Abstract
Type II toxin-antitoxin (TA) systems are the particular type of TA modules which take part in different kinds of cellular actions, such as biofilm formation, persistence, stress endurance, defense of the bacterial cell against multiple phage attacks, plasmid maintenance, and programmed cell death in favor of bacterial population. Although several bioinformatics and Pet lab studies have already been conducted to understand the functionality of already discovered TA systems, still, more work in this area is required. Rnl AB type II TA module, which is composed of RnlA toxin and RnlB antitoxin, is a newly discovered type II TA module which takes part in the defense mechanism against T4 bacteriophage attack in Escherichia coli K-12 strain MH1 that has not been widely studied in other bacteria. Because of the significant role of class Gammaproteobacteriacea in a diverse range of health problems, we chose here to focus on this class to survey the presence of the Rnl AB TA module. For better categorization and description of the distribution of this module in this class of bacteria, the corresponding phylogenetic trees are illustrated here. Neighbor-joining and the maximum parsimony methods were used in this study to take a look at the distribution of domains present in RnlA and RnlB proteins, among members of Gammaproteobacteria. Also, the possible roles of photodynamic therapy (PDT) in providing a substrate for better phage therapy are herein discussed.
Keywords: Toxin/antitoxin, Modules, type II TA module, Rnl AB system, Gammaproteobacteria, Phylogenetic tree, Antimicrobial photodynamic therapy
Introduction
The procedure employed by bacterial cells when encountering stressful situations is to reduce the rate of stable RNAs production by the alarmones, like (p)ppGpp, so it is logical to be equipped with strategies which can regulate the synthesis of DNA and protein.1 These modules are known as toxin-antitoxin (TA) systems, which usually are composed of 2 elements; the stable toxin, and the liable antitoxin, however, hybrid systems exist too.2,3 Based on this fact, the nature of the antitoxin can be of protein or antisense-RNA and, according to the recent categorization of these systems, there are six groups of TA modules, including the following:
Type I: This system is composed of 2 parts: the toxin component, which is a small hydrophobic protein; and an antitoxin part, which is either a cis-encoded antisense-RNA or a trans-encoded sRNA. The function of the toxin is to destroy the cell membrane, which leads to the inhibition of ATP synthesis as well as to important processes of cell life, like replication, transcription, and translation which result in cell death. The function of the antitoxin is to prevent this course by inhibiting toxin mRNA translation and preventing toxin production at first step (this prevention can occur in different ways). Examples of type 1 TA modules are found in different gram-positive and gram-negative bacteria; the first type 1 TA module, introduced in 1986 by Ken and Gerdes, is the Hok (host killing)-Sok (suppressor of killing) system on locus Par B of low-copy-number plasmid R1 of Escherichia coli, which is specified to take part in plasmid maintenance. Deficiency of plasmid in newborn daughter cells results in their cell death, which denotes the phenomenon of “post-segregational killing”. According to this phenomenon, the cells which do not acquire the plasmid carrying TA module are condemned to die. The reason is a pause that occurs in transcription, so after this, the ratio of a remaining toxin to antitoxin starts to be unequal and toxin reaches higher concentration. It is the unstable structure of antitoxin that leads to its destruction by cellular proteases like RNase III and in this way, free toxins start to manage their ruining effects on cells. Also, one example of type 1 TA module in Gram-positive bacteria is the fstI-RNAII system.4-8
Type II: This type of system is the best described TA module to date. There have been many bioinformatical and experimental researches in this area. The first described type II TA module, Ccd AB, with plasmid maintenance performance discovered by Ogura and Hiraga in 1983 on Mini-F plasmid of E. coli K-12, and the other was on the Sok-Hok system, resident at Par B locus of E. coli K-12 strain, introduced by Bravo, Torrontegui, and Diaz, which both take part in plasmid stability. In general, these systems are comprised of 2 protein elements, in which the antitoxin part inhibits the function of the toxin by interacting directly with the toxin. Whenever the bacterial cell is confronted with rough stressful conditions, the antitoxin protein is being degraded by proteases like Lon,9 or different kinds of CLPs found in cytoplasmic environments; the reason is the vulnerable structure of antitoxin. Their operons reside whether in plasmid or chromosome and although the proper and ultimate function of chromosomal type II TA systems is not well described,10 according to the reports of different works, we can attribute some tasks to them, of which the predominant one is stress response in which the toxin part is activated during stressful conditions.11-13 As will be mentioned further on, the functional character of type II toxin, after facing stressful conditions, is often to induce bacteriostatic term in the cells, which helps the bacteria to stop metabolism to withstand inappropriate environments, or which leads the bacteria to cell death in which the bacteria sacrifice itself for others’ sake.14,15 Different type II TA modules play important diverse roles, like Mqs RA involved in biofilm formation in E. coli,16 Hig BA influencing different virulence factors in Pseudomonas aeruginosa,17 and YefM-YoeB and some others helping bacteria to survive inside of the host cells.18 Hip A toxin in E. coli mediates the persister cell formation19 (although the type I TA module contributes in persister cell formation too),20 or involvement of systems like Maz EF system in defense against bacteriophage attack, etc.21 All are recognized as the importance of this particular TA system.
Type III: Tox IN is the first described type III TA module, discovered in Pectobacterium atrosepticum by Fineran et al in 2009, with protein toxin and antisense-RNA antitoxin-like type I system, but the difference is that antitoxin shapes a pseudoknot and interacts directly with protein toxin, which has endoribonuclease activity, and through this way the plastidic type III TA modules plays a role in defense against phage attack.22
Type IV: Both toxin and antitoxin components are protein-like type II, but the difference is in the procedure of performance. In this type, there is a competition between toxin and antitoxin, in which they never interact directly with each other but struggle on the same cellular aim. In other words, it is like antitoxin tries to fortify the cellular target, which toxin tries to demolish. An example of this system is the Yee UV system, which is identified in E. coli.23
Type V: In this type of TA module, the result of toxin translation is the destruction of the cell membrane (like type I TA systems). The function of type V antitoxin is cleavage of the toxin mRNA. An example is the Gho ST system in E. coli.24
Type VI: Newly identified, present in Caulobacter crescentus, this is an interesting module because here, instead of antitoxin, the toxin part is being degraded by the protease, and the attractive aspect of this is that the antitoxin helps the protease to degrade the toxin. Sok AB is an example of this type of system where Soc B toxin ruins the replication process.
So, the overall functions of TA systems researched before now, plasmidic or chromosomally, are to maintain the survival of the bacterial cells during stressful conditions, like the presence of antibiotics, enduring starvation, oxidative stress, pH or temperature, the stress of immune system response, and bacteriophage attack. These systems that have been categorized into 12 families are present as potential weapons in the stability of pathogenic bacteria against intra-phagosomal nutrient starvation, autophagy response, lysosomal enzymes, oxidative/nitrosative burst, acidic pH in phagosomes on the intracellular environment, and antibody/complement attack and phagocyte attack in tissues and extracellular matrix.25
Importance of Type II Toxin-Antitoxin Modules
As mentioned earlier, one of the specific types of TA modules is the type II system, which is important in virulence and in the ability of transposition by HGT, and it is capable of regulation of bacterial physiology during inappropriate, improper stressful environmental situations. Bioinformatics researches have proved that they are widely distributed among bacteria and archaea.3
RnlA-RnlB TA Module as an Interesting Type II TA System
In general, there are different kinds of RNase in E. coli, (e.g., RNase G, or RNase P, or RNase Z), in which every enzyme makes a contribution in cleavage to a special region, but their responsibilities have been proved to be limited, and they have evolved to perform their functions in different circumstances.26,27 RNase LS is also one of the bacterial enzymes which participates in cleavage of mRNAs of phage T4 when the bacteria have been attacked by an intruder phage.28 Not only does this RNase tend to cleave mRNAs of T4 phages, but also it has other functions which are not as bold as this specific duty, e.g., RNase LS takes part in total mRNA turnover of bacterial cells and also contributes to the metabolism of small RNAs. In some aspects, RNase LS is almost an equivalent purport of the protein RnlA (a protein with 357 amino acid [aa] lengths and a weight of 40 kD) 27 This is because it has been proved that RnlA protein has a role in RNase LS functionality for 2 reasons: firstly, it has endoribonuclease activity (they both cleave the socRNA almost at the same region); and secondly, whenever a phagic protein discriminates messages for degradation of dmd-binding domain (Dmd) (73), which is a protein that helps the phage to propagate after infection, and its expression increases highly after phage attack, a protein with length of 60 aa and weight of 7 kD comes around, cleaving both the RNase LS and RnlA nearly at the same region of them, and in fact halts both the RnlA and RNase LS activities. So, we can conclude that the action of Dmd is similar to the antitoxin component in common type II TA systems (an appurtenance that terminates the endoribonuclease activity) which neutralize the toxin RnlA. After the survey of DNA sequence of E. coli, researchers noticed that the ORFs of rnlA (a gene which encodes RNase LS or RnlA) and rnlB (a gene that encodes RnlB), are settled in a way like they are forming an operon and there is a 7 bp overlapping section between them, so they seem to be transcribed together and these 2 genes are likely to form an operon.29 As mentioned previously, the rnlB gene encodes a protein, RnlB (previously described as yfjO), which acts like phagic DMD and is able to suppress RnlA toxin.
The reasons for them being considered as TA modules are as follows30:
Endoribonuclease activity of RnlA and neutralization by a protein component (RnlB) are encoded from the same operon (like other type II TA systems).
The structure of RnlB is vulnerable, compared to RnlA, and can be easily degraded by cellular proteases like Lon or ClpXP. This degradation helps RnlA to be activated whenever T4 phage infection occurs.
Whenever RnlB protein is absent, a large number of mRNAs in E. coli are being demolished by RnlA.
There is an interaction between RnlA and RnlB components during the stability of bacterial cells.
In general, type 2 mRNA interferases usually preform their translation inhibition by cleavage of mRNAs, and they attend to this task in 2 different ways: ribosome-dependent (where toxin cleaves mRNAs at ribosomal A site by association directly with this part) like Hig B toxin; and ribosome-independent, like Mqs R toxin. But a toxin like RnlA has this ability to cleave mRNAs both in a ribosome-dependent and ribosome-independent manner during the translation process.
It should be mentioned that there are some exceptional instances regarding this TA module, which are discussed below:
In spite of the organization of typical type 2 TA operons, the rnlA and rnlB ORFs are located in an opposite way (rnlB is placed immediately at the downstream part of rnlA), like Mqa RA or Hic AB systems.
Another exception to this system is the structure of RnlA toxin, in comparison with the other common toxins. The architecture of this toxin is unique and it is composed of three domains usually not found in other toxins: NTD (N-terminal domain); NRD (N-repeated domain); and DBD (dmd-binding domain), which are separately organized domains. The helix, located at the C-terminus of domain DBD, is the only part of the helix which interacts with dmd phagic protein and causes RnlA to be suppressed. Practically, DBD domain is the part responsible for recognition and inhibition of phagic dmd antitoxin; also, it is a specific domain for RnlA dimerization, dmd attachment, cellular localization, and toxicity.31
Gammaproteobacteria as a Significant Pathogenic Class
A great number of significant and known pathogenic bacteria involved in different human, animal, and plant diseases are related to the class Gammaproteobacteria. So, as the deficiency of this factor can alter the process of virulence, it is worth to investigate TA systems of the pathogenic bacteria of this important class. As we know, this class is composed of orders in which we would emphasize the members which have been searched and surveyed for the presence of type 2 TA systems and, in some cases, the functionalities of the modules have been investigated.32
Although most of the type II TA systems are composed of 2 parts toxin, and antitoxin, it has to be mentioned that some of these pathogenic bacteria contain hybrid systems of TA modules.33,34
Shigella flexneri, which belongs to the family Enterobacteriaceae and induces diarrhea in humans, contains Vap BC, Maz EF, Rel BE, and Par DE type II TA modules.35,36
Salmonella typhimurium, which is also a member of this family and is a pathogen of humans because of the gastrointestinal inflammation that the bacteria induce and which eventually causes typhoid fever, is comprised of Phd-Doc, Rel BE, Vap BC, and Hig BA TA systems.37,38 Klebsiella penumoniae, another member of Enterobacteriaceae family, from Klebsiella genus, which causes ruining problems in human and animal lungs and which is problematic in nosocomial infections, contains the Rel BE type 2 TA system both in chromosome and plasmid locations.39
Some strains of E. coli, for example, O157 which causes hemorrhagic diarrhea, and K-12, which is used extensively in molecular biology, have been broadly studied and a lot of TA systems have been reported in them up to now, including Maz EF, Par DE, Rel BE, Hig BA, Hic BA, and Ccd AB, Mqs RA, Hip AB, YafQ-YoeB, PrlF-YhaV, YefM-YoeB, Hip AB, Chp BIK, DinJ-YafQ, Vap BC, Kis-Kid, Phd-Doc, Cpt IN, YafW-YkfL, Yaf NO, YfjZ-YpjF, and Rnl AB (which our focus is concentrated on in this system). Of course, other strains of this species have been investigated for the presence of TA modules, e.g., the type II systems of Pem KI, Srn BC, Vag CD, Par AB, Psi AB, Pas TI, and TomB-Hha.40-42
Yersinia pestis, which is also a member of this family (Enterobacteriaceae) and mainly is the “agent of plague” in patients, contains these functional TA modules, such as Hig BA, rel BE, Phd-Doc module and Hic AB.43,44
For the last example of a member of Enterobacteriaceae family, Proteus vulgaris should be mentioned. This opportunistic pathogen, which is especially involved in wound infections, has a widespread Hig BA type II TA system.45
Haemophilus influenza, involved in an extended range of infections including localized and invasive ones and belonging to the Pasteurellaceae family, processes vap-type systems.46,47 The TA systems of type II modules of Vibrio pathogens, the members of family Vibrionaceae, are as follows:
Vibrio cholera has five different modules of TA systems such as Par DE, Rel BE, Ccd AB, Hig BA, and Hip BA.48 V. parahaemolyticus, the bacteria which causes gastrointestinal problems in humans and another member of this group, contains type II systems of Par DE, Rel BE, Ccd AB, and Hip BA,49 and V. vulnificus, which contains Par DE, Rel BE, Ccd AB, Hic BA, and Hip BA systems, causes septicemia.50
Another important family, which belongs to the order Pseudomonadales, is Pseudomonadaceae. A very important opportunistic pathogen exists in this family, P. aeruginosa, and its association with especially hospital-acquired infections has been proved. According to researches which have been done regarding TA systems of this pathogen, it possesses YefM-yoeB, Maz EF, Hig BA, Rel BE, Par DE, Par AB, Pas TI, Tox1-Tox2, T-AT1-2, Gra TA, Vap-type system, and Hha-TomB.51-55
Acinetobacter baumannii, which is increasingly becoming a significant opportunistic pathogen in nosocomial infections, is a member of the Pseudomonadales order and Moraxellaceae family and has been observed to have the following systems: Maz EF; Hig BA; Rel BE; and also prevalent Che TA systems among their species Spl TA (or Abk AB), and Gra TA.56,57
Chasing the Chance of the Presence of the RnlA-RnlB System in Other Members of Gammaproteobacteria Class
The system Rnl AB, which was reported in the E. coli K-12 strain MH1 by Koga et al in 2011,30 has not been reported in most of the articles on pathogenic bacteria. So, it is worth preparing a comprehensive insight into the probable chance of the presence of these proteins in the genome of important pathogenic bacteria, large numbers of them of which are present in Gammaproteobacteria, by bioinformatics tools. Because it is not always possible to record the functional information about particular domain functionality, it is important to pay attention to the sequences and bioinformatics annotations.58
The process started with finding RnlA and RnlB protein sequences in protein antra in the NCBI website (https://www.ncbi.nlm.nih.gov). Among the results that were suggested by protein antra, the nearest one to the E. coli K-12 strain was CP4-57 prophage; RNase LS [E. coli str. K-12 sub.MG 1655], with 357 aa for RnlA; and CP4-57 prophage, an uncharacterized protein [E. coli str. K-12 sub.MG 1655] with 123 aa, and these results are confirmed by the UniProt website (https://www.uniprot.org),59 which contains a large amount of information about protein sequences and is freely accessible.
According to the NCBI Conserved Domain Database (CDD),60 which contains annotated biomolecular sequences with detailed information about their conserved domains, RnlA toxin possess 2 particular domains: DUF 302 like superfamily (RNLA-N-1), with length of 227 aa (Pfam15935); and the second domain, RnlA toxin, which contains 99 aa (cd14794). The aim of considering the CCD database was to gain information on the sequence of these domains. Following that, a Psi-BLAST search was used for chasing these sequences between the members of Gammaproteobacteria (taxid:1236) using a non-redundant protein sequence database as target. By considering the query coverages higher than 90%, the Psi-BLAST was iterated 2 times for both the first and second domains of RnlA (RNLA-N-1). For RNLA-N-1 domain, the number of psi-BLAST iterations was 2 times; for the second domain, this iteration was accomplished.
Phylogenetic Tree Construction to Survey the Distribution of RnlA-RnlB Module Among Gammaproteobacteria Class
Then the results of PSI-BLAST were downloaded, and they were employed for the construction of the phylogenetic tree in Mega software version 7.61 The first step toward this was ClustalW algorithm was used to perform multiple sequence alignment. The aim of inferring the phylogenetic trees here was to screen the distribution of Rnl AB systems among class Gammaproteobacteria and to study the evolutionary relationships between their protein sequences. This, in turn, could help to determine the same possible functions for this system between other bacterial species. Although, among a broad range of TA modules present in bacterial genomes, only a few are really functional and experimental approaches are required for their functional characterization.
Tree Depiction of Neighbor-Joining and Parsimony Method for Domains
Here, 2 kinds of phylogenic trees, neighbor-joining (NJ) and parsimony methods, were employed to deduce the phylogenetic trees. NJ is an agglomerative clustering method which is characterized as a progressive, distance-based method. It can be applied to a large number of sequences (> 50). It should be mentioned that NJ generates an unrooted tree.
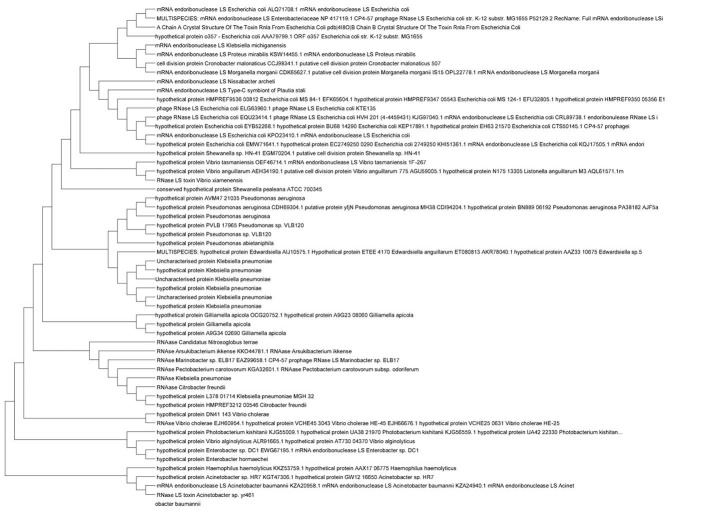
In maximum Parsimony method, the tree with the least number of changes is introduced as the most parsimonious tree, and as a result, this method is considered as one of the most precise approaches in inferring phylogenetic relationships. This method works with the “molecular clock” hypothesis, in which each family of proteins evolves with the constant rates of mutations. Figure 1 represents the maximum parsimony tree of the RNLA-N-1 superfamily of toxin RnlA.
Figure 1.
Evolutionary Relationships of RNLA-N-1 Domain-(Superfamily) in Gammaproteobacteria Via Neighbor-Joining Phylogenetic Tree.
NJ and maximum parsimony trees for the RnlA-toxin superfamily (the second domain of RnlA toxin) and RnlB-antitoxin superfamily illustrated as described here. Here, one of the trees which belongs to RNLA-N-1 domain-(superfamily) distribution among Gammaproteobacteria members is depicted to show the process (Figure 1).62
The evolutionary history was inferred using the Neighbor-Joining method in MEGA7. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (The Poisson correction for distance d assumes equal substitution rates across sites and equal amino acid frequencies. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7.
Acinetobacter baumannii and Pseudomonas aeruginosa as 2 Problematic Pathogens
Owing to their presence in a broad range of nosocomial infections like burn wounds, these 2 pathogens have been remarkable choices to study. Here according to the presence of Rnl AB type II system domains in these pathogens, we mention that they may contain phage-resistant systems, which are obstacles to T4 phage attack.
T4 Phage as a Weapon in Phage Therapy
T4-like phages have been used for therapeutic aims like phage therapy, which has been successful at least on a small scale. As for other therapeutic approaches, this procedure is not devoid of problems, and as mentioned in phage therapy articles, bacterial resistance may occur.63,64 So, through knowledge of phage-resistant systems in important bacteria like A. baumannii and P. auroginosa, it is important to consider ways to prevent any problems.
Photodynamic Therapy as a Procedure for Weakening TA Systems for Better Results of Phage Therapy
Photodynamic therapy (PDT) is a therapeutic technique which utilizes photosensitizer (PS) dye and visible light. During this process, irradiation of a specific wavelength of visible light to the photosensitizer dye leads to reactive oxygen species (ROS) production.65,66 According to previous studies, it has been proved that PDT procedure has destructive effects on the expression of particular systems in bacterial cells. For example, reduction of pmrA/pmrB 2-component signal system after PDT occurred in one study,67 and downregulation of biofilm-related genes (by conjugative PS) is another example of this effect.68 Furthermore, there have been a number of experiments that have been worked on in preparation of a substrate which can be useful for another remedy. For example, blocking efflux pumps before PS incubation through PDT approach could help the process to be more effective,69 or deletion of genes which encode components of MexAB-OprM efflux systems is useful for greater susceptibility of P. auroginosa for the PDT process (or using extracts to decrease expression of efflux pumps),70 or in another study, it was shown that the efflux pump inhibitor helps the disruption of E. fecalis biofilm by PDT.71 So, here we theorize that if the gene of the TA system which is responsible for phage resistance can be suppressed by ROS through the PDT process, it would be an appropriate substrate for phage therapy which is going to be used next.
Concluding Remarks
As described earlier, many TA systems have been introduced until now and some of them have been proved to be contributors to bacterial pathogenesis. The relationship between pathogenesis and these systems makes them an important study case for promoting new antibacterial targets in the future to use against bacterial infections, and the broad range of their distribution among important pathogenic bacteria reinforces the importance of their study. There have been a large number of studies which have focused on these systems and still more studies need to be done. Considering the important role of Rnl BA TA, and that the bioinformatics tools which have provided a background for studying more TA systems have not yet been reported, this study provides a compact report of distribution of Rnl BA TA domains among Gammaproteobacteria members, which can help in using this system in other pathogenic bacteria. Here, also the problem of phage resistance by equipping Rnl AB type II system has been discussed and suggested for utilization of PDT technique for the probable weakening of the mentioned system. By suppression of the phage-resistance TA system, bacteria would no longer be able to show resistance to the phage, so one of the problems in phage therapy procedure would be resolved.
Ethical Considerations
Not applicable.
Conflict of Interests
The authors declare no conflict of interest.
Please cite this article as follows: Hosseini N, Pourhajibagher M, Chiniforush N, Hosseinkhan N, Rezaie P, Bahador A. Modulation of toxin-antitoxin system Rnl AB type II in phage-resistant Gammaproteobacteria surviving photodynamic treatment. J Lasers Med Sci. 2019;10(1):21-28. doi:10.15171/jlms.2019.03.
References
- 1.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13(5):298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 3.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39(13):5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38(11):3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012;9(12):1488–1490. doi: 10.4161/rna.23045. [DOI] [PubMed] [Google Scholar]
- 6.Weaver KE. The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA Biol. 2012;9(12):1498–503. doi: 10.4161/rna.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rocker A, Meinhart A. Type II toxin: antitoxin systems. More than small selfish entities? Curr Genet. 2016;62(2):287–290. doi: 10.1007/s00294-015-0541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mruk I, Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 2014;42(1):70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen SK, Maenhaut-Michel G, Mine N, Gottesman S, Gerdes K, Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51(6):1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 10.Hu MX, Zhang X, Li EL, Feng YJ. Recent advancements in toxin and antitoxin systems involved in bacterial programmed cell death. Int J Microbiol. 2010;2010:781430. doi: 10.1155/2010/781430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189(17):6089–6092. doi: 10.1128/jb.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci. 2005;30(12):672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Kolodkin-Gal I, Verdiger R, Shlosberg-Fedida A, Engelberg-Kulka H. A differential effect of E coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS One. 2009;4(8):e6785. doi: 10.1371/journal.pone.0006785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Wang Q, Li M. et al. Escherichia coli toxin gene hipA affects biofilm formation and DNA release. Microbiology. 2013;159(Pt 3):633–640. doi: 10.1099/mic.0.063784-0. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi Y, Park JH, Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284(42):28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood TL, Wood TK. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen. 2016;5(3):499–511. doi: 10.1002/mbo3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan WT, Nieto C, Harikrishna JA. et al. Genetic regulation of the yefM-yoeB toxin-antitoxin locus of Streptococcus pneumoniae. J Bacteriol. 2011;193(18):4612–4625. doi: 10.1128/jb.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayaraman R. Bacterial persistence: some new insights into an old phenomenon. J Biosci. 2008;33(5):795–805. doi: 10.1007/s12038-008-0099-3. [DOI] [PubMed] [Google Scholar]
- 20.Dorr T, Vulic M, Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8(2):e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 22.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci U S A. 2009;106(3):894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan Q, Awano N, Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol. 2011;79(1):109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Lord DM, Cheng HY. et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012;8(10):855–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobato-Marquez D, Diaz-Orejas R, Garcia-Del Portillo F. Toxin-antitoxins and bacterial virulence. FEMS Microbiol Rev. 2016;40(5):592–609. doi: 10.1093/femsre/fuw022. [DOI] [PubMed] [Google Scholar]
- 26.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 27.Otsuka Y, Koga M, Iwamoto A, Yonesaki T. A role of RnlA in the RNase LS activity from Escherichia coli. Genes Genet Syst. 2007;82(4):291–299. doi: 10.1266/ggs.82.291. [DOI] [PubMed] [Google Scholar]
- 28.Otsuka Y, Yonesaki T. A novel endoribonuclease, RNase LS, in Escherichia coli. Genetics. 2005;169(1):13–20. doi: 10.1534/genetics.104.033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otsuka Y, Yonesaki T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol Microbiol. 2012;83(4):669–681. doi: 10.1111/j.1365-2958.2012.07975.x. [DOI] [PubMed] [Google Scholar]
- 30.Koga M, Otsuka Y, Lemire S, Yonesaki T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics. 2011;187(1):123–130. doi: 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naka K, Koga M, Yonesaki T, Otsuka Y. RNase HI stimulates the activity of RnlA toxin in Escherichia coli. Mol Microbiol. 2014;91(3):596–605. doi: 10.1111/mmi.12479. [DOI] [PubMed] [Google Scholar]
- 32.Gao B, Mohan R, Gupta RS. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int J Syst Evol Microbiol. 2009;59(Pt 2):234–247. doi: 10.1099/ijs.0.002741-0. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt O, Schuenemann VJ, Hand NJ. et al. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J Mol Biol. 2007;372(4):894–905. doi: 10.1016/j.jmb.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unterholzner SJ, Poppenberger B, Rozhon W. Toxin-antitoxin systems: Biology, identification, and application. Mob Genet Elements. 2013;3(5):e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dienemann C, Boggild A, Winther KS, Gerdes K, Brodersen DE. Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J Mol Biol. 2011;414(5):713–722. doi: 10.1016/j.jmb.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengupta M, Austin S. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect Immun. 2011;79(7):2502–2509. doi: 10.1128/iai.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De la Cruz MA, Zhao W, Farenc C. et al. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 2013;9(12):e1003827. doi: 10.1371/journal.ppat.1003827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Lobato-Marquez D, Moreno-Cordoba I, Figueroa V, Diaz-Orejas R, Garcia-del Portillo F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci Rep. 2015;5:9374. doi: 10.1038/srep09374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen YT, Lauderdale TL, Liao TL. et al. Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 beta-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2007;51(8):3004–3007. doi: 10.1128/aac.00167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armalyte J, Jurenaite M, Beinoraviciūte G, Teiserskas J, Suziedeliene E. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J Bacteriol. 2012;194(6):1523–1532. doi: 10.1128/JB.06104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akiyama Y, Saito E, Futai H. et al. Comprehensive study of pathogenic genes distributed in Escherichia coli isolated from cattle. Shokuhin Eiseigaku Zasshi. 2015;56(3):118–122. doi: 10.3358/shokueishi.56.118. [DOI] [PubMed] [Google Scholar]
- 42.Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence . Nat Chem Biol . 2016;12(4):208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 43.Goulard C, Langrand S, Carniel E, Chauvaux S. The Yersinia pestis chromosome encodes active addiction toxins. J Bacteriol. 2010;192(14):3669–3677. doi: 10.1128/jb.00336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bibi-Triki S, Li de la Sierra-Gallay I , Lazar N. et al. Functional and structural analysis of HicA3-HicB3, a novel toxin-antitoxin system of Yersinia pestis. J Bacteriol. 2014;196(21):3712–3723. doi: 10.1128/jb.01932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schureck MA, Maehigashi T, Miles SJ. et al. Structure of the Proteus vulgaris HigB-(HigA)2-HigB toxin-antitoxin complex. J Biol Chem. 2014;289(2):1060–1070. doi: 10.1074/jbc.M113.512095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren D, Walker AN, Daines DA. Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol. 2012;12:263. doi: 10.1186/1471-2180-12-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daines DA, Wu MH, Yuan SY. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol. 2007;189(14):5041–5048. doi: 10.1128/jb.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33(3):966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makino K, Oshima K, Kurokawa K. et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361(9359):743–749. doi: 10.1016/s0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 50.Van Melderen L. Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol. 2010;13(6):781–785. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77(16):5577–5583. doi: 10.1128/aem.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnin RA, Poirel L, Nordmann P. et al. Complete sequence of broad-host-range plasmid pNOR-2000 harbouring the metallo-beta-lactamase gene blaVIM-2 from Pseudomonas aeruginosa. J Antimicrob Chemother. 2013;68(5):1060–1065. doi: 10.1093/jac/dks526. [DOI] [PubMed] [Google Scholar]
- 53.Williams JJ, Halvorsen EM, Dwyer EM, DiFazio RM, Hergenrother PJ. Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2011;322(1):41–50. doi: 10.1111/j.1574-6968.2011.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savari M, Rostami S, Ekrami A, Bahador A. Characterization of Toxin-Antitoxin (TA) Systems in Pseudomonas aeruginosa Clinical Isolates in Iran. Jundishapur J Microbiol. 2016;9(1):e26627. doi: 10.5812/jjm.26627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez-Garcia L, Blasco L, Lopez M. et al. Toxin-Antitoxin Systems in Clinical Pathogens. Toxins (Basel) 2016;8(7) doi: 10.3390/toxins8070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurenaite M, Markuckas A, Suziedeliene E. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J Bacteriol. 2013;195(14):3165–3172. doi: 10.1128/jb.00237-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosqueda N, Gato E, Roca I. et al. Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J Antimicrob Chemother. 2014;69(10):2629–2633. doi: 10.1093/jac/dku179. [DOI] [PubMed] [Google Scholar]
- 58.Ruepp A, Zollner A, Maier D. et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32(18):5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.UniProt: UniProt:. the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–d169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchler-Bauer A, Bo Y, Han L. et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–d203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 70 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pevsner J. Bioinformatics and Functional Genomics. 3rd ed. Wiley-Blackwell; 2015.
- 63.Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013;8(6):769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 64.Donlan RM. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009;17(2):66–72. doi: 10.1016/j.tim.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 65.Hamblin MR, Hasan T. Photodynamic therapy: a new antimicrobial approach to infectious disease? Photochem Photobiol Sci. 2004;3(5):436–450. doi: 10.1039/b311900a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pourhajibagher M, Boluki E, Chiniforush N. et al. Modulation of virulence in Acinetobacter baumannii cells surviving photodynamic treatment with toluidine blue. Photodiagnosis Photodyn Ther. 2016;15:202–212. doi: 10.1016/j.pdpdt.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Boluki E, Kazemian H, Peeridogaheh H. et al. Antimicrobial activity of photodynamic therapy in combination with colistin against a pan-drug resistant Acinetobacter baumannii isolated from burn patient. Photodiagnosis Photodyn Ther. 2017;18:1–5. doi: 10.1016/j.pdpdt.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Misba L, Kulshrestha S, Khan AU. Antibiofilm action of a toluidine blue O-silver nanoparticle conjugate on Streptococcus mutans: a mechanism of type I photodynamic therapy. Biofouling. 2016;32(3):313–328. doi: 10.1080/08927014.2016.1141899. [DOI] [PubMed] [Google Scholar]
- 69.Tegos GP, Hamblin MR. Phenothiazinium antimicrobial photosensitizers are substrates of bacterial multidrug resistance pumps. Antimicrob Agents Chemother. 2006;50(1):196–203. doi: 10.1128/aac.50.1.196-203.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tegos GP, Masago K, Aziz F, Higginbotham A, Stermitz FR, Hamblin MR. Inhibitors of bacterial multidrug efflux pumps potentiate antimicrobial photoinactivation. Antimicrob Agents Chemother. 2008;52(9):3202–3209. doi: 10.1128/aac.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kishen A, Upadya M, Tegos GP, Hamblin MR. Efflux pump inhibitor potentiates antimicrobial photodynamic inactivation of Enterococcus faecalis biofilm. Photochem Photobiol. 2010;86(6):1343–1349. doi: 10.1111/j.1751-1097.2010.00792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]