Abstract
RNA helicases are a class of enzymes that unwind RNA duplexes in vitro but whose cellular functions are largely enigmatic. Here, we provide evidence that the DEAD-box protein Dbp2 remodels RNA-protein complex (RNP) structure to facilitate efficient termination of transcription in Saccharomyces cerevisiae via the Nrd1-Nab3-Sen1 (NNS) complex. First, we find that loss of DBP2 results in RNA polymerase II accumulation at the 3′ ends of small nucleolar RNAs and a subset of mRNAs. In addition, Dbp2 associates with RNA sequence motifs and regions bound by Nrd1 and can promote its recruitment to NNS-targeted regions. Using Structure-seq, we find altered RNA/RNP structures in dbp2∆ cells that correlate with inefficient termination. We also show a positive correlation between the stability of structures in the 3′ ends and a requirement for Dbp2 in termination. Taken together, these studies provide a role for RNA remodeling by Dbp2 and further suggests a mechanism whereby RNA structure is exploited for gene regulation.
Keywords: RNA helicase, RNA structure, transcription, termination, DEAD-box
RNA helicases are found in all kingdoms of life, playing central roles in all aspects of RNA metabolism (Bourgeois et al. 2016). Among them, DEAD-box proteins constitute the largest RNA helicase family. These enzymes have a conserved helicase core, which is responsible for ATP binding, hydrolysis, and RNA binding, and are characterized by the Asp-Glu-Ala-Asp (D-E-A-D) motif. Most steps in gene expression involve DEAD-box helicases, including transcription, translation and RNA decay (Linder and Jankowsky 2011). However, the detailed molecular actions of these helicases remain to be characterized.
Most DEAD-box helicases have an ATP-dependent RNA-unwinding activity in vitro (Putnam and Jankowsky 2013). This activity catalyzes a wide variety of biochemically distinct actions including nonprocessive, RNA duplex unwinding (Rogers et al. 1999; Yang and Jankowsky 2006), RNA-protein complex (RNP) remodeling activity in vitro (Fairman et al. 2004; Tran et al. 2007), and ATP-dependent “clamping” of multiprotein complexes onto RNA (Ballut et al. 2005; Nielsen et al. 2008). Studies have also shown that DEAD-box helicases can act as chaperones to promote RNA folding both in vitro and in vivo (Yang and Jankowsky 2005; Tijerina et al. 2006; Liebeg et al. 2010; Potratz et al. 2011). For example, Mss116 in Saccharomyces cerevisiae assists the folding of functional group I and II introns by unwinding misfolded RNAs to allow exchange between kinetically trapped, nonfunctional structures and functional conformations (Liebeg et al. 2010; Potratz et al. 2011). Human DEAD-box helicases, including DDX5 and DDX17, have also been reported to unwind secondary RNA structures and thereby regulate alternative splicing (Kar et al. 2011; Dardenne et al. 2014). RNA remodeling activity also appears to be critical for translation, as a recent genome-wide study of the translation factor and DEAD-box helicase Ded1 showed that this enzyme resolves structures in the 5′ ends of genes and controls translational start site choice in S. cerevisiae (Guenther et al. 2018). These examples implicate DEAD-box helicases as potential regulators of RNA metabolism and gene expression, yet we lack a thorough understanding of how RNA structure and RNP assembly affect basic molecular steps within these processes.
The basic steps in transcription include initiation, elongation, and termination. Termination by RNA polymerase II (RNAPII) is mediated mainly by two complexes in S. cerevisiae: the cleavage and polyadenylation factor (CPF) complex and the Nrd1-Nab3-Sen1 (NNS) complex. CPF-dependent 3′ end processing is the primary mode of termination for messenger RNA (mRNA) genes, whereas the NNS complex, a trimeric assembly of RNA-binding proteins Nrd1 and Nab3 with the RNA-DNA helicase Sen1, promotes termination of short, noncoding RNAPII transcripts and some mRNAs (Rondón et al. 2009; Porrua and Libri 2015). The NNS complex has also been implicated in “failsafe” termination, whereby NNS target sites can rescue defective termination from an upstream CPF-dependent site to prevent aberrant gene expression (Rondón et al. 2009).
Previous results from our laboratory showed that the ortholog of DDX5 in S. cerevisiae, Dbp2 (Xing et al. 2019), is required for efficient termination of RNAPII transcription, as loss of DBP2 results in accumulation of a 3′ extended GAL10 mRNA and GAL10s long, noncoding RNA (Cloutier et al. 2012). Both Dbp2 and DDX5 exhibit highly efficient RNA duplex unwinding in vitro, consistent with a role in altering secondary structure (Ma et al. 2013; Xing et al. 2017). Furthermore, Dbp2 associates with actively transcribed chromatin in an RNA-dependent manner (Ma et al. 2016) and is required for pre-mRNA maturation and messenger RNP assembly, as evidenced by reduced binding of export factors Nab2, Yra1, and Mex67 in dbp2∆ cells (Ma et al. 2013). As efficient termination is necessary for proper assembly of mRNA export factors (Qu et al. 2009), these two steps are likely linked through a common, upstream biochemical step mediated by Dbp2.
Several recent studies combining classic chemical mapping techniques with next-generation sequencing have shown that mRNAs are largely less structured in vivo than in vitro, that secondary structure alteration by single nucleotide polymorphisms may underlie human diseases, and that these changes in structure, when overlapping regulatory sites, have the potential to provide a new level of gene regulation (Rouskin et al. 2013; Ding et al. 2014; Wan et al. 2014; Corley et al. 2015). In addition, early biochemical studies have shown that the formation of RNA secondary structures in polyadenylation signal elements inhibits 3′ processing and termination factor binding of pre-mRNAs in mammalian cells (Chen and Wilusz 1998; Klasens et al. 1998). Genome-wide RNA structure mapping in Arabidopsis also revealed widespread formation of secondary structures in 3′ untranslated regions (UTRs) (Ding et al. 2014). Nevertheless, a role for cellular RNA structure remodeling in transcriptional termination has not been established to date. Using RNA-sequencing (RNA-seq) techniques combined with classic yeast genetics and molecular biology, we provide evidence that the DEAD-box RNA helicase Dbp2 in S. cerevisiae remodels secondary structures within the 3′ ends of a subset of mRNAs to promote efficient termination by the NNS complex. This reveals that DEAD-box RNA helicases remodel mRNA structure in vivo and that structural alteration is essential for proper gene expression.
Materials and Methods
Yeast strains and plasmids
The strains and plasmids used in this study were constructed using classical yeast genetic and/or cloning methods and are listed in Table 1 and Table 2. To construct the termination reporter plasmids, a DNA fragment containing the YOP1 3′ UTR sequence (chrXVI, 624203–624342) was generated from genomic DNA and inserted into the XhoI site of pGAC24 to make pGAC24-YOP1. The YOP1 3′UTR of the resulting plasmid was mutated by site-directed mutagenesis of the pGAC24-YOP1 construct. The primers for strain construction and cloning are listed in Table 3.
Table 1. Yeast strains.
| Strain | Genotype | Source/reference |
|---|---|---|
| Wild type (BY4741) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open biosystems |
| dbp2Δ | MATa dbp2::KanMx6 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Cloutier et al. (2012) |
| DBP2-3XFLAG | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 DBP2-3XFLAG:KanMx | Cloutier et al. (2012) |
| cup1∆ | MATa cup1∆0 his3Δ0 trp1∆0 leu2Δ0 ura3Δ0 lys2∆0 ade2∆0 | Steinmetz and Brow (1996) |
| cup1∆dbp2∆ | MATa dbp2::HygR cup1∆0 his3Δ0 trp1∆0 leu2Δ0 ura3Δ0 lys2∆0 ade2∆0 | This study |
| NRD1-3XFLAG | MATa his3D1 leu2D0 met15D0 ura3D0 NRD1-3XFLAG-KanMx | This study |
| NRD1-3XFLAG dbp2∆ | MATa his3D1 leu2D0 met15D0 ura3D0 NRD1-3XFLAG-KanMx dbp2::HygR | This study |
| RPB3-3XFLAG | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RBP3-3XFLAG-KanMx | This study |
| RPB3-3XFLAG dbp2∆ | MATa his3D1 leu2D0 met15D0 ura3D0 RPB3-3XFLAG-KanMx dbp2::HygR | This study |
Strains are all isogenic and correspond to the BY4741 or S288C background.
Table 2. Plasmids used in this study.
| Plasmid | Source/reference |
|---|---|
| p3XFLAG:KanMX | Gelbart et al. (2001) |
| pGAC24 | Lesser and Guthrie (1993) |
| pGAC24-CYC1 terminator | Steinmetz and Brow (2003) |
| pGAC24-YOP1 3′UTR | This study |
| pGAC24-YOP1 3′UTR-mut1 | This study |
| pGAC24-YOP1 3′UTR-mut1+mut2 | This study |
Plasmids were used for endogenous, 3XFLAG tagging or as the termination reporter (Figure 6).
Table 3. Primers for strain construction and cloning.
| Primer | Sequence |
|---|---|
| NRD1-3XFLAG F | 5′-GCTCAATTGAATTCTTTGATGAATATGCTTAACCAACAGCAG CAGCAACAACAACAAAGCAGGGAACAAAAGCTGGAG-3′ |
| NRD1-3XFLAG R | 5′-TATATATAGAGGTAGATTAGTTTTATGTACTATGAGCAAATA AAGGGTGGAGTAAAGATCCTATAGGGCGAATTGGGT-3′ |
| YOP1-XhoI-F | 5′-ACCGCTCGAGGAGGCAACTCACCTATATCCTC-3′ |
| YOP1-XhoI-R | 5′-ATACCTCGAGTAAACGACTCCAAAATATTTTATATGTTAAG-3′ |
| YOP1-mut F | 5′-GTTGTTATATATCGGTACTCAAGATATAAAATATTTTGGA-3′ |
| YOP1-mut R | 5′-CGACTCCAAAATATTTTATATCTTGAGTACCGATATATAAC-3′ |
| YOP1-mut2 F | 5′-TTCTCTTTGTTATCTATTCTATTCAGAGAATATAAGTACATATG-3′ |
| YOP1-mut2 R | 5′-CAACTAATAACATATGTACTTATATTCTCTGAATAGAATAGATAAC-3′ |
Primers were used for endogenous, 3XFLAG tagging or for reporter construction.
Procedures of individual-nucleotide resolution cross-linking and immunoprecipitation sequencing library construction
Purification of Dbp2-bound RNAs:
Individual-nucleotide resolution cross-linking and immunoprecipitation sequencing (iCLIP-seq) was adapted from the FAST-iCLIP protocol (Flynn et al. 2015) with the following modifications. DBP2-3XFLAG strains were grown in yeast extract and peptone with 2% glucose at 30°, to an OD600 nm of 0.5∼0.7. Then, 250 ml of cells were harvested and washed in ice-cold Tris-buffered saline with 2% glucose. After centrifugation, pellets were resuspended in 12 ml of ice-cold Tris-buffered saline with 2% glucose and irradiated on ice by Ultraviolet C (254 nm) at 180 mJ/cm2 twice for 2.5 min, with a 45-sec rest. Cells were subsequently harvested, frozen in liquid nitrogen, and lysed cryogenically using a Retsch Oscillating Mill MM400. Lysed cells were resuspended in the CLIP Lysis Buffer as previously described (Flynn et al. 2015). The soluble fraction was digested with TURBO DNase (Thermo Fisher Scientific) and RNase I (Thermo Fisher Scientific) at 37° for 10 min. RNase-treated lysates were incubated with FLAG M2 (Sigma) antibody-conjugated Protein G Dynabeads (Thermo Fisher Scientific) at 4° for 2 hr and then washed as described (Flynn et al. 2015). The labeling and purification of immunoprecipitated RNAs were performed as described (Flynn et al. 2015).
Preparation of complementary DNAs:
RNAs were mixed with barcoded 5′ phosphorylated reverse transcription (RT) primers (Table 4) and heated at 65° for 2 min. The complementary DNAs (cDNAs) were generated using TGIRT-III enzyme according to the manufacturer’s instruction. After RT, RNAs were degraded with RNase A and RNase H at 37° for 30 min. The RT products were circularized and purified as described (Flynn et al. 2015).
Table 4. Sequences used for iCLIP-seq.
| Primer | Sequence |
|---|---|
| Biotinylated 3′ adaptor | 5′-/5rApp/AGATCGGAAGAGCGGTTCAG/3Biotin/-3′ |
| RT primer 1 | 5′-/5phos/DDDNNAACCNNNNAGATCGGAAGAGCG TCGTGAT/iSp18/GG ATCC/iSp18/TACTGAACCGC-3′ |
| RT primer 2 | 5′-/5phos/DDDNNACAANNNNAGATCGGAAGAGCG TCGTGAT/iSp18/GG ATCC/iSp18/TACTGAACCGC-3′ |
| RT primer 3 | 5′-/5phos/DDDNNATTGNNNNAGATCGGAAGAGCG TCGTGAT/iSp18/GG ATCC/iSp18/TACTGAACCGC-3′ |
| P3_short | 5′-CTGAACCGCTCTTCCGATCT-3′ |
| P5_short | 5′-ACACGACGCTCTTCCGATCT-3′ |
| P3_Solexa | 5′-CAAGCAGAAGACGGCATACGAGATCGGTCTC GGCATTCCTGCTGAACCGCTCTTCCGATCT-3′ |
| P5_Solexa | 5′-AATGATACGGCGACCACCGAGATCTACACTCT TTCCCTACACGACGCTCTTCCGATCT-3′ |
Bolded letters are barcodes for different libraries.
Library amplification and sequencing:
Real-time PCR reactions were set up using 2× SYBR master mix (Applied Biosystems, Foster City, CA), P3 and P5_short primers. After 40 cycles of PCR, the products were separated by denaturing PAGE and stained with SYBR Gold (Thermo Fisher Scientific). DNAs larger than 75 nt were extracted from the gel. After elution, DNAs were ethanol-precipitated and resuspended in water. PCR products were further amplified by 10 cycles of PCR with P3 and P5_Solexa primers (Table 4). The amplified products above 140 nt were gel purified and resuspended in water. All the samples were analyzed by Agilent Bioanalyzer to determine the size distribution of the library. Sequencing was performed on the Illumina HiSeq 2500 platform for 2× 100 bp paired-end cycle run.
Processing of iCLIP-seq data:
Solexa adaptors were removed using Trimmomatic (version 0.36) (Bolger et al. 2014). The FASTQ files were demultiplexed and PCR duplicates were removed based on the random barcodes incorporated in the RT primers using scripts provided by the Chang laboratory (https://github.com/qczhang/icSHAPE/tree/master/scripts; Flynn et al. 2016). Barcode sequences (13 nt) were trimmed from the 5′ end of retained forward reads using cutadapt (version 1.9.1). The processed reads were mapped to the S288C reference genome (R64-2-1, from Saccharomyces Genome Database) using STAR (version 2.5.2b) (Dobin et al. 2013). Reads mapped to one or two sites were kept for the following analysis to include transcripts from duplicated genes, including ribosomal RNAs. The mapping rate of each replicate was >95%. The number of reads mapped to each annotated RNA transcript were counted using the summarizeOverlaps function in the Bioconductor package “GenomicAlignments” (version 1.8.4). For each of the three Dbp2-iCLIP replicates, reads that did not overlap with any read in the other two replicates were regarded as background and were discarded. Transcripts that had fewer than five counts in each library were filtered from the analysis. Only transcripts that were identified in all three replicates were regarded as binding targets. The nucleotide position before the start of each read was extracted from the forward reads as the cross-linking site in each replicate. Raw reads of Dbp2-iCLIP-seq were deposited to Gene Expression Omnibus (GEO) under the accession number GSE106479.
RNAPII chromatin immunoprecipitation sequencing
Chromatin immunoprecipitation (ChIP) was performed as described (Cloutier et al. 2013), using anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) for immunoprecipitation of endogenously 3XFLAG-tagged Rpb3. Sequencing libraries were prepared from the input and immunoprecipitated DNAs using NEXTflex ChIP-Seq Kit (BIOO Scientific, Austin, TX) according the manufacturer’s instructions. All the libraries were analyzed by Agilent Bioanalyzer to determine the size distribution. Sequencing was performed on the Illumina MiSeq platform for 2× 150 bp paired-end cycle run.
Processing of RNAPII ChIP-seq data
Adaptor sequences were removed using Trimmomatic (version 0.36) (Bolger et al. 2014). The processed reads were mapped to the S288C reference genome (R64-2-1, from Saccharomyces Genome Database) using Bowtie 2 (version 2.3.3.1) (Langmead and Salzberg 2012). Peaks of RNAPII were then determined by MACS2 (version 2.1.2) (Zhang et al. 2008). The fold enrichment of each peak over the background signal was calculated using the “bdgcmp” function in MACS2 and presented as normalized RNAPII occupancy. The overall RNAPII occupancy around termination sites of small nucleolar RNAs (snoRNAs) or mRNAs was analyzed by deepTools (version 3.1.1) (Ramírez et al. 2016).
Identification of Dbp2 binding motifs in mRNA 3′ UTRs
To identify the enriched sequence motifs in close proximity of Dbp2 binding sites in mRNA 3′ UTRs, each binding site was extended by 5 nt on each side (total 11 nt in length). All the binding regions derived from the three replicates were combined into one bed file and were analyzed by HOMER 4.7b (Heinz et al. 2010) using the findMotifsGenome.pl function.
Metagene analysis of Dbp2 binding sites in mRNAs
The coordinates of the UTRs for each mRNA were derived from two published data sets (Nagalakshmi et al. 2008; Yassour et al. 2009). If the UTR coordinates for the same transcript were different in the two data sets, the coordinates that had the widest range were used. For mRNAs without UTR annotations, 135 nt (close to the median lengths of all yeast 5′ and 3′ UTRs) were added before and after the open reading frame (ORF) region as 5′ and 3′ UTRs. The metagene plots were generated using custom R scripts with the following steps. Each 5′ UTR, ORF, and 3′ UTR, was divided into 10, 80, and 10 bins, respectively. The number of bins is proportional to the median length of each region in analyzed mRNAs. The number of cross-linking counts in each bin was then normalized to the library size of each replicate and to the expression level to obtain reads per kilobase of transcript per million mapped reads (RPKM units) of each transcript based on the Structure-seq data [wild type, no dimethyl sulfate (DMS) treatment]. The normalized Dbp2 occupancy in each bin was then divided by the total occupancy in all bins to calculate the distribution of Dbp2. The derived value for each bin in each transcript was averaged among the three replicates, and then averaged across all the transcripts analyzed. The distribution of Dbp2 occupancy across a set of mRNAs was plotted as a line graph based on the calculated value in each bin.
Genomic localization of Dbp2 and Rna15 (or Nrd1/Nab3) binding sites
The RNA binding data of Rna15, Nrd1, and Nab3 were downloaded from the GEO database (Rna15: GSM1442555, Nrd1: GSM791764, Nab3: GSM791767) (Creamer et al. 2011; Baejen et al. 2014). For each protein binding site, the distance between the position and the closest Dbp2 binding site on the same transcript was calculated. For each nearest pair of Rna15 or Nrd1 or Nab3 with Dbp2, the occupancy of the protein was normalized to the occupancy of Dbp2. The average occupancy of each protein at each distance was then calculated and plotted to demonstrate the pattern of the protein distribution near Dbp2 binding sites.
ChIP
ChIP was performed as described (Cloutier et al. 2013). Quantitative PCR was performed using Bio-Rad CFX96 Real-time system using PrimeTime Assay primers purchased from IDT (Table 5). Quantitative PCR results from 3XFLAG ChIP experiments were normalized using RNAPII ChIP from the same lysates to account for differences in transcriptional activity. The significance of the difference was tested using two-sample t-test assuming unequal variances. Strains used for ChIP analysis are listed in Table 1.
Table 5. Oligonucleotides used for ChIP.
| Primer | Sequence |
|---|---|
| NRD1-5′-ChIP-F | 5′-AGGGCAAGTGTTTCGTCC-3′ |
| NRD1-5′-ChIP-R | 5′-AAACTCGTAAAGGGAAGGAGC-3′ |
| NRD1-5′-ChIP-probe | /56-FAM/CCTCCATGT/ZEN/TCCATTCCTCGTTAGCA/3IABkFQ/ |
| PCF11-5′-ChIP-F | 5′-ATTGGATGAGAACTTGGCCT-3′ |
| PCF11-5′-ChIP-R | 5′-CTCCGAAAATTGTCCTGGTAATTC-3′ |
| PCF11-5′-ChIP-probe | /56-FAM/TTTGAAATT/ZEN/TCGCTTTCTTCCCATGCCT/3IABkFQ/ |
| YOP1-ORF-ChIP-F (1) | 5′-GGTTTTATTGGTTCCATCGTCATC-3′ |
| YOP1-ORF-ChIP-R (1) | 5′-CTTTCTTGTTAGTATAACTGCTTCGG-3′ |
| YOP1-ORF-ChIP-probe (1) | /56-FAM/TGTGTCGGG/Zen/TCATTTGGCTGTTTG/3IABkFQ/ |
| YOP1-3′UTR-ChIP-F (2) | 5′-AGACAGAAAAGGATGAAATTAGAGC-3′ |
| YOP1-3′UTR-ChIP-R (2) | 5′-TTTGAGGATATAGGTGAGTTGCC-3′ |
| YOP1-3′UTR-ChIP-probe (2) | /56-FAM/CTGTAGCCT/ZEN/TAGAAGCCTCATTGACGG/3IABkFQ/ |
| YOP1-readthrough-ChIP-F (3) | 5′-TTGGAGTCGTTTATGGTGTCC-3′ |
| YOP1-readthrough-ChIP-R (3) | 5′-TCTGTCGCGCATATCAAGAG-3′ |
| YOP1-readthrough-ChIP-probe (3) | /56-FAM/TCATCGTGT/Zen/TGTGTCGTGACGTGT/3IABkFQ/ |
| RBG1-ORF-ChIP-F (1) | 5′-CCAAAAGAACAAGGCCACATC-3′ |
| RBG1-ORF-ChIP-R (1) | 5′-GCCACATCAAAACCAATACCAG-3′ |
| RBG1-ORF-ChIP-probe (1) | /56-FAM/TGGGTCAAC/Zen/TGAAGGCCAAGCTG/3IABkFQ/ |
| RBG1-readthrough-ChIP-F (3) | 5′-CTACCGAGGGCTCTTCAAAAG-3′ |
| RBG1-readthrough-ChIP-R (3) | 5′-AGATGTGTCAATTTACCAGAAAACTC-3′ |
| RBG1-readthrough-ChIP-probe (3) | /56-FAM/CGACCTCGC/Zen/TTTGTGGTACCCAT/3IABkFQ/ |
| YNL190W- ORF-ChIP-F (1) | 5′-CTATTACTCTAGCCACCGTTGC-3′ |
| YNL190W- ORF-ChIP-R (1) | 5′-AACTACCGTCCGATGACAAAG-3′ |
| YNL190W- ORF-ChIP-probe (1) | /56-FAM/TGCCACTGC/Zen/TAAGAAGGGTGAACAT/3IABkFQ/ |
| YNL190W-3′UTR-ChIP-F (2) | 5′-GGAAGACCTAATTTTCTCCGGT-3′ |
| YNL190W-3′UTR-ChIP-R (2) | 5′-CACAAGCACACGTAAACACATAG-3′ |
| YNL190W-3′UTR-ChIP-probe (2) | /56-FAM/TCAGGATGA/ZEN/AGGGGGTAGGGGG/3IABkFQ/ |
| YNL190W-readthrough-ChIP-F (3) | 5′-GAGAAAGATCATCCAACTTTTAATCATG-3′ |
| YNL190W-readthrough-ChIP-R | 5′-AGCGTAGAAATAAGGAAAAAGAGAAAG-3′ |
| YNL190W-readthrough-ChIP-probe | /56-FAM/CGAATGTGG/ZEN/AAGGAAAATAGAGCGGAGC/3IABkFQ/ |
Preparation of Structure-seq libraries
The Structure-seq method was adapted from prior studies (Ding et al. 2015). Briefly, 50 ml of yeast cells were grown in yeast extract and peptone with 2% glucose at 30° to an OD600 nm of 0.5∼0.7. DMS was added to a final concentration of 10 mM and incubated for 10 min at 30° with vigorous shaking. The reaction was quenched with 75 ml of 4.8 M 2-mercaptoethanol (BME) and 25 ml of isoamyl alcohol. Cells were harvested by centrifugation and cell pellets were washed again with 4.8 M BME, followed by Acetate/EDTA (AE) buffer (50 mM sodium acetate, pH 5.2, 10 mM EDTA). Polyadenylated RNAs were purified and reverse transcribed by SuperScript III (Thermo Fisher Scientific) as described (Ding et al. 2015). After extraction, the cDNAs were resolved by denaturing PAGE and visualized with SYBR Gold. cDNAs longer than 30 nt were isolated and eluted from the gel in Tris-EDTA-NaCl (TEN) buffer (Ding et al. 2015) at 4° overnight. Gel-purified cDNAs were ethanol-precipitated and resuspended in water. A single-strand DNA linker (Table 6) was ligated to the cDNA 3′ ends using CircLigase I (Epicentre) as described (Ding et al. 2015). After ligation, cDNAs above 60 nt were gel purified as above and subjected to PCR amplification as described (Ding et al. 2015). PCR products were resolved by denaturing PAGE, and products above 180 bp were gel purified. After elution, the library was ethanol precipitated and resuspended in water. All the samples were analyzed by Agilent Bioanalyzer to determine the size distribution of the library. A total of 10 libraries, including three replicates of the wild type and two replicates of dbp2∆, with or without DMS treatment, were sequenced on the Illumina HiSeq 2500 platform for 2× 100 bp paired-end cycle run.
Table 6. Oligonucleotides and PCR primers for Structure-seq.
| Primer | Sequence |
|---|---|
| Random-hex RT-primer | 5′-CAGACGTGTGCTCTTCCGATCTNNNNNN-3′ |
| Single-strand DNA linker | 5′-/5Phos/NNNAGATCGGAAGAGCGTCGTGTAG/3SpC3/-3′ |
| Illumina TruSeq forward primer | 5′-AATGATACGGCGACCACCGAGATCTACA CTCTTTCCCTACACGACGCTCTTCCGATCT-3′ |
| Illumina TruSeq reverse primer_index 1 | 5′-CAAGCAGAAGACGGCATACGAGATTGGTCAG TGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′ |
| Illumina TruSeq reverse primer_index 2 | 5′-CAAGCAGAAGACGGCATACGAGATGATCTGG TGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′ |
| Illumina TruSeq reverse primer_index 3 | 5′-CAAGCAGAAGACGGCATACGAGATCGTGATG TGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′ |
Bolded letters are barcodes for multiple libraries.
Processing of Structure-seq data
Illumina adaptors were removed using Trimmomatic (version 0.36) (Bolger et al. 2014). The random trimers were trimmed from the 5′ end of forward reads using cutadapt (version 1.9.1) (Martin 2011). The processed reads were mapped to the S288C reference genome (R64-2-1, from Saccharomyces Genome Database) using STAR (version 2.5.2b) and only uniquely mapped reads (MAPQ = 255 after STAR alignment) were kept for the subsequent analysis. The transcriptome annotation was as described in the “Metagene Analysis” section. Ignoring genes with sequence overlaps with at least one other gene on the same strand, we retained 4681 mRNAs and 77 snoRNAs for differential DMS reactivity analysis. Reads were grouped according to their source RNA, and the start and end indices from genomic alignment of each read were converted to the RNA coordinates with the start of 5′ UTR or mature 5′ end as position +1.
Calculation of DMS reactivities for each transcript
The number of reads starting 1 nt downstream of each nucleotide were tallied to get the detection counts for the nucleotide. In addition, the number of reads starting −1 to +1 of each nucleotide, and ending anywhere downstream of the nucleotide were tallied as its “local coverage.” Detection rates were calculated as ratio of detection counts to local coverage for each nucleotide (Choudhary et al. 2016). Raw reactivities were calculated by combining the information from treated (+) and untreated (−) samples of the same biological replicate. For nucleotide , the raw reactivity, , for the nucleotide was obtained as follows:
where and are detection rates at nucleotide for treated and untreated samples, respectively (Aviran et al. 2011a,b). Reactivities for Gs and Us were masked as missing information. Next, raw reactivities in each replicate were normalized using a 2–8% approach (Sloma and Mathews 2015).
Differential analysis of DMS reactivity changes in the wild type and dbp2∆
Transcripts with average local coverage ≥250 were considered in the following analysis. We used a recently developed a method, dStruct, to identify differentially reactive regions of lengths 21 nt or more and assess statistical significance of differential reactivities at the level of whole transcripts (Choudhary et al. 2019). dStruct compares inherent variation in biological replicates to DBP2-dependent variation at the transcript level. The former kind of variation was calculated using the three wild-type replicates and the latter was calculated using the two replicates of dbp2∆ strain and a wild-type replicate performed in an independent batch. Results with both the raw and the false discovery rate-adjusted P-values using the Benjamini-Hochberg procedure < 0.05 were considered significant.
Identification of 3′ extended mRNAs in dbp2∆
For this analysis, only transcripts without downstream overlapping genes in the sense direction and with over five read counts were considered (3428 mRNAs). For each mRNA, reads from untreated (no DMS) Structure-seq libraries mapped to the ORF or 150 nt downstream of the 3′ UTR (referred as extended region below) were counted using the summarizeOverlaps function (IntersectionNotEmpty mode) in the Bioconductor package “GenomicAlignments” (version 1.8.4). Only transcripts meeting the following conditions were analyzed: the extended region does not overlap with the downstream transcript, with >0.97 counts per million, and detected in at least two libraries. The differential expression of ORFs and extended regions between wild type and dbp2∆ was analyzed using the Bioconductor edgeR package (version 3.14.0; Robinson et al. 2010). ORFs or extended regions with a false discovery rate ≤0.05 were considered differentially expressed in dbp2∆ compared to the wild type. For each transcript, if the reads of the ORF were not changed or down-regulated, but the extended region was upregulated in dbp2∆, then it was regarded as a 3′ extended mRNA in dbp2∆. If the ORF was upregulated in dbp2∆, the extended region in dbp2∆ should be upregulated and have a fold change twice as high as the fold change in the ORF, to be considered as a 3′ extended mRNA in dbp2∆ compared to the wild type.
Analysis of the relationship between DMS reactivity change and Dbp2 binding
Metagene plots in mRNAs were generated using custom R scripts. For each replicate, the 5′ UTR, ORF, and 3′ UTR, were divided into 10, 80, and 10 bins, respectively. DMS reactivities of As and Cs in each bin were tallied. The value in each bin of each transcript from biological replicates was averaged. For each bin, the values from all of the transcripts analyzed were averaged to represent the overall DMS reactivity. Values for all bins were then plotted as a line graph across the whole transcript. To visualize structural changes resulting from deletion of DBP2, reactivities in wild type were subtracted from reactivities in dbp2∆. The metagene-based difference in reactivity was plotted as described above. For each nucleotide in each analyzed mRNA transcript, the distance between the nucleotide and the closest Dbp2 binding site was calculated, and the reactivity value was averaged across wild type or dbp2∆ biological replicates. The average reactivity at each distance was then calculated and the DBP2-dependent changes were derived by subtracting reactivities in dbp2∆ by those in wild type. The relationship was demonstrated by plotting the change of reactivities over a range of distance to Dbp2 binding sites.
Termination reporter assay
Yeast strains cup1∆ and cup1∆dbp2∆ cells were transformed with pGAC24, pGAC24-CYC1 TER, and pGAC24-YOP1 TER. Cells were grown in SC-LEU with 2% glucose, and spotted in fivefold serial dilutions on SC-LEU with 2% glucose plates containing 0 or 1.2 mM CuSO4. Plates were incubated at 30°. For liquid growth assays, cup1∆ and cup1∆dbp2∆ cells containing pGAC24, pGAC24-YOP1 TER, pGAC24-YOP1 ter-mut1, or pGAC24-YOP1 ter-mut1+mut2 were incubated in SC-LEU with 2% glucose media containing 0 or 0.4 mM CuSO4 at 30°, with shaking in a microplate reader (Biotek epoch2, Winooski, VT). OD600 nm values were taken every 30 min. The plot was produced with the average of three replicates.
Data availability
Strains and plasmids are available upon request. Supplemental tables in Excel format are uploaded to Figshare. Raw reads and processed data of iCLIP-seq, Structure-seq, and RNAPII ChIP-seq are deposited on GEO under the accession number GSE106479. Scripts for bioinformatics analysis are deposited on Github: https://github.com/karenlai0222/Dbp2_iCLIP_Structure. Supplemental Material, Figures S1–S5, and Tables S1–S5 are available at Figshare: https://doi.org/10.25386/genetics.7791485.
Results
Dbp2 associates predominantly with snoRNAs and mRNAs
Dbp2 has been shown to function in mRNA metabolism and alter gene expression (Barta and Iggo 1995; Cloutier et al. 2012; Ma et al. 2013; Beck et al. 2014; Wang et al. 2017). Consistently, in a very recent study, Dbp2 was reported to associate with coding and noncoding RNAs, including RNAPII-transcribed snoRNAs and mRNAs (Tedeschi et al. 2018). To gain insight into the precise role of Dbp2 on snoRNAs and mRNAs, we comprehensively identified all RNA targets bound by Dbp2 in vivo. In contrast to prior studies, which utilized a denaturing protocol in conjunction with iCLIP, we used a more standard native iCLIP-seq called FAST-iCLIP (Flynn et al. 2015) using an endogenous, C-terminally 3XFLAG-tagged DBP2 strain. This method resulted in a significantly greater yield, with an average of ∼1.2 million uniquely mapped reads per Dbp2 replicate across three biological replicates.
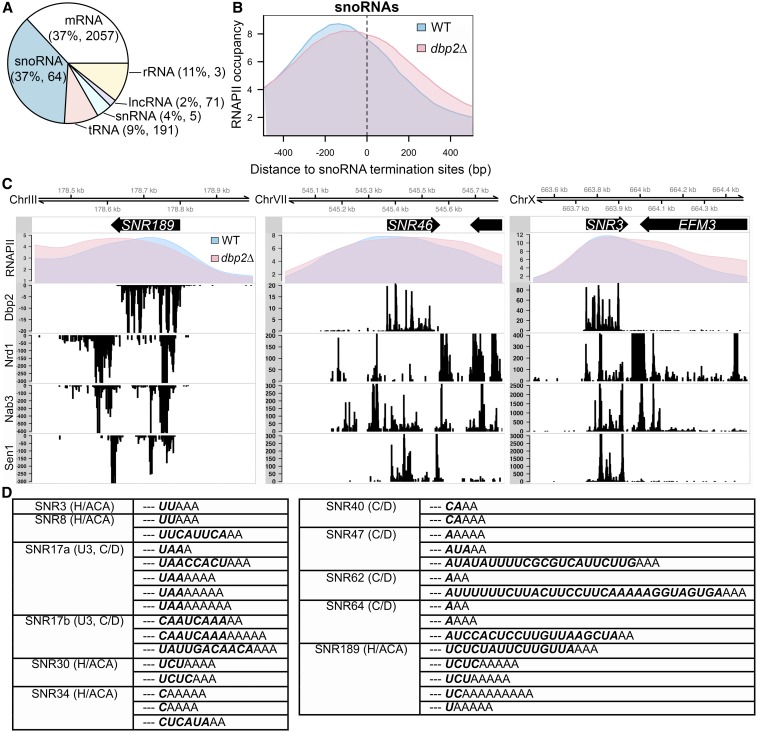
We then determined binding sites from RT stops induced at the site of cross-linking (König et al. 2010) and the binding site counts for each RNA class, which were highly reproducible across replicates (Figure S1). This resulted in a distribution of uniquely mapped reads among RNA classes similar to what was derived from the uniquely mapped reads in the prior study (data not shown) (Tedeschi et al. 2018). Among all unique mapped reads in Dbp2-iCLIP, equal proportions (37%) mapped to snoRNAs and mRNAs, and to other classes to a lesser extent (Figure 1A). The vast majority of transcribed snoRNAs and approximately one-third of protein-coding transcripts were isolated as Dbp2-bound targets. This is consistent with prior studies showing misregulation of both snoRNAs and protein-coding genes in the absence of DBP2 (Cloutier et al. 2012; Beck et al. 2014; Ma et al. 2016).
Figure 1.
Dbp2 promotes transcription termination and processing at snoRNA genes. (A) Dbp2 binds both coding and noncoding RNAPII transcripts. The composition of RNA classes identified by native iCLIP-seq of strains harboring C-terminally 3XFLAG-tagged DBP2 at the endogenous locus. Dbp2 binding sites across all RNA classes were derived from uniquely mapped reads in each replicate. The percentage of the total Dbp2-bound RNAs and number of unique transcripts in each class (shown in parentheses) were calculated and averaged from the three biological replicates. Note that the number of ribosomal RNA transcripts in Dbp2 iCLIP-seq is under-represented due to ribosomal DNA repeats. (B) RNAPII ChIP-seq reveals snoRNA gene termination defects in dbp2∆ cells. Normalized RNAPII occupancy across termination sites of monocistronic snoRNA genes from RNAPII ChIP-seq in the wild type (blue) and dbp2∆ (pink). snoRNA termination sites were obtained from Schaughency et al. (2014). (C) Comparison of RNAPII occupancy with RNA-binding sites of Dbp2 and Nrd1, Nab3, and Sen1 at individual snoRNA gene loci. Normalized RNAPII profiles across select snoRNA genes from RNAPII ChIP-seq in wild type and dbp2∆ (top panel). The binding patterns of Nrd1, Nab3, and Sen1 were reproduced from previously published data (Creamer et al. 2011). Note that the difference in scales is due to the difference in how data were generated and does not represent the absolute quantity of binding. (D) Unprocessed snoRNA sequences in Dbp2 iCLIP-seq reads. Table of all Dbp2-bound snoRNA sequences that show unprocessed nucleotides (bold italics) and nontemplated As at 3′ ends of the mature snoRNA (dashed lines). The class of snoRNA (Box C/D or Box H/ACA) are listed in parentheses next to the name. lncRNA, long noncoding RNA; rRNA, ribosomal RNA; tRNA, transfer RNA; WT, wild type.
Dbp2 promotes efficient transcription termination of snoRNAs
Dbp2 interacts physically with Sen1 (Tedeschi et al. 2018), an RNA-DNA helicase involved in transcriptional termination of snoRNAs in conjunction with RNA-binding proteins Nrd1 and Nab3 (Arndt and Reines 2015). To determine if Dbp2 plays a role in snoRNA termination, we performed RNAPII chromatin immunoprecipitation combined with high-throughput sequencing (ChIP-seq) for both wild-type and dbp2∆ cells expressing an endogenous, C-terminally 3XFLAG-tagged Rpb3 subunit. Input and immunoprecipitation libraries were prepared from three biological replicates of both wild-type and dbp2∆ cells and resulted in an average of ∼1.8 million reads per replicate. We then determined the distribution of RNAPII along genes using a package for ChIP-seq data analysis, MACS2 (Zhang et al. 2008), and asked if this occupancy is altered in dbp2∆ at snoRNA transcriptional termination sites (Schaughency et al. 2014). In wild-type cells, RNAPII shows an average occupancy profile across all monocistronic snoRNA genes that conforms to a bell-shaped curve with a peak near −200 bp, with respect to the termination site (0 bp) (Schaughency et al. 2014), that corresponds to the transcription start site (Figure 1B). In dbp2∆ cells, this peak is slightly reduced in height and shifted ∼20 bp downstream. Moreover, the average RNAPII occupancy remains higher after the annotated termination site and shows a less steep reduction in dbp2∆ cells compared to wild type. This pattern is highly similar to the RNAPII ChIP profiles following nuclear depletion of either Sen1 or Nrd1 (Schaughency et al. 2014). It has been proposed that during termination of snoRNA transcription, Nrd1 and Nab3 recognize sequence motifs in the precursor RNA and recruit Sen1, leading to termination of RNAPII elongation (Arndt and Reines 2015). Inspection of individual snoRNA gene profiles also reveals RNAPII accumulation in the absence of DBP2 after the annotated mature 3′ ends, across regions that correspond to previously identified binding sites for Nrd1, Nab3, and at the termination sites of precursor snoRNA transcripts (Figure 1C) (Jamonnak et al. 2011). Interestingly, and consistent with prior studies (Tedeschi et al. 2018), Dbp2 iCLIP sites correspond to Nrd1, Nab3, and Sen1 binding sites at some snoRNAs in the mature transcript region (SNR3, Figure 1C) but not others (SNR189 and SNR46). This binding pattern suggests that Dbp2 also binds to the mature, fully processed snoRNAs.
Because snoRNA gene termination is coupled to 3′ end processing, we then analyzed our Dbp2-iCLIP reads to detect the presence of unprocessed 3′ extensions in the Dbp2 RNA-binding data. This revealed sequences that mapped to 11 different snoRNA species that also contained 1–30 unprocessed nucleotides followed by short stretches of nontemplated As on the 3′ ends (Figure 1D). These sequences likely correspond to processing intermediates of the Trf4-Air2-Mtr4 Polyadenylation (TRAMP) complex and nuclear exosome, which are coupled with NNS-dependent termination (Arndt and Reines 2015). Similar processing intermediates have been observed in PAR-CLIP data of NNS factors (Jamonnak et al. 2011) and upon analysis of snoRNAs in rrp6∆ mutants (Grzechnik and Kufel 2008). Taken together, this suggests that Dbp2 functions in termination and maturation of snoRNAs.
Dbp2 shares RNA-binding profiles with Nrd1 and Nab3 on protein-coding transcripts and promotes loading of Nrd1 on gene 5′ ends
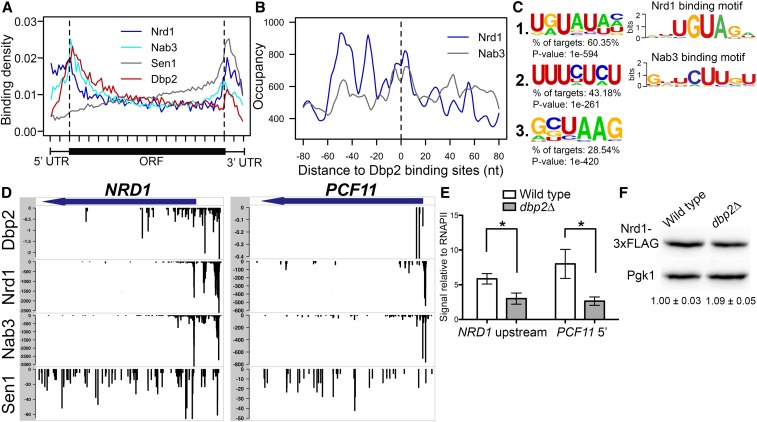
Loss of DBP2 results in accumulation of a bicistronic GAL10-GAL7 transcript (Cloutier et al. 2012), suggesting that Dbp2 may also function in termination of protein-coding genes in addition to snoRNAs. To gain insight into the role of Dbp2 binding on protein-coding genes, we then analyzed the binding pattern of Dbp2 on protein-coding transcripts using our iCLIP data set. Metagene analysis revealed Dbp2 binding across the entire length of targeted mRNAs, with highest accumulations at the 5′ end of the ORF and the 3′UTR (Figure 2A, red line). To determine if this mRNA-binding profile is similar to NNS components, we overlapped our meta-analysis of Dbp2 binding with meta-analysis profiles of Nrd1, Nab3, and Sen1 generated from previously published data sets (Creamer et al. 2011). This revealed a strikingly similar pattern of Nrd1 and Nab3 binding to Dbp2, with all three proteins exhibiting peaks at 5′ and 3′ ends of the genes (Figure 2A). Analyzing the distribution of Nrd1 and Nab3 binding sites relative to the Dbp2 cross-link site on mRNAs revealed a broad peak of Nab3 and Nrd1 binding at or near Dbp2 (within ±20 nt) (Figure 2B), suggestive of overlapping binding for all three factors. Interestingly, we also observed a periodicity in Nrd1 accumulation of ∼25 nt within the 160 nt window, with decreased levels of Nrd1 accumulation from −55 to +55 with respect to the Dbp2 cross-linking site (Figure 2B). This pattern was not observed for Nab3. We then analyzed Dbp2 iCLIP reads for the presence of enriched sequence motifs, using HOMER (Heinz et al. 2010). Despite the fact that the vast majority of DEAD-box RNA helicases exhibit sequence-independent RNA binding in vitro (Gilman et al. 2017), we found three significantly enriched motifs in Dbp2-bound 3′ UTRs (Figure 2C). The first motif with the highest enrichment (Figure 2C, motif 1) is strikingly similar to the UGUA Nrd1 RNA-binding motif (Figure 2C, top right; Creamer et al. 2011). The second most-enriched motif is U/C rich (Figure 2C, motif 2) and somewhat similar to the Nab3 sequence motif (Figure 2C, bottom right; Creamer et al. 2011). Motif 2 is also similar to the U/C-rich motif of Rna15 (UUUUCUU; Baejen et al. 2014), a component of the CPF complex that plays the predominant role in termination of protein-coding genes (Mischo and Proudfoot 2013). However, no global similarity was seen between the meta-analysis profiles of Rna15 and Dbp2 (Figure S2A), nor do the two proteins share common occupancy sites when averaged across all Dbp2-bound mRNAs (Figure S2B).
Figure 2.
Dbp2 binding in protein-coding transcripts correlates with Nrd1 and Nab3 binding sites and shares similar RNA sequence motifs. (A) Dbp2, Nrd1, Nab3, and Sen1 meta-analysis reveals similar distribution patterns on protein-coding genes. Meta-analysis of Nrd1 (blue), Nab3 (cyan), Sen1 (gray) (Creamer et al. 2011), and Dbp2 (red) binding sites across all commonly bound mRNAs. Dashed vertical lines mark boundaries between 5′ and 3′ UTRs and ORFs. (B) Nrd1 and Nab3 show enriched occupancy at Dbp2 binding sites across protein-coding transcripts. The distance between Dbp2 and Nrd1 (blue) or Nab3 (gray) binding sites in all commonly bound mRNAs. (C) Enriched sequence motifs bound by Dbp2 in protein-coding transcripts and comparison to known Nrd1 and Nab3 binding sites. Motifs were identified from Dbp2 iCLIP-seq using HOMER (Heinz et al. 2010). Web logos of sequence motifs bound by Nrd1 and Nab3 were reproduced from published reports (Creamer et al. 2011) (right panel). (D) Comparison of Dbp2 binding with Nrd1, Nab3, and Sen1 binding at previously reported protein-coding targets of the NNS complex. Meta-analysis of Nrd1, Nab3, and Sen1 RNA-binding data sets from Creamer et al. (2011) in conjunction with DBP2. (E) Loss of DBP2 reduces recruitment of Nrd1 to protein-coding genes. ChIP quantitative PCR of endogenously, 3XFLAG-tagged NRD1 strain at 5′ ends of NRD1 (left) and PCF11 (right). Signals are shown relative to input and RNAPII ChIP levels, the latter of which accounts for changes in abundance due to transcriptional activity. * indicates two-sided P-value <0.05. (F) Reduced Nrd1 association is not a result of decreased Nrd1-3XFLAG levels in dbp2∆. Representative Western blot of Nrd1-3XFLAG and Pgk1 in wild-type and dbp2∆ strains. Numbers below the blot correspond to the relative level of Nrd1-3XFLAG in dbp2∆ cells compared to wild type normalized to a Pgk1 loading control across three independent biological replicates. The signal of the wild type is set to one and SD are shown.
The common binding profiles of Dbp2 with Nrd1 and Nab3 suggest that this enzyme may function in concert with the NNS complex on protein-coding genes in addition to snoRNA genes. Association of the NNS complex with the 5′ ends of protein-coding genes has been associated with transcription attenuation (Arigo et al. 2006; Kuehner and Brow 2008; Kim and Levin 2011). Interestingly, comparison of individual RNA binding tracks on known NNS-targeted protein-coding transcripts NRD1 and PCF11 (Creamer et al. 2011) revealed strikingly similar binding patterns of Nrd1, Nab3, and Dbp2 at the 5′ ends of both genes (Figure 2D). We then asked if DBP2 is necessary for Nrd1 binding to transcribed genes by conducting ChIP of a 3XFLAG-tagged Nrd1 protein in wild-type and dbp2∆ cells. This revealed an ∼50% decrease in Nrd1 association at the 5′ end of both NRD1 and PCF11 genes in the absence of DBP2 (Figure 2E). A Western blot shows that decreased Nrd1 binding in dbp2∆ cells is not due to decreased Nrd1 protein abundance (Figure 2F). Taken together, these results suggest that Dbp2 may function in concert with the NNS complex at targeted protein-coding genes, possibly by promoting binding of Nrd1 to nascent RNA.
DBP2 facilitates proper transcription termination of a subset of mRNAs
Next, we asked if RNAPII accumulates downstream of the annotated termination site in protein-coding genes in dbp2∆ cells similar to snoRNA genes above. Importantly, this analysis was restricted to exclude overlapping genes encoded in tandem to minimize read assignment ambiguity. We also performed RNA-seq of poly(A)-selected RNAs from wild-type and dbp2∆ cells to identify gene products with putative termination defects as evidenced by 3′ extensions. Protein-coding transcripts with 3′ extensions were identified from the RNA-seq by the presence of reads mapping greater than 150 bp downstream of the annotated 3′UTR (Nagalakshmi et al. 2008; Yassour et al. 2009). This arbitrary definition was selected as 90% of RNAPII termination events occur within a 50 bp window after the polyadenylation site (Baejen et al. 2017).
Whereas RNAPII exhibited a broad peak of maximal occupancy centered at 30 bp downstream of polyadenylation site (annotated as 0 bp) in wild-type cells, loss of DBP2 resulted in a slight shift downstream, with the highest peak central at 60 bp downstream (Figure 3A). We also observed an unexpected reduction of occupancy 200 bp and further to either side of the polyadenylation site in dbp2∆ cells.
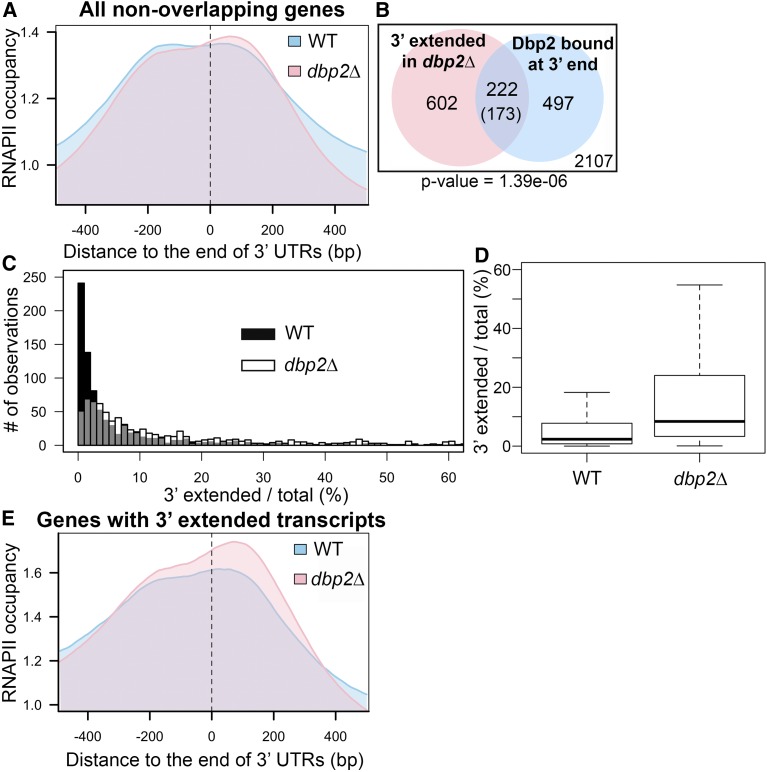
Figure 3.
Loss of DBP2 causes termination defects at a subset of protein-coding genes. (A) Meta-analysis of normalized RNAPII occupancy at the 3′ ends of all protein-coding genes in wild-type and dbp2∆ cells. Only transcripts without overlapping genes within 150 bp downstream in the sense direction were considered in the analysis. The 0 position and dotted line marks the location of the polyadenylation site (Nagalakshmi et al. 2008; Yassour et al. 2009). (B) Venn diagram showing the intersection between 3′ extended transcripts in dbp2∆ and mRNAs bound by Dbp2 at the 3′ end (50 nt of ORF 3′ end through the 3′ UTR). Aberrant transcripts were identified following RNA-seq of wild-type and dbp2∆ strains by an overaccumulation of reads mapping within 150 nt downstream of the 3′ UTR, after accounting for different expression levels between wild-type and dbp2∆ strains. The number in the parentheses is the expected value of intersection if the two groups of transcripts have no significant correlation. The number within the white square corresponds to genes that lack both detectible, putative read-through products in dbp2∆ and Dbp2 binding on mRNA 3′ ends. The P-value derived from a one-sided Fisher’s exact test is shown. (C) Loss of DBP2 results in differential accumulation of 3′ extended products for individual genes. Histograms illustrating the percentage of total transcripts corresponding to 3′ extended products in dbp2∆ or wild-type cells as determined by read counts in the extended region over the counts in the ORF in wild-type and dbp2∆ cells multiplied by 100. Gray coloring denotes overlap between the histogram of wild type and dbp2∆. (D) Loss of DBP2 results in a broader distribution of 3′extended transcripts as compared to wild-type cells. A box plot showing the quartile distribution of the ratio of extended vs. total transcript in wild-type and dbp2∆ cells. The distributions of the two strains is significantly different, as tested by the two sample Kolmogorov–Smirnov test (P-value <2.2e−16). (E) Protein-coding genes that produce 3′ extended transcripts show RNAPII accumulation in dbp2∆ cells at downstream of annotated 3′ UTRs. Meta-analysis of RNAPII occupancy across the 824 genes with 3′ extended transcripts in dbp2∆ RNA-seq. WT, wild type.
Next, we analyzed our RNA-seq data for the presence of 3′ extended mRNAs. This yielded 824 protein-coding transcripts, corresponding to ∼14% protein-coding genes, with 3′ extensions in dbp2∆ cells (Table S1). This number is likely an under-representation due to the criteria outlined above for identification of 3′ extended gene products. A Fisher’s exact test revealed a statistically significant overlap between mRNAs bound by Dbp2 at their 3′ ends and those 3′ extended isoforms, suggesting a correlation between Dbp2 binding and suppression of 3′ extension (Figure 3B and Table S2). The fact that not all aberrant transcripts in dbp2∆ cells are represented in our Dbp2 iCLIP is not surprising because iCLIP isolation depends on both RNA binding and RNA sequence context (Sugimoto et al. 2012).
By calculating the percentage of mRNAs with 3′ extensions vs. total mRNAs for a given gene product, using the formula read counts that map 150 bp downstream of polyadenylation site/read counts mapped to the ORF, we found that the percentage of 3′ extended mRNAs ranged from 0.0 to 18.0% in the wild type (median, 2.3%) and from 0.1 to 55.0% in dbp2∆ (median 8.4%) (Figure 3, C and D). The presence of 3′ extended products in wild-type cells is consistent with the recently described, heterogeneous nature of termination site choice in S. cerevisiae (Moqtaderi et al. 2018). Loss of DBP2 appears to increase the proportion of 3′ extended products per gene, as illustrated most clearly by the box plot distribution (Figure 3D). This increased abundance could be due to read-through of a predominant termination site, increased stability of the 3′ extended mRNAs in the absence of dbp2∆ (as recently described for mRNA 3′ isoforms; Moqtaderi et al. 2018), or a combination of both. To test the possibility of long 3′ isoform accumulation in dbp2∆, we compared the end positions in our analysis for read-through with the most 3′ polyadenylation sites (Pelechano et al. 2013). We found that the majority (∼77%) of the identified, extended transcripts had reads extended past the most 3′ polyadenylation site, suggestive of read-through products, whereas the other extended transcripts can be longer isoforms accumulated in dbp2∆. Future study with 3′ end-specific sequencing would be needed to determine the precise DBP2-dependent change in polyadenylation site usage. We then analyzed RNAPII occupancy for the 824 genes with 3′ extended mRNAs in dbp2∆ cells. This revealed accumulation of RNAPII in dbp2∆ across a 400-bp window centered at the polyadenylation site with the most striking accumulation 0–150 bp downstream (Figure 3E). This accumulation is centered 50 bp downstream in dbp2∆ cells as compared to wild type, and is reminiscent of nuclear depletion of Rat1 or Ysh1, nucleases required for efficient RNAPII termination (Schaughency et al. 2014; Baejen et al. 2017). In addition to genes with delayed RNAPII pausing or termination, we did observe that in a small subset of genes (∼250), the most 3′ peak summit of RNAPII in dbp2∆ is slightly upstream compared to the peak in the wild type. This may contribute to the reduction of RNAPII signal at >200 bp downstream of polyadenylation sites in dbp2∆ observed in Figure 3A. However, the resolution of this analysis is not sufficient to determine the exact mechanism contributing to the shift of peak toward upstream and will need further characterization in the future.
Dbp2 facilitates loading of termination factors at 3′ ends of protein-coding genes
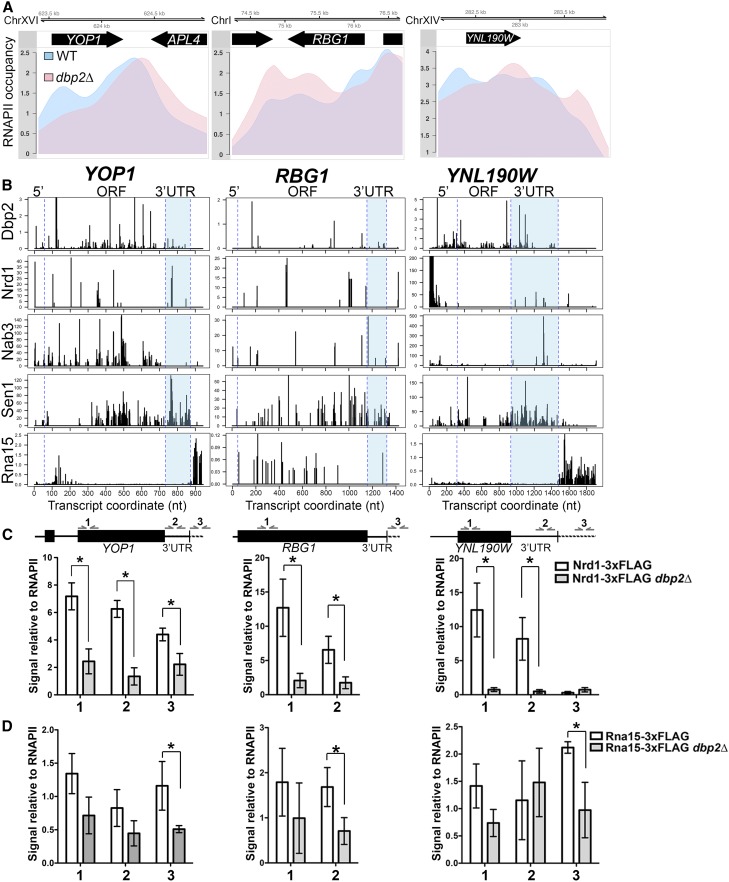
To gain insight into the mechanism for Dbp2-dependent termination of protein-coding transcripts, we then inspected the RNAPII occupancy of three genes that produce 3′ extended mRNAs in dbp2∆ that are also bound by Dbp2 in their 3′ UTRs in wild-type cells. These three genes are YOP1, an intron-containing, shorter gene with an average length 3′UTR (675 bp ORF, 140 bp 3′UTR); RBG1, an intronless, longer gene with an average 3′UTR (1100 bp ORF, 167 bp 3′UTR); and YNL190W, a shorter gene with a long 3′ UTR (614 bp ORF, 543 bp 3′UTR). Note that the average gene length and 3′UTR for all protein-coding genes are 1343 and 188 bp, respectively. All three profiles showed accumulation of RNAPII that extends further downstream in dbp2∆ than in wild-type cells, indicative of defective termination (Figure 4A). Interestingly, we also observed a decrease in RNAPII accumulation at the 5′ ends of all three genes, which likely explains the decreased accumulation toward the 5′ end of genes in our combined RNAPII profile (Figure 3A). A similar decrease was also noted for sen1 mutants on short, protein-coding genes and was speculated to be due to decreased initiation (Steinmetz et al. 2006). However, we observed no difference in the abundance between wild-type and dbp2∆ cells of transcripts of these three genes. Moreover, YNL190W shows a change in the overall pattern of RNAPII accumulation. This suggests that Dbp2 may also function in the rate of RNAPII progression along a given gene.
Figure 4.
DBP2-dependent termination of protein-coding genes correlates with efficient DBP2-dependent recruitment of Nrd1 within the gene ORF and 3′UTR. (A) RNAPII occupancy at three selected protein-coding genes shows DBP2-dependent termination. Normalized RNAPII occupancy at the three termination-defective protein-coding genes in the wild type (blue) and dbp2∆ (pink). (B) Comparison of Dbp2, Nrd1, Nab3, Sen1, and Rna15 RNA-binding sites at individual genes shows similar distributions between Dbp2 and members of the NNS complex. The published binding sites for Nrd1, Nab3, Sen1, and Rna15 were obtained from the GEO sessions GSE31764 (Creamer et al. 2011) and GSE59676 (Baejen et al. 2014). The blue dashed lines correspond to the boundaries of the coding region and the end of 3′ UTR and are aligned to the schematic representation in C. (C) Loss of DBP2 reduces recruitment of Nrd1 to protein-coding genes. ChIP quantitative PCR of endogenously, 3XFLAG-tagged NRD1 strain at 5′ ends (1), 3′UTRs (2), and downstream of polyadenylation sites (3) in YOP1, RBG1, and YNL190W. Signals are shown relative to input and RNAPII ChIP levels, the latter of which accounts for changes in abundance due to transcriptional activity. * indicates a two-sided P-value <0.05. Note that the 3′UTR (2) of RBG1 was not assayed due to a technical limitation of quantitative PCR primer binding sites. (D) Rna15 shows reduced chromatin binding in dbp2∆ cells but only downstream of the polyadenylation site. ChIP quantitative PCR was conducted as in C, but with an endogenously 3XFLAG-tagged RNA15 strain. WT, wild type. * indicates a two-sided P-value < 0.05.
Previously, the NNS complex was shown to bind the 3′ UTRs of a subset of mRNAs (Creamer et al. 2011; Webb et al. 2014); however, the precise functional role of this complex in 3′UTRs was not determined. Comparison of the RNA-binding profiles of Dbp2, Nrd1, Nab3, and Sen1 revealed that Dbp2 binding in the 3′UTRs of these transcripts overlaps with the binding of one or more factors in the NNS complex (Figure 4, D–F). Since DBP2 promotes efficient recruitment of Nrd1 to the 5′ ends of NNS-targeted protein-coding genes (Figure 2E), we then asked if this was also the case at gene 3′ ends by conducting ChIP of Nrd1-3XFLAG at the 5′ end (Figure 4C, 1), 3′UTR (Figure 4C, 2), and downstream of the polyadenylation site (Figure 4C, 3). Note that we were unable to assay 3′UTR (Figure 4C, 2) binding to RBG1 due to technical reasons (lack of efficient primer-binding sites for quantitative PCR because of AT-richness). Similar to the requirement for DBP2 at the 5′ ends of known NNS targeted genes, we also observed reduced Nrd1 association at both the 5′ and 3′ ends of our three candidate genes in the absence of DBP2, regardless of the presence of an intron or length of the 3′UTR (Figure 4C). Interestingly, loss of DBP2 also decreased the association of a 3XFLAG-tagged Rna15, but this decrease was only statistically significant downstream of the polyadenylation site, in line with the primary location of RNA binding by this protein after the 3′UTR (Baejen et al. 2014) (Figure 4B, bottom panels). This suggests that Dbp2 may also promote efficient association of Rna15 on protein-coding genes, despite the distinct RNA-binding patterns of these two proteins (Figure 4B and Figure S2, A and B). This may be due to an as-of-yet unidentified role for Dbp2 in CPF-dependent termination or to an indirect effect of reduced Nrd1 binding at sites within the 3′UTR.
Dbp2 modulates RNA/RNP structures in vivo
Given that Dbp2 is an efficient helicase in vitro (Ma et al. 2013), and DEAD-box proteins have been shown to remodel RNA structures in vivo (Guenther et al. 2018), we hypothesized that Dbp2 might remodel RNA structures and/or RNPs of RNAPII transcripts to promote termination and subsequent mRNA metabolism steps. This is likely to be true for NNS binding as both Nrd1 and Nab3 recognize RNA motifs in the context of single-strand RNA (Singh et al. 2007; Arndt and Reines 2015). Moreover, DBP2 is required for association of mRNA export factors Yra1 and Mex67 (Ma et al. 2013), which act after termination. Sequestration of polyadenylation signal sequences in secondary structures can impede CPF-dependent termination (Chen and Wilusz 1998; Klasens et al. 1998). This is likely to be true for NNS binding as both Nrd1 and Nab3 recognize RNA motifs in the context of single-strand RNA (Singh et al. 2007; Arndt and Reines 2015).
To test this, we conducted Structure-seq of total, poly(A)-selected RNA in both wild-type and dbp2∆ cells. Structure-seq uses the cell-permeable compound DMS to preferentially methylate adenosine and cytosine residues that are not involved in Watson–Crick base pairing (Peattie and Gilbert 1980) or not protected by proteins, combined with next-generation sequencing to observe RNA structure and compositional changes genome-wide (Ding et al. 2015). Methylated nucleotides were detected as RT “stops,” after library construction and RNA-seq, and then translated into nucleotide-level reactivities to DMS and a prediction of increased or decreased protection between wild type and dbp2∆ (see Materials and Methods). Total poly(A)+ RNA was used for Structure-seq to enable direct comparison with 3′ extended transcripts in our RNA-seq and because there is currently no method available to map secondary structures in nascent RNA transcripts in eukaryotes. Consistent with prior studies, we observed RT-stops indicative of DMS-dependent methylation predominantly at As and, to a lesser extent, at Cs in DMS-treated samples compared to untreated samples (Figure S3) (Rouskin et al. 2013; Ding et al. 2014). We then analyzed differential DMS reactivity in wild type and dbp2∆ for each transcript using a newly developed method, dStruct, for Structure-seq analysis (Choudhary et al. 2019). It identifies transcripts and regions that manifest significantly more variation between strains than among samples of the same strain.
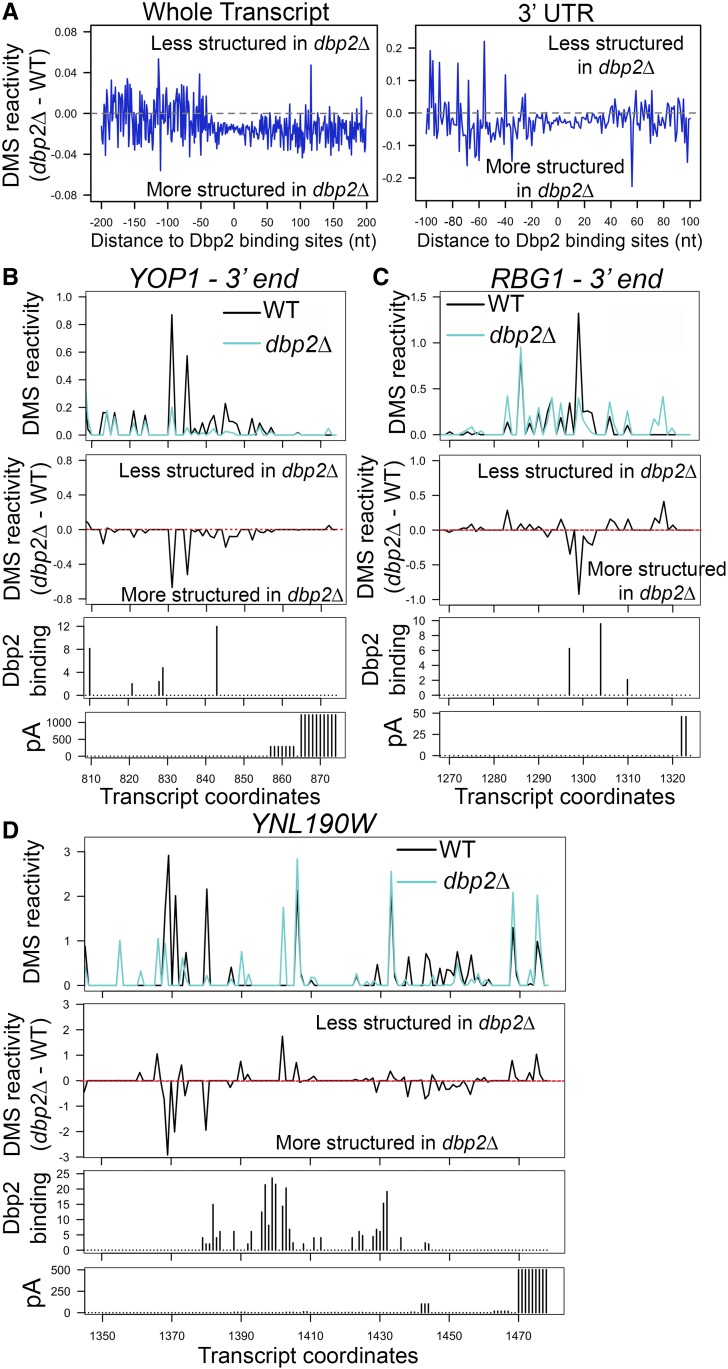
Our analysis identified 612 protein-coding transcripts with significant, DBP2-dependent changes in DMS reactivity (P-value <0.05, false discovery rate < 0.05, Table S4). Importantly, these transcripts were significantly enriched in Dbp2-bound transcripts identified in our iCLIP-seq (P-value <2.2e−16, Table S5), suggesting that these changes are directly due to Dbp2 activity. A metagene analysis of differential reactivity of these 612 transcripts revealed a lower overall DMS reactivity indicative of increased nucleotide protection in dbp2∆ as compared to wild type, centered at the Dbp2 binding site (Figure 5A, left). This trend was also observed when the analysis was restricted to 3′ UTRs (Figure 5B, right). Interestingly, the DBP2-dependent decrease in DMS accessibility spans ∼60–70 nt in length around the Dbp2 binding site, suggestive of a broad region of Dbp2-dependent RNA/RNP structural remodeling. This pattern is unlikely to be due to Dbp2 binding alone as DEAD-box proteins have a binding-site of ∼6 nt along the sugar-phosphate backbone (Andersen et al. 2006). Moreover, one would expect decreased reactivity in wild-type cells rather than dbp2∆ cells, if this pattern was due to Dbp2 binding. Instead, this is more consistent with a region of increased RNA structures and/or RNP composition.
Figure 5.
Structure-seq reveals DBP2-dependent RNA structural changes in protein-coding genes. (A) Meta-analyses of DMS reactivity at Dbp2 binding sites on mRNAs reveals 40–60 nt “trough” of DBP2-dependent structure. The relationship between Dbp2 binding sites and the changes in DMS reactivity was analyzed for both the whole transcript (left panel) and 3′ UTR (right panel). DBP2-dependent change in DMS reactivity of each nucleotide in Dbp2-bound mRNA transcripts was calculated by subtracting the average DMS reactivity in dbp2∆ by that in wild type, and was then plotted corresponding to the distance to Dbp2 binding sites identified in iCLIP-seq. (B–D) Reactivity profiles of the 3′ ends of YOP1 (B), RBG1 (C), and YNL190W (D) in wild type and dbp2∆ cells (top panels). The x-axes indicate transcript coordinates relative to the transcriptional start site. The annotated 3′ UTR region spans coordinates 733–872 for YOP1, 1156–1322 for RBG1, and 934–1476 for YNL190W as defined previously (Nagalakshmi et al. 2008; Yassour et al. 2009). Only the regions close to the end of 3′ UTR are shown (50–120 nt). DBP2-dependent changes in DMS reactivity are also presented as the values derived by subtracting the reactivity in wild type from the reactivity in dbp2∆ (middle panels). Dbp2 binding sites derived from iCLIP-seq are shown below reactivity profiles. Annotated polyadenylation sites (pA; Ozsolak et al. 2010) are shown for reference (bottom panels). Bar height in the pA histograms indicate the relative frequency of polyadenylation at each position. WT, wild type.
Next, we inspected the reactivity profiles of 3′ UTRs of YOP1, RBG1, and YNL190W (Figure 5, B–D). Although both increased and decreased reactivity was observed across all three selected 3′UTRs (Figure 5, B–D, top and middle panels), nucleotides exhibiting decreased reactivity in the absence of DBP2 gave the largest difference (Figure 5, B–D, middle panels). Interestingly, these sites appear to correspond to those that exhibit the least protection (most reactive) in wild-type cells in these three representative transcripts. This indicates that although the overall 3′UTR structure, which can include both base pairing and protein binding, is changed in the absence of DBP2, there may be a tendency for regions that are typically unprotected to be protected. Moreover, Dbp2 binding sites are found within close proximity of these regions (Figure 5B). This is consistent with RNA/RNP structural remodeling of 3′UTRs by Dbp2. Although we were able to analyze some snoRNAs for DBP2-dependent structural changes (Figure S4), we were unable to analyze regions of termination/processing due to insufficient coverage for dStruct analysis in preprocessed precursor snoRNAs. Regardless, our results above, and the known enzymatic role of Dbp2 as a DEAD-box RNA helicase, suggest that this enzyme may remodel RNA structure during termination for multiple RNAPII transcripts.
DBP2-dependent RNA structure correlates with DBP2-dependent termination
To understand the relationship between binding and structural remodeling of 3′UTRs of protein-coding transcripts with efficient termination, we qualitatively compared differential reactivity profiles across different data sets. First, we compared the DMS reactivities between wild type and dbp2∆ cells by plotting the difference in reactivity (dbp2∆ minus wild type) for all transcripts that are bound or not bound by Dbp2. This revealed reduced reactivity across Dbp2-bound transcripts in the absence of DBP2 across the length of the 5′UTR, ORF, and 3′UTR (Figure S5A). Second, we compared differential reactivities of transcripts that exhibit 3′ extensions in dbp2∆ to those that do not (Figure S5B). This revealed that transcripts from genes with DBP2-dependent termination are more protected across their 3′UTRs in the absence of DBP2 (Figure S5B). Consistent with the latter, a Fisher’s exact test showed significant correlation between the presence of DBP2-dependent differential reactivity and DBP2-dependent termination (P-value = 9.4e−16, Figure S5C and Table S6). Moreover, meta-analysis of Dbp2 binding near mRNA 3′ ends revealed a sharp peak at the immediate upstream of the polyadenylation site specifically in the group of mRNAs with 3′ extension in dbp2∆ as compared to those without 3′ extension (Figure S5D), in line with the pattern of structural change observed in Figure S5B. This suggests the Dbp2-dependent remodeling of structures and/or proteins within the 3′UTR as a likely mechanism for DBP2-dependent reactivity.
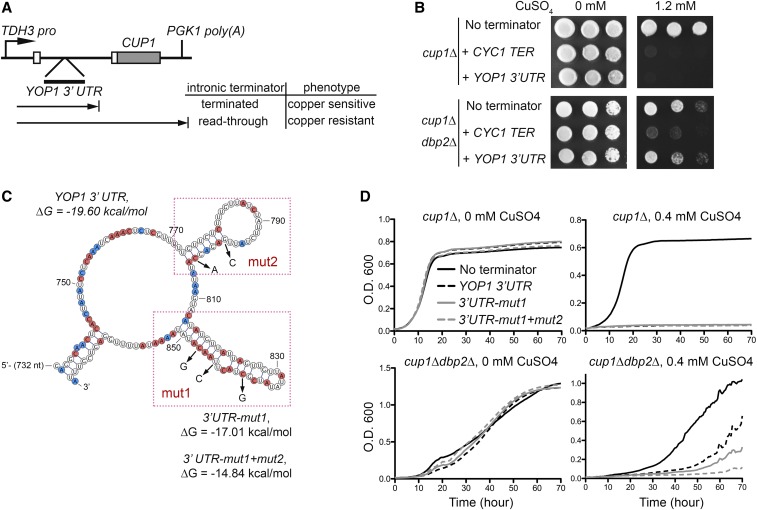
To test if the presence and stability of secondary structures in 3′ UTRs dictates a requirement for Dbp2 in termination, we utilized a transcriptional termination reporter growth assay that previously enabled identification of both cis-acting termination elements and trans-acting termination factors (Steinmetz et al. 2001; Steinmetz and Brow 2003). Briefly, the reporter encodes a chimeric actin-metallothionein (ACT1-CUP1) transcript that includes the intron of ACT1 (Figure 6A). Insertion of a terminator within the intron prevents expression of the ACT1-CUP1 chimera and results in death of cells lacking endogenous CUP1 (cup1∆ cells) on copper-containing plates. Defective termination, however, allows read-through of the internal terminator and resistant growth on different concentrations of copper, the latter of which is inversely proportional to the efficiency of termination (Steinmetz and Brow 2003).
Figure 6.
Secondary structure stability correlates with the requirement for DBP2 in termination of protein-coding genes. (A) Schematic representation the YOP1 termination reporter. The YOP1 3′ UTR region was cloned into the intron of the previously described CUP1 termination reporter plasmid (Steinmetz and Brow 2003). (B) Insertion of the YOP1 3’ UTR into the termination reporter confers DBP2-dependent copper sensitivity to cup1∆ cells. Serial dilution of cup1D∆ (top) or cup1∆ dbp2∆ cells (bottom) transformed with the reporter without terminator insertion before CUP1 (no terminator), with wild-type CYC1 terminator (CYC1 TER), or with wild-type YOP1 3’ UTR on nonselective media (0 mM) or media containing high concentrations (1.2 mM) of copper sulfate (CuSO4). (C) Predicted secondary structure of the YOP1 3’ UTR with structure-destabilizing mutations. The secondary structure of the 3′UTR of YOP1 in dbp2∆ was predicted using DMS reactivity patterns and the ViennaRNA package (Lorenz et al. 2011). Adenosines and cytosines with reduced DMS reactivity in dbp2∆ are in red and those with increased reactivity are in blue. The mutated nucleotides for the reporter assay are indicated by arrows and the folding energy (∆G) for the wild-type and mutant structures were predicted using ViennaRNA package 2.0 (temperature parameter = 30°) (Lorenz et al. 2011). (D) Destabilization of the YOP1 3’ UTR bypasses requirement for DBP2 in termination. A liquid growth assay of cup1∆ or cup1∆ dbp2∆ cells with the reporter without internal terminator insertion (no terminator), with the wild-type YOP1 3’ UTR and with the destabilized YOP1 3′ UTR containing one (3′UTR-mut1) or two (3′UTR-mut1+mut2) structural mutation sites in the presence or absence of 0.4 mM CuSO4. A reduced amount of CuSO4 was necessary compared to C, as the cells showed enhanced sensitivity in liquid culture compared to plates.
First, we generated a putative, DBP2-dependent termination reporter by inserting the 3′UTR region of YOP1 into the intron of the ACT1-CUP1 reporter (Figure 6A). To determine if the reporter recapitulates the DBP2-dependent termination defects seen at the endogenous loci, strains lacking CUP1 were first transformed with reporters either lacking a terminator (no terminator), containing an inserted CYC1 terminator (CYC1 TER), or containing the YOP1 3′UTR. CYC1 termination is independent of DBP2 and serves as a negative control (Table S1). Without an internal terminator, the transcription of the reporter is terminated only at the end of CUP1 gene (Figure 6A), allowing production of the Act1-Cup1 chimera and enabling growth of both cup1∆ and cup1∆ dbp2∆ cells on copper (Figure 6B, no terminator). Insertion of CYC1 TER resulted in a copper-sensitive phenotype in both cup1∆ and cup1∆ dbp2∆, consistent with termination in both strains prior to CUP1 (Figure 6B, + CYC1 TER). In contrast, insertion of the YOP1 3′ UTR prevented copper-resistant growth of cup1∆ cells but not cup1∆ dbp2∆ cells, indicating that the 3′UTR of YOP1 is sufficient to elicit termination in wild-type cells and that this termination is DBP2-dependent (Figure 6B, + YOP1 3′UTR).
We then generated a structure prediction of the 3′UTR of YOP1 in the absence of DBP2 using DMS reactivity data and the ViennaRNA package 2.0 (Lorenz et al. 2011) (Figure 6C). Interestingly, UA/UG-rich (Figure 6B, boxes) motifs, which are sequences recognized by Nrd1 and Nab3 (Schaughency et al. 2014), are present in predicted secondary structures. Based on the PAR-CLIP data of NNS factors (Creamer et al. 2011), Nrd1 and Nab3 cross-link to nucleotides 848 and 849, respectively, at the base of the 814–851 nt stem structure stem structure, whereas Sen1 cross-links to nucleotides 773, 774, and 776 at the base of the 773–804 stem structure located upstream.
To test if the termination defect in dbp2∆ cells is dependent on the stability of RNA secondary structures within the YOP1 3′UTR, we mutated the sequences within the reporter to destabilize one or both structures (Figure 6, C and D). We then assayed growth in liquid culture over time to increase the sensitivity of the assay. Importantly, neither mutation enabled growth of cup1∆ cells in the presence of copper, indicating that these mutations do not alter termination efficiency in wild-type cells (Figure 6D, top panels). Consistent with our plate assay, cells lacking DBP2 exhibited copper-resistant growth with the YOP1 3′UTR reporter. However, this resistance was reduced in both reporter mutants, indicative of increased termination (Figure 6D, bottom panels). Furthermore, the amount of secondary structure in the 3′ UTR correlated with the requirement for DBP2 in termination as evidenced by decreased growth of the cup1∆ dbp2∆ cells with the mut1+2 reporter over mut1 alone, indicating that the secondary structure of a given 3′UTR determines the requirement for Dbp2 in termination. This suggests that Dbp2 may promote termination efficiency by remodeling RNA/RNP structure in certain protein-coding genes. Taken together, our data point to a model whereby Dbp2 promotes efficient termination of RNPII transcription by modulating RNA/RNP structures to facilitate recruitment of NNS components. Future studies are necessary to determine the precise role of NNS complex members at the 3′ ends of protein-coding genes (see Discussion).
Discussion
Termination of RNAPII in S. cerevisiae is executed predominantly by the CPF or NNS complex, which function on protein-coding genes or noncoding RNAs, respectively. This distribution of tasks allows coupling of termination with 3′ processing steps that are appropriate for the given transcript, such as endonucleolytic cleavage and addition of a poly(A) tail to maturing mRNAs or 3′ end trimming or decay of noncoding RNAs. Although the histone modification pattern of the transcribed gene, the phosphorylation status of RNAPII, and the sequence of the nascent transcript itself all dictate the mode and efficiency of termination (Proudfoot 2016), the contribution of RNA structure and role of RNA helicases in this process has not been fully explored. By aggregating the results of multiple genome-wide analyses, we show that the RNA helicase Dbp2 promotes transcriptional termination and provide evidence that this mechanism likely involves remodeling of nascent transcripts and recruitment of Nrd1 to targeted genes.
In budding yeast, NNS-dependent termination of noncoding genes is coupled to 3′ end processing of nascent transcripts by nuclear exosome with the aid of the TRAMP complex, which adds short stretches of oligo(A) to the 3′ ends of NNS products to promote 3′ end maturation (Arndt and Reines 2015). The Sen1 RNA-DNA helicase, a component of the NNS complex, is thought to facilitate termination by unwinding the RNA-DNA hybrid produced in the wake of RNAPII (Han et al. 2017; Leonaitė et al. 2017). Our results show that loss of DBP2 results in a shift of RNAPII accumulation downstream of annotated snoRNA termination sites, well-characterized targets of the NNS complex, indicative of a role for Dbp2 in NNS-dependent termination. Dbp2 may also facilitate termination-coupled processing steps, as evidenced by the isolation of Dbp2-bound snoRNA processing intermediates with nontemplated As. These processing intermediates are highly reminiscent of semi-mature snoRNA species produced by the distributive polyadenylation polymerase activity of TRAMP in conjunction with Rrp6-dependent processing (Grzechnik and Kufel 2008) and have also been isolated by Nrd1 PAR-CLIP studies (Jamonnak et al. 2011). This role is also supported by synthetic lethal interactions between RRP6 and AIR2 with DBP2 (Wilmes et al. 2008; Cloutier et al. 2012) and the fact that loss of DBP2 results in upregulation and 3′ extension of the GAL10s long, noncoding RNA (Cloutier et al. 2012) It is interesting that Dbp2 accumulates across the snoRNA body while promoting termination (Figure 1, B and C). A likely explanation is that Dbp2 also functions in snoRNA-guided ribosomal RNA modification, a role that is supported by the nucleolar accumulation of Dbp2 and its association with ribosome subunits (Cloutier et al. 2012; Martin et al. 2013).
In addition to snoRNAs, our results also define a role for Dbp2 in termination of protein-coding genes. Although the predominant mechanism for termination of protein-coding genes involves the CPF complex, our results suggest that, like snoRNA genes, Dbp2-dependent termination of protein-coding genes may also involve the NNS complex. This is evidenced by enrichment of Nrd1 and, to a lesser extent, Nab3 consensus motifs as Dbp2-bound RNA targets (Figure 2C), DBP2-dependent binding of Nrd1 to the 5′ ends and 3′ UTRs of targeted protein-coding genes (Figure 2E and Figure 4C), and the recapitulation of DBP2-dependent termination upon insertion of a Nrd1-targeted 3′UTR into a reporter construct (Figure 6). It should be noted, however, that although our results say that Dbp2-dependent termination is mediated through Nrd1, this does not mean that this mechanism is CPF-independent.
The decision of whether to terminate transcription using CPF or NNS is largely determined by the phosphorylation state of the C-terminal domain (CTD) of RNAPII, which is recognized by CTD-interacting domains (CID) in either Pcf11 or Nrd1, respectively (Noble et al. 2005; Chinchilla et al. 2012; Eick and Geyer 2013). Specific phosphorylation patterns within the heptad repeat of the CTD mark the position of RNAPII during transcription, with high levels of serine 5 (Ser5) phosphorylation transitioning to serine 2 (Ser2) phosphorylation along the length of the gene (Harlen and Churchman 2017). Nrd1 associates with Ser5 phosphorylated CTD whereas Pcf11 associates with Ser2, confining Nrd1 to termination of short transcripts and Pcf11 to longer, protein-coding ones (Noble et al. 2005; Chinchilla et al. 2012; Eick and Geyer 2013). However, several genome-wide studies have revealed association of Nrd1 and Nab3 with the 3′ ends of mRNA transcripts, in addition to canonical snoRNAs and CUTs (Creamer et al. 2011; Wlotzka et al. 2011; Webb et al. 2014). In some cases, this association serves as a form of quality control, with NNS-directed termination serving to direct exosome recruitment to mis-processed RNAs (Gudipati et al. 2012). In line with this is the observation that NNS-targeted protein-coding transcripts are frequently upregulated upon mutation of nrd1 or components of the exosome (Gudipati et al. 2012; Webb et al. 2014). However, we observed no significant correlation between transcript upregulation and 3′ extension in dbp2∆ cells (data not shown), suggesting that DBP2-dependent termination of protein-coding genes may not be coupled with decay. Interestingly, studies have shown that the Pcf11 component of the CPF complex depends on initial binding and subsequent exchange with Nrd1 (Grzechnik et al. 2015). Although this Nrd1-Pcf11 “swap” was documented at the 5′ ends of genes, consistent with the role of NNS on shorter transcripts, evidence suggests that the converse may also occur. The most striking example is the characterization of a failsafe termination mechanism for RNAPII complexes that involves Nrd1/Nab3 sites that are downstream of a polyadenylation site (Rondón et al. 2009). Interestingly, characterization of this form of failsafe termination utilized the GAL10-GAL7 genes and associated intergenic region, which corresponds to the same region where DBP2-dependent termination of both protein-coding and noncoding genes was first described (Cloutier et al. 2012). Moreover, mounting evidence has found that the role of CTD phosphorylation in downstream processing steps may be more nuanced than the prevailing Ser5-Ser2 gradient model. Although the latter does hold true for the vast majority of genes in S. cerevisiae (Suh et al. 2016) gene-specific phosphorylation patterns and the link between Ser7 phosphorylation and Nrd1 recruitment have also been described (Vasiljeva et al. 2008; Kim et al. 2010; Kubicek et al. 2012). One interesting possibility for the role of Dbp2 stems from the observation that the Nrd1 ortholog in fission yeast, Seb1, promotes pausing of RNAPII, in addition to termination site selection and 3′ end processing (Lemay et al. 2016; Liu et al. 2017; Parsa et al. 2018). Although S. pombe does not use an NNS-like mechanism for termination of protein-coding or noncoding RNAs, with Seb1 recruiting CPF machinery to both types of RNAPII-transcribed genes (Larochelle et al. 2018), the fact that loss of DBP2 results in a change in RNAPII accumulation at Nrd1 binding sites on individual genes (Figure 2 and Figure 4) that is strikingly similar to seb1 mutation suggests that termination defects described here may be coupled to RNAPII progression. Future studies are necessary to determine if DBP2 influences RNAPII kinetics and if the kinetics are affected by aberrant RNA structure or reduced binding of Nrd1 in its absence.
Thirty years ago to date, the first DEAD-box RNA helicase was shown to have RNA duplex unwinding activity in vitro (Abramson et al. 1987; Hirling et al. 1989). Since this discovery, elegant biochemical and biophysical studies have provided an in-depth knowledge of how these enzymes function in vitro (Rudolph and Klostermeier 2015); however, their precise functions in vivo have remained elusive. Recently, the Jankowsky laboratory capitalized on the rapidly advancing technologies to view RNA-binding sites and RNA structure/composition genome-wide, providing the first evidence for widespread RNA remodeling by a DEAD-box RNA helicase that directly influences gene expression steps (Guenther et al. 2018). Although those studies provide a link between the DEAD-box RNA helicase Ded1 and translation, results herein now extend the generality of RNA remodeling events catalyzed by these enzymes to nuclear gene expression steps.
Loss of DBP2 results in decreased DMS reactivity, mostly in 3′ UTRs (Figure 5 and Figure S5). Therefore, RNA/RNP structures may contribute to regulation of specific processes. It is likely that the altered mechanism of termination and extended length of the transcripts produced in dbp2∆ cells result in different compositions and/or stoichiometries of RBPs that account for the 40–60 nt stretch of reduced DMS reactivity (Figure 5A). It has been established that efficient termination is a prerequisite for recruitment of mRNA export factors to mature mRNAs (Kessler et al. 1997; Hammell et al. 2002; Dunn et al. 2005) and our laboratory has shown that loss of DBP2 reduces association of the nuclear poly(A) binding protein, Nab2, and mRNA export factor Mex67 on poly(A)+ RNAs (Ma et al. 2013). Moreover, recent studies by the Struhl laboratory found that the protein composition of the 3′ isoform produced is influenced by the length of the transcript (Moqtaderi et al. 2018). However, the fact that the 40–60 nt stretch is centered at the Dbp2 binding site suggests that the altered folding and/or composition is directly due to loss of Dbp2 activity. These remodeling events are likely to be important for multiple steps of gene expression as recent studies have shown that Dbp2 couples translational efficiency of a given mRNA with the identity of the gene promoter (Espinar et al. 2018). Additionally, loss of this RNA remodeling activity may contribute to the cold-sensitive phenotype of dbp2∆ (Cloutier et al. 2012), due to the accumulation of stabilized, nonfunctional RNA structures as the temperature decreases in the absence of DBP2.
It is important to note that although our studies of the YOP1 3′UTR suggest that DBP2-dependent remodeling involves alteration of RNA secondary structure (Figure 6), remodeling includes both changes in RNA structure and RNA-binding protein composition that occurs either directly via RNA helicase activity or more indirectly by extending the RNA transcript length. It is well established that many RNA-binding proteins are inhibited by the presence of secondary structure in the targeted RNA, a phenomenon recently supported by genome-wide studies (Lambert et al. 2014). Consistently, formation of secondary structure has been shown to impair specific gene expression steps catalyzed by these RNA-binding proteins, including transcription termination (Chen and Wilusz 1998; Klasens et al. 1998; Mortimer et al. 2014). Because loss of DBP2 reduces recruitment of Nrd1 to individual genes and that destabilization of secondary structure in the 3′UTR of YOP1 bypasses the requirement for DBP2 in termination, our finding points to a model that Dbp2 remodels RNA structure to provide a “landing pad” for Nrd1. The precise mechanism for how this elicits termination and if that termination involves Nab3 and Sen1 are yet to be determined.
Although we have not detected a direct interaction between Dbp2 and Nrd1 or Nab3 to date, an interaction between Dbp2 and Sen1 has been reported along with a correlation of Dbp2 RNA-binding sites with locations of R-loop formation (Tedeschi et al. 2018). If Dbp2 enables recruitment and/or activation of the RNA-DNA helicase Sen1, this could explain prior observations that loss of DBP2 results in accumulation of RNA-DNA hybrids in vivo despite reduced RNA-DNA duplex unwinding activity in vitro compared to pure RNA duplexes (Ma et al. 2013; Cloutier et al. 2016). While the NNS complex is not conserved in mammalian cells, Sen1 does have an orthologous protein Senataxin that functions in transcriptional termination and resolution of R-loops (Skourti-Stathaki et al. 2011). It will be interesting to determine if DDX5, the mammalian ortholog of Dbp2, interacts with Senataxin and/or the Integrator complex, the latter of which is functionally analogous to NNS (Baillat and Wagner 2015). The fact that ectopic expression of DDX5 in dbp2∆ cells rescues defects in both growth and gene expression is highly suggestive of a conserved function in both fungi and human cells (Xing et al. 2017). Moreover, mounting evidence showing that aberrant mRNA structure may underlie select human pathophysiologies (Wan et al. 2014; Corley et al. 2015), points to a pressing need to understand the precise biochemical function of the ∼40 DEAD-box RNA helicases in mammalian cells (∼25 in budding yeast) in vivo (Linder and Jankowsky 2011).
Acknowledgments
We thank Ryan Flynn (former member of the H.Y. Chang Laboratory) at Stanford University for providing the iCLIP-seq protocol, and Yiliang Ding (John Innes Centre) for help in construction of Structure-seq libraries. We thank Mirko Ledda (Aviran laboratory at University of California, Davis) for providing guidance on DMS data-directed prediction of RNA secondary structures. We also thank Pete Pascuzzi and Nadia Atallah Lanman (Purdue University) for providing suggestions and rigorous discussion for bioinformatics analyses. This work was supported by National Institutes of Health grants R01GM097332 to E.J.T., R00-HG006860 to S.A., and grant P30CA023168 for core facilities at the Purdue University Center for Cancer Research. The authors declare that no competing interests exist.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.7791485.
Communicating editor: A. Hinnebusch
Literature Cited
- Abramson R. D., Dever T. E., Lawson T. G., Ray B. K., Thach R. E., et al. , 1987. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 262: 3826–3832. [PubMed] [Google Scholar]
- Andersen C. B., Ballut L., Johansen J. S., Chamieh H., Nielsen K. H., et al. , 2006. Structure of the exon junction core complex with a trapped DEAD-Box ATPase bound to RNA. Science 313: 1968–1972. [DOI] [PubMed] [Google Scholar]
- Arigo J. T., Carroll K. L., Ames J. M., Corden J. L., 2006. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell 21: 641–651. 10.1016/j.molcel.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Arndt K. M., Reines D., 2015. Termination of transcription of short noncoding RNAs by RNA polymerase II. Annu. Rev. Biochem. 84: 381–404. 10.1146/annurev-biochem-060614-034457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviran S., Trapnell C., Lucks J. B., Mortimer S. A., Luo S., et al. , 2011a Modeling and automation of sequencing-based characterization of RNA structure. Proc. Natl. Acad. Sci. USA 108: 11069–11074. 10.1073/pnas.1106541108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviran, S., J. B. Lucks, and L. Pachter, 2011b RNA structure characterization from chemical mapping experiments, pp. 1743–1750 in Proceedings of the 49th Allerton Conference on Communication, Control, and Computing, Institute of Electrical and Electronics Engineers, Piscataway, NJ. 10.1109/Allerton.2011.6120379 10.1109/Allerton.2011.6120379 [DOI] [Google Scholar]
- Baejen C., Torkler P., Gressel S., Essig K., Söding J., et al. , 2014. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol. Cell 55: 745–757. 10.1016/j.molcel.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Baejen C., Andreani J., Torkler P., Battaglia S., Schwalb B., et al. , 2017. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol. Cell 66: 38–49.e6. 10.1016/j.molcel.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Baillat D., Wagner E. J., 2015. Integrator: surprisingly diverse functions in gene expression. Trends Biochem. Sci. 40: 257–264. 10.1016/j.tibs.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L., Marchadier B., Baguet A., Tomasetto C., Séraphin B., et al. , 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 12: 861–869. 10.1038/nsmb990 [DOI] [PubMed] [Google Scholar]
- Barta I., Iggo R., 1995. Autoregulation of expression of the yeast Dbp2p “DEAD-box” protein is mediated by sequences in the conserved DBP2 intron. EMBO J. 14: 3800–3808. 10.1002/j.1460-2075.1995.tb00049.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck Z. T., Cloutier S. C., Schipma M. J., Petell C. J., Ma W. K., et al. , 2014. Regulation of glucose-dependent gene expression by the RNA helicase Dbp2 in Saccharomyces cerevisiae. Genetics 198: 1001–1014. 10.1534/genetics.114.170019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois C. F., Mortreux F., Auboeuf D., 2016. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat. Rev. Mol. Cell Biol. 17: 426–438. 10.1038/nrm.2016.50 [DOI] [PubMed] [Google Scholar]
- Chen F., Wilusz J., 1998. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 26: 2891–2898. 10.1093/nar/26.12.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla K., Rodriguez-Molina J. B., Ursic D., Finkel J. S., Ansari A. Z., et al. , 2012. Interactions of Sen1, Nrd1, and Nab3 with multiple phosphorylated forms of the Rpb1 C-terminal domain in Saccharomyces cerevisiae. Eukaryot. Cell 11: 417–429. 10.1128/EC.05320-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary K., Shih N. P., Deng F., Ledda M., Li B., et al. , 2016. Metrics for rapid quality control in RNA structure probing experiments. Bioinformatics 32: 3575–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary K., Lai Y.-H., Tran E. J., Aviran S., 2019. dStruct: identifying differentially reactive regions from RNA structurome profiling data. Genome Biol. 20: 40 10.1186/s13059-019-1641-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier S. C., Ma W. K., Nguyen L. T., Tran E. J., 2012. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J. Biol. Chem. 287: 26155–26166. 10.1074/jbc.M112.383075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier S. C., Wang S., Ma W. K., Petell C. J., Tran E. J., 2013. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol. 11: e1001715 10.1371/journal.pbio.1001715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier S. C., Wang S., Ma W. K., Al Husini N., Dhoondia Z., et al. , 2016. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol. Cell 61: 393–404 (erratum: Mol. Cell 62: 148). 10.1016/j.molcel.2015.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley M., Solem A., Qu K., Chang H. Y., Laederach A., 2015. Detecting riboSNitches with RNA folding algorithms: a genome-wide benchmark. Nucleic Acids Res. 43: 1859–1868. 10.1093/nar/gkv010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer T. J., Darby M. M., Jamonnak N., Schaughency P., Hao H., et al. , 2011. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 7: e1002329 10.1371/journal.pgen.1002329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardenne E., Polay Espinoza M., Fattet L., Germann S., Lambert M. P., et al. , 2014. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 7: 1900–1913. 10.1016/j.celrep.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Ding Y., Tang Y., Kwok C. K., Zhang Y., Bevilacqua P. C., et al. , 2014. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700. 10.1038/nature12756 [DOI] [PubMed] [Google Scholar]
- Ding Y., Kwok C. K., Tang Y., Bevilacqua P. C., Assmann S. M., 2015. Genome-wide profiling of in vivo RNA structure at single-nucleotide resolution using structure-seq. Nat. Protoc. 10: 1050–1066. 10.1038/nprot.2015.064 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn E. F., Hammell C. M., Hodge C. A., Cole C. N., 2005. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 19: 90–103. 10.1101/gad.1267005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Geyer M., 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113: 8456–8490. 10.1021/cr400071f [DOI] [PubMed] [Google Scholar]
- Espinar L., Tamarit M. À. S., Domingo J., Carey L. B., 2018. Promoter architecture determines cotranslational regulation of mRNA. Genome Res. 28: 509–518. 10.1101/gr.230458.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman M. E., Maroney P. A., Wang W., Bowers H. A., Gollnick P., et al. , 2004. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science 304: 730–734. 10.1126/science.1095596 [DOI] [PubMed] [Google Scholar]
- Flynn R. A., Martin L., Spitale R. C., Do B. T., Sagan S. M., et al. , 2015. Dissecting noncoding and pathogen RNA-protein interactomes. RNA 21: 135–143. 10.1261/rna.047803.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R. A., Zhang Q. C., Spitale R. C., Lee B., Mumbach M. R., et al. , 2016. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat. Protoc. 11: 273–290. 10.1038/nprot.2016.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., Rechsteiner T., Richmond T. J., Tsukiyama T., 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21: 2098–2106. 10.1128/MCB.21.6.2098-2106.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman B., Tijerina P., Russell R., 2017. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem. Soc. Trans. 45: 1313–1321. 10.1042/BST20170095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P., Kufel J., 2008. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell 32: 247–258. 10.1016/j.molcel.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P., Gdula M. R., Proudfoot N. J., 2015. Pcf11 orchestrates transcription termination pathways in yeast. Genes Dev. 29: 849–861. 10.1101/gad.251470.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati R. K., Xu Z., Lebreton A., Séraphin B., Steinmetz L. M., et al. , 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell 48: 409–421. 10.1016/j.molcel.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther U. P., Weinberg D. E., Zubradt M. M., Tedeschi F. A., Stawicki B. N., et al. , 2018. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature 559: 130–134. 10.1038/s41586-018-0258-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C. M., Gross S., Zenklusen D., Heath V., Stutz F., et al. , 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22: 6441–6457. 10.1128/MCB.22.18.6441-6457.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Libri D., Porrua O., 2017. Biochemical characterization of the helicase Sen1 provides new insights into the mechanisms of non-coding transcription termination. Nucleic Acids Res. 45: 1355–1370. 10.1093/nar/gkw1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlen K. M., Churchman L. S., 2017. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18: 263–273. 10.1038/nrm.2017.10 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirling H., Scheffner M., Restle T., Stahl H., 1989. RNA helicase activity associated with the human p68 protein. Nature 339: 562–564. 10.1038/339562a0 [DOI] [PubMed] [Google Scholar]
- Jamonnak N., Creamer T. J., Darby M. M., Schaughency P., Wheelan S. J., et al. , 2011. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17: 2011–2025. 10.1261/rna.2840711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A., Fushimi K., Zhou X., Ray P., Shi C., et al. , 2011. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell. Biol. 31: 1812–1821. 10.1128/MCB.01149-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M. M., Henry M. F., Shen E., Zhao J., Gross S., et al. , 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 11: 2545–2556. 10.1101/gad.11.19.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Y., Levin D. E., 2011. Mpk1 MAPK association with the paf1 complex blocks sen1-mediated premature transcription termination. Cell 144: 745–756. 10.1016/j.cell.2011.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Erickson B., Luo W., Seward D., Graber J. H., et al. , 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17: 1279–1286. 10.1038/nsmb.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasens B. I. F., Das A. T., Berkhout B., 1998. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 26: 1870–1876. 10.1093/nar/26.8.1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J., Zarnack K., Rot G., Curk T. T., Kayikci M., et al. , 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17: 909–915. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek K., Cerna H., Holub P., Pasulka J., Hrossova D., et al. , 2012. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 26: 1891–1896. 10.1101/gad.192781.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner J. N., Brow D. A., 2008. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell 31: 201–211. 10.1016/j.molcel.2008.05.018 [DOI] [PubMed] [Google Scholar]
- Lambert N., Robertson A., Jangi M., McGeary S., Sharp P. A., et al. , 2014. RNA bind-n-seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell 54: 887–900. 10.1016/j.molcel.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle M., Robert M. A., Hébert J. N., Liu X., Matteau D., et al. , 2018. Common mechanism of transcription termination at coding and noncoding RNA genes in fission yeast. Nat. Commun. 9: 4364 10.1038/s41467-018-06546-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J. F., Marguerat S., Larochelle M., Liu X., van Nues R., et al. , 2016. The Nrd1-like protein Seb1 coordinates cotranscriptional 3′ end processing and polyadenylation site selection. Genes Dev. 30: 1558–1572. 10.1101/gad.280222.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonaitė B., Han Z., Basquin J., Bonneau F., Libri D., et al. , 2017. Sen1 has unique structural features grafted on the architecture of the Upf1‐like helicase family. EMBO J. 36: 1590–1604. 10.15252/embj.201696174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser C. F., Guthrie C., 1993. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeg A., Mayer O., Waldsich C., 2010. DEAD-box protein facilitated RNA folding in vivo. RNA Biol. 7: 803–811. 10.4161/rna.7.6.13484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Jankowsky E., 2011. From unwinding to clamping — the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12: 505–516. 10.1038/nrm3154 [DOI] [PubMed] [Google Scholar]
- Liu X., Hoque M., Larochelle M., Lemay J. F., Yurko N., et al. , 2017. Comparative analysis of alternative polyadenylation in S. cerevisiae and S. pombe. Genome Res. 27: 1685–1695. 10.1101/gr.222331.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R., Bernhart S. H., Höner zu Siederdissen C., Tafer H., Flamm C., et al. , 2011. ViennaRNA package 2.0. Algorithms Mol. Biol. 6: 26 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. K., Cloutier S. C., Tran E. J., 2013. The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J. Mol. Biol. 425: 3824–3838. 10.1016/j.jmb.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. K., Paudel B. P., Xing Z., Sabath I. G., Rueda D., et al. , 2016. Recruitment, duplex unwinding and protein-mediated inhibition of the dead-box RNA helicase Dbp2 at actively transcribed chromatin. J. Mol. Biol. 428: 1091–1106. 10.1016/j.jmb.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M., 2011 Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17: 10–12. 10.14806/ej.17.1.200 10.14806/ej.17.1.200 [DOI]
- Martin R., Straub A. U., Doebele C., Bohnsack M. T., 2013. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 10: 4–18. 10.4161/rna.21879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo H. E., Proudfoot N. J., 2013. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim. Biophys. Acta. Gene Regul. Mech. 1829: 174–185. 10.1016/j.bbagrm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Geisberg J. V., Struhl K., 2018. Extensive structural differences of closely related 3’ mRNA isoforms: links to Pab1 binding and mRNA stability. Mol. Cell 72: 849–861.e6. 10.1016/j.molcel.2018.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer S., Kidwell M. A., Doudna J. A. 2014. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 15: 469–479. 10.1038/nrg3681 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., et al. , 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K. H., Chamieh H., Andersen C. B. F., Fredslund F., Hamborg K., et al. , 2008. Mechanism of ATP turnover inhibition in the EJC. RNA 15: 67–75. 10.1261/rna.1283109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C. G., Hollingworth D., Martin S. R., Ennis-Adeniran V., Smerdon S. J., et al. , 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 12: 144–151. 10.1038/nsmb887 [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Kapranov P., Foissac S., Kim S. W., Fishilevich E., et al. , 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029. 10.1016/j.cell.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa J. Y., Boudoukha S., Burke J., Homer C., Madhani H. D., 2018. Polymerase pausing induced by sequence-specific RNA-binding protein drives heterochromatin assembly. Genes Dev. 32: 953–964. 10.1101/gad.310136.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W., 1980. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. USA 77: 4679–4682. 10.1073/pnas.77.8.4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Wei W., Steinmetz L. M., 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497: 127–131. 10.1038/nature12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O., Libri D., 2015. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol. 16: 190–202. 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- Potratz J. P., Del Campo M., Wolf R. Z., Lambowitz A. M., Russell R., 2011. ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo. J. Mol. Biol. 411: 661–679. 10.1016/j.jmb.2011.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926 10.1126/science.aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam A. A., Jankowsky E., 2013. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim. Biophys. Acta. Gene Regul. Mech. 1829: 884–893. 10.1016/j.bbagrm.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Lykke-Andersen S., Nasser T., Saguez C., Bertrand E., et al. , 2009. Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Mol. Cell. Biol. 29: 5327–5338. 10.1128/MCB.00468-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., et al. , 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. W., Richter N. J., Merrick W. C., 1999. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274: 12236–12244. 10.1074/jbc.274.18.12236 [DOI] [PubMed] [Google Scholar]
- Rondón A. G., Mischo H. E., Kawauchi J., Proudfoot N. J., 2009. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell 36: 88–98. 10.1016/j.molcel.2009.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J. S., 2013. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. 10.1038/nature12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M. G., Klostermeier D., 2015. When core competence is not enough: functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding. Biol. Chem. 396: 849–865. 10.1515/hsz-2014-0277 [DOI] [PubMed] [Google Scholar]
- Schaughency P., Merran J., Corden J. L., 2014. Genome-wide mapping of yeast RNA polymerase II termination. PLoS Genet. 10: e1004632 10.1371/journal.pgen.1004632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. N., Singh R. N., Androphy E. J., 2007. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 35: 371–389. 10.1093/nar/gkl1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Proudfoot N. J., Gromak N., 2011. Human Senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42: 794–805. 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloma M. F., Mathews D. H., 2015. Improving RNA secondary structure prediction with structure mapping data. Methods Enzymol. 553: 91–114. 10.1016/bs.mie.2014.10.053 [DOI] [PubMed] [Google Scholar]
- Steinmetz E. J., Brow D. A., 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16: 6993–7003. 10.1128/MCB.16.12.6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Brow D. A., 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 23: 6339–6349. 10.1128/MCB.23.18.6339-6349.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Conrad N. K., Brow D. A., Corden J. L., 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331. 10.1038/35095090 [DOI] [PubMed] [Google Scholar]
- Steinmetz E. J., Warren C. L., Kuehner J. N., Panbehi B., Ansari A. Z., et al. , 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24: 735–746. 10.1016/j.molcel.2006.10.023 [DOI] [PubMed] [Google Scholar]
- Sugimoto Y., König J., Hussain S., Zupan B., Curk T., et al. , 2012. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 13: R67 10.1186/gb-2012-13-8-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H., Ficarro S. B., Kang U.-B., Chun Y., Marto J. A., et al. , 2016. Direct analysis of phosphorylation sites on the Rpb1 C-terminal domain of RNA polymerase II. Mol. Cell 61: 297–304. 10.1016/j.molcel.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi F. A., Cloutier S. C., Tran E. J., Jankowsky E., 2018. The DEAD-box protein Dbp2p is linked to noncoding RNAs, the helicase Sen1p, and R-loops. RNA 24: 1693–1705. 10.1261/rna.067249.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P., Bhaskaran H., Russell R., 2006. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc. Natl. Acad. Sci. USA 103: 16698–16703. 10.1073/pnas.0603127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E. J., Zhou Y., Corbett A. H., Wente S. R., 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA. Protein Remodeling Events. Mol. Cell 28: 850–859. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A., 2008. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 15: 795–804. 10.1038/nsmb.1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Qu K., Zhang Q. C., Flynn R. A., Manor O., et al. , 2014. Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505: 706–709. 10.1038/nature12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xing Z., Pascuzzi P. E., Tran E. J., 2017. Metabolic adaptation to nutrients involves coregulation of gene expression by the RNA helicase Dbp2 and the Cyc8 corepressor in Saccharomyces cerevisiae. G3 (Bethesda) 7: 2235–2247. 10.1534/g3.117.041814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb S., Hector R. D., Kudla G., Granneman S., 2014. PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biol. 15: R8 10.1186/gb-2014-15-1-r8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes G. M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., et al. , 2008. A genetic interaction map of RNA-processing factors reveals links between sem1/dss1-containing complexes and mRNA export and splicing. Mol. Cell 32: 735–746. 10.1016/j.molcel.2008.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlotzka W., Kudla G., Granneman S., Tollervey D., 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 30: 1790–1803. 10.1038/emboj.2011.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z., Wang S., Tran E. J., 2017. Characterization of the mammalian DEAD-box protein DDX5 reveals functional conservation with S. cerevisiae ortholog Dbp2 in transcriptional control and glucose metabolism. RNA 23: 1125–1138. 10.1261/rna.060335.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Z., Ma W. K., Tran E. J., 2019. The DDX5/Dbp2 subfamily of DEAD-box RNA helicases. Wiley Interdiscip. Rev. RNA 10: e1519 10.1002/wrna.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Jankowsky E., 2005. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 44: 13591–13601. 10.1021/bi0508946 [DOI] [PubMed] [Google Scholar]
- Yang Q., Jankowsky E., 2006. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 13: 981–986. 10.1038/nsmb1165 [DOI] [PubMed] [Google Scholar]
- Yassour M., Kaplan T., Fraser H. B., Levin J. Z., Pfiffner J., et al. , 2009. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl. Acad. Sci. USA 106: 3264–3269. 10.1073/pnas.0812841106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Supplemental tables in Excel format are uploaded to Figshare. Raw reads and processed data of iCLIP-seq, Structure-seq, and RNAPII ChIP-seq are deposited on GEO under the accession number GSE106479. Scripts for bioinformatics analysis are deposited on Github: https://github.com/karenlai0222/Dbp2_iCLIP_Structure. Supplemental Material, Figures S1–S5, and Tables S1–S5 are available at Figshare: https://doi.org/10.25386/genetics.7791485.