The effect of selection on linked neutral sites is widely used to detect selected loci. While existing models provide a good description of the patterns expected from background selection or sweeps on a small...
Keywords: diversity, background selection, selective sweeps, flycatcher
Abstract
There is an increasing awareness that selection affecting linked neutral sites strongly influences on how diversity is distributed across the genome. In particular, linked selection is likely involved in the formation of heterogenous landscapes of genetic diversity, including genomic regions with locally reduced effective population sizes that manifest as dips in diversity, and “islands” of differentiation between closely related populations or species. Linked selection can be in the form of background selection or selective sweeps, and a long-standing quest in population genetics has been to unveil the relative importance of these processes. Here, we analyzed the theoretically expected reduction of diversity caused by linked selection in the collared flycatcher (Ficedula albicollis) genome and compared this with population genomic data on the distribution of diversity across the flycatcher genome. By incorporating data on recombination rate variation and the density of target sites for selection (including both protein-coding genes and conserved noncoding elements), we found that background selection can explain most of the observed baseline variation in genetic diversity. However, positive selection was necessary to explain the pronounced local diversity dips in the collared flycatcher genome. We confirmed our analytical findings by comprehensive simulations. Therefore, our study demonstrates that even though both background selection and selective sweeps contribute to the heterogeneous diversity landscape seen in this avian system, they play different roles in shaping it.
NATURAL selection reduces genetic diversity at neutral sites mainly by two key concepts of population genetics: hitchhiking (Smith and Haigh 1974; Kaplan et al. 1989; Fay and Wu 2000) and background selection (BGS) (Charlesworth et al. 1993; Hudson and Kaplan 1995; Nordborg et al. 1996). The former process means that positive selection for an advantageous variant also selects for the genetic background (haplotype) on which the beneficial allele resides. The latter process implies that purifying selection against recurring deleterious mutations decreases the diversity at linked neutral sites.
An advantageous allele is expected to quickly increase in frequency and reach fixation in the population, thereby being a potent diversity-reducing force in large genomic regions linked to (hitchhiking with) the site under positive selection. Removal of deleterious variants is typically a slower process if the fitness effects are small and/or the effective population size is low (Crow and Kimura 1970). During the time a disadvantageous allele still segregates in the population, recombination breaks up linkage to nearby variants and thereby narrows the region in which genetic diversity will become reduced by purifying selection. Importantly, BGS only removes haplotypes on which deleterious alleles reside, leaving variants carried by other haplotypes free to segregate. Therefore, single episodes of purifying selection do not have the same diversity-reducing effect as positive selection (e.g., Stephan 2010). However, since the distribution of fitness effects is typically strongly biased toward deleterious mutations (Eyre-Walker and Keightley 2007), the combined effect of selection against many deleterious mutations means that BGS is likely to have an as strong, or even stronger, effect on genetic diversity as selective sweeps. Elucidating the relative importance of these two types of linked selection, and under which circumstances one or the other process dominates, has been a challenging question (Charlesworth 1996; Kim and Stephan 2000; McVicker et al. 2009; Comeron 2014; Elyashiv et al. 2016). The feasibility of obtaining large-scale population genomic data, together with data on recombination rate variation and detailed genome annotation, now offer exciting possibilities to understand the underlying processes behind linked selection.
If linked selection is prevalent and acts genome-wide, this should be visible as correlations between diversity and factors affecting the extent of linked selection. First, genomic regions with high recombination rates are expected to experience less linked selection since, as indicated above, recombination decouples linked loci and restricts the area of effect of a selected mutation (e.g., Kaplan et al. 1989; Nordborg et al. 1996). Thus, diversity should be positively correlated with recombination rate [see for example Mackay et al. (2012)]. Second, regions with a high density of potential targets for selection are expected to experience more linked selection simply because selection occurs more often in such regions. Thus, diversity should be negatively correlated with the density of target sites for selection. Indeed, such correlations have been found in several organisms (e.g., Begun and Aquadro 1992; Nachman 2001; Tenaillon et al. 2001; Nordborg et al. 2005; Cutter and Payseur 2013; Burri et al. 2015). Third, mutation rate variation influences the number of newly arising mutations under purifying selection and thus also the local strength of linked selection.
Given these relationships and with access to appropriate data, it should be possible to predict genetic diversity under varying influence of BGS and selective sweeps (Charlesworth 1996; McVicker et al. 2009; Comeron 2014, 2017; Elyashiv et al. 2016).
Linked selection is of particular relevance in the context of speciation (Seehausen et al. 2014; Wolf and Ellegren 2017). During speciation with gene flow, divergent selection at loci underlying ecologically relevant traits or genetic incompatibilities will hinder gene flow in the vicinity of such loci. In turn, this will lead to localized signals of increased differentiation and reduced diversity, and the formation of so-called speciation islands (Nosil 2008; Nosil et al. 2008; Feder and Nosil 2010). However, it has recently been recognized that linked selection can produce similar patterns even in the absence of gene flow (Charlesworth 1998; Noor and Bennett 2009; Turner and Hahn 2010; Cruickshank and Hahn 2014; Burri et al. 2015). Linked selection will locally reduce the effective population size (Ne), and thereby not only reduce diversity but also lead to elevated measures of relative differentiation between diverging lineages. If the extent of linked selection varies across the genome, so too will the extent of differentiation. The significance of genomic islands of differentiation in speciation is thus disputed [see Ravinet et al. (2017) for a review].
Ficedula flycatchers represent a well-studied model system for speciation research (Lundberg and Alatalo 1992) and constitute one of the most prominent examples of species with distinct differentiation islands (which fully coincide with the location of dips in genetic diversity) spread across the genome (Ellegren et al. 2012; Burri et al. 2015). Relative differentiation (FST) between these recently (<1–2 MY; Nater et al. 2015) diverged species is negatively correlated with recombination rate, indicating a role of linked selection in generating heterogeneous diversity/differentiation landscapes (Burri et al. 2015). Here, we aim to disentangle the forces responsible for shaping the genomic diversity landscape of Ficedula flycatchers by modeling the expected impact of linked selection under various scenarios of positive and purifying selection. We compare these results with extensive genome-wide resequencing data and augment the analyses with simulations. We benefit from access to recombination rate data obtained from linkage analysis (Kawakami et al. 2014) and genome annotation, including not only protein-coding genes but also conserved noncoding elements (Craig et al. 2018). While we find that BGS is generally sufficient to explain the baseline levels of genetic diversity in the flycatcher genome, our study suggests that selective sweeps are necessary to generate the most pronounced diversity dips.
Methods
Genomic and population genomic data
Estimates of nucleotide diversity (π) in nonoverlapping 200-kb windows of the collared flycatcher (Ficedula albicollis), obtained from whole-genome resequencing of extensive population samples (79 individuals from four populations), were taken from Burri et al. (2015). To estimate site frequency spectrum-based statistics, an approach was used that integrates over genotype likelihoods as implemented in the software ANGSD (Korneliussen et al. 2014). This method accounts for genotype uncertainty and considerably improves local estimates of π for low- and medium-coverage data. Additionally, the sequence data were carefully filtered to avoid biases caused by poorly aligned reads or low sequencing coverage. Briefly, repetitive regions in the reference genome were masked with a custom flycatcher-specific repeat library. Only sites fulfilling the following criteria were considered for calculation of π: minimum mapping quality of 1, minimum base quality of 20, site coverage across all individuals < 5 SD above the mean coverage, and a minimum read coverage of 5× per individual with a minimum of 10 callable genotypes per population.
We used a discrete time hidden Markov model implemented in the “HiddenMarkov” R package (https://CRAN.R-project.org/package = HiddenMarkov) to classify the window-based π estimates into background regions and diversity valleys. For the observed process dependent on the two hidden states (background and diversity valley), we assumed two normal distributions with SD fixed to the SD of the empirical distribution of π values. We then optimized the means of the distributions for the two hidden states with the Baum–Welch algorithm (Baum et al. 1970) with a maximum number of 1000 iterations. After parameter estimation, we used the Viterbi algorithm to find the most likely sequence of hidden states and identify genomic regions with a predicted diversity valley state.
Collared flycatcher recombination rate estimates in cM/Mb for 200-kb windows were taken from Kawakami et al. (2014) and were based on a high-density genetic map containing 4302 markers. To generate this map, 609 collared flycatcher individuals from a four-generation pedigree were genotyped with a custom-designed SNP array for 37,262 polymorphic loci. The use of recombination rate estimates from a pedigree-based linkage analysis instead of higher-resolution LD-based recombination maps (e.g., Kawakami et al. 2017) has the advantage that they, in contrast to the latter, are not affected by selection [i.e., it allows the direct estimation of recombination rate (r)]. Coordinates for conserved regions in the F. albicollis genome, including noncoding regions, were taken from Craig et al. (2018). Exon information was obtained from the Ensembl annotation of the collared flycatcher genome assembly version FicAlb 1.4.
Expected reduction of π due to linked selection
We compared three different models to calculate the expected deviation from neutral diversity due to selection. Model 1 was with only BGS, model 2 was model 1 plus recent sweeps, and model 3 was model 2 plus ancient sweeps. For an overview of model parameters, see Table 1.
Table 1. Model parameters.
| Parameter | Value | Reference |
|---|---|---|
| π0 | 0.0048 | This study |
| µ | 4.6 × 10−9 | Smeds et al. (2016) |
| r | Per window | Kawakami et al. (2014) |
| Ne | 450,000 | Nater et al. (2015) |
| T | 320,000 | Nater et al. (2015) |
| ud | Per window | Depends on dcs, Craig et al. (2018) |
| sd | DFE | Bolívar et al. (2018) |
| sb | 0.1/0.05/0.01 | Variable model parameter |
| α | 1/0.2 | Variable model parameter |
DFE: distribution of fitness effects. dcs: density of conserved sites.
Under neutrality, nucleotide diversity , where is the effective population size and μ the mutation rate. Mutations at neutral sites are assumed to have no effect on fitness. We used equations from Hudson and Kaplan (1995) and Nordborg et al. (1996) to calculate the expected deviation from neutral diversity due to BGS, (McVicker et al. 2009). B at a focal neutral site x can be approximated by
| (1) |
where is without selection and with free recombination, the sum is over all selected sites, and is the deleterious mutation rate per site, which is the neutral mutation rate times the proportion of selection targets. is the selection coefficient against heterozygotes, and r(x,i) is the recombination probability between the focal neutral site and the selected site i. For , the distribution of fitness effects of new mutations at nonsynonymous sites estimated for collared flycatchers was used, but with distinct values instead of intervals [13% Ns = −1, 9% Ns = −10, 14% Ns = −100, and 64% Ns = −500; compare to Bolívar et al. (2018)]. As it would be too computationally intensive to calculate B for every site along a chromosome, we implemented a window-based approach with chromosomes divided into nonoverlapping 1-kb windows. To calculate genetic distances between two sites, all sites (neutral and selected) were assumed to be located at the midpoint of their respective window. B for each window was thus obtained by considering a neutral site in the midpoint and calculating the influence of selected sites in all linked windows. The smaller the window size, the more exact this approach. To compare the model outcome with π from the data, which was available in 200-kb windows to match the recombination rate data in turn, mean B for the respective 200-kb windows was calculated from the mean of the 1-kb windows.
As a measure of the density of sites under purifying selection (subsequently dcs), we used the number of conserved sites (Craig et al. 2018) per window and assumed that all mutations occurring at these sites were deleterious. As π estimates from the data include sites under selection, the model has to include direct selection as well. For each window, the allele frequencies at conserved sites were assumed to be in mutation–selection balance. The expected frequency for additive deleterious mutations is (Crow and Kimura 1970) and for each conserved site in the window . Using the distribution of fitness effects of new mutations estimated for collared flycatchers (Bolívar et al. 2018) and a mutation rate of 4.6 × 10−9 (Smeds et al. 2016), we estimated mean π = 0.0006 for sites under purifying selection. Note that this is simply a model assumption.
Following Elyashiv et al. (2016), the effect of positive selection can be incorporated into the model. The rate of coalescence at a neutral position x due to a selective sweep at a selected position j is
| (2) |
where T is the length of the lineage for which substitutions are considered, α the fraction of substitutions that are beneficial, is the selection coefficient, and τ is the expected time to fixation of a positively selected mutation, which depends on Ne and sb [see Elyashiv et al. (2016)]. As candidates for sites that have undergone a selective sweep, we used nonsynonymous substitutions fixed in the collared flycatcher since the split from pied flycatcher (F. hypoleuca) for recent sweeps or from red-breasted flycatcher (F. parva, with F. hyperythra as an outgroup) for ancient sweeps. We set the divergence time between collared flycatcher and pied flycatcher to 320,000 generations, and between collared flycatcher and red-breasted flycatcher to 2,500,000 generations (Nater et al. 2015). We used an estimate of the long-term in collared flycatcher of 450,000 (Nater et al. 2015). In general, we used = 0.1 and α = 1 for the recent sweeps, thus treating all nonsynonymous substitutions as strong sweeps, as a contrast to the BGS-only model. For the ancient sweeps, we generally used α = 0.2 (∼0.18 estimated for flycatcher (Bolívar et al. 2018). However, we also investigated the effect on the diversity landscape when using different values for α and sb.
Following Elyashiv et al. (2016), total π is
| (3) |
To calculate the BGS parameter B from the data , we assumed a neutral (genome-wide average is 0.0041). This was obtained by averaging over all 200-kb windows with dcs < 0.05 and recombination rate >3 cM/Mb, which should represent the genomic regions least influenced by linked selection. Importantly, while the choice of affects , it does not influence the correlation between and B calculated from the model , as it is only a scaling parameter. For five chromosomes, recombination rate was not known for some windows at the chromosome ends. For chromosome 8, these were 22,200-kb windows, which were excluded from all analyses. For chromosomes 1, 6, 18, and 28, this only concerned one-to-three windows, for which recombination rates were interpolated from adjacent windows. Regions with interpolated recombination rates are marked in Supplemental Material, Figure S1.
Simulations
To validate the models, we used individual-based simulations in fwdpp 0.4 (Thornton 2014) with the following scenario. We assumed a constant population size of 2000 diploid individuals. The mutation rate was scaled accordingly to get a similar π0 as the data. The probability of a recombination event at a certain position was determined by the local recombination rate, taken from Kawakami et al. (2014). Individuals for reproduction were sampled proportionally to their fitness, with a constant population size (soft selection). Mating occurred by random pairing of gametes after mutation and recombination. As above, the density of conserved sites in a window was used to infer the probability of deleterious mutations. We simulated entire chromosomes corresponding to the sizes in the collared flycatcher reference genome and ran simulations for 6N generations. To calculate π, we drew a random sample of 1% of the individuals from the population.
Data availability
All data used for this study were previously published. See Kawakami et al. (2014), Burri et al. (2015) and Craig et al. (2018). Supplemental Material and C++ code used for the simulations and calculation of Bm are available at Figshare: https://figshare.com/s/5ee30bae0acfad563865.
Results
Correlations between π and recombination rate, and density of conserved sites
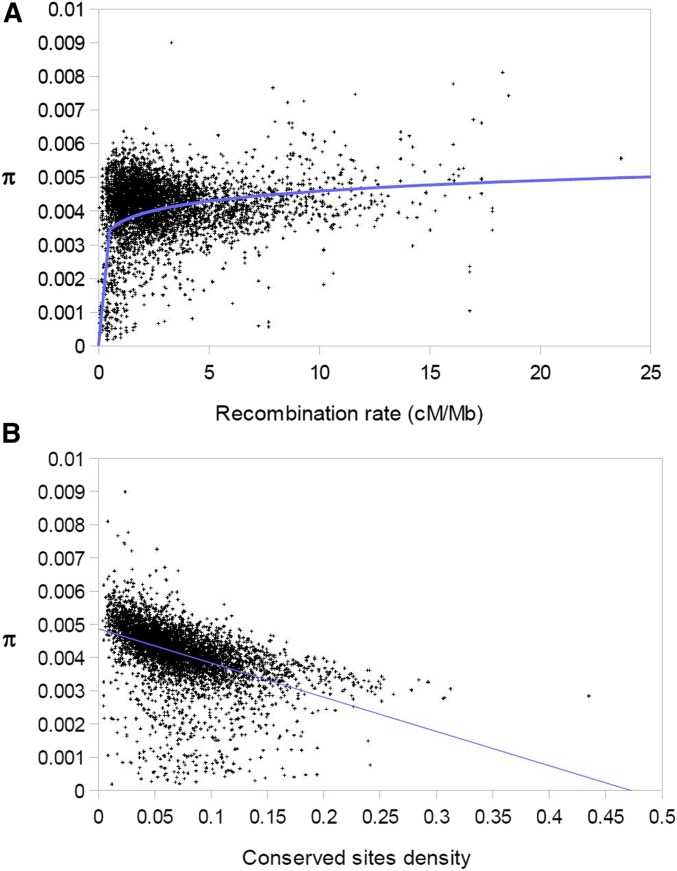
The association between nucleotide diversity and rate of recombination in the collared flycatcher is best explained by a curvilinear relationship ( with a power regression vs. r2 = 0.02 with a linear regression, Figure 1a). Nucleotide diversity increased with recombination rate up to ≈1.5 cM/Mb, but was then saturated. The variance in π among windows was higher at low than at high recombination rates. As expected, π was negatively correlated with the density of conserved sites (dcs; , linear regression, Figure 1b). The correlation was weaker when exon density was used as a proxy for the density of target sites for selection , indicating an important role of selection in noncoding regions [compare with Craig et al. (2018)]. However, note that the correlation is not exclusively due to linked selection but also due to direct selection. A multiple linear regression of π in dependence of dcs and recombination rate (linear factors) gave . Plots of the distribution of π, recombination rate, and dcs along chromosomes readily demonstrate that the overall shape of the diversity landscape resembles the recombination landscape with diversity valleys corresponding to regions with low recombination, while fine-scale differences in π tend to be due to variation in dcs (Figure S1).
Figure 1.
Relationship between mean nucleotide diversity and (a) recombination rate in cM/Mb , and (b) proportion of conserved sites (r2 = 0.22) for 200-kb windows.
Modeling of linked selection
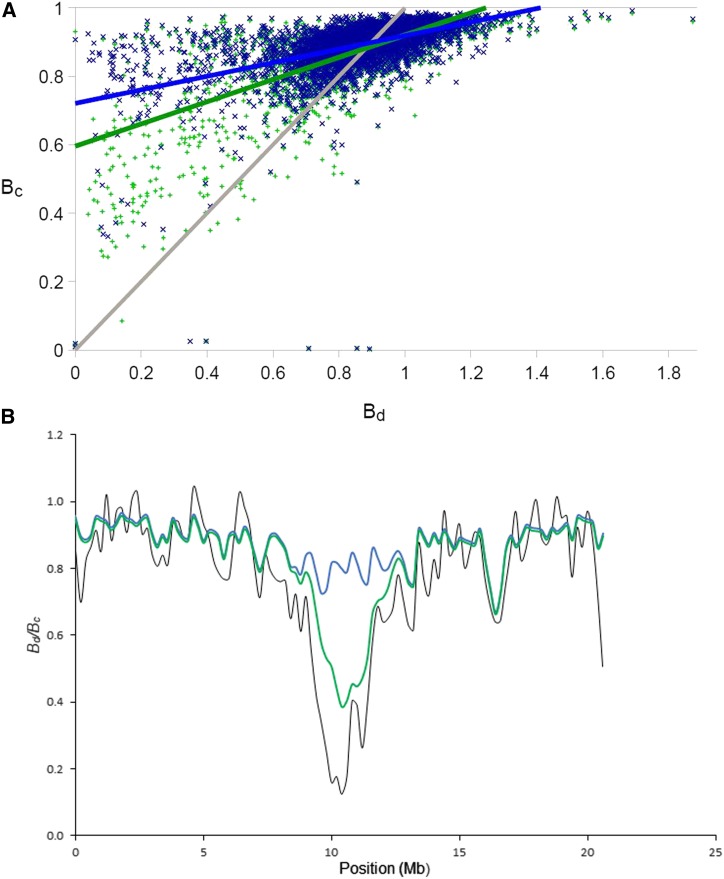
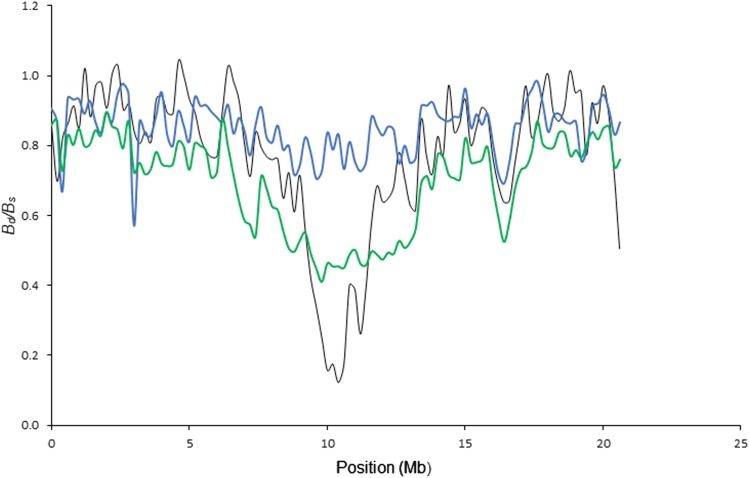
The genome-wide correlation between from model 1 (BGS) and in 200-kb windows was (Figure 2a). Correlations for individual chromosomes are given in Table S1. An example of the relationship between and using model 1 along a chromosome (chromosome 12) is shown in Figure 2b (for other chromosomes, see Figure S5). While the baseline of the modeled diversity landscape closely matches the data, the amount of reduction in the diversity valley of this chromosome deviates strongly. This was the case for basically all diversity valleys, of which there usually are one or two per chromosome [see Ellegren et al. (2012) and Burri et al. (2015)]. Higher mutation rates or lower recombination rates than the estimates used could explain some of the difference between and in valleys (see File S1).
Figure 2.
(a) Genome-wide correlation between (the modeled deviation from neutral diversity) and (B calculated from data) under two different linked-selection models. The gray line marks identity for better orientation. Colored lines are linear regressions with blue showing only background selection (model 1), and green showing BGS and recent sweeps (model 2). (b) A comparison between (black) and calculated with BGS (model 1, blue line) and BGS plus recent sweeps (model 2, green), with , α = 1 for chromosome 12. BGS, background selection.
If we include recent strong positive selection as indicated by nonsynonymous substitutions fixed in the collared flycatcher lineage since its split from pied flycatcher (model 2), the overall correlation between and for s = 0.1 was (Figure 2a). As the number of sweeps modeled is likely to be higher than the actual number, model 2 underestimates B more often and has higher variance than model 1, but the correlation is much better than with model 1. This was consistent for most chromosomes (Table S1). now showed deep and pronounced diversity valleys (Figure 2b). With lower sb, the valley in chromosome 12 is less deep and the correlation is lower, but still higher than with BGS alone (r2 = 0.55; for sb = 0.01, r2 = 0.58; for sb = 0.05, r2 = 0.81; and for sb = 0.1 r2 = 0.85.
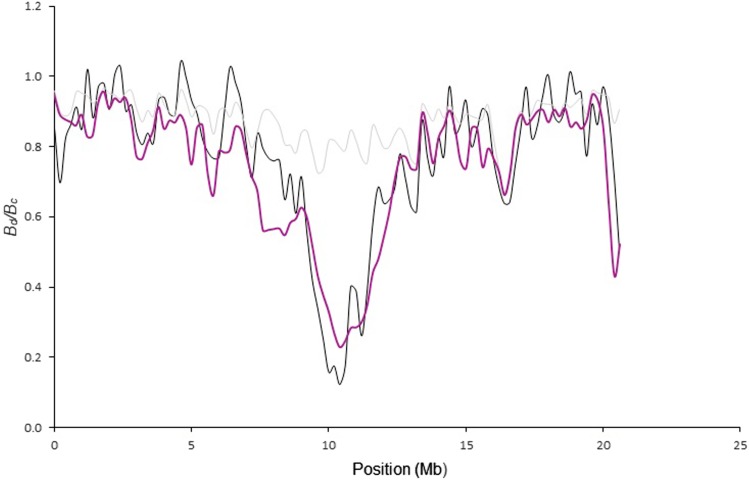
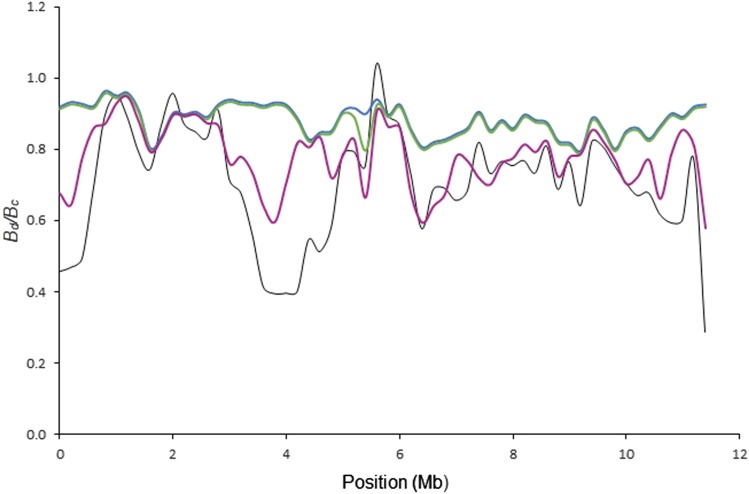
Model 3 incorporates sweeps at sites of substitutions since the split from the more distantly related red-breasted flycatcher. The overall correlation using α = 0.2 for the ancestral sweeps was weaker than with both model 1 and 2. Nevertheless, as exemplified for the terminal diversity valley of chromosome 12, some dips were only explained by model 3 (Figure 3). For some chromosomes, model 3 in fact gave the best fit (e.g., in chromosome 19 vs. r2 = 0.03 with model 2; Figure 4 and Table S1). This is important since it means that while sweeps are needed to explain diversity valleys, they do not necessarily have to be recent. Using α = 1 like for recent sweeps, the correlation becomes lower for chromosome 12 (r2 = 0.51 vs. 0.76 with α = 0.2).
Figure 3.
A comparison between (black) and calculated with background selection plus recent (, α = 1) and ancient (, α = 0.2) sweeps (model 3) (purple). The gray line represents Bc under model 1. Parameters for recent sweeps as in Figure 2.
Figure 4.
A comparison between (black) and from model 1 (blue), model 2 (green), and model 3 (purple) for chromosome 19. Parameters are as in Figure 2.
To summarize, which of the three models of linked selection best explains the observed local reductions in diversity differs along the genome? While BGS largely explains the base level of genomic diversity, the π valleys are explained better with either recent or ancient sweeps. Across all chromosomes, 56% of the π valleys were explained by model 2 (including recent sweeps), 26% by model 3 (also including ancient sweeps), and 9% by model 1 (only BGS), while 9% were not covered by any of the models. See Figure S5 for a comparison across all chromosomes.
Simulations
Individual-based simulations of diversity along one example chromosome confirmed our analytical results. Even though the general shape of the diversity landscape and also the base level of simulated B closely matched the analytical expectation under BGS , the diversity valley was not visible if we only allowed for deleterious mutations (Figure 5, blue line). If, in addition, beneficial mutations occurred, a diversity valley emerged (Figure 5, green line), though not as pronounced as in the empirical data. Importantly, the valley formed even though beneficial mutations occurred randomly all over the chromosome and not only in the valley region.
Figure 5.
Simulation of chromosome 12, population size N = 2000, run for 6N generations, selection coefficient for deleterious mutations , averaged over 40 runs. Blue line: only background selection; green line: background selection plus 1% beneficial mutations with . The black line shows B calculated from the empirical data.
Discussion
Both the simulations and the analytical results show that BGS can explain a large part of the base-level and small-scale variation of genetic diversity in the flycatcher genome. The correlation between our BGS model and π from the data lies within the range of correlations found in similar studies in human (McVicker et al. 2009) and Drosophila (Comeron 2014; Elyashiv et al. 2016). On the other hand, the deep diversity valleys present in the data cannot be explained with BGS alone. When we included selective sweeps at candidate sites in the model or when we added 1% positively selected mutations to the simulations, we generally obtained a better fit to the empirical data. However, positive selection in the model needs to be relatively high to generate valleys as pronounced as observed in the data. There are alternative explanations to why BGS alone, with the parameters used herein, does not explain the full magnitude of variation in diversity levels. Since the rate and fitness effects of newly arising deleterious mutations are key parameters in determining the strength of BGS, a major limitation of this study might be the restriction to consider only point mutations. Other mutation events, such as short insertions or deletions, might contribute a substantial part of the total input of deleterious mutations (e.g., Mills et al. 2006). Additionally, we assumed a distribution of fitness effects for deleterious mutations that was estimated based on nonsynonymous sites and might not accurately represent the distribution of effects of other mutation types. However, as discussed in File S1, while other reasons than selective sweeps, like under- or overestimation of used parameters (density of functional sites, mutation and recombination rate), or demography, may contribute to the depth of diversity valleys, we found none that would likely explain it exclusively. It is interesting to consider that while other forces than linked selection could increase the valley depth, they also would likely distort the correlation of π with the linked-selection landscape formed by recombination and the density of functional sites. Additionally, there are several modes of selection, like balancing selection, that our models did not consider. As we only used nonsynonymous substitutions as candidate sites for positive selection, it is also possible that additional sweeps affected noncoding regions, which might result in diversity valleys not explained by any of our models. These could be possible explanations for the valleys that could not be explained by our models. Our results concerning the relative contributions of BGS and hitchhiking to linked selection go in the same direction as the findings of Elyashiv et al. (2016) in Drosophila melanogaster. For 100-kb windows, they found a correlation between π and B with BGS + sweeps of 0.44, compared to 0.42 with only BGS. The difference between the models with and without sweeps was thus larger in flycatchers than Drosophila. Notably, the valleys in the heterogeneous diversity landscape so characteristic for flycatchers was better explained with sweeps at sites of nonsynonymous substitutions in the flycatcher lineage than with BGS alone. The simulations showed that even with a random location of sweeps (i.e., not restricted to sites of nonsynonymous substitutions), diversity valleys were better captured than with BGS alone (see Figure 5). This can be explained by the higher impact of positive selection in regions with low recombination. With a higher overall effect of linked selection, recombination rate has a higher impact on the diversity landscape, leading to the formation of pronounced diversity valleys in low-recombination regions.
In general, model 3 (with recent and ancient sweeps) gave a worse fit than model 2 (recent sweeps). This is because more false sweeps are introduced, producing valleys that do not fit the data and negatively affect the correlation. Although the model with recent sweeps was better than other models in explaining diversity valleys, in some cases only the model that also incorporated ancient sweeps could explain the presence of such valleys (see Figure 3 and Figure 4). That sweeps do not need to be recent or specific to the focal species to generate locally reduced diversity levels is consistent with the finding that the locations of valleys are often conserved among Ficedula species (Burri et al. 2015). Most diversity valleys are located at chromosome ends and/or at the position of presumed centromeres (Ellegren et al. 2012; Burri et al. 2015). Sweeps could occur relatively frequently in these regions due to a meiotically driven arms race in centromeres or telomeres (Henikoff et al. 2001; Malik and Henikoff 2009). A role, yet not exclusive, of positive selection behind the heterogeneous flycatcher diversity landscape is also consistent with the observation that Fay and Wu’s H statistic often shows signatures of positive selection in diversity valleys (Burri et al. 2015).
Contrary to the expectations for speciation islands, our findings indicate that diversity valleys in flycatchers are not a direct consequence of locally reduced gene flow. However, the reduced effective population size caused by the effects of linked selection in these regions might still promote the rapid build-up of drift-induced Dobzhansky–Muller incompatibilities or other isolating mechanisms during an allopatric phase, therefore reducing subsequent gene flow in secondary contact (Dobzhansky 1936; Muller 1942). Thus, although reduced gene flow can be safely rejected as cause of differentiation peaks in flycatchers, fully rejecting them as “speciation islands” might be premature.
Our results indicate that positive selection contributes to the base variation of genetic diversity across the genome rather than producing classical outliers. Recombination rate and the (spatial and temporal) abundance of selected mutations determine how selection influences nearby neutral sites. This is true for deleterious and beneficial mutations, though the impact varies. Thus, it is quite reasonable to conclude that recombination rate and functional site density form a linked-selection scaffold, which determines the impact of linked selection in a certain region of the genome. This is the shape that we can calculate with equation 1, while the total impact depends on selection strength and sign. This may be one explanation why selective sweeps are so hard to find: they are embedded in the general linked-selection landscape. But it is important to keep in mind that the reverse is also true: if there are diversity valleys in the data, it does not necessarily mean that there is something special happening, the impact of linked selection might just be stronger in this region. This is also true for summary statistics that depend on linked selection. Note that even for windows without sites under selection we do not find B = 1, which means that the whole genome is influenced by linked selection. Thus, we should be careful when using intergenic regions as neutral baseline. Our results thus add to the general demand for using BGS as a null hypothesis for diversity levels (Comeron 2017).
Acknowledgments
We thank Reto Burri and Rory Craig for sharing their data files, and David Castellano and two anonymous reviewers for helpful comments. This research was supported by the Swedish Research Council (2013-8271) and the Knut and Alice Wallenberg foundation. A.N. was financially supported by a Swiss National Science Foundation grant (P300PA_177852).
Footnotes
Supplemental material available at Figshare: https://figshare.com/s/5ee30bae0acfad563865.
Communicating editor: R. Nielsen
Literature Cited
- Baum L. E., Petrie T., Soules G., Weiss N., 1970. A maximization technique occurring in the statistical analysis of probabilistic functions of Markov chains. Ann. Math. Stat. 41: 164–171. 10.1214/aoms/1177697196 [DOI] [Google Scholar]
- Begun D. J., Aquadro C. F., 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520. 10.1038/356519a0 [DOI] [PubMed] [Google Scholar]
- Bolívar P., Mugal C. F., Rossi M., Nater A., Wang M., et al. , 2018. Biased inference of selection due to GC-biased gene conversion and the rate of protein evolution in flycatchers when accounting for it. Mol. Biol. Evol. 35: 2475–2486. 10.1093/molbev/msy149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R., Nater A., Kawakami T., Mugal C. F., Olason P. I., et al. , 2015. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res. 25: 1656–1665. 10.1101/gr.196485.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1996. Background selection and patterns of genetic diversity in Drosophila melanogaster. Genet. Res. 68: 131–149. 10.1017/S0016672300034029 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol. Biol. Evol. 15: 538–543. 10.1093/oxfordjournals.molbev.a025953 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Morgan M., Charlesworth D., 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., 2014. Background selection as baseline for nucleotide variation across the drosophila genome. PLoS Genet. 10: e1004434 10.1371/journal.pgen.1004434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., 2017. Background selection as null hypothesis in population genomics: insights and challenges from Drosophila studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160471 10.1098/rstb.2016.0471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., Suh A., Wang M., Ellegren H., 2018. Natural selection beyond genes: identification and analyses of evolutionarily conserved elements in the genome of the collared flycatcher (Ficedula albicollis). Mol. Ecol. 27: 476–492. 10.1111/mec.14462 [DOI] [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Harper & Row, New York. [Google Scholar]
- Cruickshank T. E., Hahn M. W., 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 23: 3133–3157. 10.1111/mec.12796 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Payseur B. A., 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14: 262–274. 10.1038/nrg3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. H., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Smeds L., Burri R., Olason P. I., Backström N., et al. , 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491: 756. [DOI] [PubMed] [Google Scholar]
- Elyashiv E., Sattath S., Hu T. T., Strustovsky A., McVicker G., et al. , 2016. A genomic map of the effects of linked selection in drosophila. PLoS Genet. 12: e1006130 10.1371/journal.pgen.1006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. 10.1038/nrg2146 [DOI] [PubMed] [Google Scholar]
- Fay J. C., Wu C.-I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder J. L., Nosil P., 2010. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64: 1729–1747. 10.1111/j.1558-5646.2009.00943.x [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H. S., 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1995. Deleterious background selection with recombination. Genetics 141: 1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R., Langley C., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Smeds L., Backström N., Husby A., Qvarnström A., et al. , 2014. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol. Ecol. 23: 4035–4058. 10.1111/mec.12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Mugal C. F., Suh A., Nater A., Burri R., et al. , 2017. Whole-genome patterns of linkage disequilibrium across flycatcher populations clarify the causes and consequences of fine-scale recombination rate variation in birds. Mol. Ecol. 26: 4158–4172. 10.1111/mec.14197 [DOI] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2000. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics 155: 1415–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T. S., Albrechtsen A., Nielsen R., 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15: 356 10.1186/s12859-014-0356-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg A., Alatalo R., 1992. The Pied Flycatcher. Poyser, London. [Google Scholar]
- Mackay T. F., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G., Gordon D., Davis C., Green P., 2009. Widespread genomic signatures of natural selection in hominid evolution PLoS Genet. 5: e1000471 10.1371/journal.pgen.1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. E., Luttig C. T., Larkins C. E., Beauchamp A., Tsui C., et al. , 2006. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 16: 1182–1190. 10.1101/gr.4565806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Nachman M. W., 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17: 481–485. 10.1016/S0168-9525(01)02409-X [DOI] [PubMed] [Google Scholar]
- Nater A., Burri R., Kawakami T., Smeds L., Ellegren H., 2015. Resolving evolutionary relationships in closely related species with whole-genome sequencing data. Syst. Biol. 64: 1000–1017. 10.1093/sysbio/syv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. A., Bennett S. M., 2009. Islands of speciation or mirages in the desert? examining the role of restricted recombination in maintaining species. Heredity 103: 439–444. 10.1038/hdy.2009.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996. The effect of recombination on background selection. Genet. Res. 67: 159–174. 10.1017/S0016672300033619 [DOI] [PubMed] [Google Scholar]
- Nordborg M., Hu T. T., Ishino Y., Jhaveri J., Toomajian C., et al. , 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196 10.1371/journal.pbio.0030196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P., 2008. Speciation with gene flow could be common. Mol. Ecol. 17: 2103–2106. 10.1111/j.1365-294X.2008.03715.x [DOI] [PubMed] [Google Scholar]
- Nosil P., Egan S. P., Funk D. J., 2008. Heterogeneous genomic differentiation between walking‐stick ecotypes: “isolation by adaptation” and multiple roles for divergent selection. Evolution 62: 316–336. 10.1111/j.1558-5646.2007.00299.x [DOI] [PubMed] [Google Scholar]
- Ravinet M., Faria R., Butlin R., Galindo J., Bierne N., et al. , 2017. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J. Evol. Biol. 30: 1450–1477. 10.1111/jeb.13047 [DOI] [PubMed] [Google Scholar]
- Seehausen O., Butlin R. K., Keller I., Wagner C. E., Boughman J. W., et al. , 2014. Genomics and the origin of species. Nat. Rev. Genet. 15: 176–192. 10.1038/nrg3644 [DOI] [PubMed] [Google Scholar]
- Smeds L., Qvarnström A., Ellegren H., 2016. Direct estimate of the rate of germline mutation in a bird. Genome Res. 26: 1211–1218. 10.1101/gr.204669.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. 10.1017/S0016672300014634 [DOI] [PubMed] [Google Scholar]
- Stephan W., 2010. Genetic hitchhiking vs. background selection: the controversy and its implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1245–1253. 10.1098/rstb.2009.0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon M. I., Sawkins M. C., Long A. D., Gaut R. L., Doebley J. F., et al. , 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. 10.1073/pnas.151244298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. R., 2014. A C++ template library for efficient forward-time population genetic simulation of large populations. Genetics 198: 157–166. 10.1534/genetics.114.165019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner T. L., Hahn M. W., 2010. Genomic islands of speciation or genomic islands and speciation? Mol. Ecol. 19: 848–850. 10.1111/j.1365-294X.2010.04532.x [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Ellegren H., 2017. Making sense of genomic islands of differentiation in light of speciation. Nat. Rev. Genet. 18: 87–100. 10.1038/nrg.2016.133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used for this study were previously published. See Kawakami et al. (2014), Burri et al. (2015) and Craig et al. (2018). Supplemental Material and C++ code used for the simulations and calculation of Bm are available at Figshare: https://figshare.com/s/5ee30bae0acfad563865.