167007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Jiang Z. D., Suo F., Zheng J. X., He W. Z., et al. , 2017. A large gene family in fission yeast encodes spore killers that subvert Mendel’s law. Eng. Life Sci. 6: e26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44: D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanizay L. B., Pyhäjärvi T., Lowry E. G., Hufford M. B., Peterson D. G., et al. , 2013. Diversity and abundance of the abnormal chromosome 10 meiotic drive complex in Zea mays. Heredity 110: 570–577. 10.1038/hdy.2013.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente A. M., Presgraves D. C., 2012. The selfish Segregation Distorter gene complex of Drosophila melanogaster. Genetics 192: 33–53. 10.1534/genetics.112.141390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database Collaboration 2011. The sequence read archive. Nucleic Acids Res. 39: D19–D21. 10.1093/nar/gkq1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm A. K., Dyer K. A., Firman R. C., Fishman L., Forstmeier W., et al. , 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31: 315–326. 10.1016/j.tree.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Liu H., Wang Q., He Y., Chen L., Hao C., et al. , 2016. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 26: 499–509. 10.1101/gr.199877.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Li Y., Chen D., Qi Z., Wang Q., et al. , 2017. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc. Natl. Acad. Sci. USA 114: E7756–E7765. 10.1073/pnas.1702591114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408. 10.1146/annurev.genet.37.110801.143030 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., et al. , 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43: D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin B. S., Freitag M., Selker E. U., 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44: 34–36. [Google Scholar]
- McCluskey K., Wiest A., Plamann M., 2010. The fungal genetics stock center: a repository for 50 years of fungal genetics research. J. Biosci. 35: 119–126. 10.1007/s12038-010-0014-6 [DOI] [PubMed] [Google Scholar]
- McLachlin D. T., Dunn S. D., 1997. Dimerization interactions of the b subunit of the Escherichia coli F1F0-ATPase. J. Biol. Chem. 272: 21233–21239. 10.1074/jbc.272.34.21233 [DOI] [PubMed] [Google Scholar]
- Nuckolls N. L., Núñez M. A. B., Eickbush M. T., Young J. M., Lange J. J., et al. , 2017. wtf genes are prolific dual poison-antidote meiotic drivers. Eng. Life Sci. 6: e26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., 1974. The manifestation of chromosome rearrangements in unordered asci of Neurospora. Genetics 77: 459–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju N. B., 1979. Cytogenetic behavior of Spore killer genes in Neurospora. Genetics 93: 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju N. B., 1980. Meiosis and ascospore genesis in Neurospora. Eur. J. Cell Biol. 23: 208–223. [PubMed] [Google Scholar]
- Raju N. B., 1994. Ascomycete spore killers: chromosomal elements that distort genetic ratios among the products of meiosis. Mycologia 86: 461–473. 10.1080/00275514.1994.12026437 [DOI] [Google Scholar]
- Raju N. B., Perkins D. D., Newmeyer D., 1987. Genetically determined nonselective abortion of asci in Neurospora crassa. Can. J. Bot. 65: 1539–1549. 10.1139/b87-212 [DOI] [Google Scholar]
- Raju N. B., Metzenberg R. L., Shiu P. K. T., 2007. Neurospora Spore killers Sk-2 and Sk-3 suppress meiotic silencing by unpaired DNA. Genetics 176: 43–52. 10.1534/genetics.106.069161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revington M., McLachlin D. T., Shaw G. S., Dunn S. D., 1999. The dimerization domain of the b subunit of the Escherichia coli F(1)F(0)-ATPase. J. Biol. Chem. 274: 31094–31101. 10.1074/jbc.274.43.31094 [DOI] [PubMed] [Google Scholar]
- Rhoades M., 1952. Preferential segregation in maize, pp. 66–80 in Heterosis: A Record of Researches Directed Towards Explaining and Utilizing the Vigor of Hybrids, edited by Gowen J. W. Iowa State College Press, Ames, IA. [Google Scholar]
- Samarajeewa D. A., Sauls P. A., Sharp K. J., Smith Z. J., Xiao H., et al. , 2014. Efficient detection of unpaired DNA requires a member of the Rad54-like family of homologous recombination proteins. Genetics 198: 895–904. 10.1534/genetics.114.168187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe S. J., 2011. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Semin. Cell Dev. Biol. 22: 460–468. 10.1016/j.semcdb.2011.02.019 [DOI] [PubMed] [Google Scholar]
- Shiu P. K. T., Raju N. B., Zickler D., Metzenberg R. L., 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916. 10.1016/S0092-8674(01)00609-2 [DOI] [PubMed] [Google Scholar]
- Shiu P. K. T., Zickler D., Raju N. B., Ruprich-Robert G., Metzenberg R. L., 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 103: 2243–2248. 10.1073/pnas.0508896103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A. G., Laming E. M., Sobti M., Stock D., 2014. Rotary ATPases–dynamic molecular machines. Curr. Opin. Struct. Biol. 25: 40–48. 10.1016/j.sbi.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Sugimoto M., 2014. Developmental genetics of the mouse t-complex. Genes Genet. Syst. 89: 109–120. 10.1266/ggs.89.109 [DOI] [PubMed] [Google Scholar]
- Svedberg J., Hosseini S., Chen J., Vogan A. A., Mozgova I., et al. , 2018. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nat. Commun. 9: 4242 10.1038/s41467-018-06562-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B. C., 2001. Geographic distribution of Neurospora Spore killer strains and strains resistant to killing. Fungal Genet. Biol. 32: 93–104. 10.1006/fgbi.2001.1253 [DOI] [PubMed] [Google Scholar]
- Turner B. C., Perkins D. D., 1979. Spore killer, a chromosomal factor in Neurospora that kills meiotic products not containing it. Genetics 93: 587–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora (Medium N). Microb. Genet. Bull. 13: 42–43. [Google Scholar]
- Walker J. E., Saraste M., Gay N. J., 1984. The UNC operon nucleotide sequence, regulation and structure of ATP-synthase. Biochim. Biophys. Acta 768: 164–200. 10.1016/0304-4173(84)90003-X [DOI] [PubMed] [Google Scholar]
- Wang Y., Smith K. M., Taylor J. W., Freitag M., Stajich J. E., 2015. Endogenous small RNA mediates meiotic silencing of a novel DNA transposon. G3 (Bethesda) 5: 1949–1960. 10.1534/g3.115.017921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577. 10.1002/j.1537-2197.1947.tb13032.x [DOI] [Google Scholar]
- Yu J. H., Hamari Z., Han K. H., Seo J. A., Reyes-Domínguez Y., et al. , 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41: 973–981. 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Zimmering S., Sandler L., Nicoletti B., 1970. Mechanisms of meiotic drive. Annu. Rev. Genet. 4: 409–436. 10.1146/annurev.ge.04.120170.002205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and plasmids generated during this study are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7836692.