Abstract
Levels of variability and rates of adaptive evolution may be affected by hitchhiking, the effect of selection on evolution at linked sites. Hitchhiking can be caused either by “selective sweeps” or by background selection, involving the spread of new favorable alleles or the elimination of deleterious mutations, respectively. Recent analyses of population genomic data have fitted models where both these processes act simultaneously, to infer the parameters of selection. Here, we investigate the consequences of relaxing a key assumption of some of these studies, that the time occupied by a selective sweep is negligible compared with the neutral coalescent time. We derive a new expression for the expected level of neutral variability in the presence of recurrent selective sweeps and background selection. We also derive approximate integral expressions for the effects of recurrent selective sweeps. The accuracy of the theoretical predictions was tested against multilocus simulations, with selection, recombination, and mutation parameters that are realistic for Drosophila melanogaster. In the presence of crossing over, there is approximate agreement between the theoretical and simulation results. We show that the observed relationships between the rate of crossing over, and the level of synonymous site diversity and rate of adaptive evolution in Drosophila are probably mainly caused by background selection, whereas selective sweeps and population size changes are needed to produce the observed distortions of the site frequency spectrum.
Keywords: selective sweeps, background selection, gene conversion, crossing over, neutral variability, favorable mutations, Drosophila melanogaster
THE effect of selection at a given locus on the properties of neutral variability at linked sites is a classic problem in population genetics, first studied by Sved (1968) and Ohta and Kimura (1970) in the context of associative overdominance, the apparent heterozygote advantage induced at a neutral locus by variants at linked loci that are maintained by heterozygote advantage or by mutation to partially recessive deleterious alleles. This work was followed by the paper of Maynard Smith and Haigh (1974) on the hitchhiking effect, where the spread of a favorable mutation reduces the level of neutral variability at a linked locus; this process has come to be termed a selective sweep (SSW) (Berry et al. 1991). It was later shown that selection against recurrent deleterious mutations also reduces neutral variability at linked sites by the hitchhiking process known as background selection (BGS) (Charlesworth et al. 1993). A large amount of theoretical literature on these topics has subsequently appeared, as reviewed by Barton (2010), Stephan (2010), Charlesworth (2012a), Neher (2013), and Walsh and Lynch (2018, chapter 8).
Much of the motivation for these theoretical studies came from the advent of data on genome-wide patterns of variability, which inspired attempts to infer the nature and parameters of selection from observations such as the relationships between the level of synonymous sequence diversity in a gene and the local recombination rate (Begun and Aquadro 1992), and between diversity and nonsynonymous (NS) divergence from a related species (Andolfatto 2007). Work of this type has been reviewed by Sella et al. (2009), Vitti et al. (2013), Booker et al. (2017), and Hermisson and Pennings (2017). Several recent studies have used the theory of the joint effects of recurrent SSWs and BGS—pioneered by Kaplan et al. (1989), Wiehe and Stephan (1993), and Kim and Stephan (2000)—to estimate their effects on levels of neutral diversity across the genomes of multiple species (Corbett-Detig et al. 2015), and to infer the rates of occurrence of advantageous mutations and the strength of selection acting on them (Elyashiv et al. 2016; Campos et al. 2017). These studies all concluded that the level of variability in a species is often much smaller than would be expected in the absence of selection, even in regions with relatively high rates of genetic recombination. This reduction in variability reflects the effects of both SSWs and BGS, although the estimates of the parameters involved differ substantially among the different studies.
Several important assumptions underlie the model of recurrent sweeps used in this work. One is that the effect of BGS on the probability of fixation of a linked favorable mutation is well approximated by its effect on neutral variability at a site at the same location in the genome, which is described by a factor B that multiplies the value of Ne for that site (Kim and Stephan 2000). An expression for B can be found from the standard equation for the effect of BGS in the presence of recombination (Hudson and Kaplan 1995; Nordborg et al. 1996), although this equation breaks down when the product of Ne and the selection coefficient against deleterious mutations is of the order of ≤ 1, especially when there is little or no recombination (Gordo et al. 2002; Kaiser and Charlesworth 2009; Good et al. 2014; Zhao and Charlesworth 2016). Using the formula of Kimura (1964) for an autosomal, semidominant mutation with selective advantage sA in homozygotes, the probability of fixation of a mutation with NesA >> 1 in a randomly mating, discrete-generation population of size N is then BNesA/N instead of NesA/N (Peck 1994; Barton 1995; Stephan et al. 1999; Kim and Stephan 2000).
In addition, it is usually assumed that the time occupied by an adaptive substitution is negligible compared with the coalescent time, and that Hill–Robertson interference (Hill and Robertson 1966; Felsenstein 1974) among sweeps is absent, so that the times between successive sweeps are exponentially distributed and reductions in diversity can be predicted from the formula for a single sweep. Finally, the classic theory assumes that sweeps are “hard,” such that each favorable mutation originates as a single copy in the population, as opposed to “soft” sweeps that arise from standing variation or from several independent mutational events in the same gene (Hermisson and Pennings 2005, 2017).
All of these assumptions can be questioned. The main purpose of this paper is to examine the accuracy of the assumptions concerning the effects of BGS, sweep duration, and interference among sweeps, in the context of parameter values for BGS and SSWs that appear to be fairly realistic on the basis of inferences from a Drosophila melanogaster population (Campos et al. 2017). We chose to model D. melanogaster because this species has been the basis for much of the work on the effects of hitchhiking on natural variability. We used computer simulations of multiple loci that are subject to both BGS and SSWs, together with approximations for the effects of BGS and SSWs based on replacing summations across selected sites with integration. The results indicate that the standard coalescent approach to predicting recurrent sweep effects can underestimate their magnitude. We found only small effects of interference among sweeps, so this discrepancy appears to be caused by neglecting sweep duration. To deal with this problem, we have modified the standard approach for the prediction of pairwise neutral nucleotide diversity under recurrent SSWs. We consider only hard sweeps, because these are amenable to simple analytic modeling and simulation.
Materials and Methods
We used the simulation package SLiM (Messer 2013), version 1.8. The details of the simulation methods are described in the online manual (benhaller.com/slim/SLiM.18_manual.pdf). We modeled sets of n genes separated by 2 kb of selectively neutral intergenic sequence (Figure 1), with all untranslated region (UTR) sites and 70% of NS sites subject to selection (the same selection parameters were applied to 5′and 3′ UTRs). The gene structure was chosen to represent a typical D. melanogaster gene (Campos et al. 2017). To simulate realistic parameters of selection, mutation, and recombination for a model autosome, we rescaled the values applicable to a natural population of D. melanogaster by multiplying them by the ratio of Ne for the population to the number of breeding individuals used in the simulations, N, which was usually set to 2500 (see Table 1). This conserves the products of Ne and the basic parameters of selection, recombination, and mutation, which control most aspects of evolution in finite populations if time is rescaled by a factor of N/Ne (Ewens 2004).
Figure 1.
The gene model used in the simulations.
Table 1. Parameters used in the simulations.
| Natural population | Simulations | |||
|---|---|---|---|---|
| Parameter | A | X | A | X |
| Population size (N) | 1.33 × 106 | 0.997 × 106 | 2500 | 2500 |
| Rescaling factor | — | — | 532 | 532 |
| Standard effective crossover rate | 1 × 10−8 | 1.33 × 10−8 | 5.32 × 10−6 | 5.32 × 10−6 |
| G.c. rate of initiation | 1 × 10−8 | 1.33 × 10−8 | 5.32 × 10−6 | 5.32 × 10−6 |
| G.c. tract length | 440 bp | 440 bp | 440 bp | 440 bp |
| Mutation rate per bp | 4.5 × 10−9 | 4.5 × 10−9 | 2.39 × 10−6 | 1.79 × 10−6 |
| Mean scaled selection coefficient for deleterious NS mutations (γNS) | 2,000 | 2000 | 2,000 | 2,000 |
| Mean scaled selection coefficient for deleterious UTR mutations (γUTR) | 110 | 110 | 110 | 110 |
| Scaled selection coefficient for favorable NS mutations (γa) | 250 | 375 or 500 | 250 | 375 or 500 |
| Scaled selection coefficient for favorable UTR mutations (γu) | 213 | 319.5 or 426 | 213 | 319.5 or 426 |
| Proportion of NS mutations that are favorable (pa) | 2.21 × 10−4 | 2.21 × 10−4 | 2.21 × 10−4 | 2.21 × 10−4 |
| Proportion of UTR mutations that are favorable (pu) | 9.04 × 10−4 | 9.04 × 10−4 | 9.04 × 10−4 | 9.04 × 10−4 |
| Proportion of neutral exonic mutations | 0.3 | 0.3 | 0.3 | 0.3 |
| Shape parameter of γ distribution | 0.3 | 0.3 | 0.3 | 0.3 |
| Dominance coefficient | 0.5 | 0.5 | 0.5 | 0.5 |
G.c., gene conversion; NS, nonsynonymous.
We chose an Ne/N ratio of 532, equivalent to an Ne of 1.33 million for the natural population. This value was based on the mean autosomal synonymous site diversity value of π = 0.018 for an African population and a mutation rate of μ = 4.5 × 10−9 (Campos et al. 2017), using the standard equilibrium formula π = 4Neμ for neutral variability under the infinite sites model (Kimura 1971), and assuming (rather conservatively) that mean diversity has been reduced by hitchhiking effects to 76% of its value in the absence of selection. The selection coefficients for favorable mutations, and the proportions of mutations that are favorable, were chosen to match mean values inferred from the relationship between the synonymous diversity of a gene and its rate of protein sequence evolution by Campos et al. (2017). The details of the selection parameters used here are described in Table 1. Both favorable and deleterious mutations were assumed to be semidominant.
To model recombination, we mostly used five rates of reciprocal crossing over, which were multiples of the standard autosomal recombination rate in Drosophila, adjusted by a factor of one-half to take into account the absence of recombinational exchange in males (Campos et al. 2017). These “effective rates of crossing over” span most of the observed range, and were 0.5 × 10−8, 1 × 10−8, 1.5 × 10−8, 2 × 10−8, and 2.5 × 10−8 cM/Mb, where 1 × 10−8 is the standard rate. We also ran simulations with no crossing over. The simulations were run with and without noncrossover-associated gene conversion events, using a rate of initiation of conversion events of 1 × 10−8 cM/Mb for autosomes (after correcting for the lack of gene conversion in males) and a tract length of 440 bp. Given that SLiM models gene conversion by considering only the effects of conversion events that were initiated on one side of a given nucleotide site, this rate of initiation is one-half of the values estimated from the experiments of Hilliker et al. (1994) and Miller et al. (2016), thus providing a conservative estimate of the effect of gene conversion. We did not vary the rate of initiation of gene conversion when using different rates of crossing over, since this rate appears to be fairly constant across the Drosophila genome, even in regions that lack crossing over (Langley et al. 2000; Comeron et al. 2012; Miller et al. 2016).
In addition to the simulations of autosomes, we ran simulations that were intended to represent X chromosomal mutations with equal fitness effects in the two sexes but with stronger selection than for autosomal mutations, as expected on both theoretical and empirical grounds (Charlesworth et al. 2018). X-linked loci spend two-thirds of their time in females where they can recombine, so that the effective rates of crossing over and initiation of gene conversion events for X-linked loci should be 4/3 times the autosomal values for X-linked genes that have similar parameter values in females to the autosomal ones (Campos et al. 2013). The version of SLiM that we used did not permit explicit modeling of an X chromosome. We therefore used an autosomal model with a population size of 2500, but assumed that the true Ne was three-quarters of that for the autosomes. Because N was kept constant, the autosomal rates of crossing over and initiation of gene conversion events were used in the simulations. To ensure that X-linked neutral variability in the absence of selection was three-quarters of the autosomal value, the mutation rate was multiplied by 3/4. Finally, with semidominance, and equal fitness effects of mutations in males and females, the selection coefficient for an X-linked mutation is 4/3 times that for an autosomal mutation with the same selection coefficient, implying that the scaled selection coefficients are the same. To mimic stronger selection for positively selected mutations on the X chromosome, we therefore simply multiplied the scaled selection coefficients by a given factor, either 1.5 or 2. No adjustment was made to the scaled selection coefficient for deleterious mutations.
According to the number of genes simulated, we ran four sets of simulations with genomic regions of 20 (87.4 kb), 70 (305.9 kb), 140 (610 kb), and 210 (920 kb) genes. Most of our simulations used multiples of 70 genes because this represents a genomic region with a similar number of genes to the fourth chromosome of D. melanogaster, which the simulations with zero crossing over are intended to model. Each simulation was run for 35,000 (14N) generations, which is sufficient to allow the frequency distributions of neutral and deleterious mutations to reach equilibrium (see the online Supplemental Material, File S1 and Figure S1). For the final estimates of diversity statistics (mean values of nucleotide site diversity, Tajima’s D, and the proportions of singletons at synonymous, NS, intron, and UTR sites) we used data from the final generation of each simulation. To calculate the numbers of fixations of favorable mutations at NS and UTR sites, we recorded the fixations that occurred during the last 20,000 (8N) generations. In most cases, 20 replicate simulations were run for each parameter set, but a number of cases used 9 or 10 replicates.
Four different scenarios were simulated. First, purely neutral mutations were simulated to calculate the diversity statistics for the neutral reference. Three types of scenario with hitchhiking were simulated: (i) SSWs only, (ii) BGS only, and (iii) both SSWs and BGS. Sample sizes of 20 haploid genomes [a similar size to that used by Campos et al. (2017)] were used to calculate the population genetic statistics. Mean values of each statistic over genes and replicate runs for a given model were recorded, with upper and lower 2.5 percentiles obtained by bootstrapping the mean values per gene of the chosen statistic across replicates (for brevity, we will refer to these as 95% C.I.s). The statistics generated by the simulations are presented in the online Files S2 and S3.
No new data or reagents were generated by this research. Details of the mathematical derivations are described in File S1. The detailed statistics for the results of the computer simulations are provided in Files S2 and S3. The codes for the computer programs used in the models described below are available in File S4.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7604948.
Results
BGS
The predicted effect of BGS in a multisite context can be described by the quantity B = exp(–E), where B is the ratio of expected neutral diversity at a focal neutral site under BGS to its value in the absence of BGS (which is equivalent to the corresponding ratio of mean coalescence times), and E is the sum of the effects of each selected site (Hudson and Kaplan 1995; Nordborg et al. 1996; Santiago and Caballero 1998). We assume a genomic region containing many genes, with selected sites that are continuously distributed with constant density, as in Model 3 of Charlesworth (2012b). We distinguish between NS sites and UTRs. This is, of course, a somewhat crude approximation, given that our genic model includes neutrally evolving intronic and intergenic sequences. For simplicity, we describe the case of autosomal inheritance, but parallel results hold for X-linked loci, with the appropriate changes in selection, mutation, and recombination parameters.
We model both reciprocal exchange via crossing over and noncrossover-associated gene conversion. We assume that the main contribution from gene conversion comes from sites that are sufficiently distant that gene conversion causes recombination between them at a fixed rate g = rgdg (rg is the rate of initiation of gene conversion events in females and dg is the mean tract length). This is the limiting value of the general expression for the rate of recombination due to gene conversion for sites separated by z base pairs, g[1 – exp (– z/dg)] (Langley et al. 2000; Frisse et al. 2001), after correcting for the lack of gene conversion in male meiosis.
Because SLiM assumes no crossover interference, the relationship between the frequency of crossing over and map distance in the simulations follows the Haldane mapping function (Haldane 1919), such that the frequency of crossing over between a pair of sites separated by z base pairs is given by:
| (1) |
where rc is the rate of crossing over per base pair.
The net frequency of recombination between the sites is r(z) = g + c(z). The predicted value of E for a given selection coefficient, t = hs, against heterozygous carriers of a deleterious mutation, Et, is given by Equations S1–S5 in section S1 of File S1. To obtain the final value of E, this equation is numerically integrated over the probability distribution of t values for NS and UTR sites separately, with total deleterious mutation rates UN and UU for NS and UTR sites, respectively, giving values EN and EU for the corresponding BGS effects.
To mimic the simulation results, we assume a γ distribution with a shape parameter of 0.3. As in previous studies, we ignore all deleterious mutations with a scaled selection coefficient γ = 2Nes below a critical value γc, to deal with the problem that very weakly selected mutations are subject to drift and contribute little to BGS effects (Nordborg et al. 1996). Following Nordborg et al. (1996) and Campos et al. (2017), we set γc = 5, and the γ distributions for both NS and UTR mutations were truncated accordingly. Numerical results for the integral of the kernel of the γ distribution from γc to infinity allow the proportion of mutations that exceed γc to be calculated; these are denoted by PN and PU for NS and UTR sites, respectively. With the parameters used in the simulations of autosomes, this gives PN = 0.871 and PU = 0.694. The final value for E is PNEN + PUEU, from which B can be obtained as exp(– E).
SSWs
Various methods have been used to predict the approximate effect of a single SSW on diversity statistics at a partially linked neutral site in a randomly mating population, as well as for the associated distortion of the neutral site frequency spectrum at segregating sites (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Stephan et al. 1992; Barton 1998, 2000; Gillespie 2000, 2001; Durrett and Schweinsberg 2004; Kim 2006; Pfaffelhuber et al. 2006; Coop and Ralph 2012; Bossert and Pfaffelhuber 2013). Here, we present a simple heuristic derivation of the effect of a sweep on the pairwise neutral nucleotide site diversity, π, based on a combination of coalescent process and diffusion equation approaches. Following earlier approaches, we obtain the probability that a neutral lineage associated with a favorable allele at the end of a sweep was also associated with it at the start of the sweep, rather with the wild-type allele at the selected locus.
We consider separately the deterministic and stochastic phases of the spread of a favorable mutation, which were identified early in the history of the study of sweeps (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Stephan et al. 1992; Barton 1998). The initial spread of a favorable allele A2 from a frequency of 1/(2N) is subject to large stochastic effects. With semidominance, the probability that A2 survives this effectively neutral period is approximately Q = Nes/N in a large population (Kimura 1964), assuming that the scaled selection coefficient, γ = 2Nes, is much greater than one (s is the selective advantage to homozygotes for the favorable mutation). As pointed out by Maynard Smith (1976), the overall expected frequency of A2 during this quasi-neutral phase (including losses) is approximately 1/(2N), after which it starts to behave deterministically. The expected frequency of A2 at the end of the quasi-neutral phase, conditioning on its surviving with probability Q, is thus 1/(2NQ) = γ−1. More rigorously, Martin and Lambert (2015) have used branching process theory to show that the frequency of A2 at the end of the first stochastic phase is exponentially distributed, with mean γ−1 and variance γ−2.
In the presence of BGS, we follow Kim and Stephan (2000) and assume that Ne in the formula for fixation probability is multiplied by a constant, B (see above). As shown below, this constant is somewhat different for the effect of BGS on purely neutral processes, such as the level of neutral variability, and for the effect of BGS on the fixation of favorable mutations, since selected variants are more resistant to the effects of BGS than neutral variants (Johnson and Barton 2002). We denote these two constants by B1 and B2, respectively, and write λ for the ratio B1/B2. The critical frequency at which A2 can be treated as behaving deterministically is then (B2γ)−1, using the argument in the preceding paragraph. When A2 reaches a frequency close to 1, there is a second stochastic phase in which it drifts to fixation fairly rapidly, as described below. We assume that all other effects of BGS are similar to those for neutral variability, with B1 as the factor that multiplies Ne.
The expectation of the time spent in the deterministic phase can be found as follows. As described by Ewens (2004, page 169), a semidominant favorable allele has the property that the expected time spent in a small interval of allele frequency q to q + dq is the same as the time spent in the interval 1 – q to 1 – q – dq. This implies that the expected time that A2 spends between 1/(2N) and (B2γ)−1 is the same as the expected time it spends between 1 – (B2γ)−1 and 1 – 1/(2N), so that q during the deterministic phase can conveniently be treated as lying between (B2γ)−1 and 1 – (B2γ)−1. Using the solution of the deterministic selection equation dq/dt = 1/2 spq for a semidominant allele (Haldane 1924), the expected time spent in this interval (expressed in units of coalescent time, 2Ne generations) ≈ 2γ−1 ln(B22γ2) = 4γ−1 ln(B2γ).
he expected times spent in the two stochastic phases can be found as follows. Using Equation 16 of Kimura and Ohta (1973) and the fact that Ne is multiplied by B1 to take BGS into account, the expected first passage time of a neutral allele from initial frequency 1/(2N) to a frequency q is:
| (2) |
For q << 1, this time is approximately equal to B1q, so that the additional expected time spent in the first stochastic phase is approximately λγ−1. By the above symmetry argument, the same applies to the time between 1 – (B2γ)−1 and 1–1/(2N). The total expected time to fixation of A2 when γ >> 1 is thus:
| (3) |
This expression is very close to Equation A17 of Hermisson and Pennings (2005) for the case with B1 = B2 = 1, which was derived directly from the diffusion equation for the mean sojourn time of a favorable mutation in a finite population.
As far as the effect of a substitution on neutral diversity is concerned, we note that the rate (in units of coalescent time) at which a neutral lineage that is associated with A2 at time T recombines onto a background of A1 is p(T)ρ, where p(T) is the frequency of the wild-type allele at time T and ρ = 2Ner is the scaled recombination rate. Here, T = 0 at the time of fixation of the favorable allele and T = Ts at the time when it arose in the population. From the symmetry of the selection equation, the mean frequency of A1 over the deterministic phase is 0.5, so that that ρ should be discounted by a factor of 1/2 during this part of the process (note that this argument ignores the possibility of coalescence competing with recombination during the sweep, as was pointed out to us by Matthew Hartfield; a more rigorous treatment that includes such competition will be presented elsewhere).
For sample paths in which A2 reaches the critical frequency (B2γ)−1, the expected duration of the first stochastic phase is equal to the expected value of the first passage time to this frequency, λγ−1, and its variance is λ2γ−2/3 (File S1, section S2). During this period, a single lineage recombines with A1 haplotypes at a rate close to ρ, since A1 dominates the population, thus contributing 2ρ (λγ−1 + δTs1) to the mean number of recombination events, where δTs1 is the departure of the duration of the first stochastic phase from its expectation. The final stochastic phase has effectively zero probability of contributing to recombination, due to the prevalence of the favored allele, and can be ignored for this purpose.
We exploit the fact that the frequency of A2 at the end of the first stochastic phase is exponentially distributed (Martin and Lambert 2015) to show that the variance of the time to fixation caused by fluctuations in the initial frequency of A2 at the start of the deterministic phase yields an additional variance term of 16(γ)−2 (File S1, section S2). Since this phase has a mean frequency of A2 of 0.5, the relevant product of the recombination rate and a fluctuation in deterministic sweep time (δTs2) is ρ δTs2 rather than 2ρ δTs2.
The probability Pcs that the two sampled haplotypes coalesce as a result of the sweep is equivalent to the probability that neither member of a pair of haplotypes sampled at time T = 0 recombined onto an A1 background, provided that the sweep durations are so short that no coalescence can occur among nonrecombined haplotypes during the sweep (Wiehe and Stephan 1993). This probability is given by the first term of a Poisson distribution, whose mean is equal to the expected number of recombination events over the duration of a substitution. We thus have:
| (4) |
The term (B2γ)−4r/s in the third line of Equation 4 is the deterministic phase contribution to the effect of a sweep, first derived by Barton (2000, 1998) for the case of B1 = B2 = 1, using a more rigorous approach. It has been used in several subsequent studies (Weissman and Barton 2012; Elyashiv et al. 2016; Campos et al. 2017). The last term is second-order in r/s and is likely to be of minor importance, since sweeps only have substantial effects on variability when r/s << 1. The second term has a somewhat larger effect; e.g., with no BGS, γ = 100, and r/s = 0.1, it reduces Pcs from 0.158 to 0.130. An extension to these results is described in section S3 of File S1 (Equation S20), which allows for multiple recombination events that bring a recombined lineage back onto an A2 background.
Sweeps at multiple sites:
We now consider the effects of recurrent sweeps at multiple sites. We use the standard assumption that substitutions of favorable alleles are sufficiently rare that their effects on a given site can be treated as mutually exclusive events (Kaplan et al. 1989; Wiehe and Stephan 1993; Kim and Stephan 2000; Kim 2006). We consider only a single gene, which is reasonable for favorable mutations whose selection coefficients are less than the rate of recombination between sites in different genes, as is usually the case here. These procedures are supported by our simulation results, except for cases with very low rates of recombination.
We use the expression for the probability of a sweep-induced coalescent derived above (Equation 4) to obtain an approximate expression for the net rate of coalescent events experienced at a given neutral site (in units of 2Ne generations), due to recurrent SSWs at NS and UTR sites:
| (5) |
where νa and νu are the rates (in units of coalescent time) at which substitutions of favorable mutations occur at NS and UTR sites, respectively; Pcs Ni and Pcs Uj are the rates of sweep-induced coalescent events induced by the ith NS site and jth UTR site, respectively, which can be obtained from Equation 4 and Equation S20. The summations are taken over all the sites in the gene that are under selection. The notation S−1 is used to denote the reciprocal of the expected time to coalescence due to sweeps, S, where subscripts a and u denote NS and UTR mutations, respectively.
If we assume that the fixation probability of a favorable mutation in the presence of BGS is discounted by a factor of B2 compared with the standard value (see above), we have:
| (6a) |
| (6b) |
where u is the mutation rate per nucleotide site, and pa and pu are the proportions of all new NS and UTR mutations, respectively, that are selectively favored.
Because we are confining ourselves to a single gene, a linear genetic map can be assumed. The crossing over contribution to ri is then given by rczi, where zi is the physical distance between the neutral and selected sites, and rc is the rate of crossing over per base pair. There is also a contribution from gene conversion, as described in the section on modeling BGS. The summation formula used in the sweep calculations assumes that every third base pair in an exon is a neutral site, with the other two being subject to selection (Campos et al. 2017). This differs from the SLiM procedure of randomly assigning selection status to exonic sites, with a probability ps of being under selection (ps = 0.7 in the simulations used here). To correct for this, the overall rate of NS substitutions in Equation 5 was adjusted by multiplying by 0.7 × 1.5.
Following Kaplan et al. (1989), Wiehe and Stephan (1993), and Kim and Stephan (2000), coalescent events caused by SSWs and coalescent events caused by neutral drift can be considered as competing exponential processes with rates S−1 and B1−1, respectively, on the coalescent timescale of 2Ne generations. Under the infinite sites model (Kimura 1971), the expected diversity at a neutral nucleotide site, relative to its value in the absence of selection at linked sites (θ = 4Neu), can then be written as the expected time to coalescence when time is measured in units of 2Ne generations:
| (7) |
The simulation results for synonymous site nucleotide site diversities were presented as mean values over all genes in the region simulated. Since we are modeling only a single gene, the mean of π/θ in Equation 7 over all synonymous sites in a gene should be used for comparison with the simulation results. In practice, the values obtained by substituting the mean value of S−1 across synonymous sites into Equation 7 give almost identical results, and this is used for the results described below.
The effect of sweep duration on mean coalescent time:
Equation 7 assumes that the duration of a sweep is negligible in comparison to the times between successive sweeps and to the mean neutral coalescent time 2Ne, so that sweeps can be treated as point events. However, this assumption is violated if selection is sufficiently weak. For example, with γ = 250, the deterministic component of the duration of an adaptive substitution given by Equation 3 is ∼10% of the coalescent time. Assuming that the entire time between sweep-induced coalescent events is available for neutral coalescent events can cause an underestimation of the effects of sweeps when these are sufficiently frequent.
Here, we develop an alternative approach that uses the mean diversity between successive substitutions as an estimate of the expected diversity under recurrent sweeps. This is likely to overestimate the effects of sweeps compared with the mean for randomly sampled time points, but the simulation results described below show that the resulting expressions (Equation 12) provide a good fit. We assume that adaptive substitutions occur in a gene at a constant rate ω per unit of coalescent time, given by the sum over the rates per site for the NS and UTR sites in the gene. This quantity can be found from Equation 6, by multiplying νa by 70% of the number of NS sites in a gene and νu by the number of UTR sites. We then look back in time, and evaluate the time average of the divergence of π/θ from its equilibrium value over the period since the previous substitution.
To do this, we denote the expected neutral diversity at a neutral site immediately after a substitution by π0, and the expected neutral diversity at the time of initiation of a new substitution by π1. We have:
| (8) |
where D is the probability that each member of a pair of lineages carrying the favorable mutation has failed to recombine during the substitution, conditioned on the completion of a substitution. Because the expected reduction in neutral diversity due to recurrent sweeps is S−1, we have D = (ωS) −1, thereby establishing the relationship between π0 and π1 (the assumption that the coalescence time for the pair of swept lineages is zero is relaxed below).
Under the infinite sites model (with θ << 1), the equilibrium diversity in the absence of sweeps is B1θ. In this case, the standard formula for the rate of approach of neutral diversity to its equilibrium value (Malécot 1969, p.40; Wiehe and Stephan 1993, Equation 6a) gives the following expression for the diversity at a time T after a substitution:
| (9) |
(the factor of B1−1 in the exponent reflects the reduction in Ne caused by BGS, resulting in a corresponding acceleration in the rate of approach to equilibrium).
The expected diversity over the relevant period, π, is thus given by:
| (10) |
Formulae for I(ω, B1) are derived in File S1, section 5.
Furthermore, π1 is given by:
| (11) |
where A =ω/(ω + B1−1).
In the absence of any recovery of diversity during the sweep itself, Equations 8–11 yield the final expression:
| (12a) |
In the limit as ω approaches zero, ωI and A both tend to 0, and AD tends to B1S−1. The value of π/θ for small ω is thus approximately 1/(B1 + S−1), corresponding to Equation 7.
To allow for a nonzero mean time Tcs to coalescence during the sweep, the postsweep diversity π0 is modified by adding DTcsθ to Equation 8, where Tcs is given by Equation S11 (this is an underestimation, since it ignores recombination during the sweep). This adds a small additional component to Equation 12a, giving:
| (12b) |
Equations 12a and 12b assume that the sample is taken in an interval between two successful sweeps. A correction can be applied to take into account the possibility that a sample is taken during a sweep; this effect is expected to be small unless sweep-induced coalescents are very frequent and the time occupied by a sweep is relatively large compared with the neutral coalescent time (File S1, section S6).
Continuum approximation for effects of recurrent sweeps:
A useful approximation can be obtained by treating a gene as a continuum, following Wiehe and Stephan (1993), Coop and Ralph (2012), and Weissman and Barton (2012). We correct for the effect of introns simply by reducing the density of NS sites in the coding sequence. This is done by multiplying the density within exons by the fraction of the sites that are exons among the total length of exons, introns, and UTRs. In addition, we approximate the effect of gene conversion by writing the net recombination rate between sites separated by z base pairs as (rc + gc)z when z ≤ dg, and as rc z + g (where g = gcdg) when z > dg (Andolfatto and Nordborg 1998). The resulting expressions for sweep effects are derived in File S1, section S7. These do not include any corrections for multiple recombination events, or for the variances in the first stochastic phase and deterministic phase durations, since these make the integrations analytically intractable.
Simulation results
Effects of BGS alone:
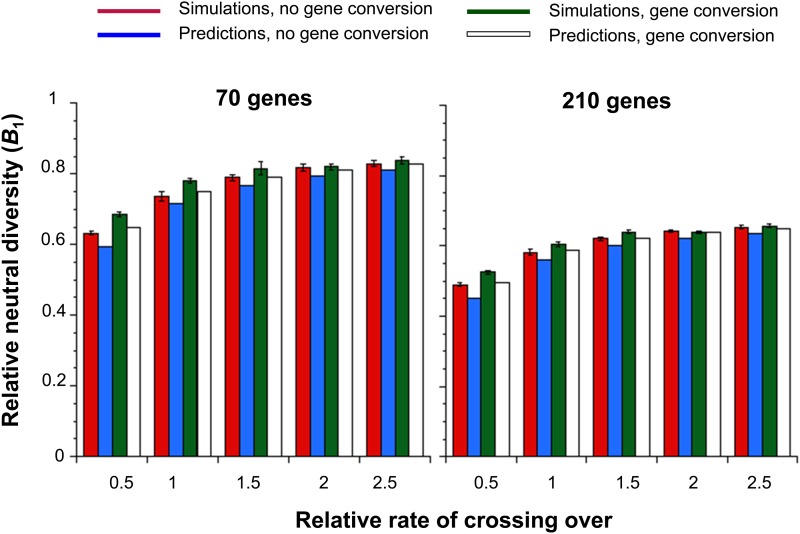
Figure 2 shows the simulation and theoretical results for B1, the ratio of the mean synonymous site nucleotide diversity (π) to its value without selection (θ) in the absence of SSWs, using the gene model in Figure 1. Chromosomal regions containing 70 and 210 genes, with and without gene conversion at the standard rate, were modeled. The mean value of θ from simulations of neutral mutations in the absence of selection at linked sites was 0.0228, with 95% C.I. (0.0227, 0.0229), which is slightly lower than the theoretical value on the infinite sites model (0.0239), presumably due to the slight deviations from the infinite sites assumption in SLiM. The ratios of the mean simulated synonymous site diversities to 0.02283 were used for the estimates of B1. Table S1 of File S1 shows more detailed results for the autosomal case, as well as for the model of X-linked loci described in Table 1.
Figure 2.
The effect of background selection on neutral diversity at autosomal loci. The bars show values of B1 = π/θ, where π is the mean diversity at synonymous sites and θ is the value in the absence of selection, in relation to the rate of crossing over relative to the standard value given in Table 1 (x-axes). Results with and without gene conversion under the standard parameters are displayed. Both the simulation results (red and green bars), with error bars indicating 95% C.I.s, and the theoretical predictions (blue and white bars) are shown.
Overall, there is a fairly good fit between the theoretical predictions and the simulation results, although the theoretical values of B1 are mostly slightly smaller than the simulation values, probably because intergenic sequences have been ignored. However, if the additional term in E contributed from neutral mutations that arise in repulsion from a linked deleterious mutation (Equations S1b, S5d and S5e) is ignored, the fits are much less good, especially with the larger numbers of genes. For example, with 210 genes and gene conversion, the predicted B1 values are 0.583, 0.696, 0.739, 0.762, and 0.776 for crossover rate factors of 0.5, 1, 1.5, 2, and 2.5, respectively; the last value is 18% larger than when the additional term is included.
Similarly, use of a linear relationship between physical distance and map distance, which has been assumed in most theoretical models of BGS, generally gives a poorer fit to the results for the higher rates of crossing over (Table S2 of File S1), except when the number of genes and the map length of the region are both small, reflecting the effect of double crossing over in reducing the net rate of recombination between distant sites. Nonetheless, the fit is surprisingly good overall; indeed, the linear map predictions often provide a better fit to the simulation results for the cases with 20 and 70 genes. The implications of these effects of the inclusion of the repulsion mutations, and the differences between the linear and Haldane maps, are considered in the Discussion.
Effects of BGS on the rate of fixation of favorable mutations:
The main goal of our work is to analyze the joint effects on neutral diversity of BGS and SSWs, and the extent to which these can be predicted by the relatively simple Equations 7 and 12. A core assumption behind these equations is that the fixation probability of a new favorable mutation is affected by BGS as though Ne is multiplied by a factor that is close to the value that applies to neutral diversity (Kim and Stephan 2000).
We have tested this assumption by comparing the mean numbers of fixations of favorable mutations observed over the last 20,000 (8N) generations of the simulations, both without BGS and with BGS. The ratio of these means provides a measure of B (B2) that can be compared to the value of B estimated from neutral diversity (B1). There are two reasons why we would not expect perfect agreement. First, a sufficiently strongly selected favorable variant could resist elimination due to its association with deleterious mutations, and instead might drag one or more of them to high frequencies or fixation (Johnson and Barton 2002; Hartfield and Otto 2011). Second, the incursion of selectively favorable mutations may perturb linked deleterious mutations away from their equilibrium, even if they do not cause their fixation.
Such Hill–Robertson interference effects (Hill and Robertson 1966; Felsenstein 1974) reduce the Ne experienced by deleterious mutations and hence their nucleotide site diversity, which is correlated with the mean number of segregating deleterious mutations. This reduction in the number of segregating deleterious mutations reduces the effects of BGS on incoming favorable mutations. For both these reasons, B1 is likely to be smaller than B2. Table S3 of File S1 provides evidence that the mean number of segregating deleterious mutations is indeed reduced by SSWs, except for the cases with no crossing over, where the rate of sweeps is greatly reduced compared with cases with crossing over.
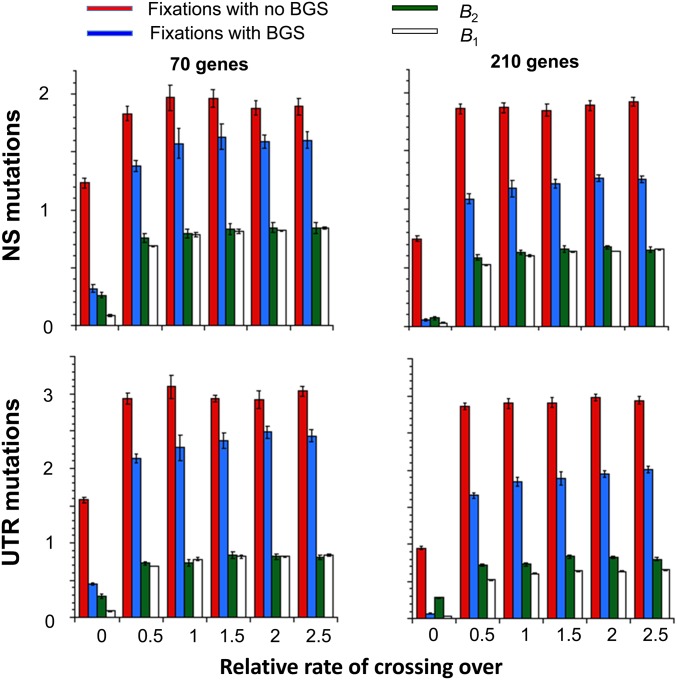
The results for autosomal loci in Figure 3 show that BGS has a substantial effect on the rate of adaptive substitutions (Table S4 of File S1 presents more detailed results for autosomal and X-linked loci). The most extreme case is when there is no crossing over, a regime in which the efficacy of BGS is undermined by Hill–Robertson interference among the deleterious mutations, so that the assumptions underlying the BGS equations tested in the previous section are violated (McVean and Charlesworth 2000; Comeron and Kreitman 2002; Kaiser and Charlesworth 2009; Seger et al. 2010; Good et al. 2014; Hough et al. 2017). For example, B1 for 70 genes with gene conversion is 0.086, close to the value found by Kaiser and Charlesworth (2009) for a similar sized region, whereas the standard BGS prediction is 0.0004. In contrast, the B2 values for favorable NS and UTR mutations are 0.26 and 0.28, respectively, around three times greater. This still represents a massive reduction in the efficacy of selection on favorable mutations, consistent with the evidence that their rates of substitution in noncrossover regions of the Drosophila genome are much lower than elsewhere (Charlesworth and Campos 2014).
Figure 3.
The effect of background selection (BGS) on the numbers of fixations of favorable nonsynonymous (NS) and UTR mutations at autosomal loci, in relation to the relative rate of crossing over (x-axes). The blue and red bars shows the numbers of fixations over the last 20,000 generations of the simulations (with 95% C.I.s), expressed as ratios of fixations to the number of genes simulated. Blue and red bars show the results for simulations with and without BGS, respectively. The corresponding ratios of numbers of fixations (B2) with and without BGS (green bars), and ratios of neutral diversities with and without BGS (white bars), obtained from the simulations (B1) are also shown. The standard gene conversion parameters are used.
For the other rates of crossing over, there is much closer agreement between the two estimations of B, although we always have B1 > B2. The discrepancy is largest for crossover rates of one-half the standard value, and seems to level off after the standard rate. As might be expected, it is smaller in the presence of gene conversion.
Effects of interference among favorable mutations on their rates of substitution:
With no recombination, Hill–Robertson interference among adaptive substitutions is likely to be important, and makes analytical models of substitution rates much harder to develop. The effects of such interference can be predicted using the approximate Equation 4 of Neher (2013), which is based on Equation 39 of Desai and Fisher (2007). When this is adapted for the case of diploids with semidominance with s >> Ub, the rate of substitution of favorable mutations, ω, is equal to 0.5s ln(Ns)/[ln(2Ub/s)]2, where s is the homozygous selection coefficient for a favorable mutation and Ub is the net mutation rate to favorable mutations for the region. Combining NS and UTR mutations (these have similar selection coefficients in our simulations), and putting s = 0.05, Ub = 0.00436, N = 2500, and ω = 0.00406, the ratio of ω to the baseline substitution rate in the absence of interference is 0.163.
The observed ratio of the rates of substitution for relative rates of crossing over of 0 and 2.5, with 70 genes, and no gene conversion and no BGS, was equal to 0.235, suggesting that the effect of interference is overpredicted by the approximation. Gene conversion increases the ratio to 0.570, so that it greatly reduces interference when crossing over is absent. BGS thus seems to play a more important role than SSWs in reducing the rate of substitution of favorable mutations when crossing over is absent, especially in the presence of gene conversion, as was suggested by Campos et al. (2014). The properties of genomic regions with very low rates of crossing over will be analyzed in more detail in a later publication.
In the absence of BGS, but with nonzero rates of crossing over, Figure 3 and Figure S4 show little effect of the crossing over rate on the rate of fixation of favorable mutations. At first sight, this suggests that there is little interference among selectively favorable mutations, with a rate of crossing over of one-half or more of the standard rate. However, there is indirect evidence for such interference effects, from estimates of the extent of underdispersion of the numbers of adaptive substitutions observed over the last 8N generations of the simulations compared with the expectation for a Poisson distribution, as described in File S1, section S8. Here, underdispersion is measured by the ratio of the variance to the mean of the number of substitutions over the period of observation (Sellers and Morris 2017).
This analysis shows that interference causes a small loss of substitutions, leading to a reduction in the extent of the reduction in diversity caused by SSWs for the cases with crossing over, with ∼5.5% of substitutions being lost due to interference. An approximate correction for interference can be made by multiplying the substitution rates for both NS and UTR mutations by the estimated proportion of substitutions that survive interference, although this ignores some of the complexities associated with the effects of interference on diversity (Kim and Stephan 2003; Chevin et al. 2008). In addition, it should be noted that the existence of underdispersion implies that the Poisson model of sweeps that is usually assumed is not exact, as pointed out by Gillespie (2001), introducing a further source of error into the predictions.
Effects of SSWs on neutral diversity:
This section is concerned with four main questions. First, to what extent does treating sweeps as point events affect the predictions of models of recurrent sweeps? Second, how well does the integral approximation for SSWs perform (Equations S24–S33) compared with the more exact summation formulae (Equations 5 and 6)? Third, how well do the competing coalescent process approximations for the joint effects of BGS and SSWs perform when the various corrections described above have been included? Finally, is less accuracy obtained by using the neutral BGS value (B1) instead of B2 in the formulae for the effect of BGS on the fixation probability of a favorable mutation?
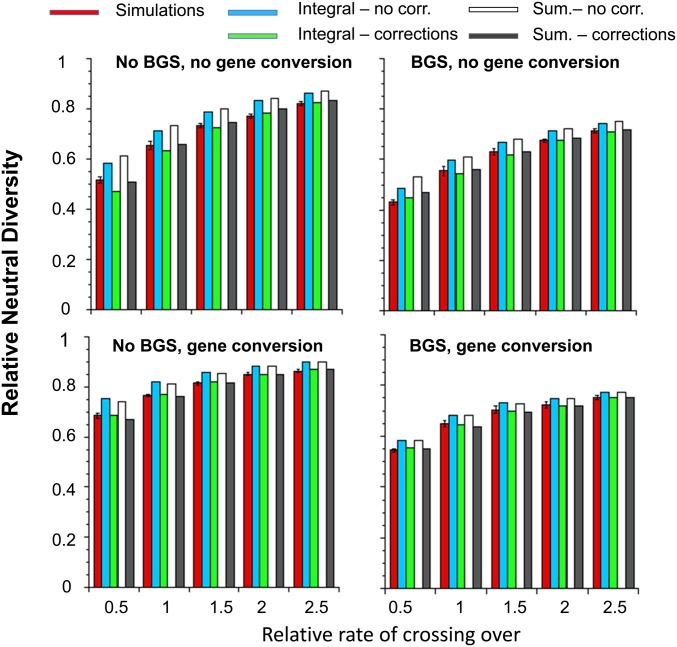
Figure 4 presents the mean values of synonymous site diversities relative to the value in the absence of selection for simulations with 70 autosomal genes, together with the predictions for the integral and summation formulae, with and without the corrections described (the correction for interference was applied to all these cases). In the case of the corrected summation formulae, all the corrections described above were applied; for the integral results, only the corrections for expected sweep duration and interference were used. More detailed results for autosomal and X-linked genes are shown in Table S6 of File S1.
Figure 4.
The effects of selective sweeps on mean diversity at autosomal synonymous sites in relation to the relative rate of crossing over (x-axes), with and without background selection (BGS), or gene conversion at the standard rate. The red bars shows the simulation values of mean synonymous diversities relative to the values without any selection (π/θ), with 95% C.I.s. The green and blue bars are the results for the integral approximations, with and without the corrections (corr.) described in the text, respectively. The gray and white bars are results for the summation formulae (sum.), with and without the corrections.
Concerning the first point, diversities are considerably overpredicted by the uncorrected values from Equation 5 (which included the correction for interference) by up to 20%, with the lowest rate of crossing over used in Figure 4 and Table S6. This shows that treating recurrent sweeps as point events can produce significant errors, especially when crossing over is infrequent and there is no gene conversion.
For the second point, the agreement between the integral and summation results is surprisingly good overall. The largest discrepancies occur when the rate of crossing over is low, and gene conversion and BGS are absent, when they are of the order of 7.6% of the lower value.
For the third point, the agreement between the simulation means and the predictions with the corrections is generally very good, although the integral results underpredict diversity by ∼20% for the autosomal case with the lowest rate of crossing over, no gene conversion, and no BGS. If the correction for interference is not applied, lower diversities are predicted, which sometimes give better agreement with the simulation results, but the effects are not major (Table S7). The main contribution to the improvements in fit from the other corrections comes from the sweep duration, as can be seen from results where one or both of the other factors (multiple recombination events and coalescence during a sweep), as well as interference, are omitted (Table S7). Omission of the correction for coalescence during sweeps usually has the next largest effect, mainly because it reduces the contribution to coalescent time from samples taken during sweeps (section S6 of File S1). Overall, omission of all the corrections except that for sweep duration produces remarkably good results.
With respect to the fourth point above, the fits with B1 alone are good, except for the lowest rate of crossing over and no gene conversion (an error of 9% in Figure 4). Overall, it seems that relatively little is to be gained by using B2.
The predictions of the effects of SSWs use a single gene model, which assumes that the effects of sweeps with the parameters assumed here are localized to single gene regions. The simulation results with sweeps alone in regions with crossing over (File S2) show that there is no noticeable effect of the numbers of genes on the mean synonymous site diversities, consistent with this assumption. This is not surprising, given that the expected reduction in diversity at a neutral site due to a single sweep at recombination distance r is approximately γ – 4r/s, where γ and s are the scaled and absolute selection coefficients for the favorable allele, respectively. With the values of γ and s for autosomal NS mutations assumed here (250 and 1 × 10−4 for natural populations, respectively), an effective crossing over rate of 1 × 10−8, and a distance of 2000 bp between sites (the minimum for sites in separate genes), the expected reduction in diversity with no gene conversion is 250 (–0.8) = 0.01, which is essentially trivial.
This conclusion does not apply in the absence of recombination, which has been studied theoretically by Kim and Stephan (2003) and Weissman and Hallatschek (2014). In this case, the simulation results displayed in File S2 show that there is a large effect of the number of genes. With no crossing over, gene conversion or BGS, the mean autosomal diversities relative to neutral expectation were 0.0819, 0.0700, and 0.0675 for 70, 140, and 210 genes, respectively. These results can be compared to the predictions from the approximate Equation 5 of Weissman and Hallatschek (2014), modified for diploidy with semidominance, which gives the absolute neutral nucleotide diversity with recurrent sweeps as 8μ ln[2ln(γ)/Ub]/s. The resulting predicted values are 0.195, 0.183, and 0.176, respectively.
As was also found by Weissman and Hallatschek (2014), the theoretical results considerably overpredict diversity. Gene conversion greatly reduces the effects of sweeps, with relative diversities of 0.130, 0.090, and 0.0832 in the absence of BGS. BGS has a much greater effect on diversity than sweeps when crossing over is absent. With gene conversion, it gives relative diversity values of 0.0867, 0.0429, and 0.0293 for 70, 140, and 210 genes, respectively. Essentially the same values are seen with both BGS and SSWs, reflecting the fact that the rate of sweeps is greatly reduced in the presence of BGS (see Figure 3). The predicted relative diversity value for a 70-gene region with no crossing over is quite close that observed for the fourth chromosome of D. melanogaster, which has a similar number of genes (Campos et al. 2014), suggesting that diversity in noncrossover regions of the genome is strongly influenced by BGS, as was also inferred by Hough et al. (2017) for the case of the newly evolved Y chromosome of Rumex.
Discussion
Accuracy of the approximations for pairwise diversity with hitchhiking
We have developed a new expression for the effect of a single substitution of a favorable allele on pairwise neutral diversity at a linked site (Equation 4). This uses an approximate formula for the duration of an adaptive substitution, which includes stochastic contributions (Equations 2 and 3). In addition, we have developed expressions for the effects of coalescence and multiple recombination events during a substitution (sections S3 and S4 of File S1), as well as a crude correction for interference among SSWs (section S8 of File S1). More importantly, we have derived new formulae to predict the effects of a constant rate of recurrent adaptive substitutions on pairwise neutral diversity (Equation 12). This approach, while admittedly somewhat heuristic, avoids the assumption made in most previous models of recurrent sweeps that the duration of an adaptive substitution can be neglected, enshrined in Equation 7 (Kaplan et al. 1989; Wiehe and Stephan 1993; Kim and Stephan 2000). This equation has been used several times for inferences about sweep parameters (Sella et al. 2009; Elyashiv et al. 2016; Campos et al. 2017), but overestimates diversities compared with the simulations, especially with high rates of adaptive substitutions and low rates of crossing over. The comparisons of the simulation results with the different types of theoretical predictions (Table S7) suggest that the corrections for coalescence and multiple recombination events during a sweep are sufficiently small that they can be ignored for most purposes.
As described at the end of the Results section, the integral approximations provide results that are quite close to the more exact results from summations, except for low rates of crossing over and no gene conversion. Similarly, applications of the reduction in diversity at neutral sites caused by BGS (B1) for predicting the reduction in rates of substitution of adaptive mutations perform nearly as well as the use of the factor B2 derived from the simulations. This suggests that inference methods can be simplified by using the integral approximations for both SSWs and BGS. As described in the section on BGS, it is relatively easy to predict BGS effects if estimates of the distribution of mutational effects on fitness are available, and the results described in Figure 3 and Figure 4 indicate that they play an important role in affecting the relationship between recombination rate, the rate of adaptive evolution, and diversity, so that it is probably unwise to ignore them when trying to infer SSW parameters
Another feature of the work presented here is the inclusion of gene conversion into sweep models, as was also done by Campos et al. (2017) but which has been ignored in previous treatments of sweeps. Gene conversion events that are not associated with crossovers are known to be a major source of recombination events at the intragenic level in Drosophila (Hilliker and Chovnick 1981; Hughes et al. 2018). With the standard autosomal effective crossing over rate for D. melanogaster of 1 × 10−8 per base pair (Campos et al. 2014), the effective rate of crossing over between two sites separated by 500 bp is 5 × 10−6. With scaled and absolute selection coefficients for NS mutations of γ = 250 and s = 10−4 used in Figure 4, the expected proportional reduction in diversity at the end of a sweep for a neutral site that is 500-bp away from the selected site is approximately γ (– 4r/s) = 250 (– 4 × 0.05) = 0.33. With the somewhat conservative gene conversion parameters assumed in the figure, equation 1 of Frisse et al. (2001) implies an additional contribution to the effective recombination rate of 4.4 × 10−6, so that the total effective recombination rate is 9.4 × 10−6. This yields a reduction in diversity of 0.13, ∼40% of the value with no gene conversion. Consistent with this result, the simulation results and theoretical predictions are significantly affected by gene conversion, such that the expected effects of sweeps on diversity are considerably reduced if gene conversion is present (Figure 4 and Table S6). Ignoring gene conversion in sweep models is likely to substantially bias estimates of sweep parameters, unless the strength of selection on favorable mutations is much larger than we have assumed. Such strongly selected mutations may, of course, contribute to SSW effects (Sella et al. 2009; Elyashiv et al. 2016). However, as discussed by Campos et al. (2017), the observed relationship between the level of diversity in a gene and its nonsynonymous divergence requires a substantial contribution from favorable mutations with local sweep effects, in which case gene conversion plays an important role.
While our models assume hard sweeps, where the new favorable mutation is introduced as a single copy, Equation 12a can also be applied to other situations, such as soft sweeps arising from standing variation or multiple mutations to the favorable allele at a locus (Hermisson and Pennings 2005, 2017). The only modification that need be made is to the expression for the reduction in diversity immediately after a sweep (D) in Equation 8.
Interference between adaptive substitutions
Our simulation results suggest that there is a minor, but noticeable, degree of interference among adaptive substitutions in the presence of crossing over (File S1, section S8) until the rate of crossing over is considerably less than one-half the standard value. Therefore, for most regions of the genome, inferences that ignore interference should not be impaired unless the effects of SSWs are much bigger than we have assumed.
A somewhat counterintuitive finding is that the proportion of substitutions eliminated by interference is nearly independent of the strength of selection and the recombination rate. Relatively weak effects of the recombination rate, provided it is not very high or low, are also evident in Table 3 of Kim and Stephan (2003) and in figure 3 of Barton (1995), for cases when the selection coefficients at the selected loci are similar. A possible explanation for this is that recombination has a dual effect on the potential for interference. If a second favorable mutation arises early enough during the substitution of a previous one, it is likely to be on the wild-type background, so that a recombination event that puts it onto the mutant background would enhance its fixation probability. The opposite would be the case if it arises late during the sweep. Similarly, the faster (lower) rate of spread of a more strongly (weakly) selected mutation would reduce (increase) the probability of either type of recombination event, so that its selective advantage might not greatly affect the opportunity for interference. Finally, the product of the rate of substitution of favorable mutations and the time taken for a substitution (ωTs) determines the chance of two substitutions overlapping in time. Because ω is proportional to the scaled selection coefficient γ and Ts is close to being inversely proportional to γ (Equation 4), ωTs is only weakly dependent on γ.
The relationship between sequence diversity and rate of crossing over
It is also of interest to ask what light the theoretical results described above shed on the observed positive relationship between DNA sequence variability at putatively neutral or nearly neutral sites within a gene in D. melanogaster, and the local rate of recombination experienced by the gene (Aguadé et al. 1989; Begun and Aquadro 1992). This observation stimulated interest in models of SSWs and BGS, and its possible causes have been a long-standing subject of debate; for reviews, see Sella et al. (2009), Stephan (2010), Cutter and Payseur (2013), and Charlesworth and Campos (2014).
Recent analyses of population genomic data have confirmed the existence of a strong relationship between synonymous nucleotide site diversity (πS) and the effective rate of crossing over, even after excluding genes in noncrossover regions. For example, figure 2 of Charlesworth and Campos (2014) shows the regressions of πS for autosomal and X-linked genes on their effective rates of crossing over, for a sample from a Rwandan population of D. melanogaster. The ratios of the value of πS for an effective rate of crossing over of 0.5 cM/Mb to the value with a crossing over rate of 2 cM/Mb (the upper limit to the autosomal rate) are K = 2.38 for autosomes and K = 1.78 for the X chromosome. The simulation results in Figure 4 for both BGS and SSWs, with 70 autosomal genes and gene conversion, give K = 1.33 and K = 1.24 with SSWs alone; K = 1.27 with BGS alone (Figure 2), a much weaker relationship than observed.
What causes this discrepancy? One possibility is that the mean scaled selection coefficients for favorable mutations used in these simulations are unrealistic. This was checked by rerunning the calculations with different γ values for the favorable mutations, using the summation method with all the corrections and B1 for the effects of BGS, as well as selection, mutation, and recombination parameter values appropriate for the natural population rather than the simulations (see Table 1). For the reasons given in the final part of the Discussion, we used a linear genetic map and no correction for the repulsion BGS terms when determining the values of B1, with selection, mutation, and recombination parameter values. We also used the rate of gene conversion indicated by experimental studies in D. melanogaster (Hilliker and Chovnick 1981, 1994; Miller et al. 2016) instead of the conservative value used previously (one-half of the empirical value). Because the effect of interference among sweeps is unknown for these parameters, and was found to be small in the presence of gene conversion and BGS, it has been ignored in the following analyses.
Because B1 is somewhat sensitive to the number of genes modeled, we used results for 210 genes, approximating the behavior of a small group of linked genes with similar recombination rates. In the absence of SSWs, but with gene conversion and BGS, K = 1.17 for the autosomal model, reflecting the greater effect of recombination with a linear map than with the Haldane mapping function. If the standard γ values for favorable mutations are used for the sweep predictions, and BGS and gene conversion are included, K = 1.27, somewhat lower than the simulation result. The ratios for γ values that are one-half and twice the standard values are 1.19 and 1.47, respectively. Thus, there is only a rather weak dependence on the strength of selection on favorable mutations. This is not surprising, in view of the fact that the effect of sweeps on diversity predicted by Equation 7 is a concave function of S−1, unless S−1 is so large that it dominates the denominator, which is not the case for these parameter values.
The effect of the proportion of mutations that are favorable can be examined in a similar way. With the standard selection coefficients, halving these proportions leads to K = 1.24 and doubling them to K = 1.34. Although this parameter has a large effect on diversity, its effect is only weakly dependent on the crossing over rate over the range considered here. Even if both the strengths of selection and the proportions of beneficial mutations are doubled, K is increased to only 1.64. To explain the observed relationship between diversity and rate of crossing, considerably larger values of both the strength of selection and proportion of favorable mutations than those used here seem to be required.
Another possibility is that intergenic and intronic sequences are subject to selection, rather than being selectively neutral. Charlesworth (2012b) used evidence on the levels of selective constraints on different types of Drosophila DNA sequences to obtain crude estimate of γ values for deleterious mutations in weakly constrained and strongly constrained noncoding sequences, as well as for deleterious NS mutations. His analysis showed that a linear genetic map provided a good approximation to the BGS predictions. We used this approach to predict the BGS parameter B1 for a genic region with a given rate of crossing over, modifying it to include the effects of gene conversion, as described in File S1, section S9. For a model of an autosome with the standard rate of gene conversion, this procedure gives B1 values of 0.379, 0.616, 0.724, 0.785, and 0.825 for relative rates of crossing over of 0.5, 1, 1.5, 2, and 2.5, respectively, yielding a ratio 2.07 for relative crossing over rates of 2 and 0.5. If these values are used in the above method for predicting π/θ for a natural population with BGS and SSWs, with the standard γ values for favorable mutations, the predictions for the different relative rates of crossing over become 0.343, 0.562, 0.652, 0.709, and 0.753, respectively, giving K = 2.05.
Both predicted values of K are still somewhat lower than the observed value of 2.38. This may reflect the fact that BGS on intergenic sequences is likely to have a weaker effect on the fixation probabilities of favorable mutations than is predicted by B1 when crossing over rates are relatively high, given their distance from positively selected mutations in coding sequences, so that the effect of increased crossing over is greater than predicted by this model.
The same procedure can also be applied to the X chromosome, as described in section S10 of File S1. If the low gene density in low crossing over regions of the D. melanogaster X chromosome is taken into account (Campos et al. 2014), moderately good fits to the data are obtained for the model with SSWs and BGS, both for the relationship between crossing over rate and diversity on the X, and for the X/autosome diversity ratios at the two extremes of the autosomal crossing over rates. The rates of substitution of favorable NS autosomal and X-linked mutations can be analyzed in a similar way, and also show a reasonable level of agreement with the observations (section S11 of File S1).
Another possibility is that a class of much more strongly selected favorable mutations (Sella et al. 2009; Elyashiv et al. 2016) is contributing to both genic and intergenic sweeps. These could cause S−1 to greatly exceed 1, so that sweep coalescents dominate over drift. In this case, a model of a neutral site embedded in a continuum of selected sites predicts that, in the absence of gene conversion, the equilibrium diversity is inversely proportional to the rate of crossing over per base pair (Coop and Ralph 2012; Weissman and Barton 2012), as can be seen from Equation S.27b when the length l of the region is very large. In the absence of BGS, this model can capture the observed ratio of π values for low- and high-crossover regions of D. melanogaster autosomes (Table S8) when γ is sufficiently large; for γ = 2000 and K = 2.48, for example. However, such a high rate of sweep coalescent events causes a large distortion of the site frequency spectrum at the neutral site; the proportion of singletons is 55% for a sample size of 20 with R = 0.5 and γ = 2000, which is over 20% greater than the observed value (Campos et al. 2014).
These analyses are very crude and require considerable refinement, but suggest that the relative values of nearly neutral variability and rates of adaptive evolution in crossover regions of the D. melanogaster genome are more strongly influenced by BGS rather than SSWs, in agreement with Comeron (2014). As discussed at the end of the Results section, this conclusion also applies to regions of the genome with zero or very low rates of crossing over, where the effects of SSWs are expected to be weak.
Distortion of the site frequency spectrum by hitchhiking
The effects of BGS and SSWs on the site frequency spectra (SFS) at the neutral loci in genomic regions with crossing over were briefly mentioned above. While it should be possible to use the theoretical frameworks developed for BGS (Zeng and Charlesworth 2011; Nicolaisen and Desai 2013) and SSWs (Durrett and Schweinsberg 2004; Kim 2006; Pfaffelhuber et al. 2006; Coop and Ralph 2012; Bossert and Pfaffelhuber 2013), this would require extensive calculations that are outside the scope of this paper. However, we note that the simulation results shown in File S2 show that recurrent SSWs have noticeable effects on the SFS, even with quite high rates of crossing over, in the direction of an excess of rare variants over neutral expectation, as expected from previous theoretical work (Kim 2006) and as has been seen in previous simulation studies, e.g., Messer and Petrov (2013). Such effects of BGS and SSWs on the SFS may bias estimates of demographic parameters when neutrality is assumed (Messer and Petrov 2013; Ewing and Jensen 2016; Schrider et al. 2016).
For example, with 70 autosomal genes and gene conversion, the mean values of synonymous site Tajima’s D per gene, with SSWs and BGS for relative rates of crossing over of 0.5, 1, 1.5, 2.0, and 2.5, were –0.209, –0.156, –0.116, –0.111, and –0.069, respectively. The corresponding mean proportions of singletons were 0.319, 0.310, 0.302, 0.299, and 0.295, compared with the neutral value from simulations of 0.275. In the presence of BGS but not SSWs, the mean values of Tajima’s D were –0.046, –0.013, –0.019, –0.036, and 0.000, respectively, compared with the neutral value of 0.042. The mean values of the proportions of singletons were 0.288, 0.286, 0.284, 0.289, and 0.282. Thus, with the parameters used here, BGS contributes very little to the distortion in the SFS, consistent with previous theoretical work on BGS with significant amounts of recombination (Zeng and Charlesworth 2011; Nicolaisen and Desai 2013). Detailed comparisons with the data are made difficult by the probable effects of demographic factors on these measures of distortion of the SFS, which tend to obscure the effects of selection at linked sites, especially their relationships with the rate of crossing over. This problem is currently under investigation.
As mentioned above, stronger selection on favorable mutations increases the extent of distortion of the SFS. For example, with the stronger of the two selection models for the X chromosome, the Tajima’s D values, and proportions of singletons for the standard rate of crossing over for 70 genes with gene conversion, SSWs and BGS were –0.434 and 0.360, respectively. The difference between X and autosomes is qualitatively similar to what is seen for the Rwandan population of D. melanogaster, shown in figure 2 of Campos et al. (2014). However, the difference between X chromosome and autosomes regarding the distortion of the SFS is much greater than is seen in the simulations. It remains to be seen whether demographic effects can explain this discrepancy.
However, the picture is very different when crossing over is absent. For 70 autosomal genes with gene conversion, the means of Tajima’s D and the proportion of singletons for synonymous sites with BGS alone were –0.880 and 0.488, respectively. With SSWs as well, the values were changed by relatively small amounts, to –1.306 and 0.563, respectively, reflecting the greatly reduced rate of fixation of favorable mutations when crossing over is absent (Figure 3). It therefore seems likely that the distorted SFSs seen in genomic regions that lack crossing over (Cutter and Payseur 2013; Campos et al. 2014) are mainly caused by BGS in the weak interference selection limit, when interference among sites subject to purifying selection causes genealogies at linked sites to have longer terminal branches relative to neutral expectation (Gordo et al. 2002; Kaiser and Charlesworth 2009; O’Fallon et al. 2010; Seger et al. 2010; Good et al. 2014).
Problems with simulating BGS
We conclude with a discussion of some technical questions concerning the modeling of BGS in SLiM. As described in the first part of the Results section, the fact that SLiM assumes a lack of crossover interference requires the modification of the standard BGS equations to model the Haldane mapping function, as described in section S1 of File S1. In addition, for accurate approximations to the simulation results, it was necessary to include an additional term in the BGS equations, caused by deleterious mutations that were in initially in repulsion with a new neutral variant (Santiago and Caballero 1998; Charlesworth 2012b); this is ignored in the equations that are usually used to model BGS.
These properties are more a reflection of the simulation procedure than of biological reality. Equation S1b implies that the extra term added to the standard BGS equation of Nordborg et al. (1996) is proportional to the sum of twice the product of the deleterious mutation rates and the mean of t = hs for deleterious mutations, multiplied by a term that is nearly independent of the factor used for rescaling. This term is exactly equal to this product when there is no recombination, and is then equal to the additive genetic variance of fitness under deterministic mutation–selection balance (Mukai et al. 1972). Since the deterministic parameters that are thought to be realistic for a Drosophila population have been multiplied by 532 for use in the simulations, the additive genetic variance in fitness is multiplied by a factor of (532)2 = 283,024 compared with its value for the real population. With 70 genes, for example, the additive variance in the simulations is 0.0542, whereas the corresponding value for the population is 1.92 × 10−7. In contrast, the Nordborg et al. (1996) equation depends largely on the ratios of deterministic parameters, except for the multiplication of the recombination rate by a factor of 1 – t, and so is largely unaffected by the rescaling. In the real population, this additional term is effectively negligible, justifying the use of the standard equation for modeling BGS (McVicker et al. 2009; Charlesworth 2012b; Comeron 2014; Elyashiv et al. 2016; Campos et al. 2017).
The use of the Haldane mapping function also means that the simulated rate of recombination for the region as a whole is affected by the rescaling, since the frequency of double crossovers is greatly increased over what would be found in a region of the same physical length in the real population. For example, with the standard rate of crossing over and 70 genes, the map length of the region with the standard rate of crossing over is 1.62. With a Poisson distribution of numbers of crossovers, as assumed in the simulations, the proportion of double crossovers among chromosomes that have experienced a crossover is 0.5 × (1.62)2 × exp(– 1.62)/[1 – exp(– 1.62)] = 0.324. For regions of the size that we have simulated, the high level of crossover interference in Drosophila (Hughes et al. 2018) means that a linear relationship between the frequency of crossing over and physical distance is close to reality for a real population (Charlesworth 2012b). Unfortunately, except for the cases with a frequency of crossing over of one-half the standard rate used here, it is impossible to simulate a linear model with 70 genes or more, since the expected number of crossovers in the region is more than one, which is inconsistent with a model that assumes that there is either a crossover or no crossover in the region.
Given that our simulation results generally support the use of the theoretical formulae for both BGS and SSWs, largely because both BGS and SSW effects extend over much smaller distances than the whole region, this implies that the use of formulae based on the BGS and SSW equations with a linear genetic map is probably justified for most analyses of population genomic data, although it would be desirable to validate this conclusion with simulations using much larger population sizes than was feasible here.
Acknowledgments
We thank Hannes Becher, Graham Coop, Matty Hartfield, and Lei Zhao for useful discussions and comments on the manuscript, and Jeff Jensen, Peter Keightley, Wolfgang Stephan, and two anonymous reviewers for their comments on the manuscript. This work was supported by grant RPG-2015-2033 from the Leverhulme Trust (to B.C.).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7604948.
Communicating editor: G. Coop
Literature Cited
- Aguadé M., Miyashita N., Langley C. H., 1989. Restriction-map variation at the zeste-tko region in natural populations of Drosophila melanogaster. Mol. Biol. Evol. 6: 123–130. [DOI] [PubMed] [Google Scholar]
- Andolfatto P., 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 17: 1755–1762. 10.1101/gr.6691007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Nordborg M., 1998. The effect of gene conversion on intralocus associations. Genetics 148: 1397–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1995. Linkage and the limits to natural selection. Genetics 65: 821–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. 10.1017/S0016672398003462 [DOI] [Google Scholar]
- Barton N. H., 2000. Genetic hitchhiking. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1553–1562. 10.1098/rstb.2000.0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 2010. Genetic linkage and natural selection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 2559–2569. 10.1098/rstb.2010.0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rate in D. melanogaster. Nature 356: 519–520. 10.1038/356519a0 [DOI] [PubMed] [Google Scholar]
- Berry A. J., Ajioka J. W., Kreitman M., 1991. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics 129: 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker T. R., Jackson B. C., Keightley P. D., 2017. Detecting positive selection in the genome. BMC Biol. 15: 98 10.1186/s12915-017-0434-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert S., Pfaffelhuber P., 2013. The Yule approximation for the site frequency spectrum after a selective sweep. PLoS One 8: e81738 10.1371/journal.pone.0081738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. L., Zeng K., Parker D. J., Charlesworth B., Haddrill P. R., 2013. Codon usage bias and effective population sizes on the X chromosome versus the autosomes in Drosophila melanogaster. Mol. Biol. Evol. 30: 811–823. 10.1093/molbev/mss222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. L., Halligan D. L., Haddrill P. R., Charlesworth B., 2014. The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster. Mol. Biol. Evol. 31: 1010–1028. 10.1093/molbev/msu056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. L., Zhao L., Charlesworth B., 2017. Estimating the parameters of background selection and selective sweeps in Drosophila in the presence of gene conversion. Proc. Natl. Acad. Sci. USA 114: E4762–E47771. 10.1073/pnas.1619434114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2012a The effects of deleterious mutations on evolution at linked sites. Genetics 190: 5–22. 10.1534/genetics.111.134288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2012b The role of background selection in shaping patterns of molecular evolution and variation: evidence from the Drosophila X chromosome. Genetics 191: 233–246. 10.1534/genetics.111.138073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Campos J. L., 2014. The relations between recombination rate and patterns of molecular evolution and variation in Drosophila. Annu. Rev. Genet. 48: 383–403. 10.1146/annurev-genet-120213-092525 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M. T., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Campos J. L., Jackson B. C., 2018. Faster-X evolution: theory and evidence from Drosophila. Mol. Ecol. 27: 3753–3771. 10.1111/mec.14534 [DOI] [PubMed] [Google Scholar]
- Chevin L.-M., Billiard S., Hospital F., 2008. Hitchhiking both ways: effect of two interfering selective sweeps on linked neutral variation. Genetics 180: 301–316. 10.1534/genetics.108.089706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., 2014. Background selection as baseline for nucleotide variation across the Drosophila genome. PLoS Genet. 10: e1004434 10.1371/journal.pgen.1004434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Kreitman M., 2002. Population, evolutionary and genomic consequences of interference selection. Genetics 161: 389–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G., Ralph P., 2012. Patterns of neutral diversity under general models of selective sweeps. Genetics 192: 205–224. 10.1534/genetics.112.141861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig R. B., Hartl D. L., Sackton T. B., 2015. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 13: e1002112 10.1371/journal.pbio.1002112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Payseur B. A., 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14: 262–274. 10.1038/nrg3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Fisher D. S., 2007. Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics 176: 1759–1798. 10.1534/genetics.106.067678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett R., Schweinsberg J., 2004. Approximating selective sweeps. Theor. Popul. Biol. 66: 129–138. 10.1016/j.tpb.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Elyashiv E., Sattah S., Hu T. T., Strutovsky A., McVicker G., et al. , 2016. A genomic map of the effects of linked selection in Drosophila. PLoS Genet. 12: e1006130 10.1371/journal.pgen.1006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. 1. Theoretical Introduction. Springer-Verlag, New York: 10.1007/978-0-387-21822-9 [DOI] [Google Scholar]
- Ewing G. B., Jensen J. D., 2016. The consequences of not accounting for background selection in demographic inference. Mol. Ecol. 25: 135–141. 10.1111/mec.13390 [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L., Hudson R. R., Bartoszewicz A., Wall J. D., Donfack J., et al. , 2001. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69: 831–843. 10.1086/323612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 2000. Genetic drift in an infinite population. The pseudohitchhiking model. Genetics 155: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 2001. Is the population size of a species relevant to its evolution? Evolution 55: 2161–2169. 10.1111/j.0014-3820.2001.tb00732.x [DOI] [PubMed] [Google Scholar]
- Good B. H., Walczak A. M., Neher R. A., Desai M. M., 2014. Genetic diversity in the interference selection limit. PLoS Genet. 10: e1004222 10.1371/journal.pgen.1004222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I., Navarro A., Charlesworth B., 2002. Muller’s ratchet and the pattern of variation at a neutral locus. Genetics 161: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1919. The combination of linkage values and the calculation of distance between loci of linked factors. J. Genet. 8: 299–309. [Google Scholar]
- Haldane J. B. S., 1924. A mathematical theory of natural and artificial selection. Part I. Trans. Camb. Philos. Soc. 23: 19–41. [Google Scholar]
- Hartfield M., Otto S. P., 2011. Recombination and the hitchhiking of deleterious alleles. Evolution 65: 2421–2434. 10.1111/j.1558-5646.2011.01311.x [DOI] [PubMed] [Google Scholar]
- Hermisson J., Pennings P. S., 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. 10.1534/genetics.104.036947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J., Pennings P. S., 2017. Soft sweeps and beyond: understanding the patterns and probabilities of selection footprints under rapid adaptation. Methods Ecol. Evol. 8: 700–716. 10.1111/2041-210X.12808 [DOI] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. 10.1017/S0016672300010156 [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Chovnick A., 1981. Further observations on intragenic recombination in Drosophila melanogaster. Genet. Res. 38: 281–296. 10.1017/S0016672300020619 [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Harauz G., Reaume A. G., Gray M., Clark S. H., et al. , 1994. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough J., Wang W., Barrett S. C. H., Wright S. I., 2017. Hill-Robertson interference reduces genetic diversity on a young plant Y-chromosome. Genetics 207: 685–695. 10.1534/genetics.117.300142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1995. Deleterious background selection with recombination. Genetics 141: 1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Miller D. E., Miller A. L., Hawley R. S., 2018. Female meiosis: synapsis, recombination, and segregation in Drosophila melanogaster. Genetics 208: 875–908. 10.1534/genetics.117.300081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., Barton N. H., 2002. The effect of deleterious alleles on adaptation in asexual populations. Genetics 162: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser V. B., Charlesworth B., 2009. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 25: 9–12. 10.1016/j.tig.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitch-hiking” effect revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., 2006. Allele frequency distribution under recurrent selective sweeps. Genetics 172: 1967–1978. 10.1534/genetics.105.048447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2000. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics 155: 1415–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2003. Selective sweeps in the presence of interference among selected loci. Genetics 164: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1964. Diffusion models in population genetics. J. Appl. Probab. 1: 177–223. 10.2307/3211856 [DOI] [Google Scholar]
- Kimura M., 1971. Theoretical foundations of population genetics at the molecular level. Theor. Popul. Biol. 2: 174–208. 10.1016/0040-5809(71)90014-1 [DOI] [PubMed] [Google Scholar]
- Kimura M., Ohta T., 1973. The age of a neutral mutation in a finite population. Genetics 75: 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Lazzaro B. P., Phillips W., Heikkinen E., Braverman J. M., 2000. Linkage disequilibria and the site frequency spectra in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics 156: 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malécot G., 1969. The Mathematics of Heredity. W. H. Freeman, San Francisco. [Google Scholar]
- Martin G., Lambert A., 2015. A simple, semi-deterministic approximation to the distribution of selective sweeps in large populations. Theor. Popul. Biol. 101: 40–46. 10.1016/j.tpb.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. 10.1017/S0016672300014634 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., 1976. What determines the rate of evolution? Am. Nat. 110: 331–338. 10.1086/283071 [DOI] [Google Scholar]
- McVean G. A. T., Charlesworth B., 2000. The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G., Gordon D., Davis C., Green P., 2009. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5: e1000471 10.1371/journal.pgen.1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., 2013. SLiM: simulating evolution with selection and linkage. Genetics 194: 1037–1039. 10.1534/genetics.113.152181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., Petrov D. A., 2013. Frequent adaptation and the McDonald-Kreitman test. Proc. Natl. Acad. Sci. USA 110: 8615–8620. 10.1073/pnas.1220835110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Smith C. B., Kazemi N. Y., Cockrell A. J., Arvanitakas A. V., et al. , 2016. Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 203: 159–171. 10.1534/genetics.115.186486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Chigusa S. I., Mettler L. E., Crow J. F., 1972. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics 72: 335–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R. A., 2013. Genetic draft, selective interference and population genetics of rapid adaptation. Annu. Rev. Ecol. Evol. Syst. 44: 195–215. 10.1146/annurev-ecolsys-110512-135920 [DOI] [Google Scholar]
- Nicolaisen L. E., Desai M., 2013. Distortions in genealogies due to purifying selection and recombination. Genetics 195: 221–230. 10.1534/genetics.113.152983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996. The effect of recombination on background selection. Genet. Res. 67: 159–174. 10.1017/S0016672300033619 [DOI] [PubMed] [Google Scholar]
- O’Fallon B. D., Seger J., Adler F. R., 2010. A continuous-state coalescent and the impact of weak selection on the structure of gene genealogies. Mol. Biol. Evol. 27: 1162–1172. 10.1093/molbev/msq006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1970. Development of associative overdominance through linkage disequilibrium in finite populations. Genet. Res. 16: 165–177. 10.1017/S0016672300012684 [DOI] [PubMed] [Google Scholar]
- Peck J., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations, and the evolution of sex. Genetics 137: 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffelhuber P., Haubold B., Wakolbinger A., 2006. Approximate genealogies under genetic hitchhiking. Genetics 174: 1995–2008. 10.1534/genetics.106.061887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago E., Caballero A., 1998. Effective size and polymorphism of linked neutral loci in populations under selection. Genetics 149: 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Houle D., Lynch M., Hahn M. W., 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194: 937–954. 10.1534/genetics.113.151670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Shanku A. G., Kern A. D., 2016. Effects of linked selective sweeps on demographic inference and model selection. Genetics 204: 1207–1223. 10.1534/genetics.116.190223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger J., Smith W. A., Perry J. J., Hunn K., Kaliszewska Z. A., et al. , 2010. Gene genealogies strongly distorted by weakly interfering mutations in constant environments. Genetics 184: 529–545. 10.1534/genetics.109.103556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., Petrov D. A., Przeworski M., Andolfatto P., 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5: e1000495 10.1371/journal.pgen.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers K. F., Morris D. S., 2017. Underdispersion models: models that are “under the radar”. Commun. Stat. Theory Methods 46: 12075–12086. 10.1080/03610926.2017.1291976 [DOI] [Google Scholar]
- Stephan W., 2010. Genetic hitchhiking vs. background selection: the controversy and its implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1245–1253. 10.1098/rstb.2009.0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., Wiehe T. H. E., Lenz M. W., 1992. The effect of strongly selected substitutions on neutral polymorphism: analytical results based on diffusion theory. Theor. Popul. Biol. 41: 237–254. 10.1016/0040-5809(92)90045-U [DOI] [Google Scholar]
- Stephan W., Charlesworth B., McVean G. A. T., 1999. The effect of background selection at a single locus on weakly selected, partially linked variants. Genet. Res. 73: 133–146. 10.1017/S0016672399003705 [DOI] [Google Scholar]
- Sved J. A., 1968. The stability of linked systems of loci with a small population size. Genetics 59: 543–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti J., Grossman S. R., Sabeti P. C., 2013. Detecting natural selection in genomic data. Annu. Rev. Genet. 47: 97–120. 10.1146/annurev-genet-111212-133526 [DOI] [PubMed] [Google Scholar]
- Walsh B., Lynch M., 2018. Evolution and Selection of Quantitative Traits. Oxford University Press, Oxford: 10.1093/oso/9780198830870.001.0001 [DOI] [Google Scholar]
- Weissman D. B., Barton N. H., 2012. Limits to the rate of adaptive substitution in sexual populations. PLoS Genet. 8: e1002740 10.1371/journal.pgen.1002740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D. B., Hallatschek O., 2014. The rate of adaptation in large sexual populations with linear chromosomes. Genetics 196: 1167–1183. 10.1534/genetics.113.160705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe T. H. E., Stephan W., 1993. Analysis of a genetic hitchhiking model and its application to DNA polymorphism data. Mol. Biol. Evol. 10: 842–854. [DOI] [PubMed] [Google Scholar]
- Zeng K., Charlesworth B., 2011. The joint effects of background selection and genetic recombination on local gene genealogies. Genetics 189: 251–266. 10.1534/genetics.111.130575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Charlesworth B., 2016. Resolving the conflict between associative overdominance and background selection. Genetics 203: 1315–1334. 10.1534/genetics.116.188912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7604948.