Abstract
Non-small cell lung cancer (NSCLC) is the most prevalent form of lung cancer and its molecular landscape has been extensively studied. The most common genetic alterations in NSCLC are mutations within the epidermal growth factor receptor (EGFR) gene, with frequencies between 10-40%. There are several molecular targeted therapies for patients harboring these mutations.
Liquid biopsies constitute a flexible approach to monitor these mutations in real time as opposed to tissue biopsies that represent a single snap-shot in time. However, interrogating cell free DNA (cfDNA) has inherent biological limitations, especially at early or localized disease stages, where there is not enough tumor material released into the patient’s circulation.
We developed a qPCR- based test (ExoDx EGFR) that interrogates mutations within EGFR using Exosomal RNA/DNA and cfDNA (ExoNA) derived from plasma in a cohort of 110 NSCLC patients.
The performance of the assay yielded an overall sensitivity of 90% for L858R, 83% for T790M and 73% for exon 19 indels with specificities of 100%, 100%, and 96% respectively. In a subcohort of patients with extrathoracic disease (M1b and MX) the sensitivities were 92% (L858R), 95% (T790M), and 86% (exon 19 indels) with specificity of 100%, 100% and 94% respectively.
Keywords: liquid biopsy, exosomes, ctDNA, exoNA, NSCLC
INTRODUCTION
Lung cancer represents one of every five cases of cancer-related death worldwide [1]. Non-small cell lung cancer (NSCLC) is one of the most common types of lung cancer and is divided into three main histological sub-types: squamous cell carcinoma, large cell carcinoma, and adenocarcinoma. NSCLC is very heterogeneous at the molecular level and between 10-40% of patients’ tumors harbor mutations within the epidermal growth factor receptor (EGFR) [2, 3].
There are now several molecular targeted therapies approved for this group of patients. Included in this group are the anti- EGFR first- and second-generation tyrosine kinase inhibitors (TKIs), which bind reversibly and irreversibly to the tyrosine kinase receptor. These are classified as evidence-based first-line treatments for NSCLC patients that harbor activating EGFR mutations (within exon 19 and the L858R missense mutation within exon 21) [4]. However, during treatment with these 1st- and 2nd- generation TKIs, there is an acquired resistance mechanism that arises through a missense driver mutation within exon 20 of EGFR (T790M) [5, 6]. This has led to the development and approval of a third-generation inhibitor, osimertinib, an oral, irreversible EGFR-TKI that is selective for both activating and resistance mutations [7].
Given the availability of these targeted therapies and higher survival rates of patients with early stage disease [8], it is critical to be able to monitor the tumor mutation profile with a sensitive and specific assay as early as possible [9].
The field of liquid biopsies has evolved greatly, and there are now several studies showing that longitudinal monitoring of cfDNA can capture tumor dynamics in NSCLC patients [10–13]. However, liquid biopsies that utilize the cell-free DNA fraction (cfDNA) face biological challenges. The majority of cfDNA is from normal cells and the level of cell-free tumor DNA (ctDNA) in that fraction can sometimes be below the limit of detection even with the most sensitive assay platforms [14, 15]. To address this limitation, three recent studies showed the benefits of combining the mutations found in exosomal nucleic acids with cfDNA (exoNA) for mutation detection [16, 17]. In one of the studies a CLIA validated qPCR test for EGFR T790M mutations in NSCLC patients (n = 210) achieved a clinical sensitivity and specificity of 92% and 89%, respectively [18].
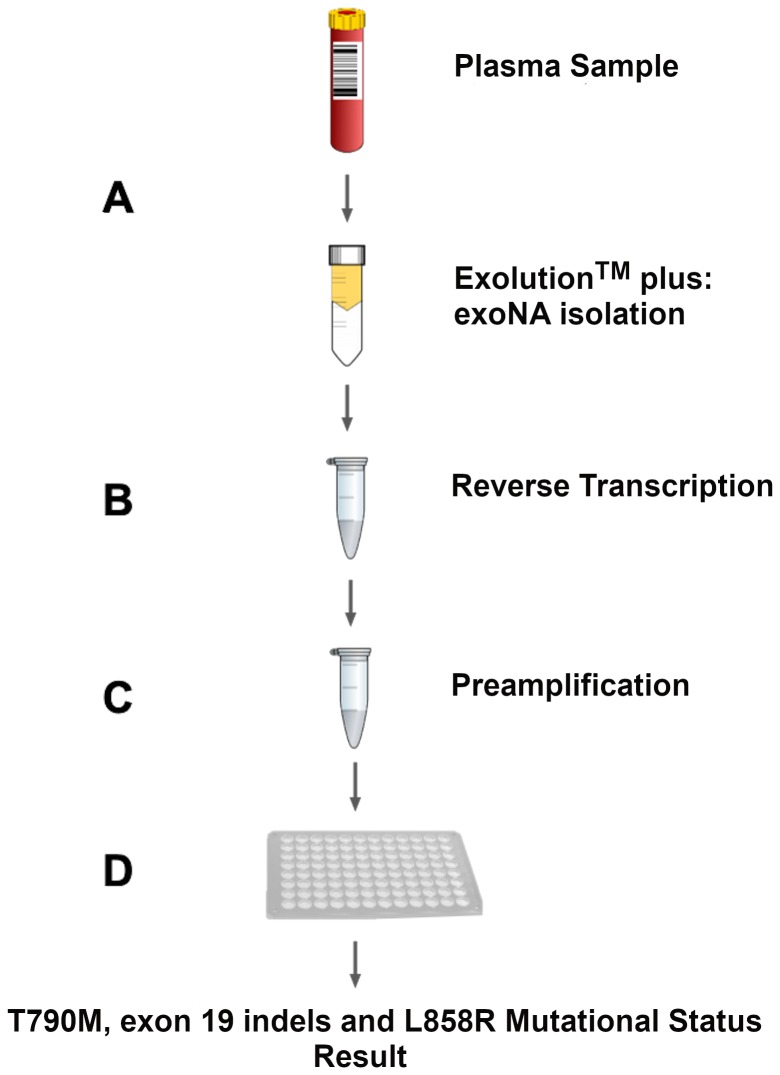
The goal of this study was to develop a qPCR-based test that interrogates a panel of 29 mutations in the EGFR gene, including the activating and resistance mutations (Table 1) that predict response to first line EGFR inhibitors and osimertinib. To mitigate the biological limitation of cfDNA assays we used the same approach as previously [16-18] and developed the assay on a combination of exosomal RNA/DNA and cfDNA (Figure 1).
Table 1. List of single point mutations, insertions and deletions interrogated with the ExoDx EGFR assay.
| Variant name | Variant type | Exon location in EGFR | Cosmic ID |
|---|---|---|---|
| c.2235_2249del15 | 15 bp deletion | Exon 19 | COSM6223 |
| c.2235_2248>AATTC | 5 bp insertion | COSM13550 | |
| c.2235_2251>AATTC | COSM13552 | ||
| c.2235_2252>AAT | 15 bp deletion | COSM13551 | |
| c.2236_2250del15 | COSM6225 | ||
| c.2236_2253del18 | 18 bp deletion | COSM12728 | |
| c.2236_2253del18 | 5 bp insertion | COSM12416 | |
| c.2237_2253>TTGCT | COSM12367 | ||
| c.2237_2254del18 | 1 bp insertion | COSM12384 | |
| c.2237_2255>T | 3 bp insertion | COSM18427 | |
| c.2238_2248>GC | 2 bp insertion | COSM12422 | |
| c.2238_2248>GC | 3 bp insertion | COSM12419 | |
| c.2238_2252del15 | 15 bp deletion | COSM23571 | |
| c.2238_2255del18 | 18 bp deletion | COSM6220 | |
| c.2239_2247delTTAAGAGAA | 9 bp deletion | COSM6218 | |
| c.2239_2248TTAAGAGAAG>C | 10 bp insertion | COSM12382 | |
| c.2239_2251>C | 1 bp insertion | COSM12383 | |
| c.2239_2253del15 | 15 bp deletion | COSM6254 | |
| c.2237_2253>TTGCT | 3 bp insertion | COSM12403 | |
| c.2239_2256del18 | 18 bp deletion | COSM6255 | |
| c.2239_2258>CA | 2 bp insertion | COSM12387 | |
| c.2240_2251del12 | 12 bp deletion | COSM6210 | |
| c.2240_2254del15 | 15 bp deletion | COSM12369 | |
| c.2240_2257del18 | 18 bp deletion | COSM12370 | |
| 2235_2255>AAT | 3 bp insertion | COSM12385 | |
| c.2237_2251del15 | 15 bp deletion | COSM12678 | |
| c.2237_2252>T | 1 bp insertion | COSM12386 | |
| T790M | Missense mutation | Exon 20 | COSM481727 |
| L858R | Missense mutation | Exon 21 | COSM6224 |
Figure 1. Assay workflow overview.
(A) Column-based exosomal RNA/DNA and cfDNA (exoNA) isolation from plasma of NSCLC patients. (B) Reverse transcription step. At this step, we added an exogenous RNA construct as control to monitor for PCR inhibition to each sample. (C) Pre-amplification step of exon 19, 20 and 21, in addition to QBeta and exon 7 (controls for sample inhibition and integrity, respectively). (D) Amplification Refractory Mutation System (ARMS) based quantitative PCR step.
RESULTS
Analytical performance
The analytical sensitivity was evaluated by spiking synthetic constructs into 2 mL of healthy pooled plasma to generate a complex background and simulate a biological sample. The results for T790M in this new test were in alignment with our previous study [18]. As shown in Table 2A, 1.25 copies of T790M/mL of plasma were detected 75% of the time (3/4 biological replicates). At ≥ 5 copies/mL, we detected T790M in all biological replicates.
Table 2. Evaluation of ExoDx EGFR on T790M (A), L858R (B) and exon 19 deletion (Δ746-750) (C).
| (A) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Copies of EGFR/mL | 4416 (Stdev±14.4) | |||||||
| Copies T790M /mL | 0 | 1.25 | 4 | 4.4 | 8 | 13 | 27 | Total |
| Fractional abundance (%)/mL | 0 | 0.03 | 0.09 | 0.10 | 0.18 | 0.30 | 0.61 | |
| Detected | 0 | 3 | 3 | 4 | 4 | 4 | 4 | 22 |
| Not detected | 4 | 1 | 1 | 0 | 0 | 0 | 0 | 6 |
| Hit Fraction* | 0 | 0.75 | 0.75 | 1.0 | 1.0 | 1.0 | 1.0 | |
*Hit fraction: Samples correctly classified.
| (B) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Copies of EGFR/mL | 4416 (Stdev±14.4) | ||||||||||
| Copies L858R /mL | 0 | 1.9 | 2.4 | 3.5 | 5.4 | 9.5 | 10.2 | 10.8 | 24.1 | 56 | Total |
| Fractional abundance (%)/mL | 0 | 0.04 | 0.05 | 0.08 | 0.12 | 0.21 | 0.23 | 0.24 | 0.54 | 1.25 | |
| Detected | 0 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 45 |
| Not detected | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Hit Fraction* | 0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| (C) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Copies of EGFR/mL | 4416 (Stdev±14.4) | ||||||||||
| Copies exon 19 deletion Δ746-750 /mL | 0 | 2 | 2.9 | 5.4 | 5.2 | 7.1 | 8.6 | 12.8 | 27.9 | 50 | Total |
| Fractional abundance (%)/mL | 0 | 0.05 | 0.07 | 0.12 | 0.12 | 0.16 | 0.19 | 0.29 | 0.63 | 1.12 | |
| Detected | 0 | 5 | 4 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 45 |
| Not detected | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Hit Fraction* | 0 | 1.0 | 0.75 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
Analytical performance was assessed by spiking mutations into healthy pooled plasma. Spike-ins were done in 2 mL plasma aliquots, but copies are depicted as copies/mL.
Table 2B and 2C summarize the assay results on activating mutations. The assay detected all five replicates at 2 copies of L858R/mL of plasma (equivalent to 0.04% allelic frequency). For exon 19 indels, 2 copies/mL (0.05% allelic frequency) were detected in all five replicates and 3 copies/mL were detected in 4/5 replicates (0.07% allelic frequency). The allelic frequencies were validated using droplet digital PCR (ddPCR). The number of mutant molecules in a plasma sample vary according to a Poisson distribution and at very low copy numbers, the variation across replicates is dominated by sampling noise rather than qPCR variation [19]. Note that the lowest theoretical limit of detection for PCR is 3 copies (assuming a Poisson distribution, 95% chance of including at least 1 copy in the PCR and single copy detection) under this assumption due to stochastic variation [20].
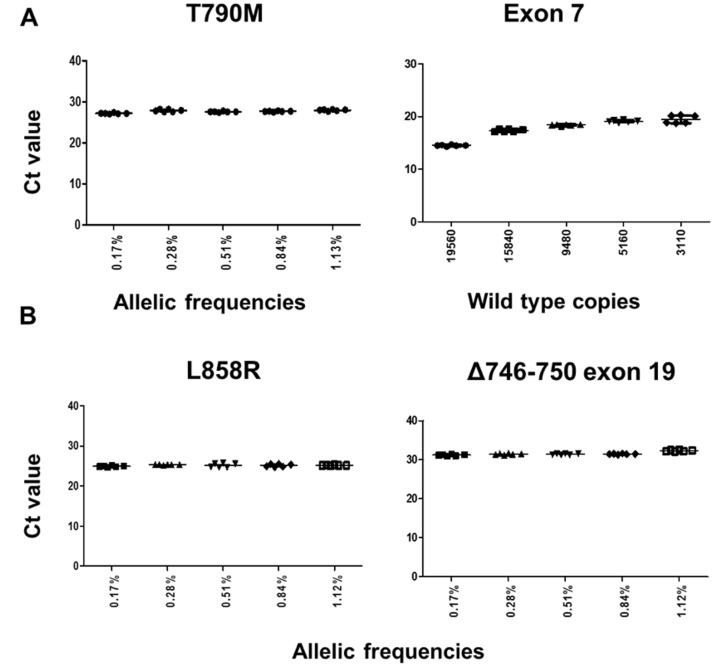
Next, we assessed the robustness of the test on mutation blends containing an average of 30 copies of mutant synthetic constructs (T790M, L858R or the most prevalent exon 19 deletion (Δ746-750) synthesized by IDT Corporation, IL, USA into wild type genomic DNA (Catalog no. G147A, Promega Corporation, WI, USA). Final allelic frequencies (Supplementary Table 1) and mutant copies in the admixtures were measured by droplet digital PCR (ddPCR) (Supplementary Table 2). The qPCR assays for all three targets were robust, mutant-specific and unaffected by the presence of wild type background (Figure 2), as we did not observe a CT delay in any of the allelic frequencies (0.17–1.13%) evaluated in a total of 35 samples, regardless of the wild type background.
Figure 2. Assay robustness on gDNA admixtures for T790M, L858R and exon 19 deletion (Δ746-750).
An average of 30 copies of mutant-containing synthetic DNA was used for the blends. (A) CT values for the T790M ARMS assay across different gDNA blends. (left). CT values for the EGFR exon 7 (control assay) (right). (B) CT values for the L858R ARMS assay (left). CT values for the exon 19 indels ARMS assay (right).
We also estimated how the assays performed on a series of commercially available cell free DNA Reference Standards (Catalog nos. HD777, HD778 and HD779, Horizon Discovery, UK). These standards contain variable amounts of mutant templates down to single copies for all three targets in the 0.1% admixture. The correlation coefficient for all three assays were greater than 0.99 on the different reference admixtures (Supplementary Figure 1).
EGFR mutation detection using exosomal nucleic acids and cfDNA on clinical samples
This patient cohort consisted of 110 clinical samples with 60 mutation-positive NSCLC patients (L858R, exon 19 indels and T790M) and 50 mutation-negative NSCLC patients and healthy donor samples with no history of familial lung cancer. 30% of NSCLC patients were diagnosed with intrathoracic disease (M0/M1a).
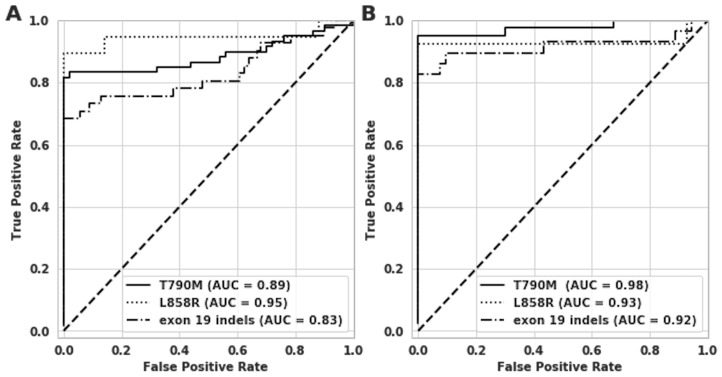
The clinical sensitivity of the assay was 83% for T790M, 90% for L858R and 73% for exon 19 indels with specificities of 100%, 100% and 96% respectively (Table 3A and 3B). The Area Under the Receiver Operator Characteristics Curve (ROC) for the T790M, L858R and exon 19 indels assays was 0.89, 0.95 and 0.83 respectively (Figure 3).
Table 3. Clinical cohort performance.
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| Plasma results using exoNA | ||||||||
| T790M | L858R* | exon 19 indels* | Total number | |||||
| Tissue result | + | - | + | - | + | - | ||
| + | 50 | 10 | 17 | 2 | 30 | 11 | 110 | |
| - | 0 | 50 | 0 | 91 | 3 | 66 | ||
*Samples are mutually exclusive for L858R and exon 19 indels
Figure 3. Receiver operating characteristic (ROC) curve analysis on clinical samples for T790M, L858R and exon 19 indels.
The x-axes show 1-Specificity or the False Positive Rate (FPR), and y-axes show the sensitivity or True Positive Rate (TPR). (A) ROC curves for all three assays for all 110 patient samples. (B) ROC curves for all three targets on M1b/MX patient samples alone.
| B | ||||||
|---|---|---|---|---|---|---|
| Parameter | Clinical Samples | |||||
| T790M | L858R | exon 19 indels | ||||
| All* | M1b | All | M1b | All | M1b | |
| AUC | 0.89 | 0.98 | 0.95 | 0.93 | 0.83 | 0.92 |
| Specificity | 100.00 | 100.00 | 100.00 | 100.00 | 95.65 | 94.33 |
| Sensitivity | 83.33 | 95.24 | 89.47 | 92.31 | 73.17 | 86.21 |
| Accuracy | 90.91 | 97.56 | 98.18 | 98.78 | 87.27 | 91.46 |
| Precision | 100.00 | 100.00 | 100.00 | 100.00 | 90.91 | 89.29 |
| NPV | 83.33 | 95.24 | 97.85 | 98.57 | 85.71 | 92.59 |
*Overall cohort includes 30% of patients with intrathoracic disease.
(A) Confusion matrix that correlates tissue results obtained in tissue with plasma exoNA for the activating and resistance mutations for all patients included in the study. (B) Assay results for the activating and resistance mutations for the entire cohort (M1b/MX/M1a/M0) and patients with disease stages M1b/MX.
The assay also demonstrated very high performance among patient samples at disease stage M1b/MX with sensitivities of 95%, 92% and 86% for T790M, L858R and exon 19 indels respectively (Table 3B). Supplementary Table 3 lists the clinical samples that were negative in plasma for at least one EGFR mutation that were classified as positive in tissue, and hence likely false negative as classified by the assay. A majority of the false negative patient samples (9/15) had intrathoracic disease (M0/M1a). ~67% of the false negative M0/M1a samples were called negative for all of EGFR mutations (L858R, exon 19 indels and T790M) tested in this assay.
DISCUSSION
The NSCLC field have been one of the pioneers for molecular therapies and personalized therapy based on individual tumor mutation status. For instance, nine superiority trials have proven the efficacy of several reversible and irreversible EGFR-TKIs versus chemotherapy alone in tumors with specific somatic variants (exon 19 indels and L858R in exon 21) [21]. Although these tumors have very good initial response rates, they eventually develop drug resistance via different mechanisms [22]. The most common mechanism of resistance is the missense mutation in codon 790 that results in an amino acid change (T790M) in exon 20 of EGFR [23]. A randomized, international, open-label, phase 3 clinical trial of osimertinib in 419 patients harboring this mutation showed a Progression Free Survival (PFS) of 10.1 months (95% CI: 8.3, 12.3) compared to 4.4 months (95% CI: 4.2, 5.6) for patients receiving doublet chemotherapy [24]. This resulted in the FDA approval of osimertinib for NSCLC patients with T790M-positive tumors.
This emphasizes the importance of assessing tumor mutational status using very accurate and sensitive methods not just prior to treatment but to track tumor dynamics over the course of therapy. In this arena, liquid biopsies have shown great potential as a tool for the clinical management of patients with NSCLC [25, 26]. However, sensitivity of liquid biopsies is limited by the amount of tumor-derived nucleic acids in circulation, especially in early stage disease. A study examining the circulating tumor DNA (ctDNA) fraction obtained from 640 subjects demonstrated that ctDNA fraction is highly variable, not only between tumor types but also within patients with the same tumor type. Only 55% of the patients with localized disease had detectable levels of ctDNA using 5 mL of plasma [27]. Sacher and collaborators recently published a prospectively validated plasma genotyping method with an overall sensitivity below 60% for patients with one metastatic site [28].
It is well known that cfDNA is not the only source of nucleic acids in biofluids but that extracellular vesicles such as exosomes also contain nucleic acids. cfDNA is typically described as coming from the dying process of the tumor (apoptosis and necrosis) whereas exosomes are released as an active metabolic process from living cells [29]. Therefore, combining these two approaches may overcome some of the inherent biological limitations of an approach based on cfDNA alone as well as assess mutation status from the living as well as the dying processes within the tumors. A recent study that compared the mutation detection rates with different liquid biopsy platforms showed a significantly higher performance when looking in the exoNA fraction compared to just cfDNA [16]. The largest added value with the exoNA over cfDNA was for patients with intrathoracic disease (M0/M1a), which is not surprising since many studies have shown limited ctDNA copies in this population. We have recently demonstrated that the exoNA approach is feasible for detection of the EGFR T790M mutation and yielded very high clinical performance (92% sensitivity and 89% specificity) for the detection of T790M in plasma of NSCLC patients [18]. The test was validated on 210 clinical samples, from which ~40% had localized disease.
There is a need for a broader EGFR panel beyond looking at the EGFR T790M mutation, so this study summarizes the results of a qPCR-based EGFR mutation panel that also includes the activating EGFR mutations. The assay had a sensitivity of 95% for T790M and 88% for activating mutations in the M1b/MX population, similar to what was achieved with BEAMing (Beads, Emulsion, Amplification, Magnetics) (95% T790M, 96% activating) and cobas® plasma test (95% T790M, 90% activating) [14]. However, the assay has a sensitivity of 56% for both T790M and activating mutations in patients with intrathoracic disease (M0/M1a) compared to 27% and 39% for BEAMing and 14% and 42% for the cobas® plasma test [14]. These results further support an increase in sensitivity that can be achieved by using exoNA in comparison to liquid biopsy tests that use only cfDNA, especially in patients with early stage disease. The increase in sensitivity does not adversely affect the specificity of the assay as demonstrated by the high specificities of 100%, 100% and 94% for T790M, L858R and exon 19 indels respectively (Table 3B). We also note that none of the EGFR mutations tested (L858R, exon 19 indels and T790M) were detected in majority of the false negative M0/M1 samples (Supplementary Table 3). The patients that remain problematic to detect with this improved liquid biopsy is likely due to the biology of the disease, releasing very low amounts of both cfDNA and exosomal RNA/DNA into circulation. A potential workaround could be to use larger plasma volumes (> 2 mL) to minimize the effects of sampling noise and improve the sensitivity of the assay. The exoNA extraction efficiency is linear, and roughly twice as much material is retained into the assay if the input volumes double.
The high sensitivity and specificity of this assay demonstrate its utility as a tool for the detection and monitoring of EGFR mutations in NSCLC patients to inform clinical management, especially in cases where tissue biopsies are not readily available.
MATERIALS AND METHODS
Assay design
The workflow for ExoDx (EGFR) is schematically represented in Figure 1. exoNA was extracted from plasma samples using a cGMP manufactured isolation kit (ExoLutionTM Plus, Exosome Diagnostics, Inc. Waltham, MA, USA) [16, 18]. ExoLution™ Plus uses a spin-column to capture both the cfDNA and extracellular vesicles. We excluded any vesicle larger than 0.8 μm in diameter from the plasma through a pre-filtration step [30]. The entire eluate was then reverse transcribed as described elsewhere [16, 30], followed by a pre-amplification reaction. This step contains EGFR exon 19 and 20 wild-type blockers as well as primers targeting EGFR exon 19, 20 and 21, exon 7 (control reaction) and a non-human control sequence (QBeta) that is spiked into every patient sample as a qPCR/inhibition control.
The final downstream detection step includes a quantitative PCR (qPCR) step using an Amplification-Refractory Mutation system (ARMS) approach to selectively amplify T790M, L858R and 27 different exon 19 insertion/deletions (indels) (Table 1). The assay includes controls for sample integrity and inhibition (EGFR exon 7 and QBeta) in each qPCR reaction as previously described [18].
Determination of qPCR CT thresholds for EGFR L858R and exon 19 indels assays
We estimated the optimal cycle thresholds (CT) for L858R and exon 19 indels detection in plasma with synthetic construct spike-ins (Table 2, Supplementary Table 4). The CT threshold for T790M was derived previously [18]. The qPCR thresholds for each assay were estimated by maximizing the Youden’s J statistic [31], which simultaneously optimizes both sensitivity and specificity.
Analytical Evaluation of ExoDx EGFR
The assay was first evaluated by spiking double-stranded synthetic DNA constructs (custom order, Integrated DNA Technologies, Coralville, IA, USA) containing T790M, L858R or exon 19 deletion Δ746-750 (c.2235_2249) into healthy pooled plasma obtained from 10 males and 10 female donors (Bioreclamation IVT, NY, USA) to simulate a patient plasma sample. The reference construct copy number determination was done by OD260 measurement from the manufacturer, and before each experiment, we performed an orthogonal PCR verification of the amplifiable copies of the reference constructs using the QX200 Droplet Digital PCR (ddPCR) System (BioRad, CA, USA) and commercially available assays (dHsaCP2000019, dHsaCP2000021 and dHsaCP2000039) following the manufacturer’s recommendations. A summary of reaction composition as well as cycling conditions can be found in Supplementary Table 5.
First, we assessed equivalency with the previously validated assay [18] to ensure that the addition of the activating mutations (L858R and exon 19 indels) did not negatively impact the assay. To do this, we tested 28 data points (four biological replicates/measurement) around the previously defined Limit of Detection (LoD). Evaluation was done using the same final assay cut-offs, QC metrics, synthetic templates and plasma samples [18] (Table 2).
The analytical conditions (thresholds) for L858R and the exon 19 deletion Δ746-750 was determined with the synthetic constructs (mutation containing sequences) spiked into healthy pooled plasma samples. The levels of EGFR in the plasma used for these experiments were also quantified by ddPCR to calculate allelic frequencies for the spike-in experiments. Allelic frequencies as low as 0.03%, 0.04% and 0.05% (estimated from ddPCR experiments) were tested for T790M, L858R and exon 19 deletion, respectively (Table 2).
The copy numbers used for the spike-in experiments for all three targets are summarized in Table 2. Input material used for these experiments were simultaneously measured by ddPCR using the commercial assays described above. Measured ddPCR copies values were used for these spike-in experiments (Supplementary Figure 2).
Next, in addition to the spike-in into plasma exoNA described above, the assay robustness for these three mutations was also assessed on a genomic DNA (gDNA) admixture. Wild-type Promega genomic DNA (125 ng/μl (37,879 genomic equivalents) (Catalog no. G147A, Promega Corporation, WI, USA), was mixed at different ratios keeping an average of 30 copies of mutant-containing DNA constant (previously quantified by ddPCR as described above) (Figure 2 and Supplementary Tables 1 and 2). The wild type gDNA background was also measured using the EGFR exon 7 assay (Figure 2).
Evaluation of ExoDx EGFR on clinical samples
The accuracy of the assay was assessed on 110 clinical samples (Table 3), see Supplementary Table 6 for the clinical description. The NSCLC patient cohort consisted of 60 mutation-positive NSCLC patients (L858R, exon 19 indels and T790M) confirmed positive by tissue biopsy using institutionally approved methods and 50 mutation-negative NSCLC patients and healthy donor samples with no history of familial lung cancer. The median plasma input volume for the study was 2 mL (standard deviation of 0.5 mL).
The 60 mutation-positive NSCLC patients included in the study represented a variety of disease stages, with M0 (n = 5), M1a (n = 13), M1b (n = 39) and MX (n = 3). All mutation-positive patients had received prior treatment with at least one first generation TKI. The 50 negative samples included in the study were from NSCLC patients with confirmed-negative biopsy (n = 25) or healthy donors and pools with no history of cancer (N = 25). Samples were provided by Clovis Oncology, Inc. (Boulder, CO, USA), ALCMI (Wilmington, NC, USA), Memorial Cancer Institute (Hollywood, FL, USA), Althia Health, S.L (Barcelona, Spain) and Biopartners, Inc. (California, USA). Mutational status from all the NSCLC patient samples included in the study was independently assessed by institutionally-approved methods. All samples were collected under clinical study protocols approved for this purpose by their respective Institutional Review Boards (IRB).
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS
We thank all the treating physicians and oncologists for providing the patient samples and clinical information from the institutions mentioned in the manuscript.
Abbreviations
- cfDNA
cell free DNA
- ctDNA
circulating tumor DNA
- exoNA
exosomal nucleic acids (DNA and RNA) and cfDNA
- NSCLC
non-small cell lung cancer
- EGFR
epidermal growth factor receptor
- TKIs
tyrosine kinase inhibitors
- ddPCR
droplet digital PCR
- ARMS
Amplification refractory mutation system
- gDNA
genomic DNA
Author contributions
JKS, ECR, VT and DG conceived and designed the study. ECR and XZ conducted the experiments. VT and DG performed the statistical analysis. LR and CK provided some of the biological materials for the study. ECR wrote the manuscript. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST
E. Castellanos-Rizaldos, Xuan Zhang, D.G. Grimm, V. Tadigotla and J.K. Skog are employees of Exosome Diagnostics, Inc.
REFERENCES
- 1.Cancer Facts and Figures 2017 American Cancer Society. 2017. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2017/cancer-facts-and-figures-2017.pdf.
- 2.Lee CK, Kim S, Lee JS, Lee JE, Kim SM, Yang IS, Kim HR, Lee JH, Kim S, Cho BC. Next-generation sequencing reveals novel resistance mechanisms and molecular heterogeneity in EGFR-mutant non-small cell lung cancer with acquired resistance to EGFR-TKIs. Lung Cancer. 2017; 113:106–14. 10.1016/j.lungcan.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Schuette W, Schirmacher P, Eberhardt WE, Dietel M, Zirrgiebel U, Muehlenhoff L, Thomas M. Treatment decisions, clinical outcomes, and pharmacoeconomics in the treatment of patients with EGFR mutated stage III/IV NSCLC in Germany: an observational study. BMC Cancer. 2018; 18:135 10.1186/s12885-018-4032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H. Osimertinib making a breakthrough in lung cancer targeted therapy. OncoTargets Ther. 2016; 9:5489–93. 10.2147/OTT.S114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka K, Nosaki K, Otsubo K, Azuma K, Sakata S, Ouchi H, Morinaga R, Wataya H, Fujii A, Nakagaki N, Tsuruta N, Takeshita M, Iwama E, et al. Acquisition of the T790M resistance mutation during afatinib treatment in EGFR tyrosine kinase inhibitor-naïve patients with non-small cell lung cancer harboring EGFR mutations. Oncotarget. 2017; 8:68123–30. 10.18632/oncotarget.19243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong L, Cui S, Ding J, Sun Y, Zhang L, Zhao Y, Gu A, Chu T, Wang H, Zhong H, Ye X, Gu Y, Zhang X, et al. Dynamics of EGFR mutations in plasma recapitulates the clinical response to EGFR-TKIs in NSCLC patients. Oncotarget. 2017; 8:63846–56. 10.18632/oncotarget.19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arriola E, García Gómez R, Diz P, Majem M, Martínez Aguillo M, Valdivia J, Paredes A, Sánchez-Torres JM, Peralta Muñoz S, Barneto I, Gutierrez V, Andrade Santiago JM, Aparisi F, et al. Clinical management and outcome of patients with advanced NSCLC carrying EGFR mutations in Spain. BMC Cancer. 2018; 18:106 10.1186/s12885-018-4004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias M, Antunes A, Campainha S, Conde S, Barroso A. Prognostic impact of M descriptors of the 8th edition of TNM classification of lung cancer. J Thorac Dis. 2017; 9:685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faehling M, Schwenk B, Kramberg S, Eckert R, Volckmar AL, Stenzinger A, Sträter J. Oncogenic driver mutations, treatment, and EGFR-TKI resistance in a Caucasian population with non-small cell lung cancer: survival in clinical practice. Oncotarget. 2017; 8:77897–914. 10.18632/oncotarget.20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyun MH, Sung JS, Kang EJ, Choi YJ, Park KH, Shin SW, Lee SY, Kim YH. Quantification of circulating cell-free DNA to predict patient survival in non-small-cell lung cancer. Oncotarget. 2017; 8:94417–30. 10.18632/oncotarget.21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Xu Y, Zhong W, Zhao J, Chen M, Zhang L, Li L, Wang M. Total DNA input is a crucial determinant of the sensitivity of plasma cell-free DNA EGFR mutation detection using droplet digital PCR. Oncotarget. 2017; 8:5861–73. 10.18632/oncotarget.14390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett CW, Berchem G, Kim YJ, El-Khoury V. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget. 2016; 7:71013–35. 10.18632/oncotarget.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ai B, Liu H, Huang Y, Peng P. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget. 2016; 7:44583–95. 10.18632/oncotarget.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlovich C, Goldman JW, Sun JM, Mann E, Sequist LV, Konopa K, Wen W, Angenendt P, Horn L, Spigel D, Soria JC, Solomon B, Camidge DR, et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin Cancer Res. 2016; 22:2386–95. 10.1158/1078-0432.CCR-15-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, Hammett T, Cantarini M, Barrett JC. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015; 90:509–15. 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, Brinkmann K, Emenegger J, Grimm DG, Castellanos-Rizaldos E, Goldman JW, Sequist LV, Soria JC, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018; 29:700–06. 10.1093/annonc/mdx765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Möhrmann L, Huang HJ, Hong DS, Tsimberidou AM, Fu S, Piha-Paul SA, Subbiah V, Karp DD, Naing A, Krug A, Enderle D, Priewasser T, Noerholm M, et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin Cancer Res. 2018; 24:181–88. 10.1158/1078-0432.CCR-17-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellanos-Rizaldos E, Grimm DG, Tadigotla V, Hurley J, Healy J, Neal PL, Sher M, Venkatesan R, Karlovich C, Raponi M, Krug A, Noerholm M, Tannous J, et al. Exosome-Based Detection of EGFR T790M in Plasma from Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2018; 24:2944–50. 10.1158/1078-0432.CCR-17-3369. [DOI] [PubMed] [Google Scholar]
- 19.Forootan A, Sjöback R, Björkman J, Sjögreen B, Linz L, Kubista M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR). Biomol Detect Quantif. 2017; 12:1–6. 10.1016/j.bdq.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009; 55:611–22. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 21.González-Larriba JL, Lázaro-Quintela M, Cobo M, Dómine M, Majem M, García-Campelo R. Clinical management of epidermal growth factor receptor mutation-positive non-small cell lung cancer patients after progression on previous epidermal growth factor receptor tyrosine kinase inhibitors: the necessity of repeated molecular analysis. Transl Lung Cancer Res. 2017. (Suppl 1); 6:S21–34. 10.21037/tlcr.2017.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol. 2018. (suppl_1); 29:i10–19. 10.1093/annonc/mdx703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ZF, Ren SX, Li W, Gao GH. Frequency of the acquired resistant mutation T790 M in non-small cell lung cancer patients with active exon 19Del and exon 21 L858R: a systematic review and meta-analysis. BMC Cancer. 2018; 18:148 10.1186/s12885-018-4075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, Lee CK, Sebastian M, Templeton A, et al. , and AURA3 Investigators . Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. 2017; 376:629–40. 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilie M, Hofman V, Leroy S, Cohen C, Heeke S, Cattet F, Bence C, Lalvée S, Mouroux J, Marquette CH, Hofman P, and STALKLUNG01 and AIR Study Consortium Investigators . Use of circulating tumor cells in prospective clinical trials for NSCLC patients - standardization of the pre-analytical conditions. Clin Chem Lab Med. 2018; 56:980–89. 10.1515/cclm-2017-0764. [DOI] [PubMed] [Google Scholar]
- 26.Veldore VH, Choughule A, Routhu T, Mandloi N, Noronha V, Joshi A, Dutt A, Gupta R, Vedam R, Prabhash K. Validation of liquid biopsy: plasma cell-free DNA testing in clinical management of advanced non-small cell lung cancer. Lung Cancer (Auckl). 2018; 9:1–11. 10.2147/LCTT.S147841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014; 6:224ra24 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, Mach SL, Jänne PA, Oxnard GR. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016; 2:1014–22. 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008; 10:1470–76. 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM, Lader E, Skog J, Noerholm M. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One. 2015; 10:e0136133 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youden WJ. Index for rating diagnostic tests. Cancer. 1950; 3:32–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.