This study demonstrates the utility of a custom targeted sequencing panel for identifying spontaneous suppressors of a mutation in the Saccharomyces cerevisiae spliceosome. The panel screens 112 genes encoding known or suspected splicing factors...
Keywords: suppressor selection, U4 snRNA, spliceosome activation, targeted sequencing, pre-mRNA splicing
Abstract
Selection of suppressor mutations that correct growth defects caused by substitutions in an RNA or protein can reveal functionally important molecular structures and interactions in living cells. This approach is particularly useful for the study of complex biological pathways involving many macromolecules, such as premessenger RNA (pre-mRNA) splicing. When a sufficiently large number of suppressor mutations is obtained and structural information is available, it is possible to generate detailed models of molecular function. However, the laborious and expensive task of identifying suppressor mutations in whole-genome selections limits the utility of this approach. Here I show that a custom targeted sequencing panel can greatly accelerate the identification of suppressor mutations in the Saccharomyces cerevisiae genome. Using a panel that targets 112 genes encoding pre-mRNA splicing factors, I identified 27 unique mutations in six protein-coding genes that each overcome the cold-sensitive block to spliceosome activation caused by a substitution in U4 small nuclear RNA. When mapped to existing structures of spliceosomal complexes, the identified suppressors implicate specific molecular contacts between the proteins Brr2, Prp6, Prp8, Prp31, Sad1, and Snu114 as functionally important in an early step of catalytic activation of the spliceosome. This approach shows great promise for elucidating the allosteric cascade of molecular interactions that direct accurate and efficient pre-mRNA splicing and should be broadly useful for understanding the dynamics of other complex biological assemblies or pathways.
INTRONS are removed from premessenger RNAs (pre-mRNAs) by the spliceosome, which comprises an assemblage of five small nuclear ribonucleoprotein particles (snRNPs) and a number of free proteins and protein complexes. Each snRNP contains one small nuclear RNA (snRNA) (U1, U2, U4, U5, or U6) and several proteins. The smallest snRNA, U6, is a key component of the catalytic core of the spliceosome, where it base pairs with the 5′ splice site of the intron (reviewed in Didychuk et al. 2018). U6 enters the spliceosome as part of the U4/U6.U5 tri-snRNP, in which it is extensively base paired to U4 snRNA. The tri-snRNP binds the prespliceosome or A complex, i.e., U1 and U2 snRNPs bound to a pre-mRNA, to form the B complex. Subsequent rearrangements result in unwinding of the U4/U6 duplex and expulsion of the U1 and U4 snRNPs to form the Bact complex, which rearranges further to form the B* complex that is competent for the first catalytic step of splicing (Will and Lührmann 2011; Hoskins and Moore 2012). The RNA-dependent ATPases Prp28 and Brr2 are required for the B-to-Bact transition due to their function in transferring the intron 5′ splice site interaction from U1 to U6 snRNA and unwinding U4/U6, respectively (Staley and Guthrie 1999; Absmeier et al. 2016).
The molecular mechanism of activation of the spliceosome for splicing catalysis has been challenging to elucidate due to the large number of constituent proteins and RNAs. A heuristic model for spliceosome activation was proposed whereby an allosteric cascade of RNA–RNA, RNA–protein, and protein–protein interactions causally links recognition of conserved intronic sequences with formation of the catalytic core of the spliceosome (Brow 2002). A disruption in the cascade due to the absence of correct splice site recognition would result in failure to complete the activation process, preventing mis-splicing. Such a mechanism requires the transmission of signals through proteins and RNAs across tens of nanometers. This intrinsic proofreading system interfaces with numerous extrinsic factors, including several DExD/H-box ATPases, that help guide accurate and efficient splicing (Burgess and Guthrie 1993; Semlow and Staley 2012; Chang et al. 2013). Although near-atomic structures of the spliceosome at several points in the splicing pathway have recently been determined (reviewed in Fica and Nagai 2017, Shi 2017, and Kastner et al. 2019), the paths of allosteric signals through the multi-megadalton spliceosome are largely unknown.
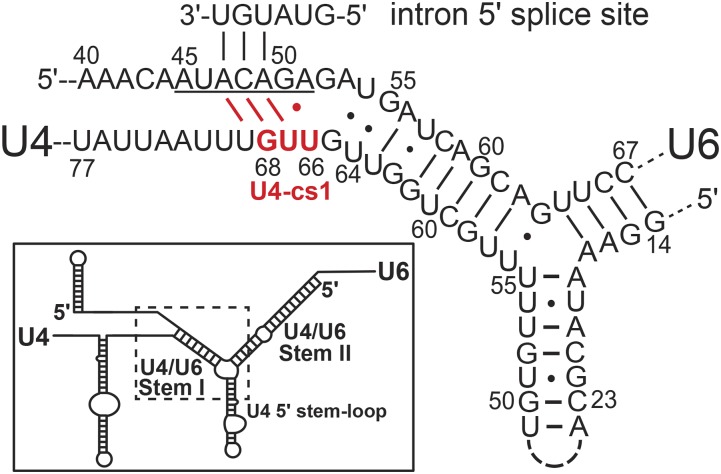
Genetic suppression analysis has proven to be a productive approach for identifying molecular interactions that drive spliceosome assembly and activation in the yeast Saccharomyces cerevisiae (Couto et al. 1987; Ares and Igel 1990; Mayerle and Guthrie 2017). When suppressors are obtained at sufficient density, they can reveal in vivo molecular interfaces that are in excellent agreement with high-resolution crystal structures (Montemayor et al. 2014). We previously created a cold-sensitive mutation in U4 snRNA, called U4-cs1, that likely extends intermolecular base pairing with U6 snRNA into the region where U6 would normally bind the intron 5′ splice site (Figure 1, Li and Brow 1996). In vitro splicing assays with U4-cs1-mutant cell extracts identified a block to activation of the B complex at 16° that could be overcome by warming the mixture to 30°, as evidenced by unwinding of U4/U6 and loss of U1 snRNA (Kuhn et al. 1999). A genome-wide selection for spontaneous mutations that allow growth of a U4-cs1 strain at 18° yielded U4-cs1-suppressor mutations in U4, U6, and Prp8 (Li and Brow 1996; Kuhn et al. 1999). Prp8 is the core regulatory protein of the spliceosome and likely evolved from the group II intron maturase protein (Galej et al. 2013; Novikova and Belfort 2017; Zhao and Pyle 2017). Targeted selections for mutations in PRP8 that suppress U4-cs1, or cold-sensitive mutations in BRR2 or PRP28, revealed multiple domains of Prp8 implicated in spliceosome activation (Kuhn and Brow 2000; Kuhn et al. 2002; Price et al. 2014). However, U4-cs1-suppressor mutations in other spliceosome proteins have not yet been identified.
Figure 1.
U4-cs1 may compete with the intron 5′ splice site for binding to the U6 ACAGA box (underlined). In the inset, the dotted box indicates the region of the U4/U6 duplex that is enlarged. The U4-cs1 substitution is shown in red; the wild-type sequence is AAA. The yeast consensus intron 5′ splice site is shown as it pairs with U6 in the catalytically competent spliceosome. Potential competing base pairs created or stabilized by the U4-cs1 mutation are shown in red.
Here I report a high-throughput method for identifying suppressor mutations that uses a custom-designed sequencing panel to scan 112 genes encoding known spliceosome components and extrinsic factors implicated in splicing. By applying this panel to a new collection of U4-cs1-suppressor strains, I identified 27 different mutations in six protein-coding genes, with PRP8 and SAD1 the most common targets. When mapped to cryogenic electron microscopy (cryo-EM) structures of the tri-snRNP and B complex, the new suppressor mutations suggest mechanisms for overcoming the cold-sensitive block to the B-to-Bact transition imposed by U4-cs1. In particular, the suppressor mutations suggest that failure of U4-cs1-arrested spliceosomes to release the assembly factor Sad1 at low temperature prevents Brr2 from engaging U4 and displacing it from U6. This finding implies that association of the intron 5′ splice site with U6 is sensed 7 nm away, at the Sad1 binding site, resulting in the release of Sad1 and repositioning of Brr2. Two potential paths for the signal from U6 to the Sad1 binding site are revealed by the U4-cs1-suppressor substitutions.
Materials and Methods
Selection for suppressors of U4-cs1
Strain DAB102 [MATa, snr6::LEU2, snr14::trp1::ADE2, trp1, ura3, lys2, his3, ade2, (pRS313-snr14-cs1), (pRS314-SNR6)], containing the U4-cs1 allele of the U4 gene (SNR14) and a wild-type U6 allele (SNR6), was constructed from CJM000 (McManus et al. 2007) by plasmid shuffle. A total of 20 colonies of DAB102 on a YPD plate were each transferred to 5 ml YPD liquid medium and grown overnight at 30°. A 200 μl volume of each culture was spread on a YPD plate and the 20 plates were placed in a 16° incubator. Over a period of 9 months, a single colony appeared on each of four plates. These four strains are not further described here. In an attempt to increase the yield of the selection, 1 ml of each of the original 20 cultures, which had been kept at 4°, was pelleted, resuspended in 200 μl fresh YPD, spread on a YPD plate, and put in an 18° incubator. Six plates yielded no colonies after 64 days. For the remaining 14 plates, colonies were picked when they grew to ∼2 mm in diameter and the plate was placed back in the incubator. All picked colonies were grown overnight in 5 ml YPD at 30° and frozen stocks were made. Five or six colonies were picked from each of the 14 plates for a total of 72 colonies. The suppressor strains selected at 18° were named DAB105 to DAB176, in the order in which they were picked.
Targeted sequencing of genomic DNA
Genomic DNA was prepared from each DAB105–176 strain using a Yeast DNA Extraction Kit (Thermo Scientific) according to the manufacturer’s recommendations except that, after isopropanol precipitation, a 10 min digestion with 50 μl of 40 μg/ml ribonuclease A at 37° was included, followed by ethanol precipitation from 1.7 M ammonium acetate. The U4 (SNR14) and U6 (SNR6) genes from each strain were PCR-amplified and Sanger sequenced to confirm the U4-cs1 mutation was still present and to identify any suppressor mutations in these snRNAs (Table 1). Genomic DNAs from the 62 strains without suppressor mutations in U4 and one strain with a suppressor mutation in U4 (DAB122) were processed with an Illumina TruSeq Custom Amplicon Kit version 1.5 along with 32 unrelated strains for a total of 95 samples. The panel was designed to generate 463 amplicons 400–425 bp in length with a cumulative coverage of ∼200,000 bp, resulting in full coverage of 112 open reading frames or snRNA-coding regions (Supplemental Material, Tables S1–S3). Indexed amplicon libraries were constructed in parallel by the University of Wisconsin Biotechnology Center DNA Sequencing Facility according to the manufacturer’s instructions (https://support.illumina.com/downloads/truseq_custom_amplicon_library_prep_guide_15027983.html) and the pooled libraries were subjected to 2 × 250 bp paired-end sequencing in an Illumina MiSeq. The number of reads per strain ranged from 111,160 to 209,078.
Table 1. Mutations identified in U4-cs1-suppressor strains selected at 18°.
| Genea | Mutationb | Substitutionc | Human residued | Strain(s) with mutatione |
|---|---|---|---|---|
| SNR14 | n.55_64delUUUGCUGGUU | 105 | ||
| n.56_71delUUGCUGGUUGUUGUUU | 109 | |||
| n.63_68delUUGUUG | 107 | |||
| n.64U > G | 112, 115 | |||
| n.64U > A | 108, 113, 114 | |||
| n.65_68delGUUG | 106 | |||
| n.69U > G | 122 | |||
| SNR6 | n.55G > U | 148 | ||
| PRP8 | n.2363A > C | p.Glu788Ala | Glu 715 | 133 |
| n.2582A > C | p.Gln861Pro | Gln788 | 139 | |
| n.3297C > A | p.Asn1099Lys | Asn1026 | 144, 147, 153, 154, 156 | |
| n.3571C > G | p.Pro1191Ala | Pro1118 | 135 | |
| n.3572C > T | p.Pro1191Leu | Pro1118 | 151 | |
| n.4900C > T | p.Leu1634Phe | Met1562 | 149 | |
| n.4919C > A | p.Thr1640Lys | Thr1568 | 159 | |
| n.5063C > G | p.Pro1688Arg | Pro1616 | 110 | |
| n.5615C > G | p.Thr1872Arg | Thr1800 | 111, 121 | |
| n.5615C > A | p.Thr1872Lys | Thr1800 | 116, 131 | |
| SAD1 | n.143G > T | p.Cys48Phe | Cys124 | 117, 118*, 119, 125†, 132*, 138†, 141 |
| n.144C > G | p.Cys48Trp | Cys124 | 120, 129 | |
| n.151A > C | p.Thr51Pro | Ser127 | 130*, 142* | |
| n.209_210insTGGACATTATTATC | p.Arg70Ser | Arg146 | 123 | |
| AGGGGAG, 211C > A | p.Gly64_Arg70dup, | Gly140_Arg146 | ||
| p.His71Asn | Gly147 | |||
| n.458C > A | p.Thr153Asn | Asn228 | 169 | |
| n.928C > A | p.Gln310Lys | Gln412 | 140 | |
| n.1085C > A | p.Thr362Lys | Thr469 | 152*, 157* | |
| n.1093G > A | p.Glu365Lys | Asn472 | 137 | |
| n.1302A > T | p.Leu434Phe | Ile542 | 155 | |
| BRR2 | n.854T > A | p.Ile285Lys | Pro260 | 150 |
| n.868G > A | p.Glu290Lys | Ala265 | 145 | |
| n.884G > T | p.Arg295Ile | Arg 270 | 126, 160 | |
| n.1075G > T | p.Gly359Cys | Cys331 | 124, 136 | |
| PRP6 | n.523G > T | p.Asp175Tyr | Asp252 | 143 |
| n.673A > G | p.Lys225Glu | Lys293 | 127 | |
| PRP31 | n.728C > T | p.Ala243Val | Gly233 | 128 |
| SNU114 | n.1834C > A | p.Pro612Thr | Pro597 | 134, 146 |
n, nucleotide; del, deletion; p, protein; ins, insertion; dup, duplication.
SNR14 is the U4 snRNA gene and SNR6 is the U6 snRNA gene; all other genes code for the protein of the same name.
SNR14 and SNR6 mutations are shown as RNA sequence. The U4-cs1 substitution is in bold. Numbering is from the first nucleotide for snRNAs and the first nucleotide of the start codon for mRNAs.
Effects of the mutations on the protein sequence, where relevant.
Human residue equivalent to the substituted yeast residue when the yeast and human orthologs are aligned.
All strains have the prefix “DAB.” For a given mutation, strains marked with “*” or “†” were from the same plate and so may not have arisen independently.
Bioinformatic analysis of sequencing data
Analysis of the sequencing data to generate variant calls was done by the University of Wisconsin Biotechnology Center’s Bioinformatics Resource Center (Figure S1). Fastq sequencing reads for each genomic DNA sample were adapter and quality trimmed using the Skewer trimming program (Jiang et al. 2014). FLASH (Magoc and Salzberg 2011) was used to merge paired-end reads into amplicon sequences. Amplicons were aligned to the Saccharomyces Genome Database S288C reference genome release R64-2-1 (http://www.yeastgenome.org) using BWA-MEM (Li 2013) and local realignment was performed with GATK (McKenna et al. 2010). Between 6 and 10% of the amplicons had low coverage, likely due at least in part to strain polymorphisms. Variants were called using GATK HaplotypeCaller version 3.6 and annotated with SNPeff (Cingolani et al. 2012).
Because the CJM000 strain is not a perfect match to the S288C reference genome, 15 variants common to ≥90% of the 63 strains were classified as polymorphisms between the two strains and were subtracted from all variant calls. A total of 103 variants remained, some of which were common to several strains. The aligned reads for each variant were inspected manually in one or more strains using Integrative Genomics Viewer version 2.3.98 (Robinson et al. 2011). For each of 46 strains, including the strain with a known mutation in the U6 gene (DAB148), a single nucleotide substitution with a read frequency of 88–100% and total number of reads from 68 to 395 was identified (Table 1 and Table S4). Strain DAB132 also has a 21-bp duplication adjacent to the substitution. These mutations are highly likely to be causative for suppression (see Results and Discussion). The remaining variants had a read frequency of 20% or less and/or 10 or fewer reads, or were associated with a truncated read in PRP28 or 11 consecutive A residues in MUD1 (see Table S7). These variants were considered to be sequencing errors or spurious. Two exceptions are a silent mutation in SYF2 (213T > A,Thr71Thr), which is likely a true polymorphism that just missed the 90% cutoff by being identified in 56 of 63 strains, and a missense mutation in PRP31 (295A > T, Asn99Tyr) in strain DAB175, which is present in only 29% of 137 reads and was not investigated further. The SNR14 (U4 RNA gene) variants (Table 1) are based solely on Sanger sequencing since the read depth for this amplicon was very poor.
Data availability
Yeast strains are available upon request. Sequence read. bam files for each of the 63 strains subjected to amplicon sequencing have been deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with BioProject ID PRJNA526548. The supplemental material includes Figures S1 and S2, Tables S1–S7, File S1 (which describes the supplemental materials in detail), and Files S2–S9 (the PyMOL files used to create Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, and Figure 9). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7865240.
Figure 3.
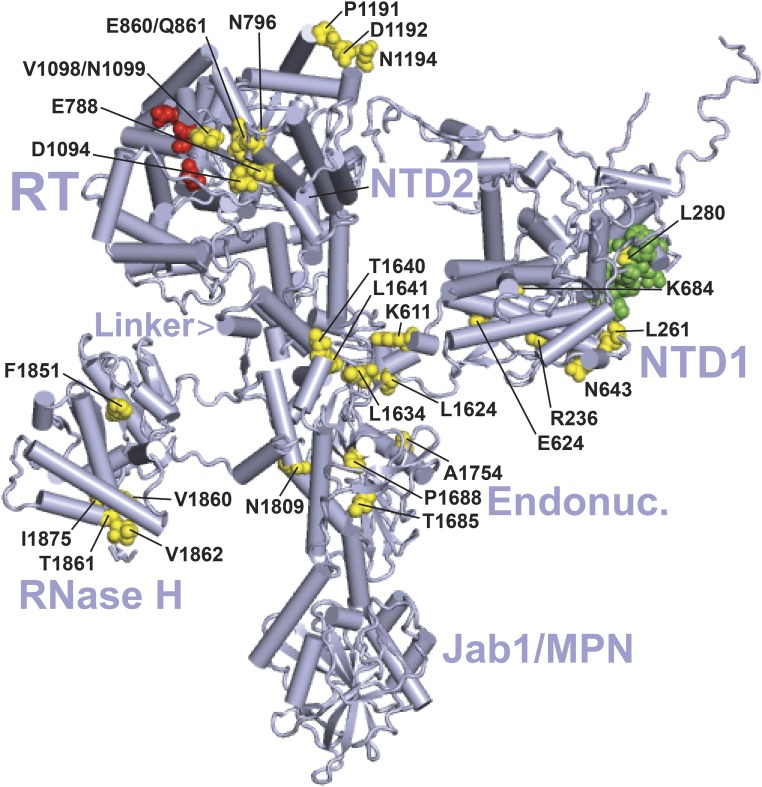
Location of suppressor substitutions in Prp8 in the yeast B complex. The cryo-EM structure is from Plaschka et al. (2017) (PDB: 5nrl). The major domains of Prp8 are indicated as follows: NTD1 and NTD2, RT, Linker, Endonuclease-like (Endonuc.), RNase H-like (RNase H), and Jab1/MPN (Galej et al. 2013; Nguyen et al. 2016; Bertram et al. 2017). Residues altered by U4-cs1-suppressors identified in Kuhn and Brow (2000) or this study (yellow, Table S6) are labeled if clearly visible. Sites of identified prp28-1 suppressors (Price et al. 2014) and brr2-1 suppressors (Kuhn et al. 2002) are colored green and red, respectively. See Table S6 for substitutions. This and subsequent figures were created using the PyMOL Molecular Graphics System, version 1.8.2.1 (Schrödinger, LLC).
Figure 4.
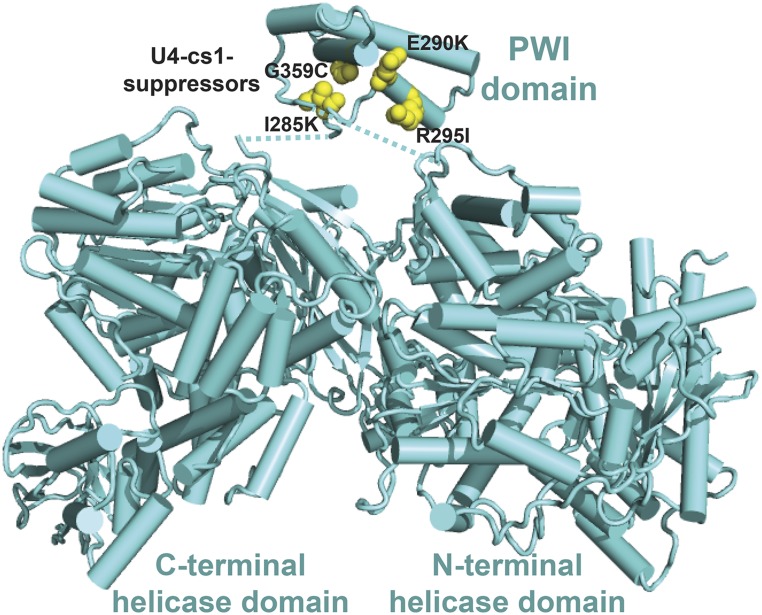
U4-cs1-suppressors in Brr2 cluster together in its N-terminal PWI domain. The U4-cs1-suppressor sites in Brr2’s PWI domain (yellow spheres) face, but do not contact, its catalytic N-terminal helicase domain and regulatory C-terminal helicase domain. Linkers that join the PWI domain to the rest of Brr2 are disordered; dotted lines indicate the connectivity. The structure is from Absmeier et al. (2017) (PDB: 5m52).
Figure 5.
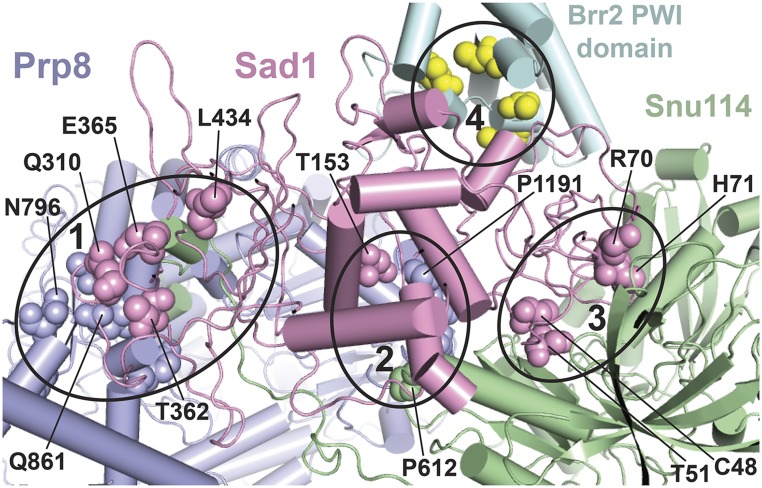
A subset of U4-cs1-suppressor mutations map to the Sad1-Brr2-Snu114-Prp8 interface in a model of the human U4/U6.U5 tri-snRNP. Amino acid residues changed in certain U4-cs1-suppressor strains are shown in spheres of the same color as the parent protein, except for Brr2 where the residues are yellow. Some are labeled with the residue number and wild-type identity. Four interfaces that harbor suppressor substitutions are marked by ellipses and comprise portions of the following protein domains: (1) Prp8(HB/RT1)-Sad1(CTD)-Snu114(NTD), (2) Prp8(RT2)-Sad1(mid)-Snu114(D3), (3) Sad1(ZnF-UBP)-Snu114(D2/3/4a), and (4) Sad1(ZnF-UBP)-Brr2(PWI). The model is based on a 7-Å cryo-EM structure of the human tri-snRNP (Agafonov et al. 2016) and was provided by Holger Stark and Reinhard Lührmann. Equivalent residues in yeast and human were assigned based on sequence alignment. Portions of the proteins that do not directly participate in the Sad1-Brr2-Snu114-Prp8 interface are not shown.
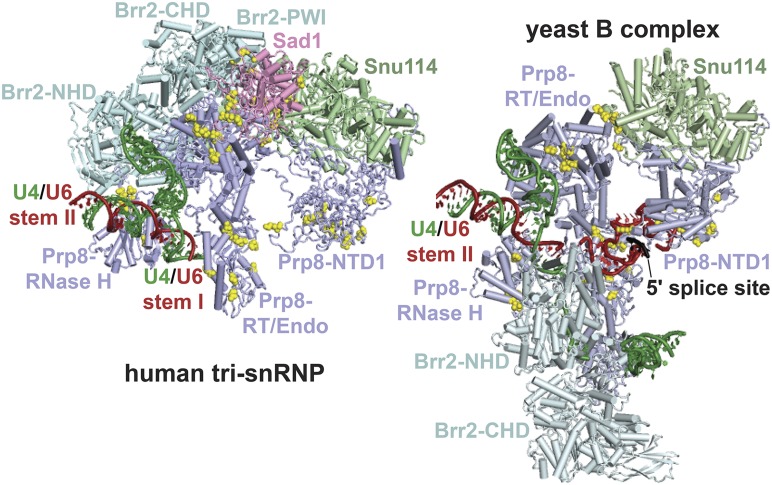
Figure 6.
Retention of Sad1 in the B complex is expected to prevent engagement of U4 by the Brr2 N-terminal helicase domain. Shown are cryo-EM structures of selected components of the human tri-snRNP (left, as in Figure 5) and the yeast B complex (right; Plaschka et al. 2017; PDB: 5nrl). Residues in which U4-cs1-suppressor substitutions were obtained (Table 1 and Table S6) are indicated by yellow spheres. The C-terminal Jab1/MPN domain of Prp8 is omitted from the human tri-snRNP structure. In the human tri-snRNP, the catalytic N-terminal helicase domain (NHD) of Brr2 is far from U4/U6 Stem I (green and red), presumably due to stable contacts between its PWI domain and Sad1. In the yeast B complex, Sad1 is absent and Brr2 has rotated ∼180°, along with the Prp8 RNase H-like domain, and has engaged the single-stranded region of U4 downstream of U4/U6 Stem I in the Brr2-NHD active site. The Brr2 PWI domain is not modeled in the yeast B complex. The intron 5′ splice site is shown paired with the U6 ACAGA stem-loop (Plaschka et al. 2017), upstream of its final location at the U6 ACAGA box. CHD, C-terminal helicase domain; Endo, endonuclease-like.
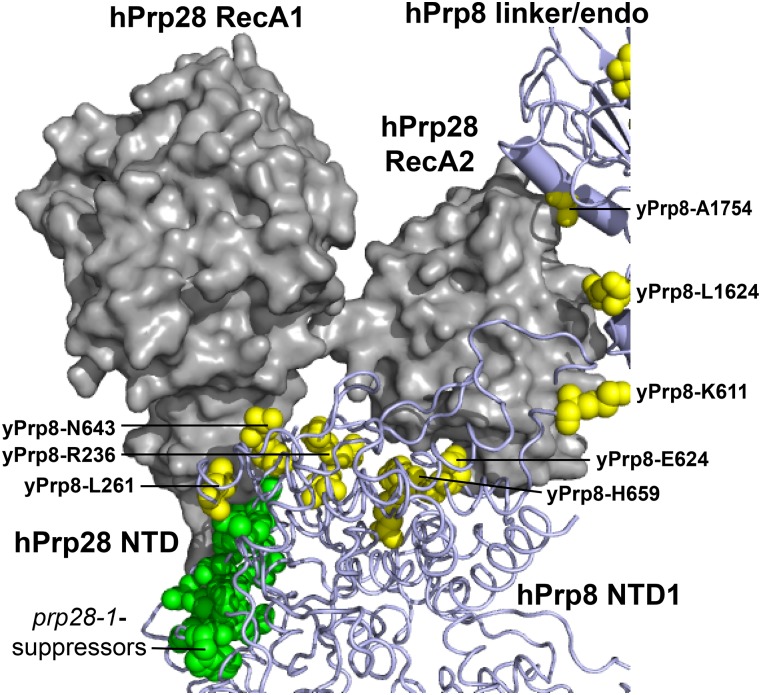
Figure 7.
A subset of U4-cs1-suppressors in Prp8 colocalize with Prp28 in the human tri-snRNP. The two RecA domains of human (h) Prp28 (gray) contact residues in Prp8 that, when mutated in yeast, suppress the cold sensitivity of prp28-1 (green) or U4-cs1 (yellow) strains. Parts of the hPrp8 NTD1 and linker/endonuclease-like (linker/endo) domains are shown, and residues are indicated with yeast (y) numbering. The N-terminal 351 residues of hPrp28 are not modeled, but the C-terminal end of the Prp28-NTD (labeled) is adjacent to prp28-1-suppressor substitutions in Prp8. Model coordinates were kindly provided by Holger Stark and Reinhard Lührmann.
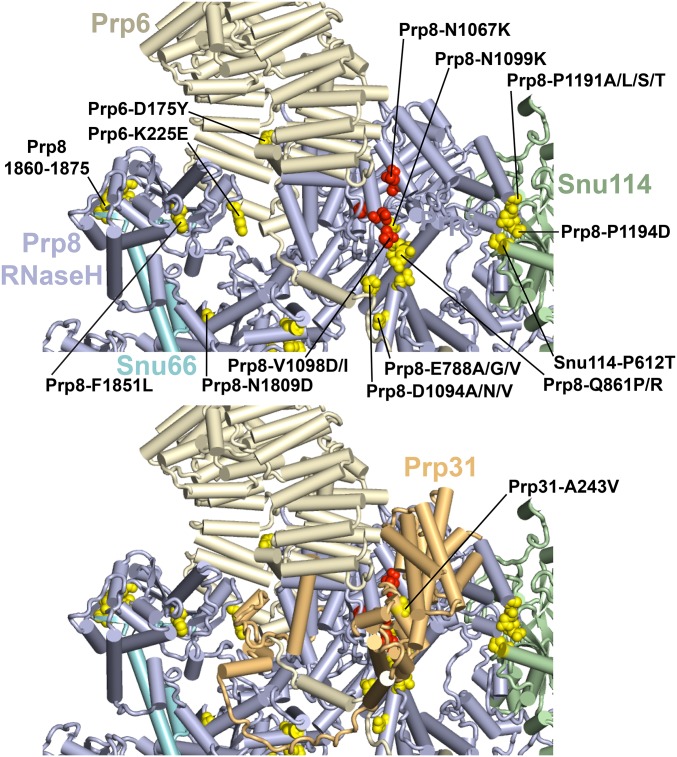
Figure 8.
A subset of U4-cs1-suppressors colocalize with brr2-1 suppressors in the RT domain of Prp8, at an interface with Prp6, and Prp31. (Top) The view is similar to that of the top of the yeast B complex structure shown in Figure 6, with the removal of U4/U6 and addition of Prp6 and Snu66. Selected suppressors of U4-cs1 (yellow) and brr2-1 (red) are labeled with their wild-type and mutant identities. Prp8-V1098D suppress both mutations (Kuhn et al. 2002). U4-cs1-suppressors in the β-hairpin of the Prp8 RNase H-like domain are in residues 1860–1862, 1872, and 1875, and appear to contact Snu66. (Bottom) Same as figure above, but with Prp31 added. The sole U4-cs1-suppressor in Prp31 is labeled. Note that Prp31 appears to contact several of the U4-cs1 and brr2-1 suppressors.
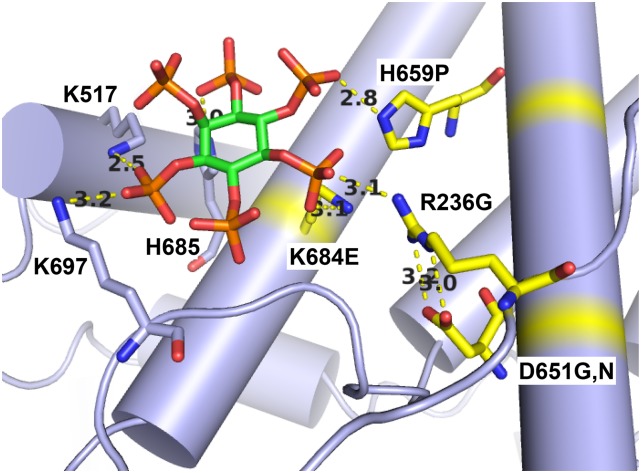
Figure 9.
Several U4-cs1-suppressor substitutions in Prp8 map to a potential IP6-binding site. The model shown is of the yeast C* spliceosome (Fica et al. 2017; PDB: 5mq0). Prp8 is lavender with U4-cs1-suppressor sites in yellow. The inositol group is green and the attached phosphates are orange and red. Modeled distances between atoms indicated by yellow dotted lines are in Ångstroms and are consistent with hydrogen bonds.
Results and Discussion
Efficient selection of spontaneous suppressors of U4-cs1
The U4-cs1 mutation is a three-nucleotide substitution (AAA to UUG) in U4 snRNA adjacent to U4/U6 Stem I (Figure 1) that blocks conversion of the B complex to the Bact complex at low temperature. Initially, we suspected that this conserved sequence in U4 might be an entry site for a helicase that unwinds U4/U6 during spliceosome activation. Indeed, Brr2, a 3′-to-5′ RNA translocase that is a component of the U5 snRNP and has this activity (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998; Kim and Rossi 1999), binds downstream of U4/U6 Stem I in U4 (Hahn et al. 2012; Mozaffari-Jovin et al. 2012). However, after analyzing spontaneous suppressors of U4-cs1-induced cold-sensitivity in U4 and U6, we proposed that extended base pairing between U4-cs1 and U6 prevents the intron 5′ splice site from pairing with the U6 “ACAGA box,” and that such pairing is required for U4/U6 unwinding (Li and Brow 1996). For example, we obtained a tandem duplication of the U6 ACAGA box as a spontaneous suppressor of U4-cs1 (Figure 1, underlined sequence), consistent with occlusion of this sequence by U4-cs1. In addition, other U4-cs1-suppressor mutations in U4 and U6 decrease potential intermolecular base pairing around the ACAGA box, while mutations that further increase potential stability of pairing are lethal even at 30° (Li and Brow 1996).
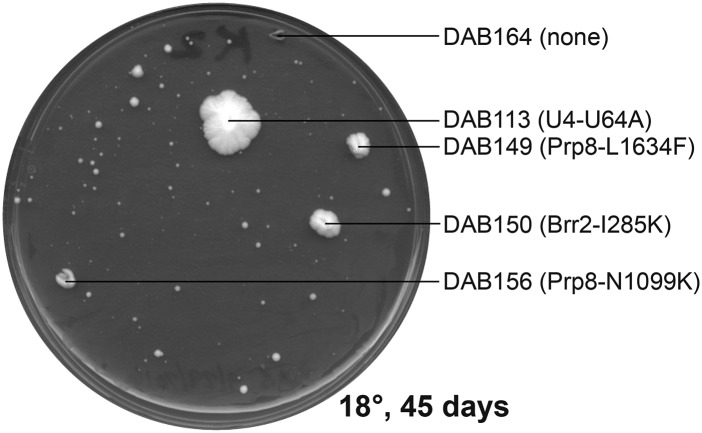
To expand the genome-wide U4-cs1-suppressor search, each of 20 plates containing rich medium (YPD) was spread with an individual saturated culture of a haploid yeast strain containing U4-cs1 in place of wild-type U4 and the plates were incubated at 18° for up to 64 days. An important advantage of selecting spontaneous suppressor mutations, rather than mutagenizing cells prior to the selection, is that all nonsilent mutations identified in genes for splicing factors are highly likely to be causative for suppression (Montemayor et al. 2014). A total of 6 plates produced no colonies; from the other 14 plates a total of 72 colonies were picked as they arose. Figure 2 shows an example of a plate after 45 days at 18°. Colonies picked from the same plate almost always had different mutations, suggesting that the spontaneous suppressors were usually acquired late in the growth of each culture when the cell population was highest. In the last few weeks of incubation, a large number of small colonies appeared (Figure 2), and most colonies picked in the last third of the selection had no mutations in splicing factor genes (see below). These slow-growing strains may suppress the splicing defect by an indirect, less-effective mechanism or could simply reflect stochastic growth of the unsuppressed mutant strain at 18°.
Figure 2.
Distinct U4-cs1-suppressor mutations arise in a single culture. A plate from the selection for cold-resistant U4-cs1 colonies is shown. Colonies that were picked are labeled with their strain designation (DAB1XX) and mutation, if any, in the 112 genes sequenced.
A prior selection using the same 20 yeast cultures plated at 16° rather than 18° yielded only four viable colonies after 9 months of incubation (see Materials and Methods). It is possible that 16° is too stringent a condition for survival of most suppressor strains. Alternatively, storing the 20 liquid cultures at 4° prior to the 18° selection, which was not done before the 16° selection, may have enriched for cold-resistant mutants.
High-throughput identification of U4-cs1-suppressor mutations
Suppressor mutations were identified with a custom-designed, targeted-amplicon sequencing panel commissioned from Illumina. The panel amplifies 112 yeast genes implicated in splicing (Table S1) in up to 95 genomic DNA samples simultaneously. Prior to applying genomic DNA samples to the panel, mutations in U4 and U6 snRNA genes were identified by PCR amplification and Sanger sequencing. Ten strains have mutations in the U4 snRNA gene, SNR14, other than the U4-cs1 mutation (Table 1). Four of these strains have deletions of 4–16 nucleotides in the highly G-/U-rich stretch encompassing the U4-cs1 substitutions, as also obtained in the previous selection (Li and Brow 1996). Five strains have a G or A substitution in U64 that is expected to disrupt a U-G wobble pair in U4/U6 Stem I (Figure 1). The final strain has a U69G substitution that would disrupt a U-A pair with the first nucleotide of the ACAGA sequence. All 10 strains were isolated in the first 19 days of the selection (Table S5), which suggests they are strong suppressors, as expected if they directly alleviate the primary defect.
Only one mutation was found in the U6 snRNA gene, SNR6: a transversion in G55 that would disrupt the same wobble pair affected by the U4-U64 substitutions (Figure 1). This result provides additional validation for a stabilizing function of the U4-U64/U6-G55 pair. The late appearance of the U6-G55U strain (32 days) suggests that mutations in U4 are better tolerated than are mutations in U6 near its ACAGA box.
The remaining 61 strains selected at 18° were analyzed using the sequencing panel. The indexed libraries made from these 61 strains were pooled and sequenced in a single Illumina MiSeq run and the reads were aligned to the S288C reference genome to identify any unique variants in each strain (see Materials and Methods). For 16 of the 61 strains selected at 18°, no variant was found in the 112 genes sequenced (Table S4 and Table S5, “none”). All of these 16 strains arose late in the selection, after at least 40 days of incubation, which is around when numerous other small colonies arose (Figure 2). Thus, the strains without identified variants most likely confer either weak or no suppression.
The 45 remaining strains each have a single nucleotide substitution in a protein-coding gene resulting in a single amino acid substitution, except one (DAB123) that acquired a 21-bp tandem duplication in the SAD1 protein-coding region with a single nucleotide substitution 2 bp downstream (Table 1). As some mutations were obtained more than once, a total of 27 different mutations in 6 different splicing proteins were recovered, all of which have previously been implicated in spliceosome activation: Brr2, Prp6, Prp8, Prp31, Sad1, and Snu114 (Table 1).
There are numerous reasons to think that the identified mutations are responsible for suppression of U4-cs1. First, not a single silent mutation was confirmed in the ∼200 kb of sequence from each of the 63 strains, demonstrating the low frequency of spontaneous mutation and thus the high probability that the recovered mutations were selected. Second, the acquisition of precisely one missense mutation in each of the 45 cold-resistant strains is highly unlikely unless each mutation confers a strong growth advantage. Third, the restriction of substitutions to a small number of the 112 genes sequenced, including one that had previously been confirmed to harbor U4-cs1 suppressors (PRP8), is consistent with a specific mechanism for overcoming the cold-sensitive block created by the U4-cs1 mutation. Indeed, of the eight residues in Prp8 that acquired substitutions in this selection, five were previously shown to harbor confirmed U4-cs1 suppressors and the other three are within one to three residues of known suppressors (Kuhn and Brow 2000; Table S6). Fourth, the temporal clustering of similar mutants is consistent with varying degrees of strength of suppression. For example, two hot spots for substitutions in Prp8 are Thr1872 to Arg or Lys, with four strains from different cultures, and Asn1099 to Lys, with five strains from different cultures (Table 1). Strains with substitutions in these two residues arose at distinct times in the selection, from 14 to 21 days for Thr1872 and 32–40 days for Asn1099 (Table S4, highlighted). Similarly, substitutions in residues 48–71 of Sad1 arose between days 17 and 24, while substitutions in residues 153–434 arose between days 24 and 54. Thus, the highly nonrandom spatial and temporal distributions of the substitutions argues strongly for their suppression of cold sensitivity conferred by U4-cs1.
When all of the past and current U4-cs1-suppressor substitutions are mapped onto the cryo-EM structure of Prp8 in the yeast B complex (Plaschka et al. 2017), they form six clusters that localize to every major domain except the C-terminal Jab1/MPN domain (Figure 3). The interactions that may be altered by the substitutions in Prp8 and other proteins are considered in turn below, starting with Brr2.
U4-cs1-suppressor mutations in Brr2 define a potential site of regulatory contact
Given that Brr2 unwinds U4/U6, I expected to recover U4-cs1 suppressors that promote its activation, most likely by disrupting negative regulation. Negative regulation of Brr2 is required to prevent premature unwinding of U4/U6 in the tri-snRNP and B complex (Absmeier et al. 2016), and loss-of-function mutations that impair such regulation are expected to be more frequent than helicase gain-of-function mutations. Strikingly, all four unique suppressor substitutions in the 2163-amino acid Brr2 map to a 110-residue noncanonical PWI domain (Figure 4). PWI domains are so named due to a conserved proline–tryptophan–isoleucine tripeptide that stabilizes a four-helix bundle; yeast Brr2 has a variant Phe-Phe-Leu tripeptide at residues 291–293 (Absmeier et al. 2015a). This tripeptide is flanked by U4-cs1-suppressor mutations at Glu290 and Arg295. The PWI domain is situated in the tertiary structure between Brr2’s active N-terminal helicase cassette and its regulatory C-terminal helicase cassette (Absmeier et al. 2017). The fold of the PWI domain brings the mutated residues into close proximity and all of the substitutions result in a substantial change in the size or charge of the side chain (Figure 4). I hypothesize that any one of these substitutions disrupts negative regulation of Brr2, allowing it to function in U4-cs1/U6 unwinding at low temperature.
Biochemical studies with purified Brr2 suggested that its N-terminal domain, which includes the PWI domain, has a negative autoregulatory function (Absmeier et al. 2015b, 2017). However, no U4-cs1-suppressor mutations were obtained in the PWI-adjacent surface of either helicase cassette. Furthermore, the PWI domain is displaced from the helicase cassettes in the assembled tri-snRNP (see below). These observations suggest that the substitutions in the PWI domain do not relieve autoregulation, rather they disrupt intermolecular contacts that confer negative regulation of Brr2. If so, then substitutions in the molecules that interact with the Brr2 PWI domain may also be present among the selected suppressor mutations. To identify interfaces that might mediate this regulation, I mapped the U4-cs1-suppressor substitutions present in other splicing factors with which Brr2 interacts in the tri-snRNP.
The locations of U4-cs1-suppressor mutations in the tri-snRNP suggest that Sad1 is retained in the U4-cs1 B complex
Previous studies identified three proteins that appear to regulate Brr2 activity: Prp8 (Kuhn et al. 2002; Maeder et al. 2009; Mozaffari-Jovin et al. 2012), Snu114 (Small et al. 2006), and Sad1 (Huang et al. 2014; Absmeier et al. 2015b). In this study, U4-cs1-suppressor substitutions were recovered in all three proteins; I will discuss those in Sad1 first.
Sad1 (snRNP-assembly defective 1; Lygerou et al. 1999) is only weakly associated with the U4/U6.U5 tri-snRNP in yeast (Huang et al. 2014; Nguyen et al. 2016) and was not identified in the yeast B complex (Fabrizio et al. 2009; Plaschka et al. 2017; Bai et al. 2018). However, yeast Sad1 was detected in a functional penta-snRNP complex isolated from whole-cell extract under low-salt conditions (Stevens et al. 2002). In contrast, Sad1 is a stable component of the human tri-snRNP (Makarova et al. 2001; Agafonov et al. 2016). It is also found in the human pre-B complex, which has a tightly bound U1 snRNP due to a dominant-negative mutation in Prp28 (Boesler et al. 2016), although no structure of this complex is available. Sad1 is not present in the yeast pre-B complex (Bai et al. 2018) or the human B complex (Bertram et al. 2017). In vitro biochemical studies indicate that Sad1 inhibits Brr2 activity via Brr2’s N-terminal domain (including the PWI domain), preventing dissociation of the tri-snRNP prior to its incorporation into the B spliceosome (Huang et al. 2014; Absmeier et al. 2015b). Sad1 presumably leaves the spliceosome in the pre-B to B transition, thus allowing Brr2 to catalyze the B-to-Bact transition.
Half of the 18 U4-cs1-suppressor strains with a mutation in SAD1 have a substitution of cysteine 48; 7 of these 9 are demonstrably independent, i.e., came from different cultures (Table 1). This Cys residue is conserved in human Sad1 (Makarova et al. 2001). Sad1 contains an N-terminal C2H2 zinc finger, but the zinc atom is bound to Cys60 and Cys63, not Cys48 (Hadjivassiliou et al. 2014). Thus, the function of Cys48 is not apparent, nor are the functions of the other residues at which substitutions were identified.
To gain insight into the possible mechanism of U4-cs1 suppression by substitutions in Sad1 and associated proteins, I used sequence alignments (see Figure S2 and Table S6 for Prp8) to map the human residues corresponding to the yeast substitutions onto a model of the human tri-snRNP kindly provided by Holger Stark and Reinhard Lührmann. Since the human tri-snRNP model is based on a 7 Å-resolution cryo-EM structure (Agafonov et al. 2016), the positions of amino acid side chains are uncertain, but the general proximity of residues is apparent. This is the only available model that includes Sad1, and it reveals a striking correspondence between the locations of U4-cs1-suppressor sites and contacts between Sad1, Prp8, Snu114, and Brr2 (Figure 5).
One Prp8-Sad1 interface (Figure 5, region 1) contains U4-cs1-suppressor residues in the Prp8 N-terminal domain 2 (NTD2) and reverse transcriptase-like (RT) domains (Figure 3) and Sad1 residues Q310, T362, E365, and L434 (Table 1 and Table S6). This interface includes the N terminus of Snu114; although no suppressor mutations were isolated in that part of Snu114, deletion of its 128 N-terminal residues blocks the B-to-Bact transition in vitro (Bartels et al. 2002). A second Prp8-Sad1-Snu114 interface (Figure 5, region 2) contains three yeast suppressor residues in the Prp8 RT domain (P1191, D1192, N1194) bracketed by substitutions in Sad1-T153 and Snu114-P612. The remaining suppressor substitutions in Sad1, in residues C48, T51, R70, and H71, along with the seven-residue insertion in strain DAB123, face a region in Snu114 where no suppressor substitutions have yet been recovered (Figure 5, region 3). The suppressor substitutions in the Brr2 PWI domain are adjacent to the N terminus of Sad1 and close to a conserved pentapeptide, residues 31–35 in yeast Sad1 (Figure 5, region 4). No suppressor mutations were obtained in this region of Sad1.
Based on these results, I propose that, in a U4-cs1 strain at low temperature, Sad1 does not dissociate from the tri-snRNP upon its incorporation into the B spliceosome, resulting in a strong arrest to spliceosome activation due to inactive Brr2. I propose further that any of the identified substitutions in the Sad1/tri-snRNP interface weakens Sad1 binding and allows release of Sad1 even at low temperature, permitting spliceosome activation to proceed in a U4-cs1 strain. My hypothesis is consistent with the observation that, in the human tri-snRNP containing Sad1, the N-terminal helicase domain of Brr2 is >10 nm away from its binding site on U4, downstream of U4/U6 Stem I (Agafonov et al. 2016; Figure 6, left). In contrast, in the yeast B complex, which lacks Sad1, the active N-terminal helicase domain of Brr2 is bound to U4 adjacent to U4/U6 Stem I, poised to unwind U4/U6 (Plaschka et al. 2017; Figure 6, right). These results support a model for wild-type B complex activation in which binding of the intron 5′ splice site to the U6 ACAGA box transmits an allosteric signal to the Sad1 binding site, resulting in release of Sad1 and repositioning of Brr2 such that it can engage U4 in its active site. This model predicts that other U4-cs1-suppressor substitutions may act by eliciting transmission of such a signal even in the absence of 5′ splice site/U6 ACAGA box pairing. With this prediction in mind, I investigated the substitutions that lie outside the Sad1 binding site.
U4-cs1 suppressors in Prp8 NTD1 map to the Prp28 binding site and are adjacent to suppressors of prp28-1
One cluster of U4-cs1 suppressors maps to NTD1 of Prp8 (Figure 3), adjacent to suppressors of the cold-sensitive mutation prp28-1 in the DEAD-box ATPase Prp28, which we isolated previously (Price et al. 2014; Table S6). Prp28 assists spliceosome activation by displacing the U1 snRNP from the 5′ splice site, allowing the 5′ splice site to pair with the U6 ACAGA box (Staley and Guthrie 1999; Chen et al. 2001). The direct target of Prp28 is not known, but stabilizing U1/5′ splice site base pairing antagonizes Prp28 function in spliceosome activation.
Prp28 is not present in the cryo-EM structure of either the yeast or human B complex (Bertram et al. 2017; Plaschka et al. 2017) and is not clearly resolved in the yeast pre-B complex (Bai et al. 2018). However, it is present in the human tri-snRNP structure (Agafonov et al. 2016). Figure 7 shows that, when mapped to this structure, the prp28-1-suppressor substitutions in Prp8 contact the N-terminal end of Prp28’s RecA1 domain and likely part of its ∼350 residue NTD, which was mostly unresolved. In contrast, the U4-cs1 suppressors span the gap between the RecA1 and RecA2 domains of Prp28. A few U4-cs1-suppressor sites in the linker (L1624) and endonuclease-like (A1754) domains of Prp8 appear to contact Prp28’s RecA2 domain as well.
Only one identified substitution, Prp8-L280P, suppresses both prp28-1 and U4-cs1 (Kuhn et al. 2002; Price et al. 2014), so the mechanisms of suppression of prp28-1 and U4-cs1 appear to differ. Yang et al. (2013) provided evidence that Prp28 performs two distinct functions: unwinding the U1/5′ splice site duplex and chaperoning pairing of the 5′ splice site with U6. Perhaps the prp28-1 suppressors in Prp8 define a surface of Prp8-NTD1 that mediates U1 snRNP displacement, while the U4-cs1 suppressors in Prp8-NTD1 define a surface that mediates 5′ splice site/U6 pairing. Alternatively or in addition, these U4-cs1 suppressors may act via other interactions after Prp28 has been released. In the yeast B complex, the face of Prp8-NTD1 containing these suppressors is close to Prp38 and Spp381 (Plaschka et al. 2017), both of which have been implicated in the B-to-Bact transition (Xie et al. 1998; Lybarger et al. 1999).
U4-cs1 suppressors in the Prp8 RT domain may influence signaling from Snu114
Three U4-cs1-suppressor residues in the Prp8 RT domain, P1191, D1192, and N1194, that contact Domain III of Snu114 in the tri-snRNP (Figure 5, region 2), continue to do so in the B complex (Figure 8). Snu114 is a GTP-binding protein that is homologous to translation elongation factor G (EF-G, EF-2 in eukaryotes) and regulates Brr2 function in spliceosome activation and disassembly (Bartels et al. 2002; Brenner and Guthrie 2005; Small et al. 2006). In EF-G, Domain III is a mobile element that couples GTP hydrolysis to conformational changes (Macé et al. 2018). Snu114 connects the NTD1 and RT domains of Prp8 (Figure 6), and so could potentially transmit an allosteric signal from Prp28 to Brr2. Intriguingly, the sole U4-cs1-suppressor mutation obtained in Snu114, which arose twice in independent cultures, is in Domain III adjacent to these U4-cs1 suppressors in the RT domain of Prp8 (Figure 8). Thus, these substitutions in Snu114 and Prp8 might mimic a conformational change that signals 5′ splice site/U6 ACAGA box pairing to Brr2.
U4-cs1-suppressors in the Prp8 NTD2/RT domain interface are adjacent to brr2-1 suppressors
Release of Sad1 from the tri-snRNP upon formation of the B complex exposes residues within the RT and NTD2 domains of Prp8 (Figure 5, region 1) that then interact with other proteins, including Prp6 and Prp31. Thus, U4-cs1-suppressor mutations in these Prp8 residues may alter multiple interactions. Prp6 comprises 19 tetratricopeptide repeats preceded by a 240-residue NTD (Legrain et al. 1991). The Prp6-NTD is essential for spliceosome activation in Schizosaccharomyces pombe (Lützelberger et al. 2010) and two U4-cs1 suppressors were recovered in this domain, D175Y and K225E (Figure 8, top). In addition, Prp6 appears to contact at least two U4-cs1-suppressor residues in Prp8, E788 and D1094. Three different amino acid substitutions were isolated in each of these Prp8 residues, and four in P1191 at the Snu114 interface (Figure 8 and Table S6). Furthermore, substitutions in E788 and P1191 were obtained both in the genome-wide and PRP8-targeted selections. Thus, this region of Prp8 is a common target for U4-cs1-suppressors.
Prp31 also has been implicated in spliceosome activation (Weidenhammer et al. 1997), and it appears to interact with several of the U4-cs1-suppressors in Prp8 and Prp6 (Figure 8, bottom). The sole U4-cs1-suppressor we obtained in Prp31, A243V, is in a region that does not directly contact Prp8 or Prp6, but it may alter such contacts indirectly. We previously reported a targeted selection for spontaneous suppressors of a cold-sensitive mutation in BRR2 (brr2-1) in the RT domain of Prp8 (Kuhn et al. 2002; Table S6). As shown in Figure 3 and Figure 8, these suppressors map adjacent to the U4-cs1 suppressors in the RT domain, and also appear to contact Prp31. One substitution in Prp8, V1098D, suppresses both U4-cs1 and brr2-1 (Figure 8 and Table S6). This juxtaposition of U4-cs1-suppressors with suppressors of a catalytic defect in Brr2 is strikingly similar to what we observed for the prp28-1 suppressors (Figure 7) and suggests that closely apposed yet distinct interfaces regulate the ATPases that drive changes in spliceosome conformation. The colocalization of U4-cs1 and brr2-1 suppressors in Prp8 with domains of Prp6 and Prp31 (Figure 8) suggests a nexus for allosteric signaling to Brr2 during activation of the B complex, which may allow integration of several allosteric signals for transmission to Brr2 through Prp8, U4/U6, or other splicing factors, like Snu66.
U4-cs1-suppressors in the Prp8 RNase H-like domain colocalize with Snu66
One additional well-defined cluster of U4-cs1 suppressors is in the β-hairpin of the RNase H-like domain of Prp8. The β-hairpin was previously shown to adopt two conformations and to “toggle” between these conformations in different phases of the splicing pathway (Schellenberg et al. 2013; Mayerle et al. 2017). However, some of the U4-cs1 suppressors in the β-hairpin are predicted to favor the “open loop” conformation (e.g., T1861P and V1862Y), while others are predicted to favor the “closed hairpin” conformation (e.g., V1860D and I1875T). Therefore, suppression may be due to some other effect of these substitutions. In the B complex, the β-hairpin contacts Snu66 (Figure 8). Snu66 is a nonessential tri-snRNP protein that interacts with Sad1 in a two-hybrid assay (Huang et al. 2014) and also binds Hub1, a ubiquitin-like protein that enhances recognition of noncanonical 5′ splice site sequences (Wilkinson et al. 2004; Mishra et al. 2011). Deletion of SNU66 confers cold sensitivity, suggesting that Snu66 is important for helicase function at low temperature. No U4-cs1-suppressors were obtained in Snu66, so it is not certain that alteration of the interaction with Snu66 is the mechanism of U4-cs1 suppression by substitutions in the β-hairpin.
Inositol hexakisphosphate may stabilize the U4-cs1-arrested spliceosome
Cryo-EM-based models of a yeast C* spliceosome, arrested immediately prior to exon ligation, and the human Bact spliceosome include an inositol hexakisphosphate (IP6) molecule bound to Prp8-NTD1 (Fica et al. 2017; Haselbach et al. 2018; Zhang et al. 2018). IP6 is a known cofactor of other RNA processing or modifying enzymes, such as adenosine deaminases that act on RNA (Macbeth et al. 2005). Although IP6 was not modeled in the yeast B complex, its location does not change between the human Bact and yeast C* complexes. Using the yeast C* structure, potential hydrogen bonds are present between IP6 and three Prp8 U4-cs1-suppressor sites (R236, H659, K684), as well as one indirect contact with IP6 (D651 via R236) (Figure 9). If these suppressor mutations act by disrupting binding of IP6, that would imply that IP6 stabilizes the cold-arrested U4-cs1 spliceosome and antagonizes its conversion to the Bact complex at low temperature. IP6 may normally act to stabilize the spliceosome at higher temperatures and/or link spliceosome activation to the metabolic state of the cell.
Notably, the Prp8-H659P substitution in the presence of wild-type U4 results in slow growth at 18 and 30°, and extremely slow growth at 37° (Kuhn and Brow 2000). U4-cs1 and Prp8-H659P mutually suppress each other’s cold sensitivity at 18°, consistent with having opposite effects on B-complex stability. Suppression of the heat sensitivity of Prp8-H659P by a U4-cs1-suppressor in the Linker domain, Prp8-L1634F (Kuhn and Brow 2000), is likely mediated by docking of NTD1 with the Linker domain (Figure 3).
Conclusions and future directions
This study validates the use of custom-targeted sequencing panels for identifying suppressor mutations obtained in genome-wide selections, given that the components of the biological pathway being interrogated are largely known. The cost of this approach, roughly $100 per suppressor strain, is at least 10-fold less expensive than Sanger sequencing the candidate genes and requires far less time and effort. The relatively low error frequency of Illumina sequencing and substantial read depth possible when sequencing only a small fraction of the yeast genome allow high confidence in variant calls. While the mutations identified in this selection are not yet formally proven to be responsible for suppression of U4-cs1, the very low mutation frequency observed, the fact that all substitutions recovered in Prp8 are at or adjacent to the sites of validated U4-cs1 suppressors (Table S6), and the temporal clustering of substitutions in distinct regions of the target proteins strongly suggest that the spontaneous mutations identified here are true suppressors of U4-cs1.
The U4-cs1-suppressor mutations identified in this study and Kuhn and Brow (2000), when mapped to cryo-EM structures of spliceosomal complexes, reveal at least two potential pathways for allosteric signals from the U6 snRNA ACAGA box to Brr2 (Figure 3 and Figure 6). The signal could pass through the Endonuclease-like and Linker domains of Prp8 to its NTD2 and RT domains, and on to Brr2 through Sad1 or other factors (Figure 5 and Figure 8). Alternatively, or in addition, the signal could pass through the Prp8 NTD1 domain to Snu114 and on to the Prp8 RT domain. An intriguing possibility is that this allosteric circuit functions something like a transistor, with Snu114 acting as the “base” that modulates the flow of the allosteric signal through Prp8. Activation of Brr2 by Snu114 in the tri-snRNP is stimulated by GTP binding and inhibited by GDP binding (Small et al. 2006), raising the possibility that modulation of the allosteric signal by Snu114 could be influenced by the intracellular GTP:GDP ratio. Since translation is dependent on GTP hydrolysis to GDP, such a mechanism would couple spliceosome activation to conditions required for translation. In rapidly growing S. cerevisiae cells, ∼90% of spliced mRNAs encode ribosomal proteins (Ares et al. 1999). Thus, decreasing spliceosome activation when the GTP:GDP ratio drops make physiological sense, since ribosome biosynthesis would consequently decrease.
The limited genetic selection described here just begins to illuminate the deep mystery surrounding the allosteric cascade of spliceosome assembly, activation, catalysis, and disassembly. Our targeted selection of U4-cs1-suppressor mutations in PRP8 (Kuhn and Brow 2000) generated more than four times as many unique substitutions in PRP8 as the genome-wide selection reported here, which therefore was not saturating. Gene-targeted selections for U4-cs1-suppressor mutations in BRR2, PRP28, SNU114, PRP31, PRP6, and SNU66 should provide more insight into the role of these splicing factors in the B-to-Bact transition. Use of the custom sequencing panel will greatly accelerate the identification of suppressor mutations in such targeted selections and, in addition, will capture background spontaneous mutations in other genes. By this means it should be possible to map the allosteric signals that drive the B-to-Bact transition. A similar strategy can be used to understand the molecular mechanisms of other steps in the splicing cycle by exploiting primary mutations in factors important for those steps. For example, the prp2-Q548N cold-sensitive mutation (Wlodaver and Staley 2014) could be used to probe the Bact-to-B* transition.
More near-atomic resolution cryo-EM structures of spliceosomal complexes are likely to be determined and will facilitate the further generation of hypotheses for molecular mechanisms of suppression. The suppressor alleles will be valuable tools for both genetic and biochemical experiments to validate hypothesized mechanisms of activation. Thus, the parallel advances in cryo-EM and next-generation sequencing synergize well to accelerate progress in understanding the molecular mechanisms of complex biological nanomachines.
Acknowledgments
I thank Brian Carrick for initiating the new U4-cs1-suppressor selection; Emma Goguen and Erica Schwotzer for genomic DNA preparation and PCR/Sanger sequencing; the University of Wisconsin Biotechnology Center (UWBC) DNA Sequencing Facility for preparing and sequencing the amplicon libraries; the UWBC Bioinformatics Resource Center for identifying sequence variants from fastq files; Holger Stark and Reinhard Lührmann for their model of the human tri-snRNP; members of the Brow, Butcher, and Hoskins laboratories for suggestions; Aaron Hoskins, Manny Ares, and Allyson Yake for insightful comments on the manuscript; and anonymous reviewers for constructive feedback. This work was funded by a grant from the National Institutes of Health (R35 GM-118075), an award from the University of Wisconsin–Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation, and by a University of Wisconsin Chancellor’s Distinguished Teaching Award.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7865240.
Communicating editor: C. Kaplan
Literature Cited
- Absmeier E., Rosenberger L., Apelt L., Becke C., Santos K. F., et al. , 2015a A noncanonical PWI domain in the N-terminal helicase-associated region of the spliceosomal Brr2 protein. Acta Crystallogr. D Biol. Crystallogr. 71: 762–771. 10.1107/S1399004715001005 [DOI] [PubMed] [Google Scholar]
- Absmeier E., Wollenhaupt J., Mozaffari-Jovin S., Becke C., Lee C.-T., et al. , 2015b The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev. 29: 2576–2587. 10.1101/gad.271528.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absmeier E., Santos K. F., Wahl M. C., 2016. Functions and regulation of the Brr2 RNA helicase during splicing. Cell Cycle 15: 3362–3377. 10.1080/15384101.2016.1249549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absmeier E., Becke C., Wollenhaupt J., Santos K. F., Wahl M. C., 2017. Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2. Cell Cycle 16: 100–112. 10.1080/15384101.2016.1255384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agafonov D. E., Kastner B., Dybkov O., Hoefele R. V., Liu W. T., et al. , 2016. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351: 1416–1420. 10.1126/science.aad2085 [DOI] [PubMed] [Google Scholar]
- Ares Jr., M., Igel A. H., 1990. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 4: 2132–2145. 10.1101/gad.4.12a.2132 [DOI] [PubMed] [Google Scholar]
- Ares Jr., M., Grate L., Pauling M. H., 1999. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA 5: 1138–1139. 10.1017/S1355838299991379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R., Wan R., Yan C., Lei J., Shi Y., 2018. Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. Science 360: 1423–1429. 10.1126/science.aau0325 [DOI] [PubMed] [Google Scholar]
- Bartels C., Klatt C., Lührmann R., Fabrizio P., 2002. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 3: 875–880. 10.1093/embo-reports/kvf172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K., Agafonov D. E., Dybkov O., Haselbach D., Leelaram M. N., et al. , 2017. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170: 701–713.e11. 10.1016/j.cell.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Boesler C., Rigo N., Anokhina M. M., Tauchert M. J., Agafonov D. E., et al. , 2016. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat. Commun. 7: 11997 10.1038/ncomms11997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner T. J., Guthrie C., 2005. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics 170: 1063–1080. 10.1534/genetics.105.042044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow D. A., 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. 10.1146/annurev.genet.36.043002.091635 [DOI] [PubMed] [Google Scholar]
- Burgess S. M., Guthrie C., 1993. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem. Sci. 18: 381–384. 10.1016/0968-0004(93)90094-4 [DOI] [PubMed] [Google Scholar]
- Chang T.-H., Tung L., Yeh F.-L., Chen J.-H., Chang S.-L., 2013. Functions of the DExD/H-box proteins in nuclear pre-mRNA splicing. Biochim. Biophys. Acta 1829: 764–774. 10.1016/j.bbagrm.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Stands L., Staley J. P., Jackups R. R., Latus L. J., et al. , 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7: 227–232. 10.1016/S1097-2765(01)00170-8 [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto J. R., Tamm J., Parker R., Guthrie C., 1987. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1: 445–455. 10.1101/gad.1.5.445 [DOI] [PubMed] [Google Scholar]
- Didychuk A. L., Butcher S. E., Brow D. A., 2018. The life of U6 small nuclear RNA, from cradle to grave. RNA 24: 437–460. 10.1261/rna.065136.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., et al. , 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36: 593–608. 10.1016/j.molcel.2009.09.040 [DOI] [PubMed] [Google Scholar]
- Fica S. M., Nagai K., 2017. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 24: 791–799. 10.1038/nsmb.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica S. M., Oubridge C., Galej W. P., Wilkinson M. E., Bai X.-C., et al. , 2017. Structure of a spliceosome remodeled for exon ligation. Nature 542: 377–380. 10.1038/nature21078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej W. P., Oubridge C., Newman A. J., Nagai K., 2013. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493: 638–643. 10.1038/nature11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjivassiliou H., Rosenberg O. S., Guthrie C., 2014. The crystal structure of S. cerevisiae Sad1, a catalytically inactive deubiquitinase that is broadly required for pre-mRNA splicing. RNA 20: 656–669. 10.1261/rna.042838.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D., Kudla G., Tollervey D., Beggs J. D., 2012. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 26: 2408–2421. 10.1101/gad.199307.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbach D., Komarov I., Agafonov D. E., Hartmuth K., Graf B., et al. , 2018. Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172: 454–464.e11. 10.1016/j.cell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Hoskins A. A., Moore M. J., 2012. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem. Sci. 37: 179–188. 10.1016/j.tibs.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-H., Chung C.-S., Kao D.-I., Kao T.-C., Cheng S.-C., 2014. Sad1 counteracts Brr2-mediated dissociation of U4/U6.U5 in tri-snRNP homeostasis. Mol. Cell. Biol. 34: 210–220. 10.1128/MCB.00837-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Lei R., Ding S.-W., Zhu S., 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15: 182 10.1186/1471-2105-15-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B., Will C. L., Stark H., Lührmann R., 2019. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb. Perspect. Biol.: a032417 10.1101/cshperspect.a032417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H., Rossi J. J., 1999. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 5: 959–971. 10.1017/S135583829999012X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A. N., Brow D. A., 2000. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics 155: 1667–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A. N., Li Z., Brow D. A., 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell 3: 65–75. 10.1016/S1097-2765(00)80175-6 [DOI] [PubMed] [Google Scholar]
- Kuhn A. N., Reichl E. M., Brow D. A., 2002. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. USA 99: 9145–9149. 10.1073/pnas.102304299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B., Achsel T., Lührmann R., 1998. The human U5–200kD DEXH-box protein unwinds U4/U6 duplices in vitro. Proc. Natl. Acad. Sci. USA 95: 4188–4192. 10.1073/pnas.95.8.4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P., Chapon C., Galisson F., 1991. Proteins involved in mitosis, RNA synthesis and pre-mRNA splicing share a common repeating motif. Nucleic Acids Res. 19: 2509–2510. 10.1093/nar/19.9.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997.
- Li Z., Brow D. A., 1996. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA 2: 879–894. [PMC free article] [PubMed] [Google Scholar]
- Lützelberger M., Bottner C. A., Schwelnus W., Zock-Emmenthal S., Razanau A., et al. , 2010. The N-terminus of Prp1 (Prp6/U5–102K) is essential for spliceosome activation in vivo. Nucleic Acids Res. 38: 1610–1622. 10.1093/nar/gkp1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger S., Beickman K., Brown V., Dembla-Rajpal N., Morey K., et al. , 1999. Elevated levels of a U4/U6.U5 snRNP-associated protein, Spp381p, rescue a mutant defective in spliceosome maturation. Mol. Cell. Biol. 19: 577–584. 10.1128/MCB.19.1.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Christophides G., Séraphin B., 1999. A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell. Biol. 19: 2008–2020. 10.1128/MCB.19.3.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth M. R., Schubert H. L., VanDemark A. P., Lingam A. T., Hill C. P., et al. , 2005. Inositol hexakisphophate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539. 10.1126/science.1113150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macé K., Giudice E., Chat S., Gillet R., 2018. The structure of an elongation factor G-ribosome complex captured in the absence of inhibitors. Nucleic Acids Res. 46: 3211–3217. 10.1093/nar/gky081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder C., Kutach A. K., Guthrie C., 2009. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat. Struct. Mol. Biol. 16: 42–48. 10.1038/nsmb.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T., Salzberg S. L., 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O. V., Makarov E. M., Lührmann R., 2001. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 20: 2553–2563. 10.1093/emboj/20.10.2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerle M., Guthrie C., 2017. Genetics and biochemistry remain essential in the structural era of the spliceosome. Methods 125: 3–9. 10.1016/j.ymeth.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerle M., Raghavan M., Ledoux S., Price A., Stepankiw N., et al. , 2017. Structural toggle in the RNaseH domain of Prp8 helps balance splicing fidelity and catalytic efficiency. Proc. Natl. Acad. Sci. USA 114: 4739–4744. 10.1073/pnas.1701462114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus C. J., Schwartz M. L., Butcher S. E., Brow D. A., 2007. A dynamic bulge in the U6 RNA internal stem-loop functions in spliceosome assembly and activation. RNA 13: 2252–2265. 10.1261/rna.699907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S. K., Ammon T., Popowicz G. M., Krajewski M., Nagel R. J., et al. , 2011. Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing. Nature 474: 173–178. 10.1038/nature10143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montemayor E. J., Curran E. C., Liao H. H., Andrews K. L., Treba C. N., et al. , 2014. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-Å resolution. Nat. Struct. Mol. Biol. 21: 544–551. 10.1038/nsmb.2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S., Santos K. F., Hsiao H. H., Will C. L., Urlab H., et al. , 2012. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 26: 2422–2434. 10.1101/gad.200949.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. H. D., Galej W. P., Bai X.-C., Oubridge C., Newman A. J., et al. , 2016. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 530: 298–302. 10.1038/nature16940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova O., Belfort M., 2017. Mobile group II introns as ancestral eukaryotic elements. Trends Genet. 33: 773–783. 10.1016/j.tig.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C., Lin P.-C., Nagai K., 2017. Structure of a pre-catalytic spliceosome. Nature 546: 617–621. 10.1038/nature22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. M., Görnemann J., Guthrie C., Brow D. A., 2014. An unanticipated early function of DEAD-box ATPase Prp28 during commitment to splicing is modulated by U5 snRNP protein Prp8. RNA 20: 46–60. 10.1261/rna.041970.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan P. L., Guthrie C., 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8: 847–855. 10.1016/S0960-9822(07)00345-4 [DOI] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Na"t. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg M. J., Wu T., Ritchie D. B., Fica S., Staley J. P., et al. , 2013. A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation. Nat. Struct. Mol. Biol. 20: 728–734. 10.1038/nsmb.2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow D. R., Staley J. P., 2012. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci. 37: 263–273. 10.1016/j.tibs.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., 2017. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18: 655–670. 10.1038/nrm.2017.86 [DOI] [PubMed] [Google Scholar]
- Small E. C., Leggett S. R., Winans A. A., Staley J. P., 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell 23: 389–399. 10.1016/j.molcel.2006.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J. P., Guthrie C., 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3: 55–64. 10.1016/S1097-2765(00)80174-4 [DOI] [PubMed] [Google Scholar]
- Stevens S. W., Ryan D. E., Ge H. Y., Moore R. E., Young M. K., et al. , 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9: 31–44. 10.1016/S1097-2765(02)00436-7 [DOI] [PubMed] [Google Scholar]
- Weidenhammer E. M., Ruiz-Noriega M., Woolford J. L., 1997. Prp31p promotes the association of the U4/U6.U5 tri-snRNP with pre-spliceosomes to form spliceosomes in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 3580–3588. 10.1128/MCB.17.7.3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C. R., Dittmar G. A., Ohi M. D., Uetz P., Jones N., et al. , 2004. Ubiquitin-like protein Hub1 is required for pre-mRNA splicing and localization of an essential splicing factor in fission yeast. Curr. Biol. 14: 2283–2288. 10.1016/j.cub.2004.11.058 [DOI] [PubMed] [Google Scholar]
- Will C. L., Lührmann R., 2011. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3: a003707 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodaver A. M., Staley J. P., 2014. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic core of the spliceosome. RNA 20: 282–294. 10.1261/rna.042598.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Beickman K., Otte E., Rymond B. C., 1998. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 17: 2938–2946. 10.1093/emboj/17.10.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wang X.-Y., Zhang Z.-M., Pu J., Fan Y.-J., et al. , 2013. Splicing proofreading at 5′ splice sites by ATPase Prp28p. Nucleic Acids Res. 41: 4660–4670. 10.1093/nar/gkt149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yan Z., Zhan X., Li L., Lei J., et al. , 2018. Structure of the human activated spliceosome in three conformational states. Cell Res. 28: 307–322. 10.1038/cr.2018.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Pyle A. M., 2017. The group II intron maturase: a reverse transcriptase and splicing factor go hand in hand. Curr. Opin. Struct. Biol. 47: 30–39. 10.1016/j.sbi.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Yeast strains are available upon request. Sequence read. bam files for each of the 63 strains subjected to amplicon sequencing have been deposited at the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) with BioProject ID PRJNA526548. The supplemental material includes Figures S1 and S2, Tables S1–S7, File S1 (which describes the supplemental materials in detail), and Files S2–S9 (the PyMOL files used to create Figure 3, Figure 4, Figure 5, Figure 6, Figure 7, Figure 8, and Figure 9). Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7865240.
Figure 3.
Location of suppressor substitutions in Prp8 in the yeast B complex. The cryo-EM structure is from Plaschka et al. (2017) (PDB: 5nrl). The major domains of Prp8 are indicated as follows: NTD1 and NTD2, RT, Linker, Endonuclease-like (Endonuc.), RNase H-like (RNase H), and Jab1/MPN (Galej et al. 2013; Nguyen et al. 2016; Bertram et al. 2017). Residues altered by U4-cs1-suppressors identified in Kuhn and Brow (2000) or this study (yellow, Table S6) are labeled if clearly visible. Sites of identified prp28-1 suppressors (Price et al. 2014) and brr2-1 suppressors (Kuhn et al. 2002) are colored green and red, respectively. See Table S6 for substitutions. This and subsequent figures were created using the PyMOL Molecular Graphics System, version 1.8.2.1 (Schrödinger, LLC).
Figure 4.
U4-cs1-suppressors in Brr2 cluster together in its N-terminal PWI domain. The U4-cs1-suppressor sites in Brr2’s PWI domain (yellow spheres) face, but do not contact, its catalytic N-terminal helicase domain and regulatory C-terminal helicase domain. Linkers that join the PWI domain to the rest of Brr2 are disordered; dotted lines indicate the connectivity. The structure is from Absmeier et al. (2017) (PDB: 5m52).
Figure 5.
A subset of U4-cs1-suppressor mutations map to the Sad1-Brr2-Snu114-Prp8 interface in a model of the human U4/U6.U5 tri-snRNP. Amino acid residues changed in certain U4-cs1-suppressor strains are shown in spheres of the same color as the parent protein, except for Brr2 where the residues are yellow. Some are labeled with the residue number and wild-type identity. Four interfaces that harbor suppressor substitutions are marked by ellipses and comprise portions of the following protein domains: (1) Prp8(HB/RT1)-Sad1(CTD)-Snu114(NTD), (2) Prp8(RT2)-Sad1(mid)-Snu114(D3), (3) Sad1(ZnF-UBP)-Snu114(D2/3/4a), and (4) Sad1(ZnF-UBP)-Brr2(PWI). The model is based on a 7-Å cryo-EM structure of the human tri-snRNP (Agafonov et al. 2016) and was provided by Holger Stark and Reinhard Lührmann. Equivalent residues in yeast and human were assigned based on sequence alignment. Portions of the proteins that do not directly participate in the Sad1-Brr2-Snu114-Prp8 interface are not shown.
Figure 6.
Retention of Sad1 in the B complex is expected to prevent engagement of U4 by the Brr2 N-terminal helicase domain. Shown are cryo-EM structures of selected components of the human tri-snRNP (left, as in Figure 5) and the yeast B complex (right; Plaschka et al. 2017; PDB: 5nrl). Residues in which U4-cs1-suppressor substitutions were obtained (Table 1 and Table S6) are indicated by yellow spheres. The C-terminal Jab1/MPN domain of Prp8 is omitted from the human tri-snRNP structure. In the human tri-snRNP, the catalytic N-terminal helicase domain (NHD) of Brr2 is far from U4/U6 Stem I (green and red), presumably due to stable contacts between its PWI domain and Sad1. In the yeast B complex, Sad1 is absent and Brr2 has rotated ∼180°, along with the Prp8 RNase H-like domain, and has engaged the single-stranded region of U4 downstream of U4/U6 Stem I in the Brr2-NHD active site. The Brr2 PWI domain is not modeled in the yeast B complex. The intron 5′ splice site is shown paired with the U6 ACAGA stem-loop (Plaschka et al. 2017), upstream of its final location at the U6 ACAGA box. CHD, C-terminal helicase domain; Endo, endonuclease-like.
Figure 7.
A subset of U4-cs1-suppressors in Prp8 colocalize with Prp28 in the human tri-snRNP. The two RecA domains of human (h) Prp28 (gray) contact residues in Prp8 that, when mutated in yeast, suppress the cold sensitivity of prp28-1 (green) or U4-cs1 (yellow) strains. Parts of the hPrp8 NTD1 and linker/endonuclease-like (linker/endo) domains are shown, and residues are indicated with yeast (y) numbering. The N-terminal 351 residues of hPrp28 are not modeled, but the C-terminal end of the Prp28-NTD (labeled) is adjacent to prp28-1-suppressor substitutions in Prp8. Model coordinates were kindly provided by Holger Stark and Reinhard Lührmann.
Figure 8.
A subset of U4-cs1-suppressors colocalize with brr2-1 suppressors in the RT domain of Prp8, at an interface with Prp6, and Prp31. (Top) The view is similar to that of the top of the yeast B complex structure shown in Figure 6, with the removal of U4/U6 and addition of Prp6 and Snu66. Selected suppressors of U4-cs1 (yellow) and brr2-1 (red) are labeled with their wild-type and mutant identities. Prp8-V1098D suppress both mutations (Kuhn et al. 2002). U4-cs1-suppressors in the β-hairpin of the Prp8 RNase H-like domain are in residues 1860–1862, 1872, and 1875, and appear to contact Snu66. (Bottom) Same as figure above, but with Prp31 added. The sole U4-cs1-suppressor in Prp31 is labeled. Note that Prp31 appears to contact several of the U4-cs1 and brr2-1 suppressors.
Figure 9.
Several U4-cs1-suppressor substitutions in Prp8 map to a potential IP6-binding site. The model shown is of the yeast C* spliceosome (Fica et al. 2017; PDB: 5mq0). Prp8 is lavender with U4-cs1-suppressor sites in yellow. The inositol group is green and the attached phosphates are orange and red. Modeled distances between atoms indicated by yellow dotted lines are in Ångstroms and are consistent with hydrogen bonds.