Abstract
Reduced ribosome biogenesis in response to environmental conditions is a key feature of cell adaptation to stress. For example, ribosomal genes are transcriptionally repressed when cells are exposed to tunicamycin, a protein glycosylation inhibitor that induces endoplasmic reticulum stress and blocks vesicular trafficking in the secretory pathway. Here, we describe a novel regulatory model, in which tunicamycin-mediated stress induces the accumulation of long-chain sphingoid bases and subsequent activation of Pkh1/2 signaling, which leads to decreased expression of ribosomal protein genes via the downstream effectors Pkc1 and Sch9. Target of rapamycin complex 1 (TORC1), an upstream activator of Sch9, is also required. This pathway links ribosome biogenesis to alterations in membrane lipid composition under tunicamycin-induced stress conditions. Our results suggest that sphingolipid/Pkh1/2-TORC1/Sch9 signaling is an important determinant for adaptation to tunicamycin-induced stress.
Keywords: ribosome, stress response, sphingolipid-Pkh1/2, TORC1-Sch9
TO survive, cells must respond and adapt to environmental conditions, typically by modulating gene expression (Gasch and Werner-Washburne 2002; de Nadal et al. 2011). For instance, ribosome synthesis, a process that consumes enormous amounts of energy, is tightly regulated in response to environmental cues (Warner 1999). Indeed, stresses such as heat shock, osmotic shock, and lack of nutrients reduce the expression of ribosomal proteins (Gasch et al. 2000; Causton et al. 2001) and rRNAs (Laferté et al. 2006; Xiao and Grove 2009). In the yeast Saccharomyces cerevisiae, this downregulation is attributed to the dissociation of the transcriptional activator Ifh1 from Fhl1, a forkhead-like transcription factor at the promoters of ribosomal protein genes (Martin et al. 2004; Schawalder et al. 2004; Wade et al. 2004). In turn, this dissociation is mediated by the inhibition of the AGC kinase (AGC kinase stands for cAMP-dependent protein kinase A, cGMP-dependent kinase G, and protein kinase C) Sch9, a major downstream effector of target of rapamycin complex 1 (TORC1) (Albert et al. 2016). Since this pathway can be triggered by different events (Hughes Hallett et al. 2014), it is likely to be a universal mechanism for the downregulation of ribosome synthesis under stress. In line with this notion, Ifh1 was shown to interact with Utp22, an rRNA-processing protein that coordinates the synthesis of ribosomal proteins and rRNA (Albert et al. 2016).
Ribosomal genes are also sensitive to secretion defects, such as those due to sec mutations, or to exposure to tunicamycin, an inhibitor of N-linked glycosylation (Mizuta and Warner 1994; Mizuta et al. 1998; Nierras and Warner 1999; Miyoshi et al. 2003). In this case, the silencing domain in Rap1 may be involved, as are the 60S ribosomal subunit assembly (Mizuta et al. 1998; Li et al. 2000; Miyoshi et al. 2002; Zhao et al. 2003; Horigome et al. 2008) and relocation of nuclear proteins to the cytoplasm (Nanduri and Tartakoff 2001). Cell wall integrity and stress response component (Wsc) proteins, Pkc1, glycogen synthase kinase-3, the Arp2/3 complex, and spindle pole body components such as Mps3 have also been implicated (Nierras and Warner 1999; Li et al. 2000; Yabuki et al. 2014, 2017). In contrast, the Ire1-mediated unfolded protein response, which is typically coordinated with the heat-shock response (Liu and Chang 2008; Hou et al. 2014), is not required (Nierras and Warner 1999), although it is otherwise essential for the survival of secretory mutants (Chang et al. 2004). In any case, how cells sense secretory defects, at what sites, and how signals are transmitted to the nucleus remain largely unknown.
One possibility is that alterations in membrane composition, following impaired transport of membrane proteins and lipids from the endoplasmic reticulum (ER) via the secretory pathway, may trigger a signal to repress ribosome biogenesis and relieve secretory stress. For example, sphingolipids are essential not only for membrane structure, but also function as secondary messengers and are trafficked among organelles mainly through the secretory pathway (Funato et al. 2002; Olson et al. 2016; Teixeira and Costa 2016). Sphingoid-base backbones are synthesized in the ER, acylated into ceramides, transported to the Golgi by vesicular and nonvesicular mechanisms (Funato and Riezman 2001), converted into more complex sphingolipids, and are finally delivered to the plasma membrane or vacuoles by vesicle-mediated pathways (Funato et al. 2002; Schnabl et al. 2005; Olson et al. 2016).
Here, we report that accumulation of long-chain sphingoid bases, caused by tunicamycin exposure, activates Pkh1/2, Pkc1, and Sch9, and thereby represses ribosomal protein gene expression. Our study also shows that, like Sch9, TORC1 is required to inhibit ribosomal protein gene expression upon tunicamycin-induced stress. Thus, we propose that long-chain sphingoid bases act as a sensor that inhibits ribosome biogenesis in the stress response to tunicamycin.
Materials and Methods
Plasmids, yeast strains, cultivation, and drug sensitivity
Plasmids and yeast strains used in this study are listed in Supplemental Material, Tables S1 and S2, respectively. Yeast cells were grown in yeast extract polypeptone dextrose (YPD), a synthetic complete medium containing 2% glucose, dropout supplements depending on plasmid selection markers, or synthetic dextrose medium. Growth curves and culture conditions are shown in Figure S1 for cells to be used in northern blotting. Tunicamycin sensitivity was assessed by serially diluting cells fivefold, spotting on YPD plates containing tunicamycin, and culturing at 30 or 25°.
Northern blotting
Total RNA was extracted by the hot phenol method from yeast cells lysed with glass beads and analyzed by northern blotting from 1.5% agarose gels, as described previously (Mizuta and Warner 1994).
Microscopy
Yeast cells were grown to midlog phase in YPD, fixed in 3.7% formaldehyde for 30 min, stained with 20 units/ml rhodamine-conjugated phalloidin in 0.1% Triton X-100 as described previously (Yamada et al. 2007), and imaged by fluorescence microscopy.
Western blotting
To analyze Sch9 phosphorylation, protein extracts from cells expressing SCH9-5HA were treated with 2-nitro-5-thiocyanobenzoic acid as previously described (Urban et al. 2007), resolved by SDS-PAGE, immunoblotted, and visualized using mouse anti-HA monoclonal antibody (12CA5; Roche) and sheep anti-mouse IgG (NA931; GE Healthcare) conjugated to horseradish peroxidase. The protein extracts from wild-type cells were analyzed by immunoblotting, and visualized with rabbit antibody against phospho-T570 Sch9 (Urban et al. 2007) and anti-rabbit IgG (Sigma [Sigma Chemical], St. Louis, MO) conjugated to horseradish peroxidase. Bands were quantified using ImageJ to determine the relative amounts of phosphorylated Sch9.
Sphingolipid labeling
As described previously (Kajiwara et al. 2014), cells were grown in synthetic dextrose medium at 25°, treated with 2.5 μg/ml tunicamycin, and then labeled with 10 μCi [3H]dihydrosphingosine (American Radiolabeled Chemical). The reaction was stopped by placing on ice, and adding 10 mM NaF and 10 mM NaN3. Lipids were subsequently extracted, of which one-half were hydrolyzed in mild alkali to deacylate glycophospholipids and identify base-resistant sphingolipids as described previously (Ikeda et al. 2015). Radiolabeled lipids were analyzed by thin-layer chromatography in 9:7:2 v/v chloroform:methanol:4.2 N-ammonium hydroxide, and quantified using FLA-7000 (Fujifilm).
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, supplemental figures, and supplemental tables. Supplemental material available at Figshare. Supplemental material available at https://doi.org/10.25386/genetics.7782599.
Results and Discussion
Tunicamycin-induced stress response requires long-chain sphingoid bases
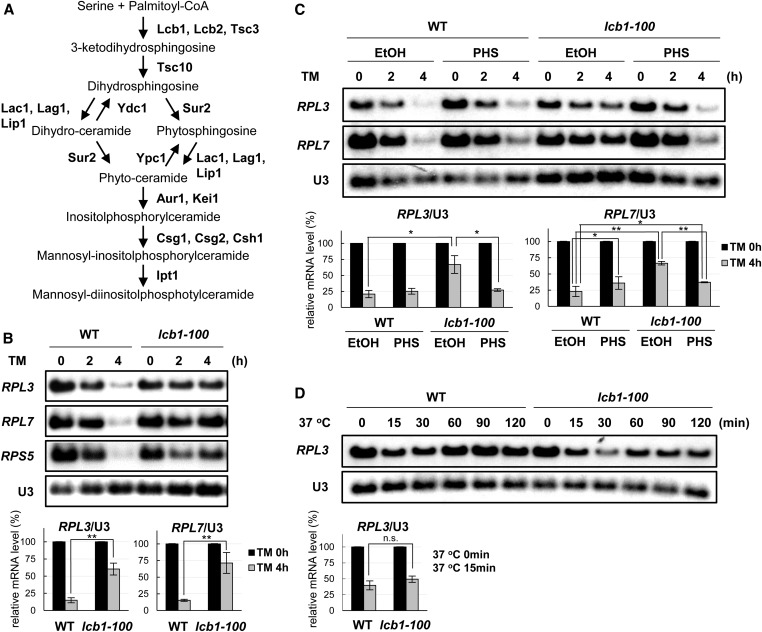
We speculated that alterations in the composition of membrane proteins and lipids following inhibition of the secretory pathway might trigger a stress response. Changes in sphingolipid content are likely among such triggers in yeast, since sec mutations or exposure to tunicamycin inhibit the biosynthesis of inositolphosphorylceramide (IPC), a complex sphingolipid (Funato and Riezman 2001; Pittet et al. 2006). Thus, we examined ribosomal protein gene expression in lcb1-100 mutant cells exposed to tunicamycin. This mutant is conditionally impaired in the first step of sphingolipid biosynthesis (Figure 1A) and is thus deficient in all sphingolipids (Zanolari et al. 2000). As shown in Figure 1B, the lcb1-100 mutant cells exhibited a significant reduction in repression of ribosomal protein gene expression in response to tunicamycin in comparison with the wild-type, suggesting that sphingolipids are required for the stress response to tunicamycin.
Figure 1.
Downregulation of ribosomal protein genes in response to TM, but not to heat stress, requires PHS. (A) Sphingolipid biosynthesis in yeast. (B) WT (FKY2736) and lcb1-100 (FKY2739) cells were cultured in YPD at 25°, shifted to 30° for 4 hr, exposed to 2.5 μg/ml TM for the indicated times, and harvested for northern analysis. Data represent one of three reproducible and independent experiments. The mRNA levels were quantified from three independent experiments, normalized to U3, and reported as mean ± SD of radioactivity relative to radioactivity at time zero. * P < 0.05 and ** P < 0.01 by Student’s t-test. (C) WT (FKY2736) and lcb1-100 (FKY2739) cells were cultured in YPD at 25°, treated for 1 hr with 5 μM PHS or an equal volume of EtOH, shifted to 30° for 4 hr, exposed to 2.5 μg/ml TM for the indicated times, and harvested for northern analysis. The mRNA levels in (C and D) were quantified as described in (B). (D) WT (FKY2736) and lcb1-100 (FKY2739) cells were cultured in YPD at 25°, shifted to 37° for the indicated times, and harvested for northern analysis. EtOH, ethanol; n.s., not significant; PHS, phytosphingosine; TM, tunicamycin; WT, wild-type.
Deletion of LIP1, a regulatory subunit of ceramide synthase that is required for its enzymatic activity (Figure 1A) (Vallée and Riezman 2005), did not affect the stress response to tunicamycin (Figure S2A). Similar results were obtained with mutants deficient in CSG1, CSG2, and CSH1 (Figure S2B), which are required to synthesize mannosyl-IPC, and with a mutant deficient in IPT1 (Figure S2C), which is required to synthesize mannosyl-di-IPC. These results indicate that long-chain sphingoid bases, but not ceramides or complex sphingolipids, are required for the stress response to tunicamycin. Exogenously added phytosphingosine restored the stress response in lcb1-100 mutant cells (Figure 1C), confirming the link to long-chain sphingoid bases. Finally, we found that the heat-stress response is intact in lcb1-100 mutant cells (Figure 1D), implying that transcriptional repression of ribosomal protein genes by long-chain sphingoid bases is specific to the stress response to tunicamycin.
Pkh1/2 and the downstream effectors Pkc1 and Sch9, but not Ypk1/2, are required for the stress response to tunicamycin
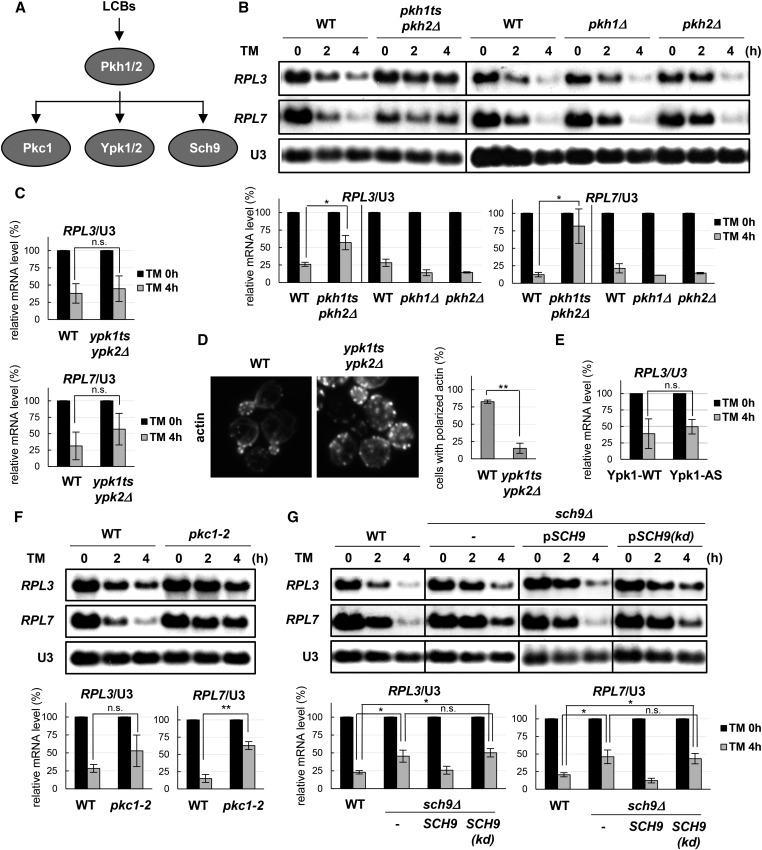
Long-chain sphingoid bases activate Pkh1 and Pkh2 (Figure 2A) (Friant et al. 2001; Liu et al. 2005; Fröhlich et al. 2009; Roelants et al. 2010; Huang et al. 2012), which are redundant yeast homologs of mammalian phosphoinositide-dependent protein kinase 1 (Casamayor et al. 1999; Zanolari et al. 2000). Notably, repression of ribosomal protein gene expression in response to tunicamycin was significantly reduced in pkh1ts pkh2Δ double-mutant cells compared with the wild type, but was unaffected in pkh1Δ or pkh2Δ deletion-mutant cells (Figure 2B), confirming that Pkh1 and Pkh2 have redundant functions in the stress response to tunicamycin.
Figure 2.
TM-induced repression of ribosomal protein genes depends on Pkh1/2 kinases, and the downstream effectors Pkc1 and Sch9, but not on Ypk1/2. (A) Signaling by LCBs via Pkh1/2. (B) WT (SEY6210) and pkh1ts pkh2Δ (MTY29) cells were cultured in YPD at 25°, and shifted to 32° for 4 hr. WT (FKY59), pkh1Δ (FKY61), and pkh2Δ (FKY58) cells were cultured in YPD at 30°. The cells were then treated with TM. The mRNA levels were quantified as described in Figure 1B. (C) WT (SEY6210) and ypk1ts ypk2Δ (MTY77) cells were cultured in YPD at 25°, shifted to 33° for 4 hr, treated with TM, and harvested for northern analysis as described in (B). (D) The same strains as in (C) were cultured in YPD at 25°, shifted to 33° for 4 hr, fixed for 30 min with 3.7% formaldehyde, stained with rhodamine-phalloidin, and visualized by fluorescence microscopy. Data are mean ± SD of the percentage of cells with polarized actin. ** P < 0.01 by Student’s t-test. (E) Ypk1-WT (PLY1083) and Ypk1-AS (PLY1098) were cultured in YPD at 25°, treated with 0.5 μM 2,3-DMB-PPI for 1 hr, and then treated with TM. (F) WT (SEY6210) and pkc1-2 (FKY2746) cells were cultured in YPD at 25°, shifted to 32° for 4 hr, and then treated with TM. (G) WT (W303-1A), sch9Δ (FKY3873), and sch9Δ cells with pRS416-SCH9 or pRS416-SCH9(kd) were cultured in YPD at 30°, and then treated with TM. The mRNA levels were determined and quantified as described in (B). 2,3-DMB-PP1, 4-amino-1-tert-butyl-3-(2,3-dimethylbenzyl)pyrazolo[3,4-d]pyrimidine; LCB, long-chain sphingoid bases; TM, tunicamycin; WT, wild-type.
The AGC kinases Pkc1, Ypk1/2, and Sch9 are downstream effectors of Pkh1/2 (Figure 2A). The ypk1ts ypk2Δ double mutation had no statistically significant changes in repression of ribosomal protein gene expression caused by tunicamycin (Figure 2C), suggesting that Ypk1/2 are not necessary for the stress response, even though the cells exhibit impaired actin polarization at a semipermissive temperature (Figure 2D), as reported previously (Roelants et al. 2002). Consistent with this, treatment of Ypk1-AS cells (ypk1Δypk2Δ + pYpk1-L424G) with the inhibitor 4-amino-1-tert-butyl-3-(2,3-dimethylbenzyl)pyrazolo[3,4-d]pyrimidine (2,3-DMB-PP1) (Niles et al. 2014) did not exhibit a significant effect on tunicamycin-induced repression of ribosomal protein RPL3 gene expression when compared with inhibitor-insensitive Ypk1-WT cells (ypk1Δypk2Δ + pYpk1-WT) (Figure 2E). In contrast, pkc1-2 cells caused a significant decrease in repression of RPL7 gene expression in response to tunicamycin (Figure 2F), as previously observed (Nierras and Warner 1999). Similarly, repression of ribosomal protein gene expression was defective in sch9Δ cells, as shown in Figure 2G. Expression of SCH9, but not a kinase-dead allele, restored the stress response in sch9Δ cells, implying that enzymatically active Sch9 is required to transcriptionally repress the expression of ribosomal protein genes in response to tunicamycin.
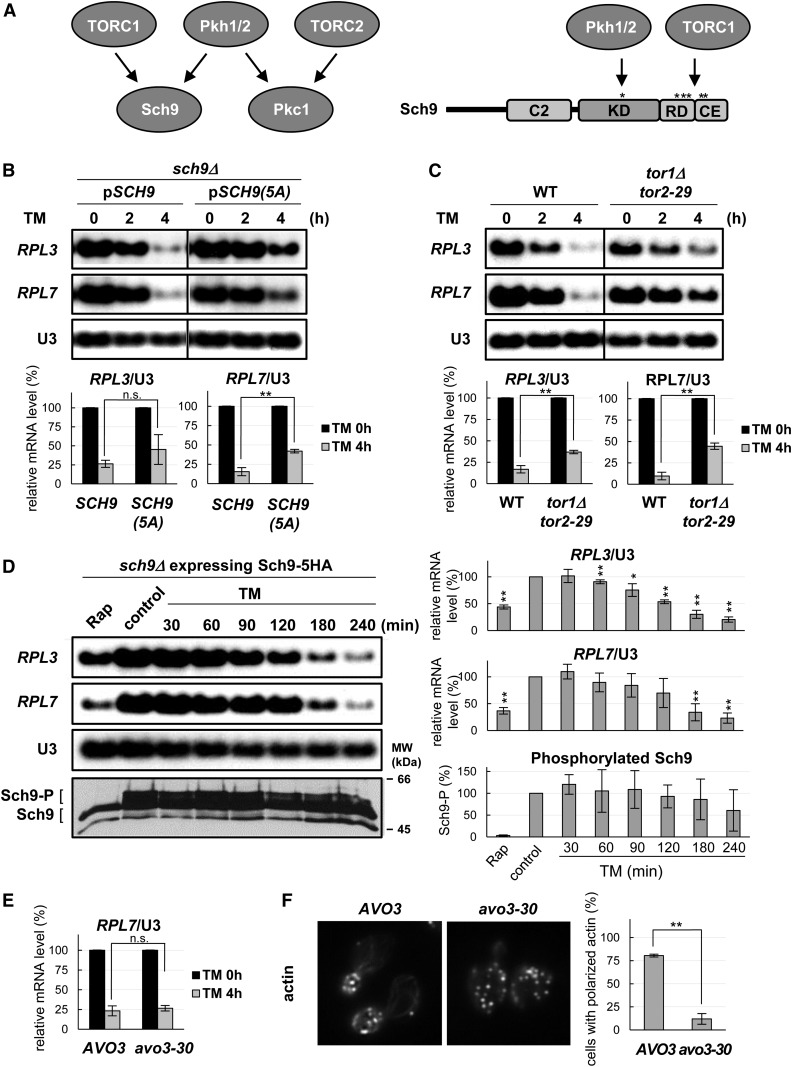
TORC1, but not TORC2, regulates the stress response to tunicamycin
Sch9 is directly phosphorylated not only by Pkh1/2 but also by TORC1 (Figure 3A) (Urban et al. 2007), although the former phosphorylates a threonine residue in the kinase domain, whereas the latter phosphorylates 5–6 serine/threonine residues in the C-terminal region. Expression of SCH9(5A), a constitutively inactive SCH9 with five point mutations at its C-terminal phosphorylation sites (T723A, S726A, T737A, S758A, and S765A) (Urban et al. 2007), did not suppress the tunicamycin-induced repression of ribosomal protein RPL7 gene expression in sch9Δ cells (Figure 3B), suggesting that Sch9 phosphorylation by TORC1 is also required for the stress response. The TORC1-deficient mutant tor1Δ tor2-29 (Helliwell et al. 1998) also exhibited a significant reduction in tunicamycin-induced repression of ribosomal protein gene expression (Figure 3C), further confirming a role for TORC1 in the stress response to tunicamycin. On the other hand, repression of ribosomal protein gene expression under heat stress was unaffected in the tor1Δ tor2-29 cells (Figure S3A). Similarly, the response to heat shock appears to be normal in sch9Δ cells expressing SCH9(kd), SCH9(5A), or pkh1ts pkh2Δ cells (Figure S3, B-E), indicating that TORC1-Sch9 and Pkh1/2 signaling plays a specific role in the stress response to tunicamycin.
Figure 3.
TORC1, but not TORC2, regulates the stress response to TM via Sch9. (A) Relationships between Pkh1/2, TORC1, TORC2, and phosphorylation sites in Sch9. (B) sch9Δ (FKY3873) cells with pRS416-SCH9 or pRS416-SCH9(5A) were cultured in YPD at 30°. The mRNA levels in (B and C) were quantified as described in Figure 1B. (C) WT (JK9-3da) and tor1Δ tor2-29 (SH229) cells were cultured in YPD at 25°, shifted to 33° for 6 hr, treated with TM, and harvested for northern analysis. (D) sch9Δ (FKY3873) cells transformed with pRS416-SCH9-5HA were cultured to log phase at 30° in synthetic complete medium without uracil, and treated with 200 ng/ml Rap for 30 min or with 2.5 μg/ml TM for the indicated times or left untreated (control), and split into two parts for RNA and protein extraction. RNA was analyzed by northern blotting and quantified as described in (B), while protein extracts were reacted with 2-nitro-5-thiocyanobenzoic acid and analyzed by immunoblotting using anti-HA. Data are mean ± SD of phosphorylated Sch9 as percentage of total Sch9 at time zero. * P < 0.05 and ** P < 0.01 by Student’s t-test. (E) AVO3 (PLY718) and avo3-30 (PLY1134) were cultured in YPD at 25°, shifted to 30° for 4 hr, treated with TM, and harvested for northern blotting as described in (B). (F) The same strains as in (E) were cultured in YPD at 25°, shifted to 30° for 4 hr, fixed with formaldehyde, stained with rhodamine-phalloidin, visualized by fluorescence microscopy, and quantified as described in Figure 2D. CE, C-terminal extension; KD, kinase domain; Rap, rapamycin; RD, regulatory domain; TM, tunicamycin; WT, wild-type.
Furthermore, there were no obvious changes in TORC1-dependent Sch9 phosphorylation within 180 min of exposure to tunicamycin (Figure 3D), although ribosomal protein gene expression diminished to ∼30% of the initial levels (Figure 3D). These results suggest that transcriptional repression of ribosomal protein genes is not due to decreases in TORC1-dependent Sch9 phosphorylation, consistent with a previous report showing that tunicamycin does not affect TORC1 activity (Stauffer and Powers 2015). These findings may suggest that TORC1 is not actively involved in the response to stress generated by tunicamycin, but rather acts passively for Sch9 activation. Finally, an avo3-30 mutant defective in TORC2, which phosphorylates Pkc1, and has distinct structure and functions from TORC1 (Figure 3A) (Loewith et al. 2002; Nomura and Inoue 2015), did not exhibit a defect in the stress response to tunicamycin despite complete loss of actin organization (Figure 3, E and F). These data suggest that TORC2 is not involved in the stress response.
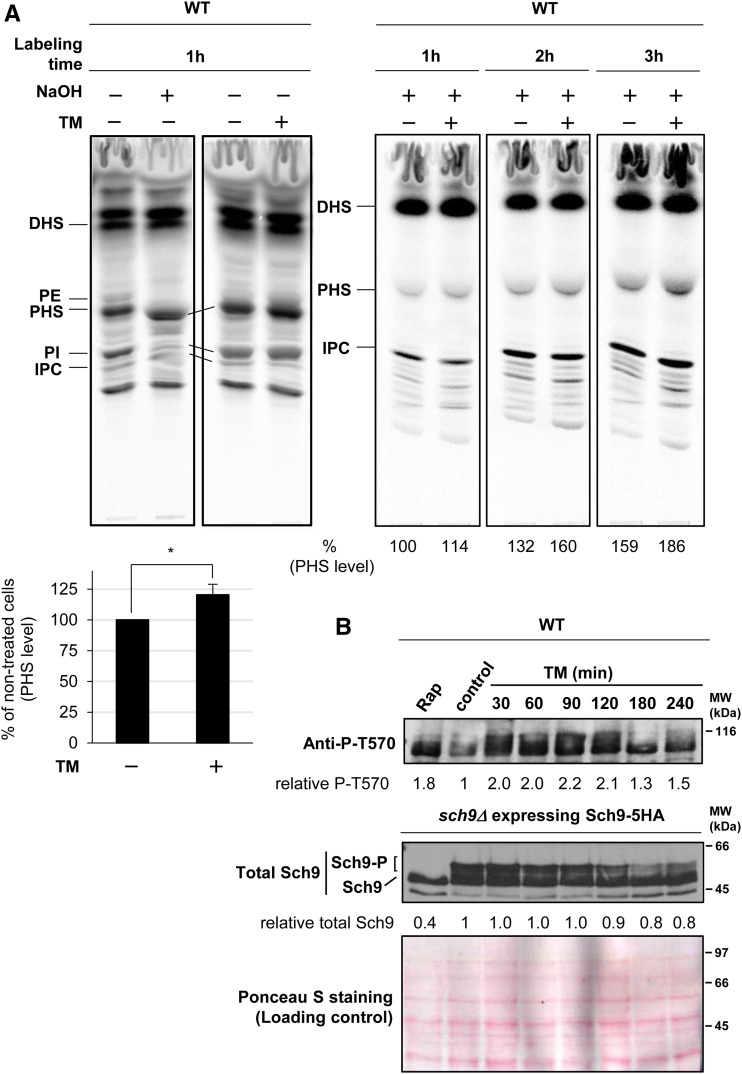
Tunicamycin treatment leads to increased levels of phytosphingosine and T570 phosphorylation of Sch9
Since TORC1 activity did not significantly change after exposure to tunicamycin (Figure 3D) (Stauffer and Powers 2015), other signaling factors may mediate the stress response. Such factors could include long-chain sphingoid bases, which may accumulate via unknown mechanisms in the presence of tunicamycin, given that tunicamycin induces a decrease of IPC levels (Pittet et al. 2006). Indeed, we found a significant increase in phytosphingosine levels in wild-type cells 3 hr after tunicamycin addition (Figure 4A, left). The increase in phytosphingosine levels occurred 1 hr after tunicamycin treatment and continued for 3 hr (Figure 4A, right). These results are consistent with the ability of phytosphingosine to rescue the tunicamycin-induced stress response in lcb1-100 cells (Figure 1C).
Figure 4.
TM treatment leads to increased levels of phytosphingosine and T570 phosphorylation of Sch9. (A) WT (FKY2736) cells were cultured to log phase at 25° in synthetic dextrose medium, treated for 2 hr with 2.5 μg/ml TM, and labeled with 10 µCi [3H]DHS for 1 hr (left panel). WT (FKY1131) cells were treated for 30 min with 5 μg/ml TM and labeled with [3H]DHS for the indicated times (right panel). Labeled lipids were (+) or were not (−) mildly hydrolyzed with NaOH to deacylate base-sensitive glycophospholipids, and only detect base-resistant sphingolipids such as PHS and IPC, and analyzed by thin-layer chromatography. Incorporation into base-resistant PHS was quantified as percentage of total radioactivity, and is reported relative to incorporation in cells not exposed to TM. Data are mean ± SD of three independent experiments (bottom left graph). * P < 0.05 by Student’s t-test. (B) WT (W303-1A) cells and sch9Δ (FKY3873) cells transformed with pRS416-SCH9-5HA were cultured in YPD, treated with 200 ng/ml Rap for 30 min or with 2.5 μg/ml TM for the indicated times or left untreated (control). The protein extracts from WT cells were prepared and analyzed by immunoblotting using anti-P-T570. The extracts from cells expressing Sch9-5HA were reacted with 2-nitro-5-thiocyanobenzoic acid and analyzed by immunoblotting using anti-HA. Data represent one of two reproducible and independent experiments. The relative amounts of T570-phosphorylated Sch9 (top panel) and total Sch9 (middle panel) compared to the levels in untreated cells were calculated and shown. Ponceau S staining (bottom panel) as a loading control is also shown. DHS, dihydrosphingosine; IPC, inositolphosphorylceramide; PE, phosphatidylethanolamine; PHS, phytosphingosine; PI, phosphatidylinositol; Rap, rapamycin; TM, tunicamycin; WT, wild-type.
The accumulation of phytosphingosine in the presence of tunicamycin could be the result of a delay in ceramide synthesis due to impaired N-glycosylation. However, a cwh8Δ mutant defective in N-glycosylation, which has a reduced level of IPC, exhibits normal ceramide synthase activity (Pittet et al. 2006). As IPC synthase activity is also normal in the cwh8Δ cells (Pittet et al. 2006), it is likely that aberrant N-glycosylation impairs the transport of ceramides between the ER and the Golgi, which in yeast occurs mainly via vesicular transport (Funato and Riezman 2001). Thus, N-glycosylation may mediate the vesicular transport of ceramide and thereby modulate levels of long-chain sphingoid bases.
As sphingolipids appear to function as upstream activators of the Pkh1/2 kinases (Huang et al. 2012) and in vitro phosphorylation of Sch9 by Pkh1/2 is stimulated by phytosphingosine (Liu et al. 2005), we next examined if tunicamycin treatment affects the phosphorylation of residue T570 in Sch9, which is catalyzed by Pkh1/2 (Urban et al. 2007). As shown in Figure 4B, tunicamycin treatment resulted in increased T570 phosphorylation, but had little effect on the protein levels of total Sch9. Thus, these results suggest that tunicamycin treatment activates the Pkh1/2 kinases. Interestingly, although rapamycin decreased Sch9 protein expression, it led to an increased level of T570-phosphorylated Sch9, implying that phosphorylation by Pkh1/2 may be negatively regulated by TORC1.
ER contacts with the plasma membrane and vacuole are not required for the stress response to tunicamycin
Although long-chain sphingoid bases are synthesized in the ER (Funato et al. 2002), those trafficked to the plasma membrane or other organelles may trigger Pkh1/2-dependent signaling instead, especially since sphingoid bases accumulate at unknown sites following tunicamycin exposure. Given that the ER forms membrane contact sites with the plasma membrane, Golgi, endosomes, mitochondria, lipid droplets, and vacuoles, and that such contact sites are required to exchange lipids between organelles (Lahiri et al. 2015; Jain and Holthuis 2017), we investigated the effect of deleting the genes involved. Deletion of all six tether proteins that form contact sites between the cortical ER and the plasma membrane (Δtether), including Ist2, Scs2/Scs22, and Tcb1/2/3, inhibited the formation of such contact sites (Manford et al. 2012) but did not affect tunicamycin-induced repression of ribosomal protein gene expression, as shown in Figure S4A. Cells separately deficient in Ist2 (ist2Δ), Scs2/Scs22 (scs2Δ22Δ), or Tcb1/Tcb2/Tcb3 (tcb1Δ2Δ3Δ) also did not display obvious defects in the stress response. Similarly, Epo1, a polarisome subunit required to tether cortical ER at bud tips (Neller et al. 2015), was not involved in the stress response (Figure S4B). Moreover, tunicamycin-induced repression of ribosomal protein gene expression occurred normally in a deletion mutant of NVJ1, which is required to tether the perinuclear ER (nuclear envelope) to the vacuole (Pan et al. 2000), or of LTC1, which regulates such nucleus–vacuole junctions (Murley et al. 2015) (Figure S4C), even though TORC1 and Sch9 are predominantly localized to the vacuole (Urban et al. 2007; Sturgill et al. 2008). Collectively, the data indicate that ER contacts with the plasma membrane or vacuole are not required for the stress response to tunicamycin.
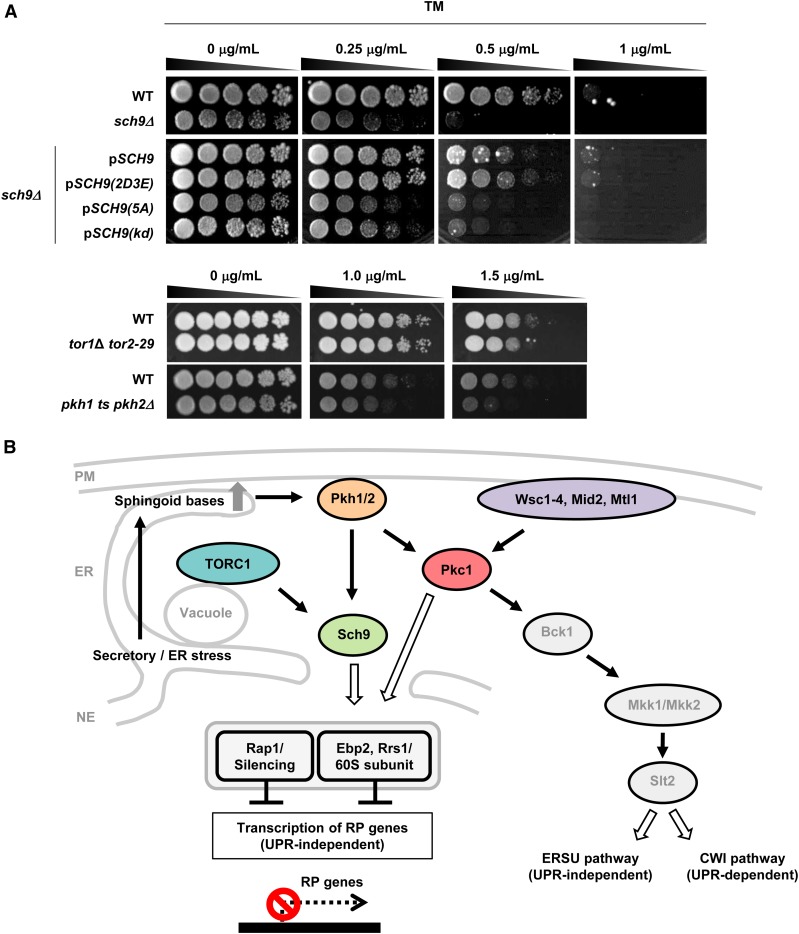
Pkh1/2 and Sch9 signaling is essential for survival to tunicamycin-induced stress
Deletion of SCH9 enhanced the sensitivity to tunicamycin, as assessed by cell growth (Figure 5A). Conversely, expression of SCH9 and SCH9(2D3E), a constitutively active allele, restored sensitivity to tunicamycin, but expression of SCH9(kd) or SCH9(5A) did not. These results suggest that Sch9 is indispensable for adaptation to tunicamycin-induced stress. In addition, tor1Δ tor2-29 and pkh1ts pkh2Δ mutant cells showed enhanced sensitivity to tunicamycin, although the tor1Δ tor2-29 cells were only slightly more sensitive than control cells. Thus, these results imply that the reduction of ribosome biogenesis plays an important role in the adaptation to stress caused by tunicamycin, consistent with the finding that ribosomal protein gene deletion mutants exhibit high resistance to tunicamycin (Steffen et al. 2012).
Figure 5.
Loss of Pkh1/2 and Sch9 signaling sensitizes cells to TM. (A) WT (W303-1A); sch9Δ (FKY3873); sch9Δ cells with pRS416-SCH9, pRS416-SCH9(2D3E), pRS416-SCH9(5A), or pRS416-SCH9(kd); WT (JK9-3da); and tor1Δ tor2-29 (SH229), WT (SEY6210), and pkh1ts pkh2Δ (MTY29) cells were cultured at 25° to OD600 = 1.0 in YPD. Fivefold serial dilutions were then spotted on YPD plates containing TM at the indicated concentrations, and incubated at 30° for 3 days (upper) or 25° for 6 days (bottom). (B) A working model for sphingolipid/Pkh1/2- and TORC1/Sch9-dependent regulation of ribosomal biogenesis under TM-induced stress. CWI, cell wall integrity; ERSU, ER stress surveillance; NE, nuclear envelope; PM, plasma membrane; RP, ribosomal protein; TM, tunicamycin; UPR, unfolded protein response; WT, wild-type.
Collectively, our results reveal that, in response to tunicamycin-induced stress, ribosomal protein genes are specifically downregulated by long-chain sphingoid bases and downstream effectors. In particular (Figure 5B), phytosphingosines accumulate following tunicamycin addition, activate Pkh1/2, and thereby promote the phosphorylation of Pkc1 and Sch9. This model is consistent with previous observations that phytosphingosine and Pkh1/2 stimulate the phosphorylation of Pkc1 and Sch9 in vitro (Friant et al. 2001; Liu et al. 2005), that the lcb1-100 mutation diminishes Pkc1 phosphorylation in vivo (Clarke et al. 2017), and that downregulation of Lcb1 reduces Sch9 phosphorylation via Pkh1/2 in vivo (Huang et al. 2012). The stress response to tunicamycin is also defective in a tor1Δ tor2-29 mutant and an SCH9(5A) mutant that lacks TORC1 phosphorylation sites, implying that Sch9 phosphorylation by TORC1 is also required. Consistent with this, Sch9 phosphorylation by TORC1 appears to be essential for adaptation to tunicamycin-induced stress, since SCH9(5A) mutant cells were sensitive to tunicamycin. However, Pkc1 may also act in parallel with Sch9 (Figure 5B), since SCH9(2D3E), a constitutively active allele, does not rescue the stress response in pkc1-2 cells (Figure S5A) or the tunicamycin sensitivity of pkh1ts pkh2Δ cells (Figure S5B). In addition, tunicamycin-induced transcriptional repression of ribosomal protein genes was not completely attenuated in the lcb1-100 mutant cells, suggesting that other sensor proteins such as Wsc1 are required for the stress response.
Tunicamycin not only causes transcriptional repression of ribosomal protein gene expression, but also activates ER stress surveillance and cell wall integrity signaling through the cell-surface sensors Wsc and Pkc1 (Babour et al. 2010). Notably, sphingolipids also activate ER stress surveillance (Piña et al. 2018). Strikingly, repression of ribosomal protein gene expression does not require Slt2 (Li et al. 2000), an otherwise important determinant of stress survival and, like ER stress surveillance and cell wall integrity signaling, a driver of Rlm1-mediated transcription (Babour et al. 2010; Levin 2011). How transcriptional repression of ribosomal protein gene expression is triggered during tunicamycin-induced stress is unknown, except that it is independent of the Ire1-mediated unfolded protein response (Nierras and Warner 1999). However, we propose that the downstream effectors of phytosphingosine-Pkh1/2, Pkc1, and Sch9 repress ribosomal protein gene expression through Rap1-mediated silencing (Mizuta et al. 1998), 60S ribosomal subunit assembly (Miyoshi et al. 2002; Horigome et al. 2008), or other as yet unknown mechanisms (Figure 5B).
The cellular response to stress caused by tunicamycin treatment is believed to be coupled to cell growth (Mizuta and Warner 1994; Mizuta et al. 1998; Li et al. 2000; Steffen et al. 2012) as such stress probably occurs in rapidly growing cells. Indeed, as TORC1-Sch9 signaling is a master regulator of cell growth and is activated under growth conditions (Loewith and Hall 2011), it can be exploited to effectively and rapidly repress ribosomal protein genes in response to tunicamycin-induced stress. TOR-Sch9 signaling negatively regulates the transcriptional repression of ribosomal protein genes under stress conditions such as heat shock or nitrogen deficiency, whereas positive regulation is seen in response to stress caused by tunicamycin. It is largely unknown how TOR-Sch9 signaling induces such opposite outputs under different conditions. We found that T570 phosphorylation of Sch9 was increased when cells were treated with rapamycin, suggesting that Sch9 phosphorylation via Pkh1/2 is negatively regulated by TORC1. This may also suggest that Sch9 phosphorylated by Pkh1/2 plays a distinct role from that phosphorylated by TORC1. Given that tunicamycin treatment leads to an increase in Sch9 phosphorylation via Pkh1/2, and that both Pkh1/2 and TORC1 are required for the stress response to tunicamycin, we propose that phosphorylation of Sch9 by both Pkh1/2 and TORC1 is required to activate unknown factor(s) that transcriptionally repress ribosomal protein genes. Since phosphorylation of the TORC1 sites in Sch9 was unaffected by tunicamycin, basal TORC1 activity may be required to maintain active Sch9.
The accumulation of phytosphingosine following tunicamycin treatment could be due to impaired ceramide transport from the ER to the Golgi and subsequent degradation of ceramide by ER-resident ceramidases. Although the exact sites where phytosphingosine activates Pkh1/2 remain to be elucidated, phytosphingosine localized in the ER may be implicated in the activation of Pkh1/2 kinases. Otherwise, phytosphingosine delivered to the eisosome, a specialized plasma membrane subdomain where a part of Pkh1/2 is present (Walther et al. 2007), or to the vacuole membrane where TORC1 and Sch9 functions appear to be regulated (Urban et al. 2007; Binda et al. 2009; Kira et al. 2016; Takeda et al. 2018), may activate Pkh1/2 to phosphorylate Sch9. In this case, the delivery of phytosphingosine to these organelles does not seem to require membrane contact sites between organelles, because transcriptional repression of ribosomal protein genes in response to tunicamycin was unaffected in deletion mutants of genes that function as tethers between the ER and plasma membrane, or vacuole membrane.
In summary, our study indicates a new role for sphingolipid/Pkh1/2-TORC1/Sch9 signaling in the transcriptional repression of ribosomal protein genes upon tunicamycin-induced stress. Further work is required to identify the downstream effector(s) of Sch9 that represses ribosomal protein gene expression. It will also be interesting to determine whether a similar mechanism for stress-induced repression of ribosomal protein genes exist in mammalian cells.
Acknowledgments
We thank M. N. Hall, R. Loewith, M. Mizunuma, T. Powers, H. Riezman, C. J. Stefan, M. Tabuchi, T. Ushimaru, and the Yeast Resource Center for providing yeast strains, antibodies, and plasmids used in this study, and Editage (www.editage.jp) for English language editing. This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (KAKENHI) (grants JP25450516 to K.M. and JP16K07693 to K.F.) and by a JSPS Research Fellowship for Young Scientists (JP15J05830 to Y.Y.). The authors declare no conflicts of interest.
Footnotes
Supplemental material available at https://doi.org/10.25386/genetics.7782599.
Communicating editor: O. Cohen-Fix
Literature Cited
- Albert B., Knight B., Merwin J., Martin V., Ottoz D., et al. , 2016. A molecular titration system coordinates ribosomal protein gene transcription with ribosomal RNA synthesis. Mol. Cell 64: 720–733. 10.1016/j.molcel.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Babour A., Bicknell A. A., Tourtellotte J., Niwa M., 2010. A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell 142: 256–269. 10.1016/j.cell.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M., Péli-Gulli M. P., Bonfils G., Panchaud N., Urban J., et al. , 2009. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35: 563–573. 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R., 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9: 186–197. 10.1016/S0960-9822(99)80088-8 [DOI] [PubMed] [Google Scholar]
- Causton H. C., Ren B., Koh S. S., Harbison C. T., Kanin E., et al. , 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12: 323–337. 10.1091/mbc.12.2.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. J., Jesch S. A., Gaspar M. L., Henry S. A., 2004. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics 168: 1899–1913. 10.1534/genetics.104.032961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J., Dephoure N., Horecka I., Gygi S., Kellogg D., 2017. A conserved signaling network monitors delivery of sphingolipids to the plasma membrane in budding yeast. Mol. Biol. Cell 28: 2589–2599. 10.1091/mbc.e17-01-0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E., Ammerer G., Posas F., 2011. Controlling gene expression in response to stress. Nat. Rev. Genet. 12: 833–845. 10.1038/nrg3055 [DOI] [PubMed] [Google Scholar]
- Friant S., Lombardi R., Schmelzle T., Hall M. N., Riezman H., 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20: 6783–6792. 10.1093/emboj/20.23.6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich F., Moreira K., Aguilar P. S., Hubner N. C., Mann M., et al. , 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 185: 1227–1242. 10.1083/jcb.200811081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K., Riezman H., 2001. Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J. Cell Biol. 155: 949–959. 10.1083/jcb.200105033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K., Vallée B., Riezman H., 2002. Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry 41: 15105–15114. 10.1021/bi026616d [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Werner-Washburne M., 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2: 181–192. 10.1007/s10142-002-0058-2 [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell S. B., Howald I., Barbet N., Hall M. N., 1998. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C., Okada T., Matsuki K., Mizuta K., 2008. A ribosome assembly factor Ebp2p, the yeast homolog of EBNA1-binding protein 2, is involved in the secretory response. Biosci. Biotechnol. Biochem. 72: 1080–1086. 10.1271/bbb.70817 [DOI] [PubMed] [Google Scholar]
- Hou N. S., Gutschmidt A., Choi D. Y., Pather K., Shi X., et al. , 2014. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc. Natl. Acad. Sci. USA 111: E2271–E2280. 10.1073/pnas.1318262111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Liu J., Dickson R. C., 2012. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet. 8: e1002493 10.1371/journal.pgen.1002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Hallett J. E., Luo X., Capaldi A. P., 2014. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 198: 773–786. 10.1534/genetics.114.168369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A., Muneoka T., Murakami S., Hirota A., Yabuki Y., et al. , 2015. Sphingolipids regulate telomere clustering by affecting the transcription of genes involved in telomere homeostasis. J. Cell Sci. 128: 2454–2467. 10.1242/jcs.164160 [DOI] [PubMed] [Google Scholar]
- Jain A., Holthuis J. C. M., 2017. Membrane contact sites, ancient and central hubs of cellular lipid logistics. Biochim. Biophys. Acta 1864: 1450–1458. 10.1016/j.bbamcr.2017.05.017 [DOI] [PubMed] [Google Scholar]
- Kajiwara K., Ikeda A., Aguilera-Romero A., Castillon G. A., Kagiwada S., et al. , 2014. Osh proteins regulate COPII-mediated vesicular transport of ceramide from the endoplasmic reticulum in budding yeast. J. Cell Sci. 127: 376–387. 10.1242/jcs.132001 [DOI] [PubMed] [Google Scholar]
- Kira S., Kumano Y., Ukai H., Takeda E., Matsuura A., et al. , 2016. Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Mol. Biol. Cell 27: 382–396. 10.1091/mbc.e15-07-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferté A., Favry E., Sentenac A., Riva M., Carles C., et al. , 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 20: 2030–2040. 10.1101/gad.386106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S., Toulmay A., Prinz W. A., 2015. Membrane contact sites, gateways for lipid homeostasis. Curr. Opin. Cell Biol. 33: 82–87. 10.1016/j.ceb.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. 10.1534/genetics.111.128264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Moir R. D., Sethy-Coraci I. K., Warner J. R., Willis I. M., 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 20: 3843–3851. 10.1128/MCB.20.11.3843-3851.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Zhang X., Lester R. L., Dickson R. C., 2005. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280: 22679–22687. 10.1074/jbc.M502972200 [DOI] [PubMed] [Google Scholar]
- Liu Y., Chang A., 2008. Heat shock response relieves ER stress. EMBO J. 27: 1049–1059. 10.1038/emboj.2008.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Hall M. N., 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., et al. , 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Manford A. G., Stefan C. J., Yuan H. L., Macgurn J. A., Emr S. D., 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23: 1129–1140. 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Martin D. E., Soulard A., Hall M. N., 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119: 969–979. 10.1016/j.cell.2004.11.047 [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Tsujii R., Yoshida H., Maki Y., Wada A., et al. , 2002. Normal assembly of 60 S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J. Biol. Chem. 277: 18334–18339. 10.1074/jbc.M201667200 [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Shirai C., Mizuta K., 2003. Transcription of genes encoding trans-acting factors required for rRNA maturation/ribosomal subunit assembly is coordinately regulated with ribosomal protein genes and involves Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 1969–1973. 10.1093/nar/gkg278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K., Warner J. R., 1994. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol. 14: 2493–2502. 10.1128/MCB.14.4.2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K., Tsujii R., Warner J. R., Nishiyama M., 1998. The C-terminal silencing domain of Rap1p is essential for the repression of ribosomal protein genes in response to a defect in the secretory pathway. Nucleic Acids Res. 26: 1063–1069. 10.1093/nar/26.4.1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Sarsam R. D., Toulmay A., Yamada J., Prinz W. A., et al. , 2015. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 209: 539–548. 10.1083/jcb.201502033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri J., Tartakoff A. M., 2001. The arrest of secretion response in yeast: signaling from the secretory path to the nucleus via Wsc proteins and Pkc1p. Mol. Cell 8: 281–289. 10.1016/S1097-2765(01)00312-4 [DOI] [PubMed] [Google Scholar]
- Neller J., Dünkler A., Rösler R., Johnsson N., 2015. A protein complex containing Epo1p anchors the cortical endoplasmic reticulum to the yeast bud tip. J. Cell Biol. 208: 71–87. 10.1083/jcb.201407126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierras C. R., Warner J. R., 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 274: 13235–13241. 10.1074/jbc.274.19.13235 [DOI] [PubMed] [Google Scholar]
- Niles B. J., Joslin A. C., Fresques T., Powers T., 2014. TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Rep. 6: 541–552. https//.org/10.1016/j.celrep.2013.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura W., Inoue Y., 2015. Methylglyoxal activates the target of rapamycin complex 2-protein kinase C signaling pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 35: 1269–1280. 10.1128/MCB.01118-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson D. K., Fröhlich F., Farese R. V., Walther T. C., 2016. Taming the sphinx: mechanisms of cellular sphingolipid homeostasis. Biochim. Biophys. Acta 1861: 784–792. 10.1016/j.bbalip.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., et al. , 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell 11: 2445–2457. 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña F., Yagisawa F., Obara K., Gregerson J. D., Kihara A., et al. , 2018. Sphingolipids activate the endoplasmic reticulum stress surveillance pathway. J. Cell Biol. 217: 495–505. 10.1083/jcb.201708068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M., Uldry D., Aebi M., Conzelmann A., 2006. The N-glycosylation defect of cwh8Delta yeast cells causes a distinct defect in sphingolipid biosynthesis. Glycobiology 16: 155–164. 10.1093/glycob/cwj043 [DOI] [PubMed] [Google Scholar]
- Roelants F. M., Torrance P. D., Bezman N., Thorner J., 2002. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13: 3005–3028. 10.1091/mbc.e02-04-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants F. M., Baltz A. G., Trott A. E., Fereres S., Thorner J., 2010. A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. USA 107: 34–39. 10.1073/pnas.0912497106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schawalder S. B., Kabani M., Howald I., Choudhury U., Werner M., et al. , 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432: 1058–1061. 10.1038/nature03200 [DOI] [PubMed] [Google Scholar]
- Schnabl M., Daum G., Pichler H., 2005. Multiple lipid transport pathways to the plasma membrane in yeast. Biochim. Biophys. Acta 1687: 130–140. 10.1016/j.bbalip.2004.11.016 [DOI] [PubMed] [Google Scholar]
- Stauffer B., Powers T., 2015. Target of rapamycin signaling mediates vacuolar fission caused by endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Biol. Cell 26: 4618–4630. 10.1091/mbc.E15-06-0344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K. K., McCormick M. A., Pham K. M., MacKay V. L., Delaney J. R., et al. , 2012. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191: 107–118. 10.1534/genetics.111.136549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill T. W., Cohen A., Diefenbacher M., Trautwein M., Martin D. E., et al. , 2008. TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell 7: 1819–1830. 10.1128/EC.00088-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda E., Jin N., Itakura E., Kira S., Kamada Y., et al. , 2018. Vacuole-mediated selective regulation of TORC1-Sch9 signaling following oxidative stress. Mol. Biol. Cell 29: 510–522. 10.1091/mbc.E17-09-0553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira V., Costa V., 2016. Unraveling the role of the target of Rapamycin signaling in sphingolipid metabolism. Prog. Lipid Res. 61: 109–133. 10.1016/j.plipres.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., et al. , 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26: 663–674. 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Vallée B., Riezman H., 2005. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 24: 730–741. 10.1038/sj.emboj.7600562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. T., Hall D. B., Struhl K., 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432: 1054–1058. 10.1038/nature03175 [DOI] [PubMed] [Google Scholar]
- Walther T. C., Aguilar P. S., Fröhlich F., Chu F., Moreira K., et al. , 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26: 4946–4955. 10.1038/sj.emboj.7601933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- Xiao L., Grove A., 2009. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr. Genomics 10: 198–205. 10.2174/138920209788185261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki Y., Kodama Y., Katayama M., Sakamoto A., Kanemaru H., et al. , 2014. Glycogen synthase kinase-3 is involved in regulation of ribosome biogenesis in yeast. Biosci. Biotechnol. Biochem. 78: 800–805. 10.1080/09168451.2014.905183 [DOI] [PubMed] [Google Scholar]
- Yabuki Y., Katayama M., Kodama Y., Sakamoto A., Yatsuhashi A., et al. , 2017. Arp2/3 complex and Mps3 are required for regulation of ribosome biosynthesis in the secretory stress response. Yeast 34: 155–163. 10.1002/yea.3221 [DOI] [PubMed] [Google Scholar]
- Yamada H., Horigome C., Okada T., Shirai C., Mizuta K., 2007. Yeast Rrp14p is a nucleolar protein involved in both ribosome biogenesis and cell polarity. RNA 13: 1977–1987. 10.1261/rna.553807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanolari B., Friant S., Funato K., Sütterlin C., Stevenson B. J., et al. , 2000. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19: 2824–2833. 10.1093/emboj/19.12.2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Sohn J. H., Warner J. R., 2003. Autoregulation in the biosynthesis of ribosomes. Mol. Cell. Biol. 23: 699–707. 10.1128/MCB.23.2.699-707.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, supplemental figures, and supplemental tables. Supplemental material available at Figshare. Supplemental material available at https://doi.org/10.25386/genetics.7782599.