Centrosomes are important mediators of mitotic spindle assembly, and loss of centrosomes can lead to mitotic errors and cell death; however, it is also clear that cells can compensate for centrosome loss through multiple mechanisms...
Keywords: centrosomes, apoptosis, JNK signaling, ROS, oxidative stress
Abstract
Centrosomes play a critical role in mitotic spindle assembly through their role in microtubule nucleation and bipolar spindle assembly. Loss of centrosomes can impair the ability of some cells to properly conduct mitotic division, leading to chromosomal instability, cell stress, and aneuploidy. Multiple aspects of the cellular response to mitotic error associated with centrosome loss appear to involve activation of JNK signaling. To further characterize the transcriptional effects of centrosome loss, we compared gene expression profiles of wild-type and acentrosomal cells from Drosophila wing imaginal discs. We found elevation of expression of JNK target genes, which we verified at the protein level. Consistent with this, the upregulated gene set showed significant enrichment for the AP-1 consensus DNA-binding sequence. We also found significant elevation in expression of genes regulating redox balance. Based on those findings, we examined oxidative stress after centrosome loss, revealing that acentrosomal wing cells have significant increases in reactive oxygen species (ROS). We then performed a candidate genetic screen and found that one of the genes upregulated in acentrosomal cells, glucose-6-phosphate dehydrogenase, plays an important role in buffering acentrosomal cells against increased ROS and helps protect those cells from cell death. Our data and other recent studies have revealed a complex network of signaling pathways, transcriptional programs, and cellular processes that epithelial cells use to respond to stressors, like mitotic errors, to help limit cell damage and maintain normal tissue development.
PROPER development requires precise spatial and temporal coordination of cell division to drive tissue growth. During cell division, chromosomes are replicated in S phase and then segregated equally into two daughter cells during mitosis. The accurate segregation of chromosomes is achieved by the action of the bipolar mitotic spindle (Walczak and Heald 2008). This microtubule (MT)-based structure is essential to generate the physical forces required to move chromosomes to opposite poles, and also has built-in checkpoints that ensure accurate segregation. The assembly of the mitotic spindle is a complex process with multiple layers of regulation to ensure its accuracy (Prosser and Pelletier 2017). Defects in mitotic spindle formation can lead to multipolar spindles or incorrect attachment of MTs to chromosomes, which in turn can lead to segregation errors that cause DNA damage and even whole-chromosome missegregation (aneuploidy). These types of defects are forms of chromosomal instability (CIN), a hallmark of many cancers that is highly correlated with tumor malignancy (Hanahan and Weinberg 2011; Nicholson and Cimini 2011).
In most animal cells, the bipolar mitotic spindle arises from the MT nucleating activity of a pair of organelles known as centrosomes, which sit at the two spindle poles (Figure 1A) (Walczak and Heald 2008; Lerit and Poulton 2016; Prosser and Pelletier 2017). As the central source of spindle MTs, the orientation of the centrosome pair also determines the geometry of mitotic spindle formation and the axis of division relative to the surrounding tissue. Centrosomes also serve a wide range of cellular functions separate from mitotic spindle assembly, including the regulation of cilia assembly, cell cycle progression, the DNA damage response (DDR), and cell signaling. Given these critical functions ascribed to centrosomes, they were long-considered essential components of most animal cells.
Figure 1.
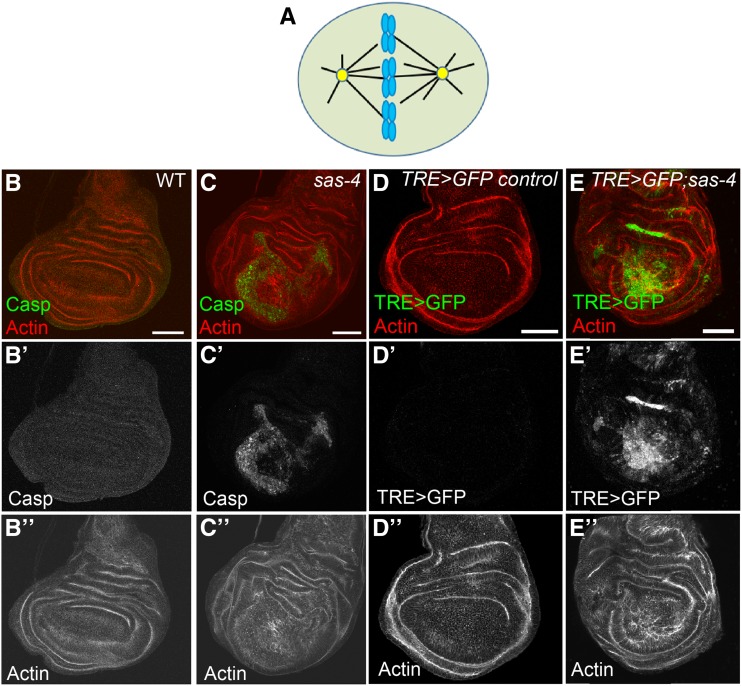
Centrosome loss dramatically increases apoptosis levels and JNK activity. (A) Model of a dividing cell. A pair of centrosomes (yellow) nucleate microtubules (black lines), some of which attach to the chromosomes (blue), to form a bipolar spindle. (B–B”) There is little to no apoptosis in wild-type (WT) third larval instar wing discs, as reported by antibodies to cleaved caspase 3. (C–C”) In sas-4 mutants, which lack centrosomes, many cells undergo apoptotic cell death. (D–D”) The JNK transcriptional reporter, TRE>GFP, has no detectable expression at this stage in control discs. (E–E”) Many of the acentrosomal cells in sas-4 mutant wing discs have highly elevated JNK activity. All images are maximum-intensity projections. Bars, 50 µm. Red channel: actin. Green channel: cleaved caspase 3 (Casp) in (B and C); TRE>GFP in (D and E).
However, more recently, it became apparent that cells possess centrosome-independent MT nucleation pathways that assist in spindle assembly (e.g., the Augmin complex and the RanGTP pathway) (Prosser and Pelletier 2017). In many cell types, these additional pathways are robust and capable of assembling a bipolar spindle even in the complete absence of centrosomes. A striking example of this occurs in Drosophila where entire animals homozygous mutant for genes essential for centrosome formation or function can develop to adulthood (Basto et al. 2006). We now know this is not unique to flies, because if p53-mediated programed cell death is blocked, mice lacking centrosomes can develop to late embryogenesis and then die because the lack of cilia impairs Hedgehog signaling (Bazzi and Anderson 2014).
In Drosophila, detailed examinations of acentrosomal cells in several tissues (e.g., brain and ovarian germline) have revealed surprisingly few mitotic errors, indicating that the noncentrosomal MT nucleation pathways are adequate for proper spindle assembly and accurate chromosome segregation in those cells (Basto et al. 2006; Stevens et al. 2007; Poulton et al. 2017). In contrast, we previously found that in the proliferative epithelial cells of the wing imaginal disc, loss of those same centrosomal proteins leads to significant defects in spindle assembly, which increases rates of aneuploidy, DNA damage, and misoriented spindles (Poulton et al. 2014). Those defects then activate a cell stress pathway, c-Jun N-terminal Kinase (JNK) signaling, which drives apoptotic cell death (Figure 1, B–E). In wing discs lacking centrosomes (mutant for sas-4 or asl) or possessing dysfunctional centrosomes (mutant for cnn), ∼15–20% of cells die (Poulton et al. 2014), suggesting that although alternative MT nucleation pathways help buffer wing disc cells against centrosome loss, they are not as effective in this tissue as they are in other tissues/cell types. Despite loss of such a substantial fraction of cells, overall wing development remains remarkably normal in most centrosome-deficient animals. Proper mitosis and subsequent wing development in acentrosomal animals are mediated by several factors. Correct spindle assembly becomes dependent on MT nucleation by the Augmin complex and the RanGTP pathway, and on delay of the cell cycle by the spindle assembly checkpoint (SAC). Cell death that does occur is buffered by compensatory proliferation of neighboring cells to replace dying cells and delayed development, which presumably allows additional time to correct tissue-level defects caused by massive cell death (Poulton et al. 2014). Together, these findings have highlighted the remarkable ability of cells and tissues to compensate not only for loss of key mitotic regulators, such as centrosomes, but also for the wide range of downstream effects of their loss, such as CIN and cell death.
The sensitivity of wing disc cells to mitotic spindle assembly errors due to centrosome loss, as well as their sensitivity to the downstream consequences of spindle assembly errors (i.e., aneuploidy and spindle misorientation), make them an excellent model to investigate the cellular response to centrosome loss, mitotic errors, and cell death, as well as the tissue-level and systemic responses to those insults. As our previous data and others have demonstrated, one important component of these complex responses to tissue damage induced by a variety of stresses is changes in gene expression, the most well characterized of which are associated with the activation of cell signaling pathways [i.e., JNK, Wnt, Dpp, and JAK-STAT (Janus Kinase-Signal Transducer and Activator of Transcription)] (Ryoo et al. 2004; Kondo et al. 2006; Pastor-Pareja et al. 2008; Perez-Garijo et al. 2009; Dekanty et al. 2012; Poulton et al. 2014). For example, JNK signaling is a central mediator of the response to multiple forms of cell stress or tissue damage (Igaki 2009). High levels of JNK activity initiate apoptosis in tissues like the wing and eye imaginal discs. To help compensate for the loss of cells due to apoptosis, lower levels of JNK in neighboring cells can help drive proliferation in the surviving cells to help maintain total cell numbers and tissue integrity, which is a central component of the regeneration process (Ryoo et al. 2004; Fan and Bergmann 2008; Martin et al. 2009; Perez-Garijo et al. 2009; Fogarty et al. 2016; Brock et al. 2017; Khan et al. 2017). Several recent studies have demonstrated the important transcriptional responses occurring in damaged/stressed cells, much of them mediated directly by JNK signaling. For example, one key pathway that helps drive compensatory proliferation is JAK-STAT signaling, whose activating ligands [the Unpaired (Upd) proteins] are themselves transcriptional targets of JNK signaling (Pastor-Pareja et al. 2008; Bunker et al. 2015; Santabárbara-Ruiz et al. 2015). Intriguingly, recent studies have also uncovered important effects of cell stress and damage on redox balance in imaginal discs, and have suggested important roles for reactive oxygen species (ROS) in mediating the activities of the relevant cell signaling pathways to control processes like cell death and compensatory proliferation (Kanda et al. 2011; Ohsawa et al. 2012; Gauron et al. 2013; Huu et al. 2015; Santabárbara-Ruiz et al. 2015; Clemente-Ruiz et al. 2016; Fogarty et al. 2016; Brock et al. 2017; Khan et al. 2017; Pérez et al. 2017). Together, these studies have begun to elucidate a regulatory network involving complex cross talk between traditional signaling pathways and ROS, which helps correct cellular damage and maintain tissue homeostasis.
We sought to define the transcriptional response to centrosome loss by performing transcriptome analysis on imaginal wing discs from wild-type (WT) animals and on two centrosome-lacking genotypes. Differential gene expression analysis identified hundreds of genes that are significantly up- or downregulated in both acentrosomal mutants relative to WT. One key finding from the transcriptional data, and our subsequent functional genetics experiments, is that centrosome loss induces significant oxidative stress; many genes upregulated in acentrosomal cells contribute to redox regulation. We then performed a reverse genetic screen in the genetically sensitized background of acentrosomal wing disc cells and identified a novel genetic interaction between sas-4, which encodes a core centrosomal protein, and the gene encoding glucose-6-phosphate dehydrogenase (g6pd), the rate-limiting enzyme in the pentose phosphate pathway and a key generator of the antioxidant reduced glutathione (Stanton 2012). We characterized the cellular defects underlying this interaction and found that G6PD upregulation is an important counterbalance to increased ROS induced by mitotic errors. Together, the current study reveals new consequences of centrosome loss (i.e., oxidative stress/redox imbalance), as well as yet another way in which acentrosomal cells buffer themselves against the deleterious effects of centrosome loss (i.e., upregulation of antioxidant promoting genes to limit ROS levels).
Materials and Methods
Drosophila genetics
The following fly stocks were used: y w [Bloomington Drosophila Stock Center (BDSC) #1495] was used as the WT control; sas-4S2214 (BDSC #12119), aslmecD (Blachon et al. 2008), UAS-sas-4 RNAi (BDSC #35049), UAS-mud RNAi (BDSC #35044), UAS-bub3 RNAi (BDSC #32989), UAS-g6pd RNAi (Vienna Drosophila Resource Center #101507), ap-Gal4 UAS-GFP (a gift from Y. Tamori, Hokkaido University), MS1096-Gal4 (BDSC #8860), en-Gal4 UAS-RFP (BDSC #30557), UAS bskDN (BDSC #6409), TRE-GFP (Chatterjee and Bohmann 2012), His2Av:eGFP (BDSC #24163), UAS-p35 (BDSC #5072), UAS-g6pd[9g] (Legan et al. 2008), GSTD1>GFP (Sykiotis and Bohmann 2008), and Ilp8:GFPMI00727 (BDSC #33079). A list of additional RNA interference (RNAi) stocks tested in the candidate gene screen can be found in Table 5.
Table 5. Candidate genes screened for interaction with sas-4.
| RNAi gene target | Stock identified | Crossed to MS>GFP alone | Crossed to MS>GFP;sas-4 RNAi |
|---|---|---|---|
| Upd2 | BDSC 33949 | Strongly dysmorphic | Strongly dysmorphic |
| BDSC 33988 | Not determined | Normal | |
| Ets21C | BDSC 39069 | Strongly dysmorphic | Strongly dysmorphic |
| Castor | VDRC 2929 | Normal | Normal |
| Cnc | VDRC 37674 | Normal | Normal |
| VDRC 108127 | Normal | Normal | |
| VDRC 101235 | Strongly dysmorphic | Strongly dysmorphic | |
| Dif | VDRC 30579 | Normal | Normal |
| VDRC 100537 | Normal | Normal | |
| G6PD | VDRC 101507 | All erect, 5% blistered and 7% furled | All erect, 29% blistered and 11% furled |
| BDSC 50667 | Strongly dysmorphic | Strongly dysmorphic | |
| ImpL3 | VDRC 110190 | Normal | Normal |
| VDRC 31192 | Normal | Normal | |
| Jra | VDRC 31595 | Normal | Necrotic spots |
| KEAP1 | VDRC 107052 | Normal | Normal |
| VDRC 330323 | Normal | Normal | |
| TRAF4 | VDRC 110766 | Normal | Normal |
| MMP1 | VDRC 31989 | Normal | Normal |
| WWOX | VDRC 108350 | Normal | Normal |
Based on cellular function, a subset of genes was selected from the list of genes that were upregulated in both sas-4 and asl mutants relative to wild-type. The stock identifiers for RNAi lines targeting the genes of interest are shown. RNAi lines were crossed to the MS1096-Gal4 UAS-GFP (MS>GFP) wing disc driver alone and in combination with sas-4 RNAi. Glucose-6-phosphate dehydrogenase knockdown alone had modest effects that were synergistically elevated in cells codepleted of centrosomes by sas-4 RNAi (see Figure 6, A and B). RNAi, RNA interference; BDSC, Bloomington Drosophila Stock Center; VDRC, Vienna Drosophila Resource Center.
Detection and quantification of ROS levels
To determine ROS levels, we stained the indicated genotypes with dihydroethidium (DHE) (Millipore, Bedford, MA) using the following protocol, modified from Owusu-Ansah et al. (2008). Wandering third-instar larvae were hemi-dissected in room temperature Schneider’s medium with penicillin/streptomycin. The inverted carcasses were immediately transferred to 1 ml of Schneider’s medium with 30 μM DHE and incubated for 10 min on a nutator at room temperature. The DHE solution was removed and the carcasses were washed three times for 2 min each with Schneider’s medium. The wing discs were then fully dissected from the carcasses, mounted in Halocarbon oil, and immediately imaged on a Zeiss ([Carl Zeiss], Thornwood, NY) LSM Pascal confocal microscope. The same protocol was used for MitoSOX staining (5 μM; Thermo Fisher Scientific).
To quantify and statistically compare ROS levels in the different genotypes, we used maximum-intensity projections of z-stack images of DHE stainings. We then isolated and extracted the GFP+ area of the wing pouch–hinge region (i.e., the dorsal region expressing ap-Gal4-driven transgenes of interest), measured the area of the selected region, and counted the number of DHE+ cells. Because of interexperimental variability associated with live sample preps and DHE staining, it is not possible to quantitatively compare DHE levels among different genotypes. To circumvent this issue, we used the ventral region of the wing disc as a control to standardize the DHE signal (the ventral compartment is homozygous sas-4 mutant but does not express any of the indicated transgenes, and thus is GFP-negative). As with the dorsal region, we measured the area of the ventral wing pouch–hinge region and counted the number of DHE+ cells. We then calculated the number of DHE+ cells/area for both the dorsal (GFP+) and ventral (GFP−) regions, and divided the dorsal by the ventral. Increased or decreased ROS levels induced by transgene expression in the dorsal region will therefore alter the ratio of dorsal:ventral DHE+ cells, with each compartment separately standardized by its area. We then used ANOVA with Dunnett’s multiple comparisons test (GraphPad Prism 7.04) to determine any significant differences in means, comparing transgenic backgrounds to the ap>GFP/+;sas-4 background.
RNA-sequencing experiment and analysis
Total RNA was isolated from wing imaginal discs from wandering larvae as described previously (McKay and Lieb 2013). RNA from 20 larval wing discs of a given genotype was isolated. This process was repeated three times per genotype to yield three biological replicates. RNA-sequencing (RNA-Seq) was performed by the University of North Carolina High-Throughput Sequencing Facility. Libraries were created using the TruSeq Stranded mRNA kit from Illumina. Reads were aligned to the annotated dm3 Drosophila genome using TopHat (v2.0.14) (Trapnell et al. 2012). Read depth for each gene was generated using the Bedtools “coverageBed” and “groupBy” tools. Differential gene expression analysis was performed with edgeR (version 3.14.0). Differentially expressed genes were defined as having a false discovery rate (FDR) ≤ 0.001 and having an average normalized counts value ≥ 10 in at least one sample. Browser shots of RNA-Seq data display read depth-normalized values. Bar plots of RNA-Seq data represent the average trimmed mean-normalized counts of three replicates (± SD). Gene ontology (GO) analysis was performed using DAVID (Database for Annotation, Visualization and Integrated Discovery version 6.8). Motif enrichment analysis was performed using analysis of motif enrichment (AME) (McLeay and Bailey 2010), in combination with transcription factor motifs from the Fly Factor Survey database. De novo motif discovery was performed using discriminative regular expression motif elicitation (DREME) (Bailey 2011). Additional details are available upon request. Graphical displays of RNA-Seq data used in figures were generated from the University of California Santa Cruz Genome Browser or using edgeR’s estimates of RNA abundance.
Immunocytochemistry, imaging, and analysis
Wing disc fixation and antibody staining were performed as previously described (Roberts et al. 2012). Briefly, third-instar larvae were fixed for 20 min in 4% paraformalydehyde (PFA), washed three times with 0.1% phosphate buffered saline with Triton X-100 (PBT), blocked with PBT plus 1% goat serum, incubated overnight in primary antibodies at 4°, washed three times with PBT, incubated with secondary antibodies (> 2 hr at room temperature or overnight at 4°), and washed three times with PBT, then mounted in Aquapolymount (Polysciences, Warrington, PA). Antibodies used were cleaved caspase 3 (1:200; Cell Signaling), MMP1 (1:50; Developmental Studies Hybridoma Bank), and Jra (1:500; Santa Cruz). Alexa secondary antibodies were used at 1:500. Phalloidin was spiked into secondary antibodies at 1:500. Confocal images were acquired on a Zeiss Pascal microscope. PhotoshopCS4 (Adobe) was used to adjust levels so that the range of signals spanned the entire output grayscale, and to adjust brightness and contrast. Adult wing images were acquired on a Samsung Galaxy S8 attached to a Unitron FS30 microscope. Cell death was quantified as the caspase 3-positive region of the disc, and then standardized to the total area of the disc or disc compartment (e.g., dorsal region). We then used ANOVA with Sidak’s or Dunnett’s multiple comparisons test (GraphPad Prism 7.04) to determine any significant differences in means.
Data availability
Drosophila stocks are available upon request from J. S. Poulton. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and supplemental figures and tables. Supplemental material available at https://doi.org/10.25386/genetics.7837943.
Results
Defining the transcriptional response to centrosome loss
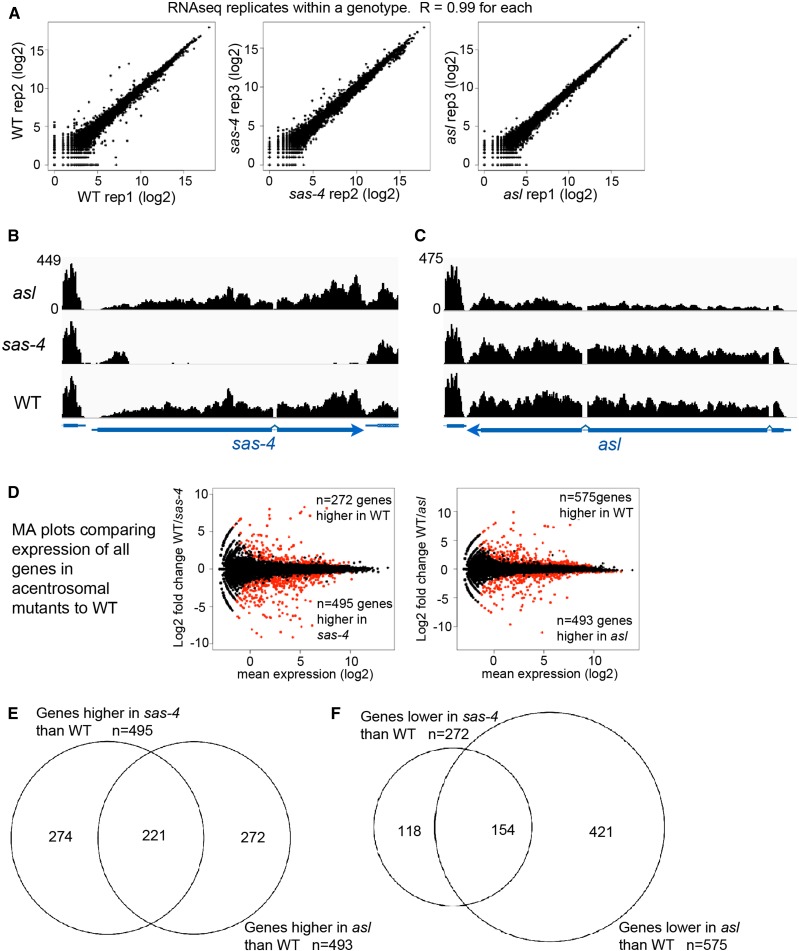
To investigate the effects of centrosome loss on the transcriptional program of a proliferating epithelial tissue, we performed RNA-Seq on Drosophila wing imaginal discs from late third-instar larvae of three genotypes: yellow white (y w; our WT control), or animals homozygous mutant for null alleles of one of two different proteins required for centriole duplication: sas-4s2214 or aslmecD. These alleles lead to complete or near-complete loss of centrosomes by third larval instar (Basto et al. 2006; Blachon et al. 2008; Poulton et al. 2014). We performed RNA-Seq on three biological replicates for each genotype. We first analyzed how well the replicates within a genotype correlated with one another, finding extremely high concordance among replicates for each genotype (Figure 2A; Pearson correlation coefficient = 0.99 for replicates within each genotype). We then examined the expression of the sas-4 and asl loci in their respective mutant backgrounds. In the sas-4 mutant, there was almost complete loss of sas-4 RNA transcripts (Figure 2B). It is worth noting that the sas-4s2214 mutant did possess transcripts of ∼200 bp arising from the 5′ end of the first exon. The sas-4s2214 allele is a P-element insertion in the first exon, previously mapped to chromosomal location 3R:2977450, which is precisely where the abrupt end of transcripts was observed in the sas-4 mutant. The aslmecD mutation is a point mutant (C1718T), leading to a premature stop codon at Q483. Consistent with a point mutant, full-length transcripts were present in the aslmecD mutant background, though overall levels were slightly reduced compared to controls (Figure 2C), which may reflect nonsense-mediated decay.
Figure 2.
Analysis of RNA-Seq data reveals highly consistent gene up- and downregulation. (A) Plots comparing RNA-Seq replicates within a genotype demonstrate extremely high concordance. Pearson’s r correlation values are shown. (B and C) Browser shots of RNA-Seq signals (normalized read depth) at the sas-4 and asl loci for the three indicated genotypes. y-axis values (shown for asl) are the same for each genotype. (D) MA plots reveal genes significantly up- (red dots below midline) or downregulated (red dots above midline), in comparisons of sas-4 to WT (left) or asl to WT (right). (E and F) Venn diagrams demonstrating the highly significant overlap of genes found to be up- (E) or downregulated (F) in both sas-4 and asl, relative to WT. RNA-Seq, RNA-sequencing; WT, wild-type.
To define changes in gene expression associated with centrosome loss, we compared RNA-Seq data from each of the acentrosomal mutants to the WT control RNA-Seq data. Plots of differential gene expression revealed many up- and down-regulated genes for each pairwise genotype comparison (Figure 2D; complete lists of gene expression data are in Supplemental Material, Table S1). To identify genes most significantly up- or downregulated in each mutant relative to WT, we filtered the comprehensive list of genes to include only those that met an FDR of P < 0.001, as well as a minimum expression threshold (fragments per kilobase of transcript per million mapped reads ≥ 10) for at least one genotype. In comparing sas-4 mutant wing discs to controls, use of these filters revealed 495 genes significantly upregulated and 272 genes downregulated in sas-4 discs (Table S2). In comparing asl mutant wing discs to controls, we found 493 upregulated and 575 downregulated genes (Table S3).
Mutations in sas-4 and asl result in centrosome loss through different mechanisms; Sas-4 is directly involved in centriole assembly (Kohlmaier et al. 2009; Schmidt et al. 2009) whereas Asl regulates daughter centriole duplication licensing (Blachon et al. 2008; Novak et al. 2014). Therefore, changes in gene expression unique to one mutant genotype might reflect the transcriptional response to centrosome-independent functions for that particular protein. To identify the common response to centrosome loss, we cross-referenced the lists of differentially expressed genes to identify genes that were significantly up- or downregulated in both acentrosomal mutants relative to WT. The two lists exhibited highly significant overlap (Figure 2, E and F): 221 genes were significantly upregulated (Table S4) and 154 genes were significantly downregulated (Table S5) in both sas-4 and asl wing discs, relative to control wing discs. This was much higher than expected by chance (hypergeometric mean test: P < 4.26e−201 for upregulated genes; P < 1.54e−91 for downregulated genes). This conservative approach should exclude unforeseen changes in gene expression associated with a particular mutant or mutant background, thus isolating only genes specifically affected by centrosome loss. Of course, for some genes, lack of concordance may simply result from experimental variability, and changes in gene expression for a particular gene might have reached the significance threshold in one genotype but have been just below that threshold in the other, thus excluding it from the shared list of significantly up- or downregulated genes.
Several categories of genes, as defined by GO term analysis via DAVID (Huang da et al. 2009), were notable in the list of jointly upregulated genes. These included genes involved in oxidation–reduction pathways, including the antioxidant and detoxifying glutathione transferase pathway, as well as genes involved in the innate immune response (Table 1, Table 2, and Table 3). Manual inspection also revealed a number of genes involved in or known to be targets of the JNK pathway (Table 4), consistent with our previous work (Poulton et al. 2014).
Table 1. GO terms enriched in genes higher in both sas-4 and asl.
| GO term | P-value |
|---|---|
| Biological Process | |
| Glutathione metabolic process | 3.90E−04 |
| Imaginal disc-derived male genitalia morphogenesis | 1.20E−03 |
| Cell adhesion | 1.60E−03 |
| Immune response | 5.80E−03 |
| Oxidation–reduction process | 9.20E−03 |
| Molecular function | |
| Glutathione peroxidase activity | 8.45E−05 |
| Signaling pattern recognition receptor activity | 3.30E−03 |
GO term analysis of genes with significantly increased expression in both sas-4 and asl mutant wing discs, relative to wild-type, suggests upregulation of several biological pathways. Notable among them are indicators of oxidative stress. Unadjusted P-values are shown. GO, gene ontology.
Table 2. Genes with known or putative roles in the oxidative stress response.
| Gene | Protein name/function | Reference |
|---|---|---|
| Zw (G6PD) | Zwischenferment. Glucose-6-phosphate dehydrogenase. Rate-limiting enzyme in the pentose phosphate pathway. Reaction generates NADPH, which is used by glutathione reductase to produce reduced glutathione, a potent antioxidant. A Nrf2 target. | Loboda et al. (2016) |
| Glutathione synthase | Second enzyme in synthesis of glutathione, the key nonenzymatic antioxidant. | Lu (2013) |
| Traf4 | TNF receptor-associated factor 4. Traf2 regulates oxidative stress with Atg9 through JNK. | Tang et al. (2013) |
| Wwox | WW domain-containing oxidoreductase. Regulates ROS and TNF-induced cell death. | O’Keefe et al. (2015) |
| Aox (CG18522) | Aldehyde oxidase 1. Potent generator of superoxides. | Kundu et al. (2012) |
| GstE8 | Glutathione S-transferase E8. Cnc and Paraquat induced. Up in hyperoxia screen. | Gruenewald et al. (2009), Misra et al. (2011) |
| GstD3 | Glutathione S-transferase D3. Cnc and Paraquat induced. Up in hyperoxia screen. | Gruenewald et al. (2009), Misra et al. (2011) |
| Men | Malic oxidoreductase. Malic enzyme knockdown modulates reductive stress. | Xie et al. (2013) |
| ImpL3 | Lactate dehydrogenase. HIF-1 target gene. Up in hyperoxia screen. Inhibition induces oxidative stress. | Gruenewald et al. (2009) |
| Naprt (CG3714) | Nicotinate phosphoribosyltransferase. Rate-limiting enzyme in NAD+ synthesis. NAD+ is a cofactor in redox reactions and helps prevent oxidative stress. | Massudi et al. (2012) |
| AcCoAS | Acetyl coenzyme A synthase. Mammalian Acss2 promotes acetylation of the stress-responsive HIF-2α subunit by the acetyltransferase/coactivator Creb-binding protein. | Chen et al. (2017) |
| CG3397 | Putative Aldo/keto reductase. Up after hyperoxia oxidative stress. | Gruenewald et al. (2009) |
A list of oxidative stress response genes that were significantly upregulated in both sas-4 and asl mutants relative to wild-type. TNF, tumor necrosis factor; ROS, reactive oxygen species.
Table 3. Genes with known or inferred roles in innate immunity or wound healing.
| Gene | Role | Reference |
|---|---|---|
| Immune response or innate immunity target genes | ||
| Dif | NF-κB transcription factor, in Toll pathway. | Buchon et al. (2014) |
| PGRP-LC | Peptidoglycan recognition protein light chain. Transmembrane receptor recognizing diaminopimelic acid-type peptidoglycan, a bacterial cell wall component. Upstream of the immune deficiency pathway. | Buchon et al. (2014) |
| PGRP-SA | See above. Other peptidoglycan recognition proteins are Cnc and Paraquat induced. Involved in the Toll pathway. Induced by LPS in S2 cells. | Boutros et al. (2002), Buchon et al. (2014) |
| GNBP2 | Gram-negative bacteria-binding protein 2. | Buchon et al. (2014) |
| Clect27 | C-type lectin. Putative roles in bacterial recognition. | O’Rourke et al. (2006) |
| Spn55B | Serpins function in Toll receptor activation. | Meekins et al. (2017) |
| upd2 | JAK/STAT ligand. Required for parasitoid wasp immune response. | Yang and Hultmark (2016) |
| upd3 | JAK/STAT ligand. Required for parasitoid wasp immune response. | Yang and Hultmark (2016) |
| cv-2 | Crossveinless 2, binds bone morphogenic proteins, is an immunity target gene in gut. | Buchon et al. (2009) |
| Ipk1 | Inositol pentakisphosphate 2-kinase. An immunity target gene in gut. | Buchon et al. (2009) |
| LamC | Lamin C. An immunity target gene in gut. | Buchon et al. (2009) |
| vir-1 | Virus-induced RNA 1. A JAK-STAT target, induced by viral infection. | Dostert et al. (2005) |
| wun2 | Wunen 2. Lipid phosphate phosphatase. An immunity target gene in gut and hemocytes. | Boutros et al. (2002), Buchon et al. (2009) |
| Pvf2 | Ligand for Pvr pathway. IMD pathway activates Pvf2 and 3 in a JNK-dependent way. Induced by LPS in Drosophila S2 cells. | Boutros et al. (2002), Bond and Foley (2009) |
| myo61f | Required for intestinal brush border integrity and resistance to bacterial pathogens. Induced by LPS. | Hegan et al. (2007), Silverman et al. (2003) |
| Jra | Jun transcription factor. Induced by LPS in Drosophila S2 cells. | Boutros et al. (2002) |
| Puc | Puckered. JNK phosphatase. Induced by LPS in Drosophila S2 cells. | Boutros et al. (2002) |
| Ets21c | Ets domain transcription factor. Induced by LPS in Drosophila S2 cells. | Boutros et al. (2002) |
| RhoL | Rho family GTPase. Induced by LPS in Drosophila S2 cells. | Boutros et al. (2002) |
| Mmp1 | Matrix metalloproteinase 1. Induced by LPS in Drosophila S2 cells. | Boutros et al. (2002) |
| Melanization and wound response | ||
| Tg | Transglutaminase. Part of cuticle cross-linking wound response pathway. | Telci and Griffin (2006) |
| amd | α methyl dopa-resistant. Part of cuticle cross-linking wound response pathway. | Tang (2009) |
| Ddc | Dopa decarboxylase. Part of cuticle cross-linking wound response pathway. | Tang (2009) |
| yellow-b | Related to Yellow, which is sufficient for the formation and deposition of melanin. | Ferguson et al. (2011) |
| yellow-c | See above. | Ferguson et al. (2011) |
| Cpr76Bc | Cuticular protein 76Bc. Three other Cprs are known to be wound inducible. | Moussian (2010) |
| mtg | Mind the gap. Chitin-binding cuticle protein. | Moussian (2010) |
| Edg78E | Insect cuticle protein. Chitin binding. | Moussian (2010) |
| Mmp1 | Matrix metalloproteinase 1. A wound response gene. | Lee and Miura (2014) |
| scb | α-integrin. JNK target required for embryonic wound repair. | Campos et al. (2010), Homsy et al. (2006) |
| Jra | Jun-related antigen (=fly Jun). Required for embryonic wound repair. | Campos et al. (2010) |
| Ets21C | Ets family transcription factor. Wound response gene. | Patterson et al. (2013) |
Genes with reported roles in the innate immune response and/or wound healing that were significantly upregulated in both sas-4 and asl mutants relative to wild-type. PVR, PDGF- and VEGF receptor-related; IMD, immune deficiency; LPS, lipopolysaccharide.
Table 4. Genes related to JNK signaling.
| Gene | Role | Reference |
|---|---|---|
| JNK pathway/regulators | ||
| Jra | Jun transcription factor, essential part of JNK pathway. Also a JNK target gene. | Ríos-Barrera and Riesgo-Escovar (2013) |
| puc | Protein phosphatase that is a feedback-negative regulator of JNK. JNK target gene. | Ríos-Barrera and Riesgo-Escovar (2013) |
| Gadd45 | Mammalian relatives are JNK activators. Fly protein genetically interacts with hep = Fly JNKK. | Peretz et al. (2007) |
| Pvf1 | PDGF- and VEGF-related factor 1. Involved in corpse removal, a role in which it and its receptor Pvr are upstream of JNK. | Ishimaru et al. (2004) |
| Pvf2 | PDGF- and VEGF-related factor 2. Activated by IMD pathway in a JNK-dependent way and acts with Pvf3 in turn as a feedback-negative regulator of JNK signaling. | Bond and Foley (2009) |
| scaf | Inactive serine protease. JNK target gene and feedback-negative regulator of the JNK pathway. | Rousset et al. (2010) |
| Traf4 | TNF receptor-associated factor 4. Traf2 regulates oxidative stress with Atg9 through the JNK pathway. | Tang et al. (2013), Ríos-Barrera and Riesgo-Escovar (2013) |
| JNK target genes | ||
| ImpL2 | Secreted insulin/IGF antagonist . | Jasper et al. (2001) |
| Ilp8 | Divergent member of the insulin/IGF/relaxin-like family. | La Fortezza et al. (2016) |
| Myo61F | Myo1C. Induction by LPS requires JNK. | Silverman et al. (2003) |
| Nlaz | Neural lazarillo. A lipocalin involved in metabolic homeostasis. | Hull-Thompson et al. (2009), Kučerová et al. (2016) |
| scb | Integrin α-chain. | Homsy et al. (2006) |
| upd2 | Unpaired 2. JAK-STAT ligand. | Pastor-Pareja et al. (2008) |
| upd3 | Unpaired 3. JAK-STAT ligand. | Santabárbara-Ruiz (2015), Pastor-Pareja et al. (2008) |
| MMP1 | Matrix metalloproteinase 1. | Uhlirova and Bohmann (2006) |
List of genes that were upregulated in both sas-4 and asl mutants relative to WT, and are either components or the JNK signaling cascade or transcriptional targets of JNK signaling. IMD, immune deficiency; TNF, tumor necrosis factor; IGF, insulin-like growth factor; LPS, lipopolysaccharide.
The transcriptional response to centrosome loss does not broadly elevate core centrosomal proteins or proteins involved in parallel pathways
Centrosomes are multiprotein organelles. Thus, we initially hypothesized that cells might sense the mitotic challenge in centrosome-deficient cells by upregulating genes encoding centrosomal proteins. However, no known centrosomal components were significantly upregulated in both asl and sas-4 mutants. Centrosome loss in wing imaginal discs is buffered by mitotic delay induced by the SAC and by noncentrosomal MT nucleation. Thus, another potential transcriptional response might be upregulation of components of the SAC, the Augmin complex, or the Ran pathway, which partially compensate for centrosome loss in wing discs and early embryos (Hayward et al. 2014; Poulton et al. 2014). Only two genes with MT or SAC connections were significantly upregulated by loss of both Asl and Sas-4: tubulin-binding cofactor A (CG1890) (Voelzmann et al. 2016) and Spindly, a protein essential for silencing the SAC via dynein recruitment to the kinetochore (Griffis et al. 2007). However, when we scanned the lists of genes upregulated by knockdown of sas4 or asl alone, a few additional genes emerged: rcd2, identified in an RNAi screen for centrosome function (Dobbelaere et al. 2008), and CP309, encoding the centrosomal protein pericentrin-like protein (Mennella et al. 2012; Lerit et al. 2015; Richens et al. 2015), were upregulated in sas-4 mutants, while mad2, a key component of the SAC (Musacchio 2015), ran, which has dual roles in nuclear import and noncentrosomal MT nucleation (Clarke and Zhang 2008), and cct5, involved in centrosome-independent spindle assembly (Moutinho-Pereira et al. 2013), were upregulated in asl mutants. Thus, coordinated transcriptional upregulation of the compensatory pathways does not appear to be a prominent response to loss of centrosomes, but it may play a minor role. Centrosome loss in the wing imaginal disc disrupts mitotic spindle assembly, leading to chromosome missegregation and DNA damage (Poulton et al. 2014). Thus, we also looked for up regulation of genes involved in the DDR, which initiates repair pathways to correct lesions or, if the damage is too severe, triggers programmed cell death (Borges et al. 2008). This response includes the upregulation of genes involved in DNA damage detection or repair (Christmann and Kaina 2013). Interestingly, we did not detect significant up- or downregulation of known DDR genes in our analysis. Perhaps the extent of DNA damage is not sufficient for it to be detected by analysis of the entire tissue; alternately, changes in gene expression may not be not a primary component of the DDR in this tissue.
Validating differential gene expression associated with centrosome loss reveals significant upregulation of genes regulated by the JNK pathway
In wing discs lacking centrosomes (sas-4 mutants), significant defects in efficient spindle assembly, accurate chromosome segregation, and proper spindle orientation lead to increased apoptosis (Figure 1, B and C) (Poulton et al. 2014). These mitotic defects appear to drive the apoptosis of affected cells by activating JNK signaling, since blocking JNK signaling prevents apoptosis in acentrosomal wing discs (Poulton et al. 2014). JNK signaling regulates gene expression, at least in part through the key transcription factor AP-1, a heterodimer of Jun [Jun-related antigen (Jra) in flies] and Fos. Consistent with this, we previously found that a JNK-signaling transcriptional reporter [TRE (tetradecanoylphorbol acetate response element)>GFP; GFP under the control of a promoter containing Jun-binding sites] is activated by centrosome loss (sas-4 RNAi), both in cells undergoing apoptosis and also in other cells in the disc (Figure 1, D and E) (Poulton et al. 2014). These data suggested that we would see elevated expression of JNK target genes in centrosome-deficient discs, both at the transcript and protein levels.
We first compared our list of genes that were significantly up- or downregulated, in both the sas-4 and asl mutant backgrounds, to the lists of genes up- or downregulated in two other transcriptomic studies in wing disc models with demonstrated activation of JNK signaling: one induced damage to the disc through misexpression of proapoptotic genes (Khan et al. 2017), and the other examined the transcriptional response to tumor formation through the loss of cell polarity proteins Scribble (Scrib) or Discs-large (Dlg) (Bunker et al. 2015). For both up- and downregulated genes, there were significantly more genes in common between our studies than one would expect by chance (Figure S1, A and B). Thus, the significant concordance with our findings and theirs suggests that, regardless of the means of insult or injury, there are inherent, shared aspects of the JNK-related transcriptional response. It will be interesting to examine the lists of genes that are common to all of our data, and functionally test those genes that have not been previously implicated in the response to tissue injury or tumor formation (Figure S1, A and B).
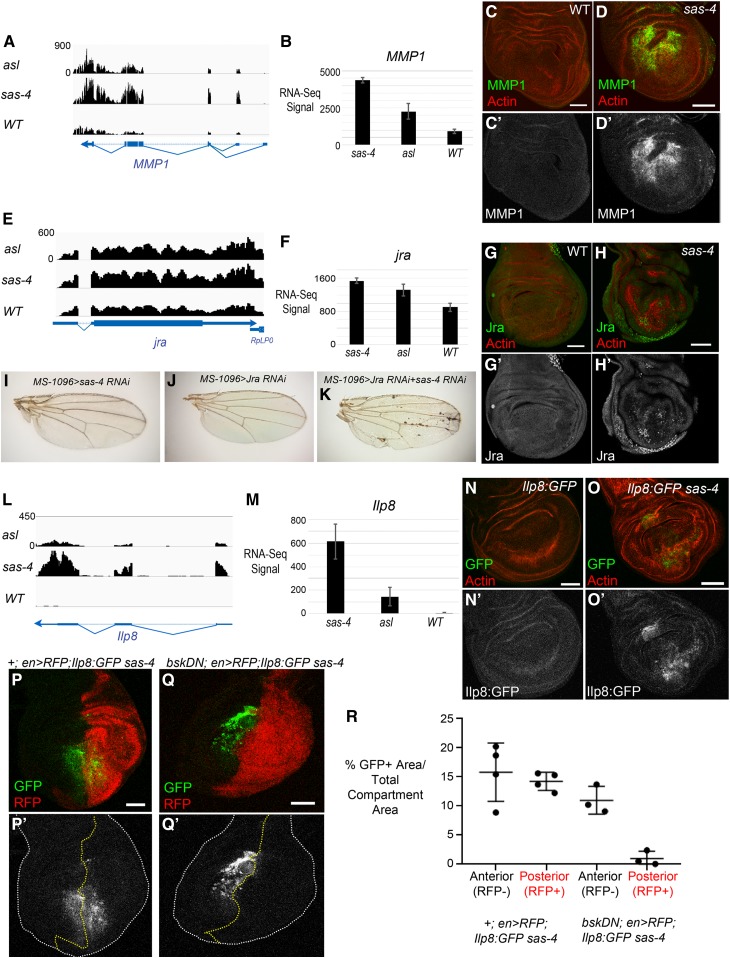
We next wished to more directly examine the hypothesis that centrosome loss activates a JNK-dependent transcriptional response. For these analyses, we focused on the sas-4 mutant because sas-4 loss elicited a stronger transcriptional response than asl for many of the genes on the shared list of differentially expressed genes. Many positively regulated transcriptional targets of JNK signaling have been identified in Drosophila. Consistently, mRNA levels of many of these, including Jra itself, puckered (puc), a feedback-negative regulator of the JNK pathway, Insulin-like peptide 8 (Ilp8), Reaper (rpr), and Matrix Metalloproteinase1 (MMP1) were elevated in both the sas-4 and asl mutant backgrounds relative to WT controls (Figure 3, A, B, E, F, L, and M and Table S4).
Figure 3.
Centrosome loss leads to upregulation of expression of JNK target genes. (A and E) Browser shots of RNA-Seq signals (normalized read depth) of two known JNK target genes, MMP1 (A) and Jra (E), for asl, sas-4, and WT genotypes. Transcription direction indicated by arrowheads. (B and F) Bar plots of average RNA-Seq signals (normalized counts ± SD) of the three biological replicates for MMP1 (B) and Jra (F). (C-C’) MMP1 protein, as visualized by antibody, has minimal expression in WT wing discs. (D-D’) MMP1 protein levels are dramatically increased in acentrosomal sas-4 wing discs. (G-G’) Jra is weakly expressed in control discs, though there is significant expression in the peripodial cells (not shown in this single-slice image). (H-H’) Jra protein is increased in sas-4 discs. (I–K) Representative adult wings from the indicated genotypes. Neither knockdown of sas-4 alone (I) nor jra alone (J) perturbs wing development. However, knockdown of both sas-4 and jra together produces necrotic spots in the adult wing. (L) Browser shot of RNA-Seq signal (normalized read depth) of the ilp8 locus for asl, sas-4, and WT genotypes. (M) Bar plot of average RNA-Seq signal (normalized counts ± SD) for ilp8 in the three genotypes. (N-N’) ilp8 expression, as assessed using a protein trap line expressing GFP-tagged Ilp8 under control of the ilp8 promoter, is low in control discs. (O-O’) ilp8 is upregulated in sas-4 discs. (P) A sas-4 mutant wing disc expressing Ilp8:GFP and en>RFP with no transgene, is a control for the experiment in Q (P’ shows the Ilp8:GFP channel alone). (Q) The upregulation of ilp8 associated with centrosome loss is JNK-dependent because misexpression of BskDN in the posterior portion of sas-4 homozygous mutant wing discs inhibits Ilp8:GFP upregulation. BskDN is driven by en>RFP (red in Q; grayscale in Q’ is the Ilp8:GFP channel alone). White dashed line marks the outer edge of the wing disc. (R) Quantification of Ilp8:GFP positive area standardized to the total area of the anterior (RFP negative) or posterior (RFP positive) area. Note that expression of en>RFP alone does not alter Ilp8:GFP levels induced by centrosome loss, whereas en>RFP driving bskDN noticeably reduces Ilp8:GFP levels. Bar, 50 µm. Images are maximum-intensity projections, except in (G and H) where single slices were used to limit the Jra signal from the peripodial cells. RFP, red fluorescent protein; RNA-Seq, RNA-sequencing; WT, wild-type.
We next examined whether the changes in RNA transcript levels observed in RNA-Seq analysis led to changes in protein levels of JNK transcriptional targets, by using antibodies to MMP1 and Jra. Control WT wing discs express little to no MMP1 or Jra protein (Figure 3, C and G; with the exception of peripodial cells, which express moderate levels of Jra). Consistent with the RNA-Seq data, we found that sas-4 mutant discs had noticeable increases in both proteins (Figure 3, D and H). Interestingly, the level of protein increase indicated by antibody staining appeared to be more dramatic than the corresponding increase in RNA levels measured in our RNA-Seq data. We suspect that these apparent differences in magnitude may result from the uneven nature of the increase within a disc (many cells do not show increased signal), thus, by a pooled sample method such as RNA-Seq, the overall increase in transcripts in the cells that are upregulating JNK targets is buffered. It is also possible that there are post-transcriptional differences between mutant and WT cells that lead to increased protein levels beyond that associated with increased transcription (e.g., release of some inhibitory mechanisms affecting RNA translation). Because Jra is an essential component of the JNK signaling pathway, we tested the importance of Jra expression in control and acentrosomal cells. While knocking down either Jra or Sas-4 alone did not perturb wing development, knocking down both led to significant morphological defects (Figure 3, I–K). This is consistent with our previous data that demonstrated a role for JNK itself in maintaining tissue homeostasis in acentrosomal wing discs (Poulton et al. 2014). This interaction likely occurs through JNK’s positive roles in apoptosis and/or compensatory proliferation stemming from centrosome loss.
Another interesting hit from our RNA-Seq data was Insulin-like peptide 8 (Ilp8) (Figure 3, L and M and Table S4). Ilp8 mediates delays in developmental timing caused by abnormal tissue growth during larval stages (Colombani et al. 2012; Garelli et al. 2012). Larvae mutant for centrosomal proteins such as Sas-4 exhibit a significant delay in larval development, taking ∼24-hr longer than controls to enter pupation (Poulton et al. 2014). Thus, we examined whether the increase in Ilp8 transcripts in centrosome-deficient animals leads to increased Ilp8 protein expression, using Ilp8:GFP, a GFP protein trap of the endogenous Ilp8 locus (Garelli et al. 2012). In control WT animals, there is minimal expression of Ilp8 in third-instar wing imaginal discs (Figure 3N). However, in Ilp8:GFP sas-4 animals, we noted a significant increase in Ilp8:GFP expression (Figure 3O). Ilp8 upregulation in response to imaginal disc growth defects induced by knockdown of endocytic or ribosomal proteins (i.e., Avl or Rpl7) requires JNK signaling (Colombani et al. 2012). Thus, we tested whether JNK signaling mediated the upregulation of Ilp8 after centrosome loss. Indeed, when we used the Gal4-UAS system to ectopically express a dominant negative form of the fly homolog of JNK (BasketDN; BskDN) in the posterior portion of sas-4 homozygous mutant wing discs, this led to a clear reduction in Ilp8:GFP in the region of the disc where JNK was inhibited (Figure 3Q; compare to en>RFP; sas-4 control in Figure 3P; quantified in Figure 3R). The increased Ilp8 expression in sas-4 mutants, along with the known developmental delay and JNK activation experienced by these animals, suggests that Ilp8 upregulation via JNK is likely an important mediator of prolonged development in acentrosomal animals.
Thus, in the wing imaginal disc, centrosome loss leads to increased JNK activity with concomitant changes in expression of JNK target genes, and also validates the accuracy of our RNA-Seq data. It will be interesting to determine if some of the other genes in our RNA-Seq data elevated after centrosome loss are previously unknown JNK signaling targets; a growing number of transcriptomic studies from Drosophila models with active JNK signaling will provide valuable data for cross-referencing (Rousset et al. 2010; Bunker et al. 2015; Clemente-Ruiz et al. 2016; Khan et al. 2017).
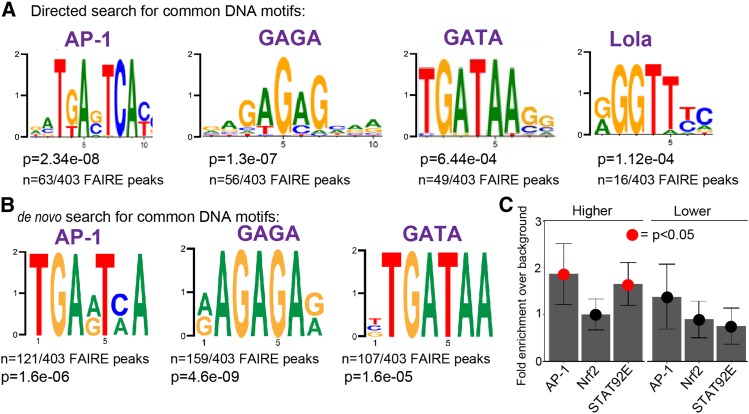
To examine this possibility in our own data, we performed transcription factor-binding motif analysis of the shared genes upregulated in acentrosomal cells. We first looked for enrichment of known transcription factor-binding motifs in open chromatin sites of third-instar wing imaginal discs within 2 kb of the 221 genes upregulated in both mutant backgrounds [= 403 FAIRE peaks (FAIRE=Formaldehyde-Assisted Isolation of Regulatory Elements)] (Uyehara et al. 2017). Remarkably, a consensus sequence significantly matching the AP-1-binding site was found in 63 of the 403 FAIRE peaks (Figure 4A; P = 2.34e−8). This suggests that there may be many additional genes directly upregulated by JNK signaling in centrosome-deficient cells. To further test this possibility, we performed de novo motif discovery in those open chromatin regions within 2 kb of the upregulated genes. This analysis revealed the presence of an AP-1-binding site in 121/403 FAIRE peaks (Figure 4B; P = 1.6e−6). Notably, the 63 AP-1 motifs identified in the directed search are a subset of the 121 motifs identified by the de novo motif search due to increased statistical stringency of the directed search analysis. Intriguingly, these analyses also revealed additional motifs unrelated to JNK signaling, including GAGA, GATA, and Lola-binding sites (Figure 4, A and B). Notably, GATA proteins, including the primary fly GATA protein Serpent, are regulated by ROS (Gao et al. 2014; Indo et al. 2017) and help promote the innate immune response (Senger et al. 2006); as noted above, centrosome loss increases expression of genes involved in oxidative stress and innate immune responses (Table 1). It will be interesting in the future to test possible roles for these transcription factors in the response to centrosome loss. It is also worth noting that neither de novo nor motif enrichment analyses applied to genes downregulated in acentrosomal cells revealed any significant support for particular transcription factor-binding sites near to those genes.
Figure 4.
Enrichment of genes near AP-1, GAGA, GATA, Lola, and STAT92E transcription factor-binding sites. (A) Transcription factor DNA-binding motifs significantly enriched in open chromatin sites near to genes upregulated in both sas-4 and asl. P-values (rank sum test) and fraction of open chromatin peaks containing a designated motif are shown. (B) A de novo motif discovery of open chromatin peaks also uncovered enrichment of AP-1-, GAGA-, and GATA-binding sites. (C) Directed analysis of open chromatin regions near the up- and downregulated genes common to sas-4 and asl for enrichment of binding sites for AP-1, Nrf2, and STAT92E. Data are plotted as the enrichment over genomic background for each motif. Error bars represent 95% C.I.s. Red dots indicate a P-value (Z-test) < 0.05. AP-1 was included as a control for this approach, since we knew from our library-based directed search and the de novo search that AP-1-binding sites should be significantly enriched in our upregulated set of genes. We did not detect significant enrichment for Nrf2 sites using this approach. However, we did detect significant enrichment of the STAT92E-binding motif in our upregulated genes [this motif was not in the library for our directed search, represented in (A)].
Based on the upregulation of the JAK-STAT ligands Upd2 and Upd3 in acentrosomal cells (Table S4), it was curious that the STAT92E-binding motif was not found in our de novo search (Figure 4B; the STAT92E consensus sequence was absent from the library of motifs used in our directed search). Therefore, we conducted a third analysis of potential transcription factor-binding sites in the open chromatin around the genes up- or downregulated in sas-4 and asl, this time specifically looking for enrichment of sequences aligning to the STAT92E-binding motif. We used the consensus AP-1-binding motif as a positive control since it was significantly enriched in the upregulated genes based on both of our other motif search approaches (Figure 4, A and B). This analysis revealed significant enrichment of STAT92E-binding sites in the open chromatin regions of genes upregulated by centrosome loss (Figure 4C). In contrast, there was no significant enrichment of STAT92E-binding sites in genes downregulated by centrosome loss. To more directly determine whether sas-4 knockdown leads to upregulation of JAK-STAT activity in acentrosomal cells, we examined expression of the JAK-STAT transcriptional reporter 10xSTAT:GFP in sas-4 mutant wing discs. While, we did not detect obvious changes in JAK-STAT activity in sas-4 mutants, when we blocked apoptosis with p35 we did find increased JAK-STAT activity (Figure S2), suggesting that centrosome loss leads to increased JAK-STAT activation, likely through JNK-induced upregulation of Upd ligands. Intriguingly, the strongest upregulation was in neighboring WT cells (Figure S2C, arrows), which may reflect JAK-STAT’s involvement in the compensatory proliferation response.
Centrosome loss leads to oxidative stress
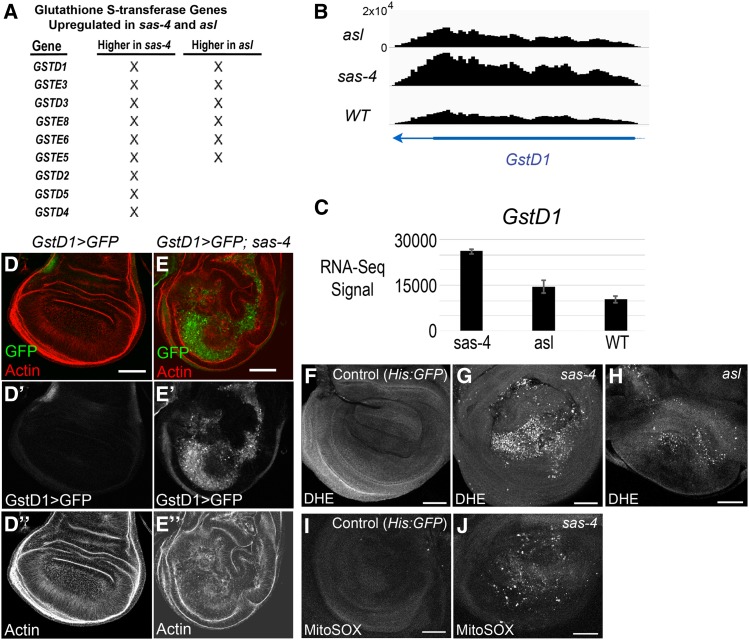
One of the most striking features of our RNA-Seq data were increased expression of genes associated with the response to oxidative stress (Table 2). These ranged from signaling proteins like tumor necrosis factor (TNF)-associated Factor 4 (TRAF4), which acts upstream of the JNK pathway to regulate the oxidative stress response (Tang et al. 2013), to enzymes like WW domain-containing oxidoreductase (WWOX), which regulates ROS and TNF-induced cell death (O’Keefe et al. 2015), or CG3714, a nicotinate phosphoribosyltransferase family member essential for the increase in cellular NAD levels to prevent oxidative stress (Hara et al. 2007). Among these, multiple glutathione S-transferase (GST) genes were upregulated in both sas-4 and asl mutants, and sas-4 loss led to the upregulation of three additional GST genes (Figure 4A). These enzymes mitigate oxidative stress by conjugating glutathione to toxic electrophilic substrates, reducing their reactivity and increasing their solubility, thus facilitating their excretion from cells and tissues (Chatterjee and Gupta 2018).
We followed this lead, using GSTD1 as an example. GSTD1 is a known target of KEAP1/Nrf2 signaling, which regulates the response to oxidative stress (Sykiotis and Bohmann 2008). In the eye imaginal disc, GSTD1 can also be upregulated by JNK signaling (Kanda et al. 2011). Our RNA-Seq data suggested that GSTD1 is upregulated in acentrosomal cells (Figure 5, A–C). To confirm this, we took advantage of a GSTD1>GFP reporter, in which the promoter region of GSTD1 drives GFP expression (Sykiotis and Bohmann 2008). In WT discs, GSTD>GFP expression is very low, while in sas-4 mutant discs there is strong upregulation of GFP driven by the GSTD1 promoter (Figure 5, D and E). Interestingly, the consensus binding site for Nrf2 did not show up in our de novo transcription factor motif analysis (Figure 4B), nor was it significantly enriched in our directed motif search of genes up- or downregulated by centrosome loss (Figure 4C). Thus, despite significant upregulation of several oxidative stress response genes, including GSTD1, the number of direct targets of KEAP1/Nrf2 signaling in our upregulated gene set may be rather small.
Figure 5.
Centrosome loss induces oxidative stress and upregulation of GST genes. (A) List of GST genes significantly upregulated in sas-4 and/or asl mutants. (B) Browser shot of RNA-Seq signal (normalized read depth) of the GstD1 locus in the three genotypes. Transcription direction indicated by arrowhead. (C) Bar plot of average RNA-Seq signal (normalized counts ± SD) for GstD1. (D-D”) Control WT wing discs have minimal expression of GstD1, as indicated by the reporter GSTD1>GFP. (E-E”) sas-4 wing discs have elevated levels of GSTD1>GFP. (F) In WT wing discs, ROS levels are essentially undetectable. His:GFP flies were used as WT in this experiment, and were mixed with sas-4 mutant discs to provide an “in tube” control. (G) sas-4 and (H) asl homozygous mutant wing discs showed strongly elevated ROS levels. (I) WT wing discs do not stain for MitoSOX, a marker of mitochondrially-derived superoxide. (J) MitoSOX staining is elevated in sas-4 mutant discs. Bar, 50 µm. DHE, dihydroethidium; RNA-Seq, RNA-sequencing; ROS, reactive oxygen species; WT, wild-type.
Increased expression of GSTD genes, and in particular the upregulation of GSTD1>GFP, can reflect the presence of ROS (Sykiotis and Bohmann 2008). As noted above, GO term analysis of our lists of genes significantly up- or downregulated in acentrosomal cells revealed changes in expression of proteins, involved in both redox metabolism and detoxification, associated with xenobiotic factors and oxidative stress. This could reflect an increase in ROS in cells lacking centrosomes. To test this hypothesis, we incubated sas-4 and asl mutant wing discs with the ROS probe DHE (Bindokas et al. 1996). Strikingly, both sas-4 and asl mutant wing discs had a dramatic increase in DHE staining (Figure 5, F–H). We also observed increased signal using the MitoSOX probe, indicating that at least some ROS production occurs in mitochondria (Figure 5, I and J). Interestingly, the vast majority of ROS+ cells are concentrated in the pouch region of the wing disc, which is the same pattern we observed in the distribution of apoptotic cells in acentrosomal wing discs (Figure 1C). This pattern similarity suggests a correlation between apoptosis and ROS production (see below). The cause of this preferential clustering of phenotypes within the wing pouch is unclear; potential explanations may include higher rates of proliferation in the pouch resulting in more mitotic errors and subsequent cell death, differences in cell signaling pathway activity that affect cell survival/sensitivity, differences in expression levels of pro- or antiapoptotic regulators, or the topography of the wing disc leading to the concentration of basally extruded apoptotic cells in the central, basal region of the pouch. Additional studies will be required to determine the mechanisms underlying the apparent propensity of pouch cells to succumb to centrosome loss.
Centrosomes regulate several cellular processes that could conceivably affect redox balance (e.g., the DDR) (Lerit and Poulton 2016). Therefore, we sought to determine if the increase in ROS was a specific effect of centrosome loss or a potential consequence of the mitotic errors induced by centrosome loss in the wing disc. Knockdown of key mitotic regulators and resulting mitotic errors were recently reported to lead to increased GSTD>GFP expression (Clemente-Ruiz et al. 2016). Therefore, we used our ROS assays to examine ROS levels following knockdown of two other mitotic regulators, Mud and Bub3. Mud is important for spindle orientation in the wing disc (Nakajima et al. 2013), while Bub3 contributes to the SAC and the attachment of MTs to kinetochores (Logarinho et al. 2008). Thus, defects caused by knockdown of these proteins should be independent of centrosome function. As previously reported (Dekanty et al. 2012; Morais da Silva et al. 2013; Poulton et al. 2014), knockdown of each of these proteins leads to significant increases in apoptosis (Figure 6, A and C). We found that Mud or Bub3 knockdown also increased ROS levels as measured by DHE staining (Figure 6, B and D), similar to that observed after disruption of centrosome function. Together, these data are consistent with the possibility that ROS production increases in acentrosomal cells as a direct or indirect result of subsequent mitotic errors.
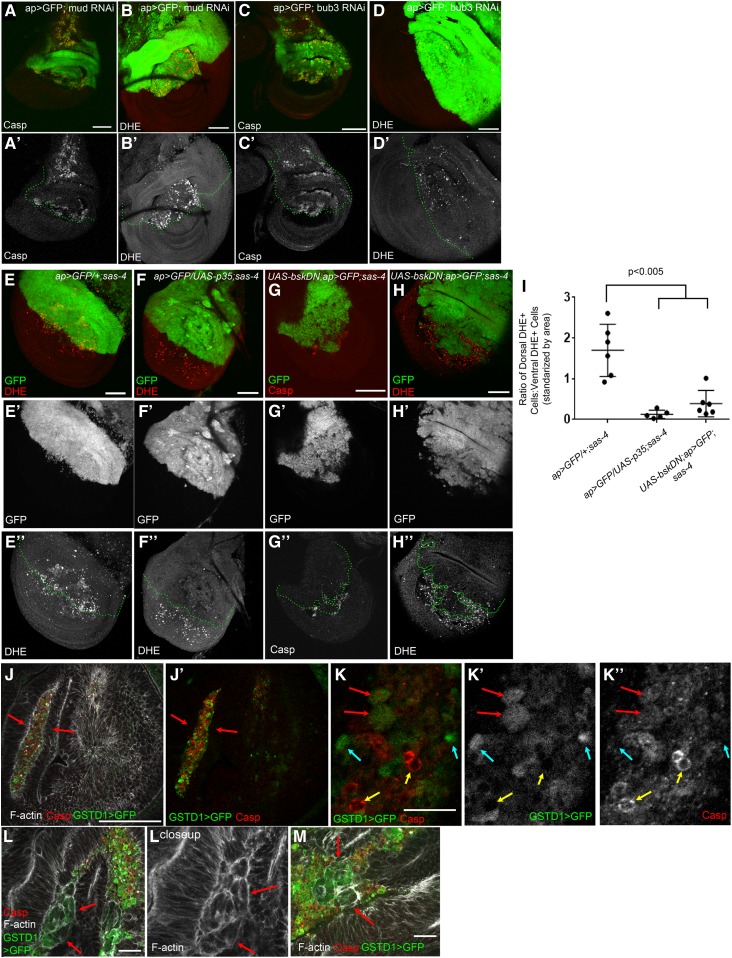
Figure 6.
Mitotic errors induced by other stimuli also elevate apoptosis and ROS levels, and blockade of either apoptosis or JNK reduces ROS production in acentrosomal cells. (A and B) Knockdown of the mitotic spindle anchoring protein, Mud, leads to both increased apoptosis (A-A’) and ROS (B-B’). (C-C’ and D-D’) Knockdown of the mitotic fidelity factor, Bub3, also increases both apoptosis (C) and ROS (D) levels. (E-E”) sas-4 homozygous mutant wing discs have elevated ROS levels, with slightly more DHE+ cells present in the dorsal compartment than the ventral [quantified in (I)]; dorsal compartment marked by ap>GFP expression. This genotype also serves as a control for the subsequent experiments. (F-F”) ROS production associated with centrosome loss (entire disc is sas-4 mutant) is reduced by inhibiting apoptosis via p35 misexpression in the GFP+ dorsal area. (G-G”) Misexpressing the JNK signaling inhibitor BskDN (a dominant negative form of JNK) prevents apoptosis caused by centrosome loss. (H-H”) JNK blockade also reduces ROS levels in sas-4 mutant discs. (I) Quantification of ROS levels in relevant genetic backgrounds (± SD). Blocking apoptosis through misexpression of p35 or bskDN in the dorsal region of sas-4 mutant discs reduces ROS levels, relative to the ventral portion, which is sas-4 mutant but does not express the indicated transgene. Bar, 50 µm. (J–M) Higher-magnification images of sas-4 homozygous mutant wing discs expressing the ROS reporter GSTD1>GFP (green channel), and stained for the apoptotic marker cleaved caspase 3 (red channel) and phalloidin to label F-actin (white channel). (J-J’) Many cells with highly elevated levels of GSTD1>GFP and/or cleaved caspase were outside of the wing disc epithelium (arrows). (K–K”) At even higher magnification, some cells displayed high levels of GSTD1>GFP and low levels of caspase (blue arrows), some with elevated levels of both (red arrows), and some with very high caspase levels and low levels of GSTD1>GFP (yellow arrows). (L and M) We also observed cells that had segregated in groups from the disc epithelium and had moderate expression of GSTD1>GFP with little or no caspase accumulation (red arrows). Scale bars in (K–M), 10 μm. Casp, caspase; DHE, dihydroethidium; RNAi, RNA interference; RNA-Seq, RNA-sequencing; ROS, reactive oxygen species; WT, wild-type.
Recent studies in the wing disc suggest that apoptosis can induce ROS production, though this effect may be indirect (see Discussion) (Santabárbara-Ruiz et al. 2015; Clemente-Ruiz et al. 2016; Fogarty et al. 2016). To test the hypothesis that the ROS induction that we observed in centrosome-deficient discs is a result of the apoptosis triggered by mitotic errors, we blocked apoptosis in sas-4 mutant wing discs using the caspase inhibitor p35 and measured ROS levels. Blocking apoptosis significantly reduced ROS levels in sas-4 mutant cells (Figure 6, E, F, and I). p35 blocks apoptosis by inhibiting the activity of the downstream caspase DrICE. However, it does not block activity of the upstream caspase Dronc, and this promotes continuous JNK activity in the resulting undead cells (Kondo et al. 2006; Martin et al. 2009). Thus, the ability of p35 to block ROS elevation in sas-4 mutant cells suggests that JNK activation alone is not sufficient to elevate ROS.
The relationship between JNK signaling, apoptosis, and ROS is complex. Activating JNK signaling can activate antioxidant pathways and can also induce cell death (Wang et al. 2003; Dhanasekaran and Reddy 2017). Thus, we tested an alternate hypothesis: JNK signaling, while not sufficient, is necessary for the elevation of ROS in centrosome-deficient wing discs. To test this, we examined sas-4 mutant wing discs in which JNK signaling was blocked through misexpression of BSK-DN. As we saw previously, JNK inhibition via BSK-DN expression suppresses virtually all of the apoptosis that is normally caused by centrosome loss (Figure 6G) (Poulton et al. 2014). DHE staining in these discs revealed a significant reduction in ROS levels after JNK blockade (Figure 6, H and I), consistent with the idea that JNK signaling is necessary for elevated ROS production in centrosome-deficient wing discs. Together, these data suggest that the highly elevated ROS levels in a subset of sas-4 mutant cells depend on the completion of apoptosis, regardless of whether JNK is hyperactivated (as in the p35+ cells) or blocked (as in the BSK-DN cells). These results are considered further in the Discussion.
As a final examination of the causal relationships between apoptosis and ROS, we examined at cellular resolution the accumulation of the ROS reporter GSTD1>GFP and the apoptotic marker cleaved caspase 3 in sas-4 mutant wing discs. Strikingly, the majority of cells with highly elevated levels of GSTD1>GFP and/or cleaved caspase were outside of the wing disc epithelium (Figure 6J, arrows), having presumably been basally extruded. At higher magnification, different populations of cells were seen, some with high levels of GSTD1>GFP and low levels of caspase (Figure 6K, blue arrows), some with elevated levels of both (Figure 6K, red arrows), and some with very high caspase levels and low levels of GSTD1>GFP (Figure 6K, yellow arrows); this final category may represent cells in which advanced progression of apoptosis eventually released the cytoplasmic GFP. In sectioning through the discs, we also observed cells that had segregated in groups from the disc epithelium and had moderate expression of GSTD1>GFP, with little or no caspase accumulation (Figure 6, L and M). These latter data are consistent with previous observations that apoptotic caspase activation in the wing disc can trigger low-level, nonautonomous elevation of ROS in neighboring cells (Santabárbara-Ruiz et al. 2015). Alternatively, elevation of ROS may occur before the activation of caspases. The precise mechanism by which this occurs remains to be determined.
G6PD expression buffers ROS production and G6PD knockdown elevates apoptosis caused by centrosome loss
Our earlier work revealed that mitotic errors induced by the absence of centrosomes trigger apoptosis in the aneuploid cells (Poulton et al. 2014). However, these data also revealed that many cells that are challenged with centrosome loss evade death, though their cell cycle is lengthened. One possibility is that some of genes upregulated in sas-4 mutant wing cells help centrosome-deficient cells survive in the presence of mitotic stress. To test this hypothesis, we performed a small candidate RNAi screen testing for genetic interactions with sas-4, predicting that knockdown of genes encoding proteins that helped cells cope with centrosome loss would enhance the sas-4 knockdown phenotype. The candidate genes were chosen based on their roles as key players in redox balance or related signaling pathways (Table 5). From this screen, we identified a significant interaction between sas-4 and glucose-6-phosphate dehydrogenase (g6pd), using a g6pd RNAi line previously shown to significantly reduce G6PD levels and activity (Teesalu et al. 2017). While knockdown of either gene alone had minimal effects on adult wing blade morphology, knocking down both genes significantly increased wing blistering (Figure 7, A and B). G6PD is the rate-limiting enzyme in the pentose phosphate pathway, converting glucose-6-phosphate to 6-phosphoglucono-s-lactone. This reaction also generates NADPH, which is used by glutathione reductase to produce reduced glutathione, a potent antioxidant. Thus, in many cell types, G6PD is a central player in the ability to limit ROS levels (Stanton 2012).
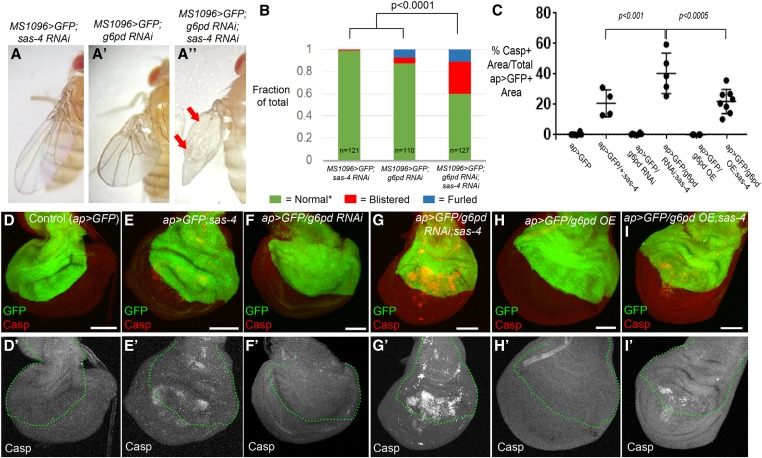
Figure 7.
G6PD helps prevent apoptosis in acentrosomal cells. (A–C) Representative adult wing from the indicated genotypes. (A) Knockdown of sas-4 with MS1096-Gal4 has no observable effect on adult wing morphology. (A’) g6pd knockdown has only minor effects on wing blade morphology, although the wings of these flies are held erect relative to the body. (A”) Simultaneous knockdown of both sas-4 and g6pd leads to significantly more wing blisters and furled wing phenotypes. (B) Quantification of effects on wing blade morphology. * The “Normal” wing category includes both the WT appearance seen in the sas-4 RNAi wings, as well as the erect wings with normal wing blades observed in g6pd RNAi flies. “Furled” refers to wings that never expanded (unfurled) after eclosion. (C) Quantification of the effects of sas-4 and g6pd manipulations on apoptosis levels. In addition to the P-values shown for the two comparisons to ap>GFP/g6pd RNAi;sas-4, a one-way ANOVA with Dunnett’s multiple comparisons test of all genotypes to the ap>GFP control indicates that neither g6pd RNAi nor g6pd OE are significantly different from ap>GFP, whereas expression of either transgene in the sas-4 mutant background is significantly different from ap>GFP alone (P < 0.001). Overexpression of G6PD did not reduce cell death associated with centrosome loss (comparison of ap>GFP/g6pd OE;sas-4 to ap>GFP/+;sas-4) (P = 0.82 by Welch’s t-test). (D–I) Assessment of apoptosis levels in third-instar wing discs of indicated genotypes. (D-D’) Control ap-Gal4 UAS-GFP (ap>GFP) wing discs have minimal apoptosis. (E-E’) The elevated apoptosis throughout the wing pouch characteristic of sas-4 homozygous mutants is not altered by expression of GFP in the dorsal region via ap>GFP. (F-F’) Expression of g6pd RNAi using ap-Gal4 does not increase apoptosis in a WT background. (G-G’) Expression of g6pd RNAi using ap-Gal4 in the sas-4 mutant background significantly increases the incidence of apoptosis associated with centrosome loss in the cells where g6pd is knocked down. (H-H’) G6PD OE does not increase apoptosis in a WT background. (I-I’) G6PD OE is not sufficient to reduce apoptosis caused by centrosome loss. Bar, 50 µm. All images are maximum intensity projections. Casp, caspase; G6PD, glucose-6-phosphate dehydrogenase; OE, overexpression; RNAi, RNA interference; WT, wild-type.
In our previous analysis of other sas-4 genetic interactions (Poulton et al. 2014), increased adult wing blistering correlated with increased levels of apoptosis during larval stages. Therefore, we examined apoptosis levels, using activated caspase staining, to determine if there is any enhancement or suppression of the sas-4 apoptotic phenotype after knockdown of G6PD. While g6pd RNAi alone led to no detectable increase in apoptosis (Figure 7, D vs. F, quantified in Figure 7C), apoptosis was significantly increased when g6pd was knocked down in acentrosomal cells relative to that observed in acentrosomal cells alone (Figure 7, E vs. G, quantified in Figure 7C). This was consistent with the hypothesis that G6PD buffers the elevated ROS production induced by centrosome loss. To directly test this, we stained for DHE in g6pd knockdown cells in the sas-4 mutant background. Consistent with the hypothesis, we observed an even greater increase in ROS levels in acentrosomal cells that also lack G6PD (Figure 8, B vs. D, quantified in Figure 8G; note, the ap>GFP/+;sas-4 data in Figure 8G are the same as those presented in Figure 6I), while G6PD knockdown alone did not elevate ROS (Figure 8, C and G). These data suggest that elevated G6PD expression in centrosome-deficient cells helps prevent the death of some of the cells attempting to cope with the loss of centrosomes, presumably by limiting the amount of ROS.
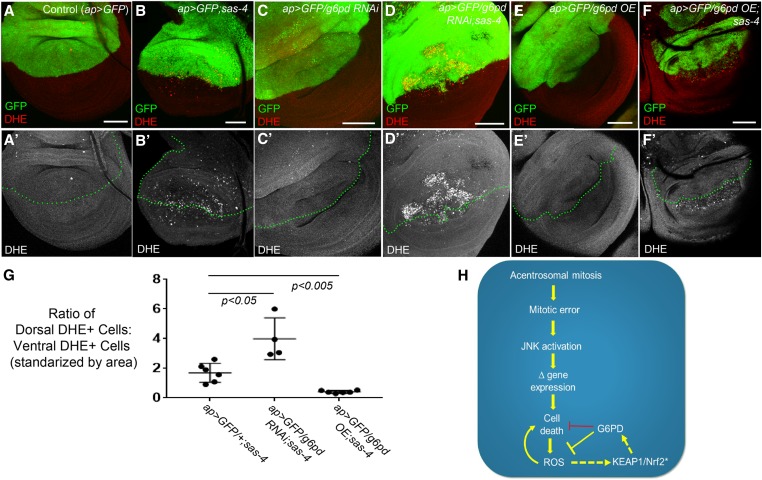
Figure 8.
G6PD buffers acentrosomal cells against ROS production. (A–F) Assessment of ROS levels in third-instar wing using DHE. (A-A’) ap>GFP alone does not induce ROS. (B-B’) ap>GFP also does not affect levels of ROS associated with centrosome loss (sas-4 homozygous mutant disc). (C-C’) g6pd knockdown in otherwise normal cells does not induce ROS production. (D-D’) However, g6pd knockdown does elevate ROS in sas-4 mutant cells above levels caused by centrosome loss alone. (E-E’) G6PD OE does not affect baseline ROS production in WT. (F-F’) G6PD OE significantly reduces levels of ROS in acentrosomal cells. Bar, 50 µm. All images are maximum-intensity projections. (G) Quantification of the effects of sas-4 and g6pd manipulations on ROS levels; see the Materials and Methods for a detailed description of these calculations. (H) Speculative model of the relationships between the relevant pathways, processes, and genes. Because G6PD buffers ROS levels in some cells and because ROS can contribute to cell death, G6PD can indirectly inhibit cell death (this indirect relationship is indicated by the red repression symbol). The activation of KEAP1/Nrf2 signaling indicated by “*” is inferred from increased ROS levels, which are known to activate KEAP1/Nrf2, and from increased expression of GstD1>GFP, which is regulated by KEAP1/Nrf2 (this inferred activity is indicated by the dashed arrow). Similarly, we infer that G6PD is upregulated by KEAP1/Nrf2 activity in acentrosomal cells because g6pd is a previously identified transcriptional target of that pathway. DHE, dihydroethidium; G6PD, glucose-6-phosphate dehydrogenase; OE, overexpression; RNAi, RNA interference; ROS, reactive oxygen species; WT, wild-type.
These observations indicated that the increased expression of G6PD we observed in sas-4 mutant cells, as revealed by our RNA-Seq data, might serve as a feedback response to limit apoptosis and ROS production. To test this, we overexpressed G6PD in sas-4 mutants. Consistent with the role of G6PD in antioxidant generation, ROS levels were significantly reduced in the sas-4 mutant cells overexpressing G6PD (Figure 8, F vs. B and G). Interestingly, overexpression of G6PD did not detectably affect the levels of apoptosis caused by centrosome loss (Figure 7, E vs. I, quantified in Figure 7C). Together, these data indicate that the upregulation of G6PD is important in limiting ROS production, and reveal that although its basal level of expression helps prevent apoptosis in mitotically stressed acentrosomal cells that have not already entered the apoptotic path, increasing G6PD levels alone is not sufficient to eliminate apoptosis caused by centrosome loss (Figure 7, I and C). This interpretation fits well with our observations above, suggesting that increased ROS levels are largely downstream of apoptosis, and thus decreasing ROS in cells already firmly committed to the path to apoptosis would not necessarily be expected to decrease apoptosis in this context. It would also be useful in the future to generate G6PD mutant clones in the sas-4 mutant background to determine if its effects are cell autonomous.
Discussion
Transcriptional responses to cellular and tissue injury are major determinants of cell behavior and homeostasis. Drosophila imaginal discs provide powerful models to identify primary signaling pathways and biological processes governing these responses, and to dissect their relationships to one another (Beira and Paro 2016). JNK signaling is now well established as a central player in these events. JNK serves multiple roles, including sensing the initiating cell stress, activating pathways that alleviate cellular stress (e.g., DNA repair) (Hayakawa et al. 2004; Picco and Pages 2013), triggering apoptosis when damage is severe (Igaki 2009), and promoting the activation of secondary, mitogenic signaling pathways by upregulating their ligands, which ultimately drives large-scale processes of tissue repair such as compensatory proliferation (Ryoo et al. 2004). More recently, in the context of cell/tissue damage, JNK has also been implicated in regulating redox balance (Clemente-Ruiz et al. 2016; Fogarty et al. 2016; Khan et al. 2017), which appears to be an important aspect of the cellular and tissue-level response to that damage.
We are interested in the response to centrosome loss, and in the pathways that buffer and compensate for the mitotic defects and resultant apoptosis that centrosome loss causes in some tissues (Poulton et al. 2014, 2017). To extend this analysis, we examined the transcriptional response to centrosome loss. This revealed that mitotic errors induced by centrosome loss trigger a complex transcriptional response in wing imaginal discs, including JNK-dependent changes in gene expression. Interestingly, a previous microarray-based study of transcriptome profiles in acentrosomal fly cells did not detect major changes in gene expression of multiple components of particular cellular processes (e.g., no upregulation of JNK or redox pathways) (Baumbach et al. 2012). We believe that the most likely reason for this difference was that the previous study pooled RNA from larval wing discs and brains. As we and others have found, larval fly brains are quite robust to centrosome loss and thus do not noticeably activate cell stress responses like JNK signaling (Basto et al. 2006; Poulton et al. 2017). Thus, it is likely that changes in gene expression occurring in acentrosomal wing discs were diluted out by the inclusion of RNA from brains. It is also possible that technical differences between RNA-Seq and microarray platforms contributed to our ability to detect expression changes in numerous genes.
In our RNA-Seq analysis, multiple regulators of redox balance were significantly upregulated. This spurred us to investigate oxidative stress levels in acentrosomal cells, revealing that a significant fraction of these cells have high levels of ROS. We went on to identify upregulation of G6PD as an important component of the ability of acentrosomal cells to buffer themselves against oxidative stress. Together, our data demonstrate that, in wing imaginal discs, error-prone, acentrosomal mitosis activates JNK signaling, leading to both the induction of apoptosis and increased ROS production (Figure 8H). Our data also reveal an important mechanism to deal with this threat: transcriptional changes in redox regulators, including G6PD, that can then feed back into the process, limiting the extent of both ROS production and cell death, and thus potentially giving cells more time to fix mitotic errors without losing the affected cells.
ROS, JNK, apoptosis, and proliferation/repair: a complex network
Recent studies have begun to reveal the complexities of the signaling network linking ROS, JNK, apoptosis, and proliferation/repair, casting doubt on the idea that they form a simple linear pathway, and instead suggesting that the responses vary depending on the tissue and damaging agent. It is well established that high levels of JNK signaling in imaginal discs can induce apoptosis (Igaki 2009) and that ROS production increases rapidly following cell stresses such as aneuploidy (Clemente-Ruiz et al. 2016). Indeed, in both wing and eye imaginal discs, initiation of the apoptotic pathway by overexpression of proapoptotic proteins (Hid or Rpr) is sufficient to significantly increase ROS levels (Santabárbara-Ruiz et al. 2015; Fogarty et al. 2016; Brock et al. 2017; Khan et al. 2017). In addition, ROS is elevated in tumor-forming genetic models (Ohsawa et al. 2012; Pérez et al. 2017) or following physical tissue damage (Santabárbara-Ruiz et al. 2015). However, trying to fit all the data into a single linear pathway is difficult. Directly inducing apoptosis or triggering caspase activity without death can lead to ROS elevation (Huu et al. 2015; Santabárbara-Ruiz et al. 2015; Fogarty et al. 2016; Khan et al. 2017; Pérez et al. 2017), but it is also well established that elevating ROS can trigger apoptosis (Camhi et al. 1995; Martindale and Holbrook 2002; Redza-Dutordoir and Averill-Bates 2016) and that reducing ROS can limit the apoptotic response (O’Keefe et al. 2015; Clemente-Ruiz et al. 2016; Fogarty et al. 2016). There is evidence that ROS production is induced by JNK signaling (Kanda et al. 2011; Khan et al. 2017; Pérez et al. 2017), but also evidence that ROS play a role in activating JNK (Wang et al. 2003; Ohsawa et al. 2012; Santabárbara-Ruiz et al. 2015; Clemente-Ruiz et al. 2016; Fogarty et al. 2016; Khan et al. 2017; Pérez et al. 2017) and that JNK signaling can induce an antioxidant response (Wang et al. 2003). Finally, in some situations, elevating ROS appears to reduce JNK signaling (Brock et al. 2017).
The studies most relevant to our work are from the Milan laboratory, which examined the consequences of aneuploidy induced by disrupting the spindle assembly checkpoint or spindle assembly in eye and wing disc epithelia. The parallels with centrosome loss are striking. As with centrosome loss (Poulton et al. 2014), disruption of spindle assembly by reducing levels of mitotic regulators such as Rod or Bub3 elevated the incidence of mitotic defects like lagging chromosomes and elevated DNA damage, and the resulting aneuploid cells were removed by apoptosis (Dekanty et al. 2012; Clemente-Ruiz et al. 2016). However, if apoptosis is blocked, highly aneuploid cells accumulate and a JNK-dependent transcriptional response is triggered. The “undead cells” elevate expression of the morphogen Wingless and drive tissue overgrowth, paralleling the effects of centrosome loss (Poulton et al. 2014). In a follow-up study, two more parallels were identified (Clemente-Ruiz et al. 2016). First, dying cells accumulate elevated levels of ROS, as measured by GSTD1>GFP. Second, they also induce a transcriptional response that includes a series of JNK target genes and a set of genes involved in buffering ROS.
In other cases, the apoptosis–JNK–ROS connections are even more complex. For example, in eye imaginal discs, when apoptosis is induced by expression of Hid but death is blocked by p35 expression, the production and release of extracellular ROS is triggered (Fogarty et al. 2016). This leads to the recruitment of hemocytes that secrete the TNFα relative Eiger, which in turn activates lower and thus nonapoptotic levels of JNK activation in neighboring cells (Fogarty et al. 2016). This promotes the expression of mitogenic signals that contribute to compensatory proliferation/regeneration and apoptosis-induced proliferation. The generation of extracellular ROS appears to rely on plasma membrane-targeted ROS generators like Dual Oxidase (DUOX) (Fogarty et al. 2016; Khan et al. 2017). Intriguingly, the DUOX maturation factor NIP, encoded by moladietz (mol), is itself upregulated by JNK activity (Khan et al. 2017). JNK-induced extracellular ROS (produced by the NIP-DUOX mechanism) then help to maintain JNK activity in neighboring cells to drive tissue repair (Khan et al. 2017).
Comparison of our findings with these and other studies reveals interesting similarities and differences. First, consistent with these studies, induction of apoptosis caused by loss of centrosomes or other mitotic regulators triggers ROS production. However, our p35 data suggest that completion of apoptosis is important for ROS production in acentrosomal cells, which contrasts with data indicating that blocking apoptosis did not reduce ROS levels associated with Hid misexpression (Fogarty et al. 2016) or CIN induction (Clemente-Ruiz et al. 2016). Interestingly, in scrib rasV12 eye disc clones (a tumor model), blocking caspase activity with p35 did significantly reduce ROS levels (Pérez et al. 2017). These differences suggest that the circuitry of these interconnected processes may be wired differently in acentrosomal cells, leading to different experimental outcomes. For example, different tissues might have different responses downstream of Dronc, which remains active after p35-mediated apoptosis blockade. Further studies will be also required to elucidate the cellular mechanisms generating ROS in these systems; NIP-DUOX is an interesting candidate (see below).
It is also worth considering what our data suggest about the relationship between JNK activity and ROS levels. As described above, JNK can drive ROS production through NIP-DUOX (Khan et al. 2017). However, others have found that JNK is not essential for ROS production in the wing disc in response to aneuploidy (Clemente-Ruiz et al. 2016) and that JNK activation is not sufficient for ROS elevation in the eye disc (Fogarty et al. 2016). In contrast, we found that blocking JNK signaling in acentrosomal cells reduced ROS levels, which is very similar to recent findings from the Bergmann laboratory, which found that blocking JNK in scrib rasv12 clones prevents ROS production (Pérez et al. 2017). From our experiments, we speculate that reduction in ROS after JNK blockade is due to the subsequent inhibition of apoptosis, because when we blocked acentrosomal cell death via p35 misexpression, which leads to hyperactive JNK signaling, we did not detect increased ROS levels, instead finding decreased ROS in the p35+ cells. Perhaps the sustained, high-level JNK signaling in these undead cells somehow subverts the NIP-DUOX mechanism of ROS production, either directly or indirectly, through massive upregulation of ROS antagonists like G6PD. Alternatively, the mechanism driving NIP-DUOX-mediated ROS levels may not be active in acentrosomal cells. Indeed, despite clearly identifying the upregulation of numerous JNK targets, our RNA-Seq data did not detect any significant upregulation of mol, even in individual mutant gene lists (i.e., sas-4 vs. control or asl vs. control). Thus, ROS production caused by centrosome loss may occur independently of NIP-DUOX. Of course, differential sensitivities associated with different RNA-Seq protocols or downstream analyses may explain the absence of mol upregulation in our data. Thus, our data and those of others clearly suggest important links between cellular damage, JNK activation, initiation of the apoptotic cascade, and ROS production. However, the circuitry connecting these events and the mechanisms underlying these relationships appears to vary depending on the nature or severity of the damage. This would benefit from further exploration.
JAK-STAT and KEAP1-Nrf2 signaling
Another signaling pathway implicated by our RNA-Seq data was the JAK-STAT pathway. Our list of upregulated genes shared by both sas-4 and asl included Upd2 and Upd3; both are ligands of the pathway and are previously identified JNK targets (Pastor-Pareja et al. 2008; Bunker et al. 2015; Santabárbara-Ruiz et al. 2015). This list also included inositol pentakisphosphate 2-kinase (Ipk1), which can regulate JAK-STAT signaling (Seeds et al. 2015), and the JAK-STAT target gene vir1 (Dostert et al. 2005). Consistent with this, we found enrichment of STAT92E-binding sites in our set of genes upregulated in both sas-4 and asl loss (Figure 4C), and saw upregulation of a JAK-STAT reporter in acentrosomal cells, though only when apoptosis was inhibited (Figure S2, A–C). We attempted to test for genetic interactions by inducing simultaneous upd2 and sas-4 knockdown, but the severe phenotypes (massive apoptosis and abnormal adult wings) caused by knockdown of Upd2 alone precluded analysis of the interaction. Other studies demonstrate a role for JAK-STAT in regulating compensatory proliferation and regeneration in damaged imaginal discs (Ohsawa et al. 2012; Katsuyama et al. 2015; Santabárbara-Ruiz et al. 2015; Clemente-Ruiz et al. 2016). We speculate that the increased levels of JAK-STAT ligands that we detected are likely involved in the compensatory proliferation response that we previously observed in acentrosomal wing discs (Poulton et al. 2014).
We also looked for connections with the KEAP1-Nrf2 pathway, an important regulator of cytoprotective responses to oxidative stress (Sykiotis and Bohmann 2008; Loboda et al. 2016; Sies et al. 2017). Nrf2 is a bZIP transcription factor that positively regulates antioxidant response proteins. KEAP1 keeps it inactive by anchoring it in the cytoplasm and targeting it for proteasomal destruction, but oxidative stress relieves this inhibition (Figure 8H). G6PD is a target of KEAP1-Nrf2 signaling (Loboda et al. 2016). Many of the GST family proteins that were upregulated in acentrosomal cells have previously been found to be upregulated when KEAP1-Nrf2 signaling is experimentally activated (Loboda et al. 2016). Indeed, the Drosophila Nrf2 homolog, Cap-n-collar (Cnc), was recently proposed to help limit ROS levels in the wing disc following tissue damage induced by Rpr misexpression (Brock et al. 2017). In the model of Brock et al. (2017), increased ROS levels activate Cnc, which negatively regulates ROS levels via increased transcription of ROS suppressors. The authors also suggested that this Cnc-mediated reduction in ROS levels helps maintain an optimal level of JNK signaling needed for tissue repair and development. Our attempts to disrupt KEAP1-Nrf2 signaling in our RNAi-based genetic interaction screen did not reveal significant genetic interactions with sas-4 knockdown (Table 5). However, whether that is due to a true lack of functional interaction or a simple lack of effective knockdown remains unclear. Nevertheless, our data suggest that G6PD upregulation is an important buffer against excessive ROS levels: reducing G6PD levels in acentrosomal cells significantly increases ROS levels above those induced by centrosome loss alone. Furthermore, apoptosis is elevated after loss of G6PD in acentrosomal cells, perhaps due to the increased levels of ROS. Induction of antioxidant-promoting proteins like G6PD and the GST proteins may be part of a response through which lower levels of JNK, induced in not yet apoptotic cells, may allow them to survive and correct minor mitotic errors, similar to the response in neighboring cells induced by extracellular ROS (Santabárbara-Ruiz et al. 2015; Fogarty et al. 2016). In contrast, the higher levels of ROS associated with loss of G6PD in acentrosomal cells may lead to higher, intolerable levels of JNK activity that push them down the apoptotic pathway.
Centrosome loss, immunity, and xenobiotic detoxification
Some mysteries remain in the gene sets upregulated after centrosome loss. While JNK pathway target genes, and genes involved in the oxidative stress response and apoptosis, now make sense, two other prominent upregulated gene sets remain more puzzling: genes involved in innate immunity and in the xenobiotic response to toxic compounds (Table 3 and Table 6). Work by the Ruvkun laboratory in Caenorhabditis elegans suggests an intriguing interpretation of these data. In the course of an unrelated RNAi screen, a common response to knockdown of genes involved in several core biological processes, including protein translation, proteasome or mitochondrial function, and mRNA processing was noted (Melo and Ruvkun 2012). All led to the activation of both the innate immune and xenobiotic detoxification responses. Similar effects were induced by pathogens, or by pathogen-derived or natural toxins (Dunbar et al. 2012; McEwan et al. 2012; Govindan et al . 2015). Further, responses to the knockdown of genes involved in core biological processes were mediated through the JNK pathway (Melo and Ruvkun 2012). Based on these data, they have developed the intriguing hypothesis that animals have evolved a conserved mechanism to detect and respond to pathogen attack on key cellular machines via small-molecule or protein effectors that inhibit their function. They suggest that cells evolved to respond to defects in the function of key cellular machines by upregulating genes mediating innate immune and xenobiotic detoxification responses. Our data fit well with this hypothesis. We saw upregulation of many components of the xenobiotic detoxification response, and those involved in or activated by the innate immune response (Table 3 and Table 6). Others have seen similar changes in response to other stresses, including the response to xenobiotic drugs (Misra et al. 2011) or to pathogenic protein aggregation (Dialynas et al. 2015; Zhan et al. 2015). Genes upregulated after SAC inactivation also include xenobiotic response genes (Clemente-Ruiz et al. 2016). Intriguingly, oxidative stress also induces many of these genes (Misra et al. 2011; Kucinski et al. 2017), including ones with known antioxidant roles and those, like cytochrome p450s, without known roles in this process.
Table 6. Known or suspected xenobiotic response genes.
| Gene | Role | Reference |
|---|---|---|
| Phase 1: cytochrome p450s | ||
| Cyp18a1 | Cytochrome P450-18a1. Part of a superfamily of heme-containing microsomal oxidase enzymes that metabolize both xenobiotic compounds for detoxification and endogenous substrates. | Feyereisen (2005) |
| Cyp6a17 | Cytochrome P450-6a17. See above. | Feyereisen (2005) |
| Cyp6a23 | Cytochrome p450-6a23. See above. | Feyereisen (2005) |
| CG12224 | Cytochrome P450 reductase/NADPH:ferrihemoprotein oxidoreductase. Membrane-bound enzyme required for electron transfer to cytochrome P450 in the microsome from NADPH | Pandey and Fluck (2013) |
| Phase 2: modify xenobiotics to increase hydrophilicity | ||
| AcCoAS | Acetyl coenzyme A synthase. Functions in glycine conjugation of xenobiotics. Cofactor of NATs. | Montooth et al. (2006) |
| St3 | Sulfotransferase 3. | James and Ambadapadi (2013) |
| CG15661 | UDP-glucuronosyl/UDP-glucosyltransferase involved in detoxification. | Gruenewald et al. (2009) |
| Aox (CG18522) | Aldehyde oxidase 1. In addition to metabolic roles, catalyzes the oxidation of both cytochrome P450 (CYP450) and monoamine oxidase intermediate products. | |
| GSS | Glutathione synthase | |
| GstD1 | Glutathione S-transferase D1. JNK and Nrf2 target gene. | Board and Menon (2013), Misra et al. (2011) |
| GstD3 | Glutathione S-transferase D3. Nrf2 target. | Misra et al. (2011) |
| GstE3 | Glutathione S-transferase E3. Nrf2 target. | Misra et al. (2011) |
| GstE5 | Glutathione S-transferase E5. | Misra et al. (2011) |
| GstE6 | Glutathione S-transferase E6. Nrf2 target. | Misra et al. (2011) |
| GstE8 | Glutathione S-transferase E8. Nrf2 target. | Misra et al. (2011) |
| sda | Slamdance or aminopeptidase N. Modifies glutathione-S-conjugate. γ-glutamyl transpeptidase, aminopeptidase N, and cysteine-conjugate-β-lyase may comprise a multienzyme pathway that acts on xenobiotic–glutathione conjugates. | Hausheer et al. (2011) |
| Men-b | Malic enzyme b. Induced by xenobiotics. May provide NADPH for glutathione reductase and NADPH-cytochrome c reductase. | Oda et al. (1999) |
| Zw (G6PD) | Zwischenferment. Glucose-6-phosphate dehydrogenase. A NRF2 target gene. Provides NADPH to glutathione reductase. | Loboda et al. (2016) |
| CG10365 | Glutathione-specific γ-glutamylcyclotransferase 1. Similar to mouse Chac1. May destroy glutathione. | Bachhawat and Yadav (2018) |
| CG18641 | Triacylglycerol lipase family. May function in metabolism of 3-phenoxybenzoic acid-containing xenobiotic triacylglycerols. | Lian et al. (2018) |
| CG4267 | Triacylglycerol lipase family. See above. | Lian et al. (2018) |
| CG7367 | Triacylglycerol lipase family. See above. | Lian et al. (2018) |
| Phase 3: export xenobiotics | ||
| l(2)03659 | MDR4/MOAT-B. ABC transporter-like. | Hoffmann and Kroemer (2004) |
A list of genes that have increased expression in sas-4 and asl mutants, relative to wild-type, and that are also proposed to function in the response to xenobiotics. (NATs)=N-acetyltransferases.
Our upregulated gene list also overlapped with genes upregulated after wounding and during imaginal disc regeneration. Imaginal discs have a remarkable ability to regenerate after surgical or radiation damage (Haynie and Bryant 1977), by a process known as compensatory proliferation (Fan and Bergmann 2008). This response also requires the JNK pathway. Strikingly, 9 of the top 20 genes upregulated during the early stages of regeneration (Katsuyama et al. 2015) are also significantly upregulated after knockdown of both sas-4 or asl. Many of these upregulated genes are part of the generalized wound response, such as MMP1 and PGRP-SA (Patterson et al. 2013), suggesting that at least some parts of these transcriptional responses are parallel ones. Genes upregulated after disc regeneration also included xenobiotic response genes (e.g., three Cytochrome p450 genes and seven GST family members), suggesting the possibility that animals are genetically programmed to interpret tissue injury as a pathogen attack.
Together, our previous and current data demonstrate the robust and multilayered mechanisms that help cells, and tissues, compensate for the absence of centrosomes. In proliferating epithelia, such as the wing imaginal disc, alternative MT nucleation pathways help build mitotic spindles and the SAC slows mitotic progression to facilitate these less-efficient pathways (Poulton et al. 2014). Although many cells appear to divide without obvious mitotic errors, a significant number suffer chromosome segregation errors (e.g., lagging chromosomes, aneuploidy, and DNA damage). Our data and those of others suggest that it is these cells that activate high levels of JNK signaling, which drives those cells into the apoptotic pathway (Dekanty et al. 2012; Poulton et al. 2014). JNK also acts as a homeostatic buffer in the tissue by promoting compensatory proliferation in the neighboring cells (Igaki 2009). In the present study, we uncovered an additional consequence of centrosome loss, increased ROS, and another compensatory process, upregulation of oxidative stress response genes like g6pd. A growing body of recent work has begun to reveal a complex network of signaling pathways and cellular processes that link cellular defects, such as mitotic errors, and subsequent activation of JNK signaling and apoptosis, to oxidative stress and compensatory proliferation/regeneration. The relationships among these players are not yet fully understood, in part due to the complexity that has emerged from recent studies, including bidirectionality (e.g., ROS ←→ JNK), tissue-specific mechanisms (e.g., eye disc vs. wing disc), and threshold-dependent responses (e.g., JNK promoting apoptosis vs. survival/proliferation). Future studies will help further characterize the circuitry defining these relationships and the underlying causes for the apparent specificity of some mechanisms.
Acknowledgments
We thank D. Bohmann, W. Orr, Y. Tamori, L. Wallrath, N. Rusan, T. Avidor-Reiss, the Bloomington Drosophila Stock Center, the Vienna Drosophila Resource Center, and the Developmental Studies Hybridoma Bank for reagents; J. Cuningham and J. Pearson for technical assistance; S. Nystrom for assistance with motif analysis; T. Perdue for assistance with microscopy; the University of North Carolina (UNC) High-Throughput Sequencing Facility; and our UNC colleagues for excellent suggestions. The work was supported by National Institutes of Health grant R35 GM1-18096 to M.P., a Research Scholar grant (17-164-01-DDC) from the American Cancer Society to D.J.M., and start-up funds from UNC’s Department of Medicine to J.S.P. The authors declare no competing financial interests.
Footnotes
Supplemental material available at https://doi.org/10.25386/genetics.7837943.
Communicating editor: H. Bellen
Literature Cited
- Bachhawat A. K., Yadav S., 2018. The glutathione cycle: glutathione metabolism beyond the gamma-glutamyl cycle. IUBMB Life 70: 585–592. 10.1002/iub.1756 [DOI] [PubMed] [Google Scholar]
- Bailey T. L., 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. 10.1093/bioinformatics/btr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., et al. , 2006. Flies without centrioles. Cell 125: 1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Baumbach J., Levesque M. P., Raff J. W., 2012. Centrosome loss or amplification does not dramatically perturb global gene expression in Drosophila. Biol. Open 1: 983–993. 10.1242/bio.20122238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H., Anderson K. V., 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl. Acad. Sci. USA 111: E1491–E1500. 10.1073/pnas.1400568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beira J. V., Paro R., 2016. The legacy of Drosophila imaginal discs. Chromosoma 125: 573–592. 10.1007/s00412-016-0595-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindokas V. P., Jordan J., Lee C. C., Miller R. J., 1996. Superoxide production in rat hippocampal neurons: selective imaging with hydroethidine. J. Neurosci. 16: 1324–1336. 10.1523/JNEUROSCI.16-04-01324.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., et al. , 2008. Drosophila asterless and vertebrate Cep152 are orthologs essential for centriole duplication. Genetics 180: 2081–2094. 10.1534/genetics.108.095141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board P. G., Menon D., 2013. Glutathione transferases, regulators of cellular metabolism and physiology. Biochim. Biophys. Acta 1830: 3267–3288. 10.1016/j.bbagen.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Bond D., Foley E., 2009. A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 5: e1000655 10.1371/journal.ppat.1000655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges H. L., Linden R., Wang J. Y., 2008. DNA damage-induced cell death: lessons from the central nervous system. Cell Res. 18: 17–26. 10.1038/cr.2007.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., Agaisse H., Perrimon N., 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3: 711–722. 10.1016/S1534-5807(02)00325-8 [DOI] [PubMed] [Google Scholar]
- Brock A. R., Seto M., Smith-Bolton R. K., 2017. Cap-n-collar promotes tissue regeneration by regulating ROS and JNK signaling in the Drosophila melanogaster wing imaginal disc. Genetics 206: 1505–1520. 10.1534/genetics.116.196832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Poidevin M., Pradervand S., Lemaitre B., 2009. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5: 200–211. 10.1016/j.chom.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Buchon N., Silverman N., Cherry S., 2014. Immunity in Drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 14: 796–810. 10.1038/nri3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker B. D., Nellimoottil T. T., Boileau R. M., Classen A. K., Bilder D., 2015. The transcriptional response to tumorigenic polarity loss in Drosophila. Elife 4: e03189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camhi S. L., Lee P., Choi A. M., 1995. The oxidative stress response. New Horiz. 3: 170–182. [PubMed] [Google Scholar]
- Campos I., Geiger J. A., Santos A. C., Carlos V., Jacinto A., 2010. Genetic screen in Drosophila melanogaster uncovers a novel set of genes required for embryonic epithelial repair. Genetics 184: 129–140. 10.1534/genetics.109.110288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Gupta S., 2018. The multifaceted role of glutathione S-transferases in cancer. Cancer Lett. 433: 33–42. 10.1016/j.canlet.2018.06.028 [DOI] [PubMed] [Google Scholar]
- Chatterjee N., Bohmann D., 2012. A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One 7: e34063 10.1371/journal.pone.0034063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Xu M., Nagati J., Garcia J. A., 2017. Coordinate regulation of stress signaling and epigenetic events by Acss2 and HIF-2 in cancer cells. PLoS One 12: e0190241 10.1371/journal.pone.0190241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann M., Kaina B., 2013. Transcriptional regulation of human DNA repair genes following genotoxic stress: trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Res. 41: 8403–8420. 10.1093/nar/gkt635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P. R., Zhang C., 2008. Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 9: 464–477. 10.1038/nrm2410 [DOI] [PubMed] [Google Scholar]
- Clemente-Ruiz M., Murillo-Maldonado J. M., Benhra N., Barrio L., Perez L., et al. , 2016. Gene dosage imbalance contributes to chromosomal instability-induced tumorigenesis. Dev. Cell 36: 290–302. 10.1016/j.devcel.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., Leopold P., 2012. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336: 582–585. 10.1126/science.1216689 [DOI] [PubMed] [Google Scholar]
- Dekanty A., Barrio L., Muzzopappa M., Auer H., Milan M., 2012. Aneuploidy-induced delaminating cells drive tumorigenesis in Drosophila epithelia. Proc. Natl. Acad. Sci. USA 109: 20549–20554. 10.1073/pnas.1206675109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran D. N., Reddy E. P., 2017. JNK-signaling: a multiplexing hub in programmed cell death. Genes Cancer 8: 682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G., Shrestha O. K., Ponce J. M., Zwerger M., Thiemann D. A., et al. , 2015. Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet. 11: e1005231 10.1371/journal.pgen.1005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Josue F., Suijkerbuijk S., Baum B., Tapon N., et al. , 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6: e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C., Jouanguy E., Irving P., Troxler L., Galiana-Arnoux D., et al. , 2005. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 6: 946–953. 10.1038/ni1237 [DOI] [PubMed] [Google Scholar]
- Dunbar T. L., Yan Z., Balla K. M., Smelkinson M. G., Troemel E. R., 2012. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11: 375–386. 10.1016/j.chom.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Bergmann A., 2008. Apoptosis-induced compensatory proliferation. The cell is dead. Long live the cell! Trends Cell Biol. 18: 467–473. 10.1016/j.tcb.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L. C., Green J., Surridge A., Jiggins C. D., 2011. Evolution of the insect yellow gene family. Mol. Biol. Evol. 28: 257–272. 10.1093/molbev/msq192 [DOI] [PubMed] [Google Scholar]
- Feyereisen R., 2005. Insect cytochrome, pp. 1–77 in Comprehensive Molecular Insect Science: Biochemistry and Molecular Biology, Vol. 4, edited by Gilbert L. I., Iatrou K., Gill S. S. Elsevier, Oxford. [Google Scholar]
- Fogarty C. E., Diwanji N., Lindblad J. L., Tare M., Amcheslavsky A., et al. , 2016. Extracellular reactive oxygen species drive apoptosis-induced proliferation via Drosophila macrophages. Curr. Biol. 26: 575–584. 10.1016/j.cub.2015.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Wu X., Simon L., Fossett N., 2014. Antioxidants maintain E-cadherin levels to limit Drosophila prohemocyte differentiation. PLoS One 9: e107768 10.1371/journal.pone.0107768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garelli A., Gontijo A. M., Miguela V., Caparros E., Dominguez M., 2012. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336: 579–582. 10.1126/science.1216735 [DOI] [PubMed] [Google Scholar]
- Gauron C., Rampon C., Bouzaffour M., Ipendey E., Teillon J., et al. , 2013. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci. Rep. 3: 2084 10.1038/srep02084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan J. A., Jayamani E., Zhang X., Breen P., Larkins-Ford J., et al. , 2015. Lipid signalling couples translational surveillance to systemic detoxification in Caenorhabditis elegans. Nat. Cell Biol. 17: 1294–1303. 10.1038/ncb3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E. R., Stuurman N., Vale R. D., 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177: 1005–1015. 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenewald C., Botella J. A., Bayersdorfer F., Navarro J. A., Schneuwly S., 2009. Hyperoxia-induced neurodegeneration as a tool to identify neuroprotective genes in Drosophila melanogaster. Free Radic. Biol. Med. 46: 1668–1676. 10.1016/j.freeradbiomed.2009.03.025 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A., 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hara N., Yamada K., Shibata T., Osago H., Hashimoto T., et al. , 2007. Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J. Biol. Chem. 282: 24574–24582. 10.1074/jbc.M610357200 [DOI] [PubMed] [Google Scholar]
- Hausheer F. H., Parker A. R., Petluru P. N., Jair K. W., Chen S., et al. , 2011. Mechanistic study of BNP7787-mediated cisplatin nephroprotection: modulation of human aminopeptidase N. Cancer Chemother. Pharmacol. 67: 381–391. 10.1007/s00280-010-1333-x [DOI] [PubMed] [Google Scholar]
- Hayakawa J., Mittal S., Wang Y., Korkmaz K. S., Adamson E., et al. , 2004. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell 16: 521–535. 10.1016/j.molcel.2004.10.024 [DOI] [PubMed] [Google Scholar]
- Haynie J. L., Bryant P. J., 1977. The effects of X-rays on the proliferation dynamics of cells in the imaginal wing disc ofDrosophila melanogaster. Wilehm Roux Arch Dev Biol 183: 85–100. 10.1007/BF00848779 [DOI] [PubMed] [Google Scholar]
- Hayward D., Metz J., Pellacani C., Wakefield J. G., 2014. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell 28: 81–93. 10.1016/j.devcel.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegan P. S., Mermall V., Tilney L. G., Mooseker M. S., 2007. Roles for Drosophila melanogaster myosin IB in maintenance of enterocyte brush-border structure and resistance to the bacterial pathogen Pseudomonas entomophila. Mol. Biol. Cell 18: 4625–4636. 10.1091/mbc.e07-02-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann U., Kroemer H. K., 2004. The ABC transporters MDR1 and MRP2: multiple functions in disposition of xenobiotics and drug resistance. Drug Metab. Rev. 36: 669–701. 10.1081/DMR-200033473 [DOI] [PubMed] [Google Scholar]
- Homsy J. G., Jasper H., Peralta X. G., Wu H., Kiehart D. P., et al. , 2006. JNK signaling coordinates integrin and actin functions during Drosophila embryogenesis. Dev. Dyn. 235: 427–434. 10.1002/dvdy.20649 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hull-Thompson J., Muffat J., Sanchez D., Walker D. W., Benzer S., et al. , 2009. Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. PLoS Genet. 5: e1000460 10.1371/journal.pgen.1000460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huu N. T., Yoshida H., Yamaguchi M., 2015. Tumor suppressor gene OSCP1/NOR1 regulates apoptosis, proliferation, differentiation, and ROS generation during eye development of Drosophila melanogaster. FEBS J. 282: 4727–4746. 10.1111/febs.13528 [DOI] [PubMed] [Google Scholar]
- Igaki T., 2009. Correcting developmental errors by apoptosis: lessons from Drosophila JNK signaling. Apoptosis 14: 1021–1028. 10.1007/s10495-009-0361-7 [DOI] [PubMed] [Google Scholar]
- Indo H. P., Hawkins C. L., Nakanishi I., Matsumoto K. I., Matsui H., et al. , 2017. Role of mitochondrial reactive oxygen species in the activation of cellular signals, molecules, and function. Handb. Exp. Pharmacol. 240: 439–456. 10.1007/164_2016_117 [DOI] [PubMed] [Google Scholar]
- Ishimaru S., Ueda R., Hinohara Y., Ohtani M., Hanafusa H., 2004. PVR plays a critical role via JNK activation in thorax closure during Drosophila metamorphosis. EMBO J. 23: 3984–3994. 10.1038/sj.emboj.7600417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James M. O., Ambadapadi S., 2013. Interactions of cytosolic sulfotransferases with xenobiotics. Drug Metab. Rev. 45: 401–414. 10.3109/03602532.2013.835613 [DOI] [PubMed] [Google Scholar]
- Jasper H., Benes V., Schwager C., Sauer S., Clauder-Munster S., et al. , 2001. The genomic response of the Drosophila embryo to JNK signaling. Dev. Cell 1: 579–586. 10.1016/S1534-5807(01)00045-4 [DOI] [PubMed] [Google Scholar]
- Kanda H., Igaki T., Okano H., Miura M., 2011. Conserved metabolic energy production pathways govern Eiger/TNF-induced nonapoptotic cell death. Proc. Natl. Acad. Sci. USA 108: 18977–18982. 10.1073/pnas.1103242108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama T., Comoglio F., Seimiya M., Cabuy E., Paro R., 2015. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc. Natl. Acad. Sci. USA 112: E2327–E2336. 10.1073/pnas.1423074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. J., Abidi S. N. F., Skinner A., Tian Y., Smith-Bolton R. K., 2017. The Drosophila Duox maturation factor is a key component of a positive feedback loop that sustains regeneration signaling. PLoS Genet. 13: e1006937 10.1371/journal.pgen.1006937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B. F., Mogensen M. M., et al. , 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19: 1012–1018. 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Senoo-Matsuda N., Hiromi Y., Miura M., 2006. DRONC coordinates cell death and compensatory proliferation. Mol. Cell. Biol. 26: 7258–7268. 10.1128/MCB.00183-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerová L., Kubrak O. I., Bengtsson J. M., Strnad H., Nylin S., et al. , 2016. Slowed aging during reproductive dormancy is reflected in genome-wide transcriptome changes in Drosophila melanogaster. BMC Genomics 17: 50 10.1186/s12864-016-2383-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucinski I., Dinan M., Kolahgar G., Piddini E., 2017. Chronic activation of JNK JAK/STAT and oxidative stress signalling causes the loser cell status. Nat. Commun. 8: 136 10.1038/s41467-017-00145-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu T. K., Velayutham M., Zweier J. L., 2012. Aldehyde oxidase functions as a superoxide generating NADH oxidase: an important redox regulated pathway of cellular oxygen radical formation. Biochemistry 51: 2930–2939. 10.1021/bi3000879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fortezza M., Schenk M., Cosolo A., Kolybaba A., Grass I., et al. , 2016. JAK/STAT signalling mediates cell survival in response to tissue stress. Development 143: 2907–2919. 10.1242/dev.132340 [DOI] [PubMed] [Google Scholar]
- Lee W. J., Miura M., 2014. Mechanisms of systemic wound response in Drosophila. Curr. Top. Dev. Biol. 108: 153–183. 10.1016/B978-0-12-391498-9.00001-2 [DOI] [PubMed] [Google Scholar]
- Legan S. K., Rebrin I., Mockett R. J., Radyuk S. N., Klichko V. I., et al. , 2008. Overexpression of glucose-6-phosphate dehydrogenase extends the life span of Drosophila melanogaster. J. Biol. Chem. 283: 32492–32499. 10.1074/jbc.M805832200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit D. A., Poulton J. S., 2016. Centrosomes are multifunctional regulators of genome stability. Chromosome Res. 24: 5–17. 10.1007/s10577-015-9506-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit D. A., Jordan H. A., Poulton J. S., Fagerstrom C. J., Galletta B. J., et al. , 2015. Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J. Cell Biol. 210: 79–97. 10.1083/jcb.201503117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J., Nelson R., Lehner R., 2018. Carboxylesterases in lipid metabolism: from mouse to human. Protein Cell 9: 178–195. 10.1007/s13238-017-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A., Damulewicz M., Pyza E., Jozkowicz A., Dulak J., 2016. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell. Mol. Life Sci. 73: 3221–3247. 10.1007/s00018-016-2223-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E., Resende T., Torres C., Bousbaa H., 2008. The human spindle assembly checkpoint protein Bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol. Biol. Cell 19: 1798–1813. 10.1091/mbc.e07-07-0633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. C., 2013. Glutathione synthesis. Biochim. Biophys. Acta 1830: 3143–3153. 10.1016/j.bbagen.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. A., Perez-Garijo A., Morata G., 2009. Apoptosis in Drosophila: compensatory proliferation and undead cells. Int. J. Dev. Biol. 53: 1341–1347. 10.1387/ijdb.072447fm [DOI] [PubMed] [Google Scholar]
- Martindale J. L., Holbrook N. J., 2002. Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol. 192: 1–15. 10.1002/jcp.10119 [DOI] [PubMed] [Google Scholar]
- Massudi H., Grant R., Braidy N., Guest J., Farnsworth B., et al. , 2012. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One 7: e42357 10.1371/journal.pone.0042357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D. L., Kirienko N. V., Ausubel F. M., 2012. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11: 364–374. 10.1016/j.chom.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. J., Lieb J. D., 2013. A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27: 306–318. 10.1016/j.devcel.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeay R. C., Bailey T. L., 2010. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11: 165 10.1186/1471-2105-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meekins D. A., Kanost M. R., Michel K., 2017. Serpins in arthropod biology. Semin. Cell Dev. Biol. 62: 105–119. 10.1016/j.semcdb.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A., Ruvkun G., 2012. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149: 452–466. 10.1016/j.cell.2012.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K. L., Chhun B., Kan F., et al. , 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14: 1159–1168. 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R., Horner M. A., Lam G., Thummel C. S., 2011. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25: 1796–1806. 10.1101/gad.17280911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Siebenthall K. T., Clark A. G., 2006. Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J. Exp. Biol. 209: 3837–3850. 10.1242/jeb.02448 [DOI] [PubMed] [Google Scholar]
- Morais da Silva S., Moutinho-Santos T., Sunkel C. E., 2013. A tumor suppressor role of the Bub3 spindle checkpoint protein after apoptosis inhibition. J. Cell Biol. 201: 385–393. 10.1083/jcb.201210018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B., 2010. Recent advances in understanding mechanisms of insect cuticle differentiation. Insect Biochem. Mol. Biol. 40: 363–375. 10.1016/j.ibmb.2010.03.003 [DOI] [PubMed] [Google Scholar]
- Moutinho-Pereira S., Stuurman N., Afonso O., Hornsveld M., Aguiar P., et al. , 2013. Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc. Natl. Acad. Sci. USA 110: 19808–19813. 10.1073/pnas.1320013110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25: R1002–R1018 (erratum: Curr. Biol. 25: 3017) 10.1016/j.cub.2015.08.051 [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Meyer E. J., Kroesen A., McKinney S. A., Gibson M. C., 2013. Epithelial junctions maintain tissue architecture by directing planar spindle orientation. Nature 500: 359–362. 10.1038/nature12335 [DOI] [PubMed] [Google Scholar]
- Nicholson J. M., Cimini D., 2011. How mitotic errors contribute to karyotypic diversity in cancer. Adv. Cancer Res. 112: 43–75. 10.1016/B978-0-12-387688-1.00003-X [DOI] [PubMed] [Google Scholar]
- Novak Z. A., Conduit P. T., Wainman A., Raff J. W., 2014. Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos. Curr. Biol. 24: 1276–1282. 10.1016/j.cub.2014.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H., Suzuki Y., Shibata T., Yoshida A., 1999. Glucocorticoid-dependent induction of HMG-CoA reductase and malic enzyme gene expression by polychlorinated biphenyls in rat hepatocytes. J. Nutr. Biochem. 10: 644–653. 10.1016/S0955-2863(99)00055-8 [DOI] [PubMed] [Google Scholar]
- Ohsawa S., Sato Y., Enomoto M., Nakamura M., Betsumiya A., et al. , 2012. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature 490: 547–551. 10.1038/nature11452 [DOI] [PubMed] [Google Scholar]
- O’Keefe L. V., Lee C. S., Choo A., Richards R. I., 2015. Tumor suppressor WWOX contributes to the elimination of tumorigenic cells in Drosophila melanogaster. PLoS One 10: e0136356 10.1371/journal.pone.0136356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke D., Baban D., Demidova M., Mott R., Hodgkin J., 2006. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16: 1005–1016. 10.1101/gr.50823006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Yavari A., Mandal S., Banerjee U., 2008. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 40: 356–361. 10.1038/ng.2007.50 [DOI] [PubMed] [Google Scholar]
- Pandey A. V., Fluck C. E., 2013. NADPH P450 oxidoreductase: structure, function, and pathology of diseases. Pharmacol. Ther. 138: 229–254. 10.1016/j.pharmthera.2013.01.010 [DOI] [PubMed] [Google Scholar]
- Pastor-Pareja J. C., Wu M., Xu T., 2008. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis. Model. Mech. 1: 144–154, discussion 153 10.1242/dmm.000950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson R. A., Juarez M. T., Hermann A., Sasik R., Hardiman G., et al. , 2013. Serine proteolytic pathway activation reveals an expanded ensemble of wound response genes in Drosophila. PLoS One 8: e61773 10.1371/journal.pone.0061773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz G., Bakhrat A., Abdu U., 2007. Expression of the Drosophila melanogaster GADD45 homolog (CG11086) affects egg asymmetric development that is mediated by the c-Jun N-terminal kinase pathway. Genetics 177: 1691–1702. 10.1534/genetics.107.079517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez E., Lindblad J. L., Bergmann A., 2017. Tumor-promoting function of apoptotic caspases by an amplification loop involving ROS, macrophages and JNK in Drosophila. Elife 6: e26747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Garijo A., Shlevkov E., Morata G., 2009. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 136: 1169–1177. 10.1242/dev.034017 [DOI] [PubMed] [Google Scholar]
- Picco V., Pages G., 2013. Linking JNK activity to the DNA damage response. Genes Cancer 4: 360–368. 10.1177/1947601913486347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J. S., Cuningham J. C., Peifer M., 2014. Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation. Dev. Cell 30: 731–745. 10.1016/j.devcel.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J. S., Cuningham J. C., Peifer M., 2017. Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size. J. Cell Biol. 216: 1255–1265. 10.1083/jcb.201607022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser S. L., Pelletier L., 2017. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18: 187–201. 10.1038/nrm.2016.162 [DOI] [PubMed] [Google Scholar]
- Redza-Dutordoir M., Averill-Bates D. A., 2016. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta 1863: 2977–2992. 10.1016/j.bbamcr.2016.09.012 [DOI] [PubMed] [Google Scholar]
- Richens J. H., Barros T. P., Lucas E. P., Peel N., Pinto D. M., et al. , 2015. The Drosophila Pericentrin-like-protein (PLP) cooperates with Cnn to maintain the integrity of the outer PCM. Biol. Open 4: 1052–1061. 10.1242/bio.012914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Barrera L. D., Riesgo-Escovar J. R., 2013. Regulating cell morphogenesis: the Drosophila Jun N-terminal kinase pathway. Genesis 51: 147–162. 10.1002/dvg.22354 [DOI] [PubMed] [Google Scholar]
- Roberts D. M., Pronobis M. I., Alexandre K. M., Rogers G. C., Poulton J. S., et al. , 2012. Defining components of the ss-catenin destruction complex and exploring its regulation and mechanisms of action during development. PLoS One 7: e31284 10.1371/journal.pone.0031284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset R., Bono-Lauriol S., Gettings M., Suzanne M., Speder P., et al. , 2010. The Drosophila serine protease homologue Scarface regulates JNK signalling in a negative-feedback loop during epithelial morphogenesis. Development 137: 2177–2186. 10.1242/dev.050781 [DOI] [PubMed] [Google Scholar]
- Ryoo H. D., Gorenc T., Steller H., 2004. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell 7: 491–501. 10.1016/j.devcel.2004.08.019 [DOI] [PubMed] [Google Scholar]
- Santabárbara-Ruiz P., López-Santillán M., Martínez-Rodríguez I., Binagui-Casas A., Pérez L., et al. , 2015. ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet. 11: e1005595 10.1371/journal.pgen.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S. B., et al. , 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19: 1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Seeds A. M., Tsui M. M., Sunu C., Spana E. P., York J. D., 2015. Inositol phosphate kinase 2 is required for imaginal disc development in Drosophila. Proc. Natl. Acad. Sci. USA 112: 15660–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger K., Harris K., Levine M., 2006. GATA factors participate in tissue-specific immune responses in Drosophila larvae. Proc. Natl. Acad. Sci. USA 103: 15957–15962. 10.1073/pnas.0607608103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H., Berndt C., Jones D. P., 2017. Oxidative stress. Annu. Rev. Biochem. 86: 715–748. 10.1146/annurev-biochem-061516-045037 [DOI] [PubMed] [Google Scholar]
- Silverman N., Zhou R., Erlich R. L., Hunter M., Bernstein E., et al. , 2003. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem. 278: 48928–48934. 10.1074/jbc.M304802200 [DOI] [PubMed] [Google Scholar]
- Stanton R. C., 2012. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64: 362–369. 10.1002/iub.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N. R., Raposo A. A., Basto R., St Johnston D., Raff J. W., 2007. From stem cell to embryo without centrioles. Curr. Biol. 17: 1498–1503. 10.1016/j.cub.2007.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis G. P., Bohmann D., 2008. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 14: 76–85. 10.1016/j.devcel.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., 2009. Regulation and function of the melanization reaction in Drosophila. Fly (Austin) 3: 105–111. 10.4161/fly.3.1.7747 [DOI] [PubMed] [Google Scholar]
- Tang H. W., Liao H. M., Peng W. H., Lin H. R., Chen C. H., et al. , 2013. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev. Cell 27: 489–503. 10.1016/j.devcel.2013.10.017 [DOI] [PubMed] [Google Scholar]
- Teesalu M., Rovenko B. M., Hietakangas V., 2017. Salt-inducible kinase 3 provides sugar tolerance by regulating NADPH/NADP(+) redox balance. Curr. Biol. 27: 458–464. 10.1016/j.cub.2016.12.032 [DOI] [PubMed] [Google Scholar]
- Telci D., Griffin M., 2006. Tissue transglutaminase (TG2)–a wound response enzyme. Front. Biosci. 11: 867–882. 10.2741/1843 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [corrigenda: Nat. Protoc. 9: 2513 (2014)] 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlirova M., Bohmann D., 2006. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 25: 5294–5304. 10.1038/sj.emboj.7601401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyehara C. M., Nystrom S. L., Niederhuber M. J., Leatham-Jensen M., Ma Y., et al. , 2017. Hormone-dependent control of developmental timing through regulation of chromatin accessibility. Genes Dev. 31: 862–875. 10.1101/gad.298182.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelzmann A., Hahn I., Pearce S. P., Sanchez-Soriano N., Prokop A., 2016. A conceptual view at microtubule plus end dynamics in neuronal axons. Brain Res. Bull. 126: 226–237. 10.1016/j.brainresbull.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C. E., Heald R., 2008. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 265: 111–158. 10.1016/S0074-7696(07)65003-7 [DOI] [PubMed] [Google Scholar]
- Wang M. C., Bohmann D., Jasper H., 2003. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell 5: 811–816. 10.1016/S1534-5807(03)00323-X [DOI] [PubMed] [Google Scholar]
- Xie H. B., Cammarato A., Rajasekaran N. S., Zhang H., Suggs J. A., et al. , 2013. The NADPH metabolic network regulates human alphaB-crystallin cardiomyopathy and reductive stress in Drosophila melanogaster. PLoS Genet. 9: e1003544 10.1371/journal.pgen.1003544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Hultmark D., 2016. Tissue communication in a systemic immune response of Drosophila. Fly (Austin) 10: 115–122. 10.1080/19336934.2016.1182269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Xie Q., Tibbetts R. S., 2015. Opposing roles of p38 and JNK in a Drosophila model of TDP-43 proteinopathy reveal oxidative stress and innate immunity as pathogenic components of neurodegeneration. Hum. Mol. Genet. 24: 757–772. 10.1093/hmg/ddu493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Drosophila stocks are available upon request from J. S. Poulton. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and supplemental figures and tables. Supplemental material available at https://doi.org/10.25386/genetics.7837943.