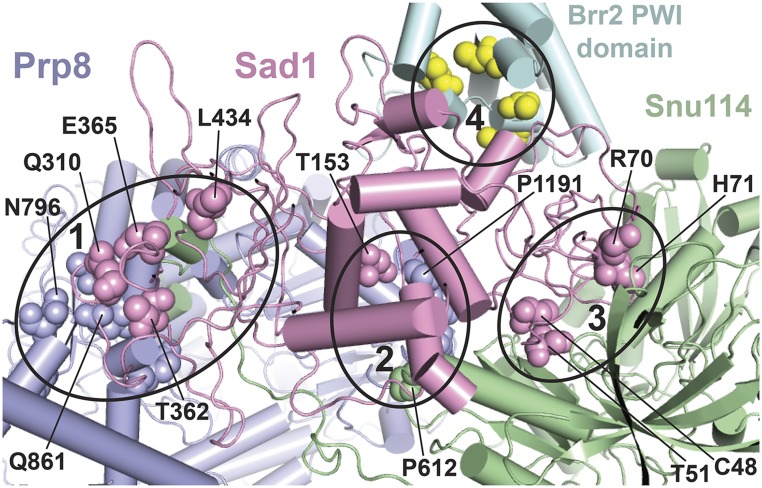
Figure 5.
A subset of U4-cs1-suppressor mutations map to the Sad1-Brr2-Snu114-Prp8 interface in a model of the human U4/U6.U5 tri-snRNP. Amino acid residues changed in certain U4-cs1-suppressor strains are shown in spheres of the same color as the parent protein, except for Brr2 where the residues are yellow. Some are labeled with the residue number and wild-type identity. Four interfaces that harbor suppressor substitutions are marked by ellipses and comprise portions of the following protein domains: (1) Prp8(HB/RT1)-Sad1(CTD)-Snu114(NTD), (2) Prp8(RT2)-Sad1(mid)-Snu114(D3), (3) Sad1(ZnF-UBP)-Snu114(D2/3/4a), and (4) Sad1(ZnF-UBP)-Brr2(PWI). The model is based on a 7-Å cryo-EM structure of the human tri-snRNP (Agafonov et al. 2016) and was provided by Holger Stark and Reinhard Lührmann. Equivalent residues in yeast and human were assigned based on sequence alignment. Portions of the proteins that do not directly participate in the Sad1-Brr2-Snu114-Prp8 interface are not shown.