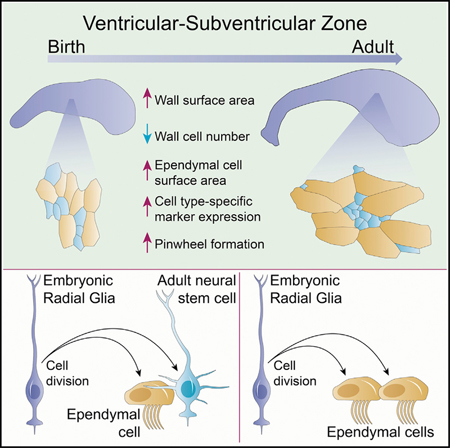
SUMMARY
The adult mouse brain contains an extensive neurogenic niche in the lateral walls of the lateral ventricles. This epithelium, which has a unique pinwheel organization, contains multiciliated ependymal (E1) cells and neural stem cells (B1). This postnatal germinal epithelium develops from the embryonic ventricular zone, but the lineage relationship between E1 and B1 cells remains unknown. Distinct subpopulations of radial glia (RG) cells in late embryonic and early postnatal development either expand their apical domain >11-fold to form E1 cells or retain small apical domains that coalesce into the centers of pin-wheels to form B1 cells. Using independent methods of lineage tracing, we show that individual RG cells can give rise to clones containing E1 and B1 cells. This study reveals key developmental steps in the formation of the postnatal germinal niche and the shared cellular origin of E1 and B1 cells.
In Brief
Redmond et al. examine the transformation of the embryonic ventricular zone into the largest germinal niche of the adult rodent forebrain. They trace the origin of ependymal and neural stem cells to a common embryonic progenitor and follow the postnatal apical maturation that results in this niche’s characteristic pinwheels.
Graphical abstract
INTRODUCTION
Ependymal cells (E1 cells) are postmitotic epithelial cells in the walls of the brain ventricles (Spassky et al., 2005). A key function of E1 cells is the propulsion of cerebrospinal fluid (CSF) along the ventricular system through the coordinated beating of their cilia (Sawamoto et al., 2006; Del Bigio, 2010). E1 cells are part of the neurogenic niche in the adult ventricular-subventricular zone (V-SVZ) of the lateral ventricles. This germinal ventricular wall contains adult neural stem cells (NSCs); a subpopulation of glial fibrillary acidic protein (GFAP)-expressing astrocytes known as B1 cells that function as primary progenitors of neurons and oligodendrocytes (Doetsch et al., 1999; Menn et al., 2006; Mirzadeh et al., 2008; Ortega et al., 2013; Tong et al., 2015). From their birth in the embryo until the time when they produce olfactory bulb (OB) interneurons later in life, B1 cells remain largely quiescent (Fuentealba et al., 2015; Furutachi et al., 2015). The apical domains of B1 cells share the surface of the ventricular wall with E1 cells. Both cell types are organized into polarized structures called pin-wheels, where the small apical endings of B1 cells form the center of pinwheels and are surrounded by a rosette of ependymal cells with large apical surfaces (Mirzadeh et al., 2008; Kokovay et al., 2012; Hu et al., 2017). This organization is unique to regions of the ventricular walls where neurogenesis continues throughout adulthood.
In the embryonic brain, the walls of the lateral ventricles are composed of a pseudostratified ventricular zone (VZ). This epithelium transforms into a mixed epithelium in the adult that in mouse includes the cuboidal E1 cells and pseudostratified B1 cells. It has been proposed that the adult V-SVZ niche forms by the progressive transformation of the embryonic VZ (Tramontin et al., 2003), but the cellular dynamics underlying this transformation are poorly understood. It has been shown that radial glial (RG) cells, the NSCs during embryogenesis, give rise to B1 cells and to multiciliated E1 cells (Merkle et al., 2004, 2007; Spassky et al., 2005). However, it remains unknown whether single RG cells have the potential to give rise to both B1 and E1 cells or whether distinct RG populations generate exclusively B1 or E1 cells. Recent findings suggest that mouse RG cells that divide around embryonic day 15.5 (E15.5) generate B1 cells; a subpopulation of these NSCs divide symmetrically to self-renew in postnatal life (Fuentealba et al., 2015; Obernier et al., 2018). Interestingly, the lineages for postnatal and embryonic NSCs also separate around the same time (E15.5) (Fuentealba et al., 2015). It is intriguing that the time when RG cells divide to become prospective postnatal B1 cells closely matches the time when E1 cells are born (Spassky et al., 2005). However, the lineage relationship between E1 and B1 cells remains unknown.
Here, we investigated how the lateral wall of the lateral ventricles transitions from an embryonic VZ into the adult V-SVZ, how E1 and B1 cells acquire their unique apical domains and marker expression, and how these cells become organized into pinwheels. Given the similar times of birth of B1 and E1 cells in the embryo, we also ask whether E1 and B1 are generated from single RG progenitors. This work reveals the shared cellular origins and maturation of E1 and B1 cells, two cells with key functions for brain homeostasis and persistent neurogenesis.
RESULTS
Postnatal Growth of the Lateral Walls Occurs Despite Net Loss of Epithelial Cells
How the epithelial cells of the brain lateral ventricles contribute to the postnatal growth of its walls remains unknown. The principal cell populations that inhabit the lateral wall of the lateral ventricle, E1 and B1 cells, are postmitotic (E1 cells) or largely quiescent (B1 cells) after they are specified in mid-fetal development (Spassky et al., 2005; Fuentealba et al., 2015; Furutachi et al., 2015). We therefore hypothesized that the apical domain of VZ cells must grow to explain the expansion of this wall.
We measured the total area of the lateral wall of the lateral ventricle from embryonic and postnatal stages by tracing whole mounts immunostained for β-catenin, which labels the lateral membranes of ventricular epithelial cells (Figure 1A). The area containing brightly stained β-catenin+ apical surfaces was considered the lateral wall and was distinct from the surrounding tissue that was largely β-catenin-negative (Figures 1B and 1C). We found that the surface area of the lateral wall of the lateral ventricle grew from an average of 3.19 mm2 (n = 3; SD, 0.50 mm2) to 5.97 mm2 (n = 3; SD, 0.50 mm2) from E17.5 to postnatal day 15 (P15) (p = 0.000016, one-way ANOVA with Tukey honest significant difference [HSD] correction for multiple comparisons; see Table 1 for statistics details) but did not significantly change in size between P15 and P40 (n = 4; mean, 6.58 mm2; SD, 0.40 mm2; p = 0.51). Because of the inherent variability in microscope-assisted dissections of whole mounts, we also prepared serial frontal sections of mouse brains and reconstructed the lateral wall from cross-sections (Figures 1D and 1E). A similar growth pattern was observed by this serial section analysis (Figure 1E).
Figure 1. Development of the Lateral Wall of the Lateral Ventricle.
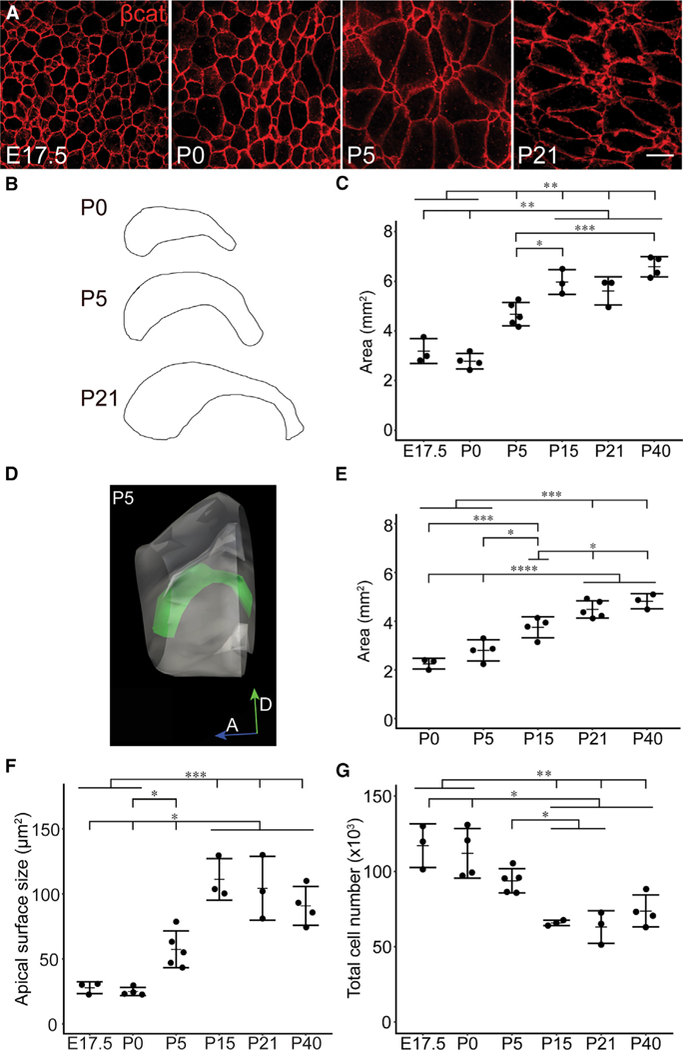
(A) Confocal images taken at the surface of whole mounts of the lateral wall of the lateral ventricle at different ages. In red, β-catenin delineates the apical profiles of ventricular epithelial cells.
(B) Camera lucida drawings of whole mounts of the lateral ventricular wall illustrate the increases in the size of the lateral wall over postnatal development.
(C) Area of the lateral wall of the lateral ventricle, measured from whole-mount preparations collected at different ages of development.
(D) Three-dimensional reconstruction of a P5 lateral wall measured from serial frontal crosssections.
(E) Lateral wall areas measured by 3D reconstruction of frontal sections at progressive stages of development.
(F) Mean apical surface sizes of ventricular epithelial cells.
(G) Number of ventricular epithelial cells per lateral wall at different ages.
Each point represents one quantified whole mount from one brain hemisphere, plotted with mean (dash) and SD (error bars). Between 237 and 538 cells per whole mount were analyzed across 200 sampling sites in (F) and (G) (totaling 939–2,004 cells/age; see STAR Methods). One-way ANOVA was used in (C) and (E)-(G) with Tukey HSD correction for multiple comparison, where p < 0.05 was considered significant (see Table 1 for all p values and summary of statistical tests). Horizontal lines above graphs indicate unidirectional significant differences between groups; groups located under a tick plus horizontal bar are significantly different than the groups indicated by a tick mark, with a p value < 0.05 (*), 0.01 (**), 0.001 (***),or0.0001 (****).If groupsare connected by only tick marks (no horizontal bars in the comparison), the significance relationship is bidirectional. Example: in (C), E17.5 or P0 are significantly different than P5, P15, P21, and P40, with p < 0.01, but P21 is not necessarily significantly different than P40. Scale bar: 10 μm.
Table 1.
Summary of Statistics Applied to Data Presented in Figure 1
| One-Way ANOVA | Tukey HSD Adjusted p Values, 95% Confidence Interval | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Figure | Experiment | Degrees of Freedom, Between |
Degrees of Freedom, Within |
F Statistic | p Value | Age | E17.5 | P0 | P5 | P15 | P21 | P40 |
| 1C | traced whole- | 5 | 16 | 41.59 | 1.30E-08* | E17.5 | ||||||
| mount area | P0 | 0.8462807 | ||||||||||
| P5 | 0.0042380* | 0.0001550* | ||||||||||
| P15 | 0.0000161* | 0.0000011* | 0.0132359* | |||||||||
| P21 | 0.0000840* | 0.0000055* | 0.1021914 | 0.9248499 | ||||||||
| P40 | 0.0000005* | 0.0000000* | 0.0001395* | 0.5127735 | 0.1110083 | |||||||
| 1E | serial | 4 | 14 | 29.80 | 1.03E-06* | P0 | NA | |||||
| reconstruction | P5 | NA | 0.3403408 | |||||||||
| area | P15 | NA | 0.0009096* | 0.0198807* | ||||||||
| P21 | NA | 0.0000081* | 0.0000746* | 0.0657414 | ||||||||
| P40 | NA | 0.0000059* | 0.0000430* | 0.0146639* | 0.7350948 | |||||||
| 1F | average apical | 5 | 16 | 24.02 | 6.54E-07* | E17.5 | ||||||
| surface size | P0 | 0.9997739 | ||||||||||
| P5 | 0.1017804 | 0.0360687* | ||||||||||
| P15 | 0.0000287* | 0.0000081* | 0.0011026* | |||||||||
| P21 | 0.0000804* | 0.0000229* | 0.0040271* | 0.9903036 | ||||||||
| P40 | 0.0003412* | 0.0000859* | 0.0299407* | 0.4516533 | 0.8103080 | |||||||
| 1G | total cell | 5 | 16 | 14.10 | 2.21E-05* | E17.5 | ||||||
| number | P0 | 0.9904301 | ||||||||||
| P5 | 0.1078104 | 0.2191231 | ||||||||||
| P15 | 0.0005639* | 0.0008395* | 0.0385908* | |||||||||
| P21 | 0.0003220* | 0.0004581* | 0.0202644* | 0.9996279 | ||||||||
| P40 | 0.0015663* | 0.0024889* | 0.1484919 | 0.9385329 | 0.8183208 | |||||||
Asterisk (*) indicates p < 0.05.
We next measured the apical surface areas of cells delineated by β-catenin staining (Figure 1F). We found that the average apical surface area of ventricular epithelial cells increased significantly between E17.5 (27.84 μm2; SD, 4.51 μm2; n = 3) and P15 (110.26 μm2; SD, 16.00 μm2; n = 3; 3.96-fold change; p = 0.000029) (Figure 1F).
Next, we determined how the number of apical profiles in the lateral wall changes over time using systematic random sampling-based stereology to estimate the total population of cells (Figure 1G). We found that the lateral wall had an average of 117,148 cells at E17.5 (n = 3; SD, 14,535.15). Interestingly, this number began to decrease directly after birth and declined by nearly 44% at P15 (65,879 cells; SD, 1,804.51; n = 3; p = 0.00056) (Figure 1G). This loss of cells in the walls of the lateral ventricles is likely due to the symmetric consumption of B1 cells as they produce neurons destined for the OB (Obernier et al., 2018). The growth of the lateral wall during this time of cell loss is explained by the nearly 4-fold increase in the average apical surface size of ventricular epithelial cells (E17.5 to P15; see above) (Figure 1F). E1 cells have very large apical domain sizes compared to B1 cells (Mirzadeh et al., 2008; Figure 2A; see below), and the population of B1 cells declines after birth (Obernier et al., 2018). Thus, we infer that the postnatal growth of the ventricles is overwhelmingly mediated by the expansion of E1 cells.
Figure 2. Development of Marker Expression in Ventricular Epithelial Cells within Four Quadrants of the Lateral Wall ofthe Lateral Ventricle.
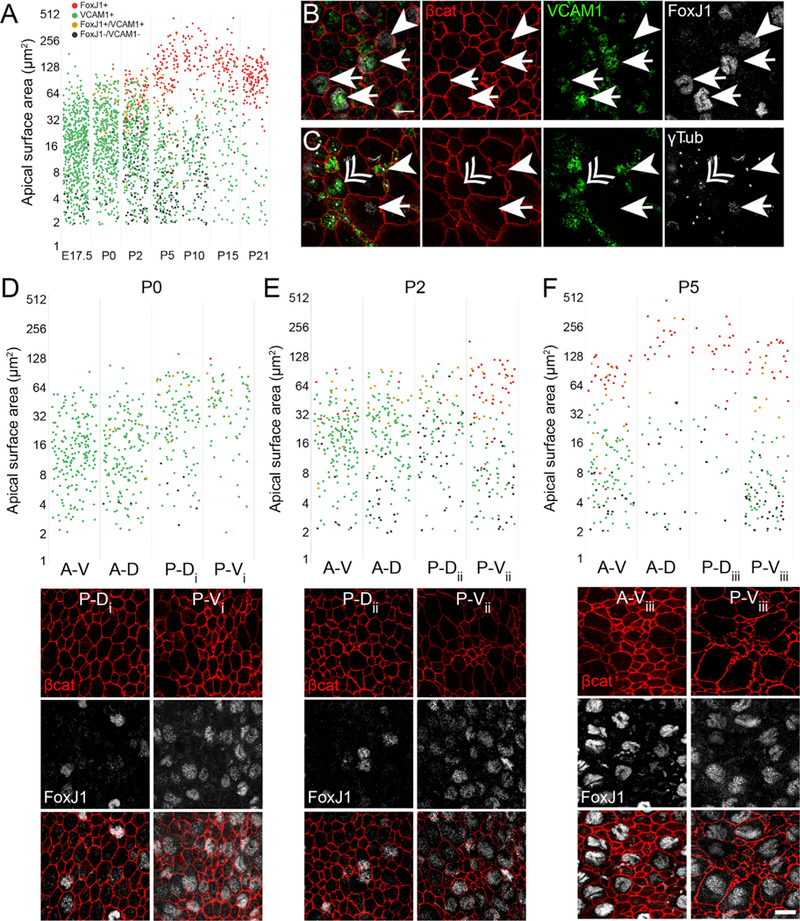
(A) Quantification of the apical domain areas from all four quadrants during development. Cells expressing the marker proteins FoxJ1 (E1 cells) and VCAM1 (adult B1 cells) are color-coded as follows: FoxJ1+/VCAM1−, red; FoxJ1−/VCAM1+, green; FoxJ1+/VCAM1+, yellow; and FoxJ1− /VCAM1−, black. Note that the apical surface sizes of FoxJ1+/VCAM1− cells segregate from FoxJ1−/VCAM1+ cells within the first postnatal week.
(B) Lateral-wall whole mounts from P2 mice immunostained for β-catenin (red), VCAM1 (green), and FoxJ1 (white) show apical domains of cells that are VCAM1+/FoxJ1+ (arrows) or VCAM1-/FoxJ1+ (arrowheads).
(C) Lateral-wall whole mounts from P2 mice immunostained for β-catenin (red), VCAM1 (green), and gamma-tubulin (white) show apical domains of cells that are VCAM1+ with one basal body (arrowhead), deuterosomes (double arrowhead), and VCAM1− with multiple basal bodies (arrow).
(D–F) Quantification of apical domain sizes of by region at progressive perinatal ages (D, P0; E, P2; and F, P5), color-coded by marker expression (see A). Below each graph are examples of images taken from the whole-mount quadrants indicated by the subscripts i-iii. Numbers of cells plotted are as follows: (A) 888 (E17.5), 572 (P0), 562 (P0), 346 (P5), 227 (P10), 153 (P15), and 171 (P21).
(D) 173 (A-D), 213 (A-V), 115 (P-D), and 71 (P-V).
(E) 162 (A-D), 173 (A-V), 102 (P-D), and 125 (P-V).
(F) 51 (A-D), 136 (A-V), 37 (P-D), and 122 (P-V).
A-D, anterior-dorsal; A-V, anterior-ventral; P-D, posterior-dorsal; P-V, posterior-ventral. See Mirzadeh et al. (2008). Scale bars: 10 μm.
Maturation of E1 and B1 Cells and the Formation of Pinwheels
We next analyzed how apical domain size correlated with the expression of E1 and B1 cell-specific markers over perinatal and postnatal development (Figure 2). The apical domain of mature B1 cells has been shown to be positive for vascular cell adhesion molecule-1 (VCAM1) (Kokovay et al., 2012) and was also recently described in a subset of embryonic RG cells (Hu et al., 2017). In contrast, mature E1 cells do not express VCAM1 but are positive for the forkhead transcription factor J1 (FoxJ1), which is also required for ependymal cell differentiation (Jacquet et al., 2009).
Whole mounts were dissected and immunostained for cell-type-specific proteins and β-catenin to analyze the molecular differentiation of ventricular epithelial cells over time. We analyzed the molecular marker expression and apical surface sizes of 153–888 ventricular epithelial cells per time point (Figure 2A). In agreement with previous findings, we found that VCAM1 was expressed in the apical domain of many embryonic VZ cells (Figure 2A; Hu et al., 2017). During postnatal development, VCAM1 expression decreased in cells that continued to expand their apical domains (Figures 2A–2C). By P5, the majority of cells with large apical domains were negative for VCAM1, while cells that maintained VCAM1 expression from P5 onward had smaller apical domains (Figure 2A). These results suggest that RG cells that become either E1 or B1 cells express VCAM1 around the time of birth, but E1 cells rapidly downregulate the expression of VCAM1 while B1 cells maintain VCAM1 expression.
Given the expression pattern of VCAM1 in the majority of RG cells shortly before birth and the lack of VCAM1 protein in mature E1 cells, we wanted to understand the relationship between RG VCAM1 expression and ependymal cell differentiation. To follow RG transformation into E1 cells, we determined the expression pattern of the E1 cell marker FoxJ1 in cells with increasing apical domain size (Figures 2A and 2D-2F). We subdivided the lateral wall into four quadrants based on anterior-posterior and dorsal-ventral axes (Mirzadeh et al., 2008) to determine whether there was a spatiotemporal pattern of E1 cell differentiation. We did not detect FoxJ1 expression in whole-mount preparations of the lateral wall before E17.5 (data not shown). At E17.5, we found FoxJ1+ cells in the lateral wall, consistent with the previously reported wave of ependymal cell differentiation (Spassky et al., 2005) (Figure 2A). By P0, the density of FoxJ1+ cells in the P-V region had increased and FoxJ1+ cells were also detected in the posterior-dorsal (P-D) and anterior-dorsal (A-D) regions (Figure 2D). At P2, FoxJ1+ cells were detected in all regions of the lateral wall (Figure 2E). At P5, the expression of FoxJ1 was clearly correlated with larger apical domains, consistent with E1 cell differentiation (Figure 2F). Interestingly, at P0, P2, and P5, we detected many FoxJ1+ cells that expressed VCAM1 (Figures 2A, 2B, and 2D-2F). Moreover, we observed VCAM1+ cells that contained γ-tubulin+ deuterosomes, organelles associated with the formation of basal bodies in multiciliated cells and the differentiation of E1 cells (Figure 2C) (Spassky et al., 2005; Klos Dehring et al., 2013; Al Jord et al., 2014). This suggests that El-producing RG progressively acquire E1 identity while VCAM1 continues to be present. However, this transitional stage of differentiation was confined to a short period of time. Double-labeled VCAM1+/FoxJ1+ cells were rare in the lateral wall after P5 (Figure 2A).
As ventricular epithelial cells structurally and molecularly differentiated, we also observed the emergence of pinwheel structures. Beginning at P5, pinwheels could be observed in the P-V region of the lateral wall but were absent from the other three regions (Figure 3A). Within the pinwheels, the majority of cells with smaller apical domains expressed VCAM1, while the cells with larger apical domains expressed FoxJ1. At P7, well-defined pinwheels could also be seen in the P-D V-SVZ (Figure 3B, top row). We did not observe clear pinwheel structures in the anterior-dorsal (A-D) or anterior-ventral (A-V) regions at P7, although FoxJ1+ cells with large apical domains and numerous VCAM1+ cells with small apical domains were present (Figure 3B, bottom row). After P10, pinwheels became evident in A-V and A-D regions (Figure 3C).
Figure 3. Emergence of Pinwheels during Early Postnatal Development.
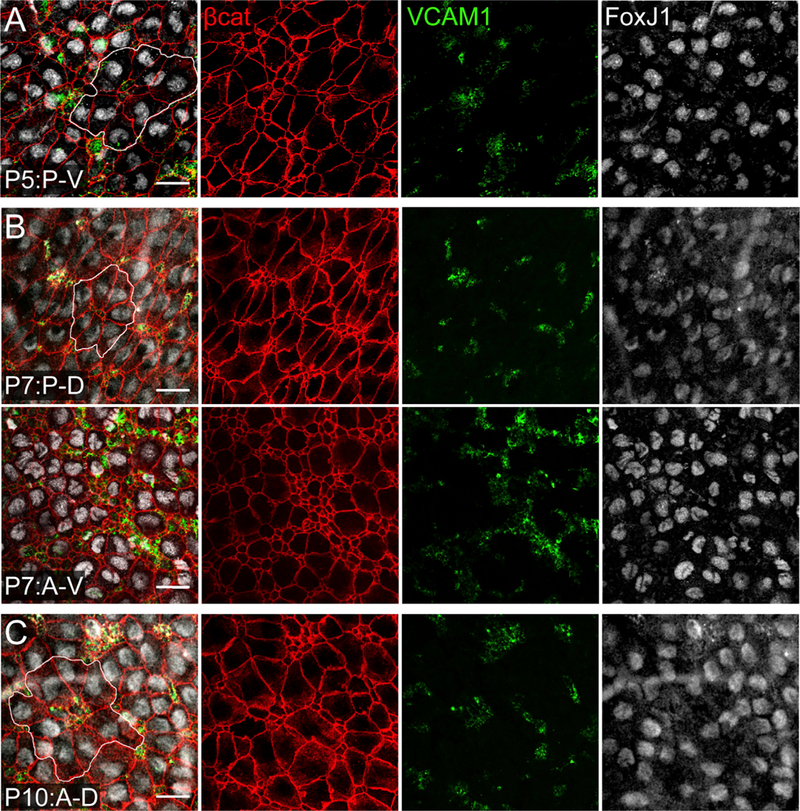
Confocal images were taken at the surface of whole mounts of the lateral wall of the LV.
(A) At P5, pinwheels could be observed in the P-V region of the lateral wall (white outline in merged image). Cells with a smaller apical domain expressed VCAM1, while the cells with a larger apical domains expressed FoxJ1.
(B) At P7, pinwheels structures could be identified in dorsal (P-D) regions of the V-SVZ (white outline in merged image; top row). Clear pinwheel structures were not observed in the A-V region (bottom row) or A-D region (data not shown).
(C) By P10, pinwheels had also emerged in the anterior regions of the lateral wall: A-D, white outline in merged image, top row; A-V, data not shown. Scale bars: 20 μm.
Taken together, these data suggest that the formation of pinwheels is associated with the enlargement of RG apical domains as they differentiate into E1 cells. During RG differentiation, we found that VCAM1 was transiently expressed in nearly all V-SVZ cells while apical domains expanded (Figure 2). VCAM1 expression was maintained as cells became B1 cells (Figures 2D–2F), but was downregulated as FoxJ1 expression increased in young E1 cells that continued their apical expansion (Figures 2A and 2D–2F).
Do Ependymal Cells and Postnatal NSCs Share a Common Origin?
Embryonic VZ RG cells give rise to both B1 and E1 cells (Merkle et al., 2004, 2007; Spassky et al., 2005). Intriguingly, the time of birth of E1 cells closely matches the time when RG cells give rise to B1 cells (Spassky et al., 2005; Fuentealba et al., 2015; Furutachi et al., 2015). These observations suggest that B1 and E1 cells could share a common cell of origin in the embryonic VZ.
To study the lineage of single RG cells, we have previously used a library of retroviruses (QmGFP-OL), each encoding a unique 24-base pair DNAsequence, or”barcode” (Cepkoetal., 1995,1998; Golden et al., 1995; Fuentealba et al., 2015). We analyzed the barcode data from this study and found one E1 cell that shared the same barcode as a group 3 granule cells in the OB (clone Q056R-C3) from an animal injected with the retroviral library at E13.5. Because E1 cells do not divide in the adult brain (Spassky et al., 2005), we infer that the adult E1 cell was clonally related to a B1 cell that generated the OB neurons. We therefore laser-microdissected and analyzed additional OB neurons and ependymal cells from P34 (n = 5) or P56 (n = 5) mice that received intraventricular QmGFP-OL injections at E12.5, E13.5, or E15.5, and found five additional E1 cells that shared a barcode with OB neurons (for experimental details, see Fuentealba et al., 2015). These observations suggested that E1 cells are lineage related to B1 cells, which generate OB interneurons postnatally. However, since it is not possible to laser-microdissect neighboring E1 and B1 cells separately in the walls of lateral ventricles, with this lineage- tracing method we could not directly observe possible E1-B1 cells clones in the V-SVZ.
A Subpopulation of RG Gives Rise to E1 Cells and B1 Cells
In order to directly observe clonal relationships among cells in the ventricular epithelium of the adult brain, we used the UbC-StarTrack lineage-tracing method (Figure 4) (Figueres-Oñate et al., 2016). This clonal analysis is based on the electroporation of 12 transposon constructs (encoding six fluorophores with either nuclear or cytoplasmic localization), as well as plasmids encoding hyperactive piggyBac transposase (hyPBase) and tamoxifen-inducible Cre recombinase (CreERT2). Following hyPBase-mediated transposition, fluorophores are randomly integrated into the cell genome. Tamoxifen (Tmx) induces Cre-mediated recombination at loxP sites of non-integrated constructs, removes terminal repeat sequences required for genomic integration, and halts additional transpositions (Figure 4A). This results in the continuous expression of a heritable color code with very high complexity (16 × 106 possible combinations). We electroporated VZ cells at E14.5, injected Tmx at P1 to halt further integration events, and analyzed whole mounts at P60 (Figure 4B). UbC-StarTrack-labeled cells were present along the entire surface of the whole mount (Figure 4C). Clonal relationships among labeled cells on the lateral wall of the lateral ventricle were determined by analyzing the combination of the six different fluorescent markers (analyzed in separate fluorescent channels) for each individual labeled cell. Thus, specific color codes were generated by the presence or absence of the six fluorophores in the cytoplasm and/or nucleus of each cell (Figures 4C–4I). B1 cells were identified by their morphology and E1 cell identity was confirmed by FoxJ1 staining (Figures 4D–4F, 4H, and 4I, asterisks). Consistent with the above results, we found 8 E1-B1 cell pairs (Figures 4D–4F and 4I [lower clone]), 20 pairs of E1-E1 cells (Figures 4H and 4I [upper clone]), and 125 single E1 cells (Figure 4J). We also observed six other StarTrack clones containing more than two cells, indicating additional divisions (data not shown): three consisted of E1 cells only, and three had B1 and E1 cells.
Figure 4. StarTrack LineageTracing of Embryonic Radial Glia at E14.5.
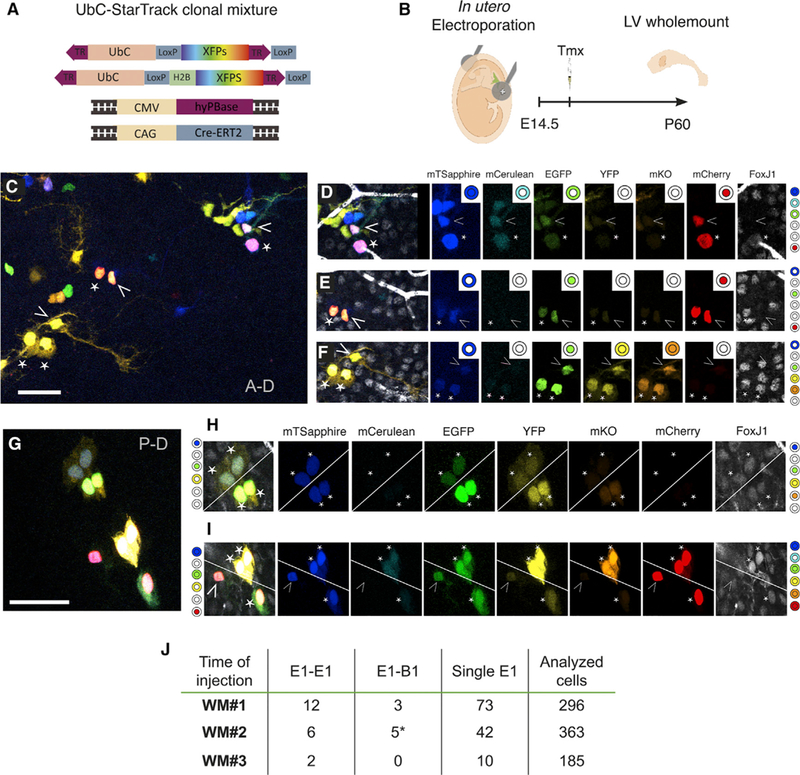
(A) UbC-StarTrack clonal mixture contains six transposable fluorescent reporters (nuclear or cytoplasmic mTsapphire, mCerulean, EGFP, YFP, mKO, and mCherry), as well as hyPBase- and Cre-ERT2-encoding plasmids, all under ubiquitous promoters.
(B) In utero electroporations with the UbC-StarTrack clonal mixture at E14.5. Whole mounts were dissected at P60 and immunostained for FoxJ1.
(C) Whole mount of the lateral wall of the lateral ventricle with multiple clones of labeled cells.
(D–F) Three different clones of E1 and B1 cells from (C) expressing five (D), three (E), or six (F) fluorescent reporters. Clonal relationships were assessed by identifying the unique combination of fluorescent reporters expressed in cell nuclei orcytoplasm in each fluorescent channel. Clone color codes are illustrated to the right as filled or unfilled concentric circles: the center circle denotes nuclear localization, and the outer ring denotes cytoplasmic localization.
(G) Four clones, three made up of E1-E1 pairs, and a fourth with an E1 and B1 cell.
(H) Two E1-E1 clonal pairs are separated by the diagonal white line (left color code, upper clone; right color code, lower clone).
(I) Two nearby clones are separated by a diagonal line. The upper clone is E1-E1 (right color code), and the lower clone is made up ofan E1 cell with a B1 cell (left color code).
(J) Numbers of E1-E1, E1-B1, and single E1 clones identified by analyzing the color codes of 844 cells in three whole mounts injected and electroporated at E14.5. The asterisk in the E1-B1 column indicates that one clone had two E1 cells and one B1 cell (shown in F). Arrowheads correspond to B1 cells, and the other asterisks correspond to FoxJ1+ E1 cells. Scale bars: 50 μm.
The barcode and StarTrack lineage-tracing experiments suggests that B1 cells and E1 cells can originate from a common progenitor in the embryonic forebrain. However, given the large number of fluorophores needed for the StarTrack approach, we could not use additional markers for cell identification or to demarcate apical cell borders. We therefore used a third approach for lineage tracing: Nestin::CreER;Ai14 pregnant female mice were given a single low dose of Tmx to induce labeling at clonal density (at E11.5, E14.5, or E17.5; Figure 5A) (Bonaguidi et al., 2011; DeCarolis et al., 2013; Obernier et al., 2018). Because it is possible that two cells next to each other could have undergone Cre recombination independently, or that active Cre recombinase protein in one cell was transferred to a neighbor (Zomer et al., 2015), we also administered two pulses of BrdU: the first, immediately, and the second, 6 h after Tmx administration (Figure 5A; Obernier et al., 2018). Whole mounts of the V-SVZ were analyzed at P10 (Figure 5A). E1 and B1 cells were identified based on morphology; β-catenin staining was used to identify cell borders and to confirm E1 or B1 cell identity based on the size of their apical domain (Figure 5B). We quantified the number of tdT+/BrdU+ pairs in whole mounts at P10 and reasoned that pairs of double-labeled cells must be derived from a recombined progenitor cell that divided soon after bromodeoxyuridine (BrdU) administration (Figure 5C). Tdt+/BrdU+ E1-E1 cell pairs were found in animals injected at E11.5 and E14.5 (E1-E1 pairs: 2 at E11.5; 1 at E14.5; 0 at E17.5; n = 5 whole mounts per age). Interestingly, we also found 6 tdT+/BrdU+ E1-B1 cell pairs (E1-B1 pairs: 2 at E11.5; 4 at E14.5; 0 at E17.5). We also found 32 single tdT+/BrdU+ E1 cells. We cannot exclude that, in the intervening time between the initial labeling and when whole mounts were analyzed, additional members of these clones were lost by cell death, migration, or consumption. This double-labeling experiment further supports the hypothesis that a subpopulation of RGs that divide in the embryo can give rise to pairs of E1-E1 and B1-E1 cells. The number of tdT+/BrdU+ E1-B1 clones was higher at E14.5 compared to E17.5, indicating that the majority of these E1-B1 progenitors divide predominantly around E14.5 (Figure 5C). Interestingly, we also found single tdT+B1 cells (data not shown), with no other labeled cell in their proximity. These cells are likely derived from RGs that divided in the embryo, but one of the two daughter cells may have died or, in the case of a B1 cell, could have been consumed by the generation of OB interneurons or other glia (Calzolari et al., 2015; Obernier et al., 2018).
Figure 5. Clonally Related V-SVZ Cells Derived from Nestin+ RG.
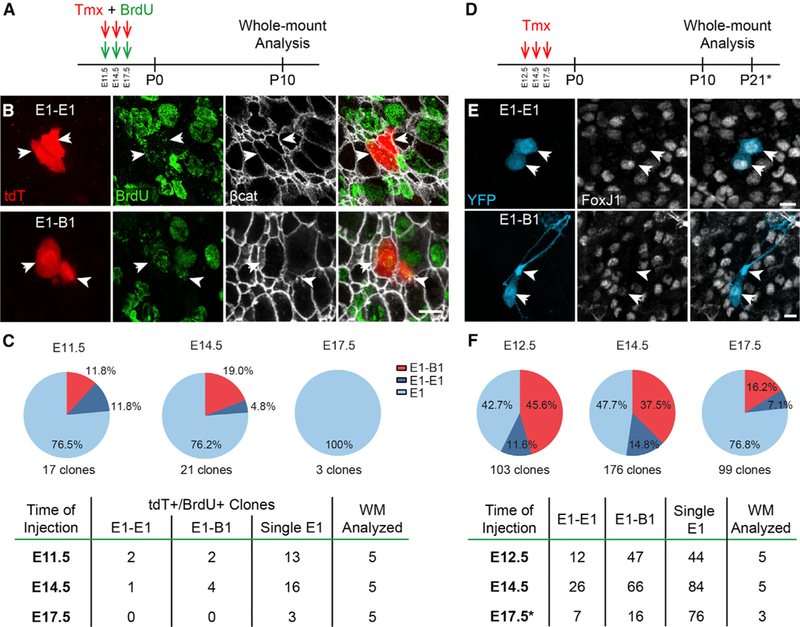
(A) Low doses of tamoxifen were given to Nestin::CreER;Ai14 timed-pregnant female mice at E11.5, E14.5, or E17.5. BrdU was injected simultaneously and 6 h later. Whole mounts were analyzed at P10 for double-labeled tdT+/BrdU+ cells.
(B) Confocal images of an isolated tdT+/BrdU+ clone of E1 cells (arrows, top row) and a clone containing a B1 cell (arrowhead, bottom row) and an E1 cell (arrow, bottom row).
(C) Quantification of tdT+/BrdU+ clones observed at different ages of injection, from a total of 5 analyzed whole mounts (WMs) per age.
(D) Sixty-fold higher doses of Tmx were administered to Nestin::CreER;Confetti timed-pregnant female mice at E12.5, E14.5, and E17.5. Whole mounts were analyzed at P10 (E12.5, E14.5) or P21 (E17.5) for single-fluorophore-labeled pairs of cells that included E1 cells (arrows) and/or B1 cells (arrowheads).
(E) Confocal images of isolated single-fluorophore-expressing clones (YFP shown here) of E1 cells (arrows, top row) and of a B1 cell (arrowhead) with an E1 cell (arrow, bottom row).
(F) Quantification of clones observed using the Nestin::CreER;Confetti mice at different ages of injection, from a total of 3–5 WMs analyzed per age.
Reporter-based lineage-tracing techniques necessitate sparse labeling of cell populations to ensure the correct identification of single clones. However, sparse labeling may not reveal rare events. Our UbC-StarTrack experiments overcome the problem of clonal density with high-complexity fluorophore combinational codes, but electroporation does not allow for cell type specificity of reporter expression. To test whether our findings using sparse labeling in Nestin::CreER;Ai14 were representative of Nestin+ RG clones, we used Nestin::CreER;Confetti mice (Figures 5D–5F). The Confetti reporter allows for four possible recombination events, leading to expression of different reporters: nuclear GFP, membrane-bound CFP, cytoplasmic YFP, or cytoplasmic RFP (Snippert et al., 2010).
A cluster of cells labeled by one of the four reporters is likely derived from a single progenitor that underwent one of the four Cre-induced recombination events. We injected a single dose of Tmx into timed-pregnant Nestin::CreER;Confetti mice (at E12.5, E14.5, or E17.5) to induce recombination in RG cells (Figure 5D). The amount of Tmx was 60-fold higher than that administered to Nestin::CreER;Ai14 mice (100 mg/kg versus 1.67 mg/kg), resulting in a greater number of recombination events per whole mount; the presence of multiple reporters allowed us to resolve single clones. At P10 or P21, the presence of E1-E1 and/or E1-B1 cell pairs expressing one of the four fluorescent proteins was analyzed in whole mounts of the lateral wall (Figures 5E and 5F). Similar to our previous results (Figures 5A–5C), we found 45 E1-E1 cell pairs (12 pairs at E12.5; 26 pairs at E14.5; 7 pairs at E17.5) and 129 E1-B1 cell pairs (47 pairs at E12.5; 66 pairs at E14.5; 16 pairs at E17.5) in animals injected at different stages of development (Figure 5F). These findings are in line with the hypothesis that a common progenitor can give rise to both E1 and B1 cells. In this analysis too, the number of E1-B1 cell pairs was highest at E14.5, suggesting that E1 cells and B1 cells with a common origin are mostly produced around E14.5 (Figures 5C and 5F). We conclude that progenitor cells in the embryo can produce pairs of E1 cells, as well as mixed pairs containing both B1 and E1 cells.
DISCUSSION
Here, we describe the changes the murine embryonic VZ undergoes as it transforms into the ependyma and germinal epithelium of the adult V-SVZ (Figures 1, 2, and 3). Furthermore, we show that the two main cell types in this adult epithelium, E1 and B1 cells, can have a common origin from RG cells that divide during mid-fetal development (Figures 4 and 5). First, we show how the surface area of the lateral wall expands nearly two times to accommodate the growth of the lateral ventricle (Figures 1B and 1C). This expansion occurs despite a net loss in epithelial cell number as postnatal NSCs, through symmetric consuming divisions, generate OB neurons (Obernier et al., 2018), a process that is particularly active during juvenile development. The ventricular growth, despite this net loss of epithelial cells, is explained by the increasing size of the apical domains of differentiating E1 cells (Figures 1F, 1G, and 2A). During differentiation from RG cells at E17.5to morphologically mature cells at P21, E1 cells expand their apical domain up to 11-fold (average sizes, 16.5 μm2 [E17.5 RG cells] and 185.6 μm2 [P10 FoxJ1+cells]; Figure 2A). The expansion and differentiation of E1 cells in the lateral wall occur in a region-specific manner, with the P-V region being first to show mature E1 cells and pinwheel structures, followed by P-D, A-D, and A-V regions (Figures 2D–2F and 3). This gradient of differentiation is similar to that observed for the time of birth of E1 cells (Spassky et al., 2005).
The growth of E1 cell apical domains provides a cellular platform for the organization of their planar polarity. The basal body of the RG primary cilium generates a deuterosome, which in turn generates the many basal bodies that will anchor each E1 cell motile cilium (Spassky et al., 2005). Planar polarity is established as RG cells differentiate into E1 cells, a process that requires the primary cilium of RG and hydrodynamic forces (Mirzadeh et al., 2008, 2010b; Guirao et al., 2010), and is essential for E1 cells to generate proper CSF flow and prevent hydrocephalus (Ohata et al., 2014, 2015; Foerster et al., 2017). In the absence of a RG primary cilium, RG apical domains prematurely expand in an mTORC-dependent process, leading to embryonic ventriculomegaly and postnatal hydrocephalus (Foerster et al., 2017). Interestingly, normal apical surface expansion coincides with E1 cells acquiring FoxJ1 expression but occurs before multiciliation. At P2, FoxJ1 is co-expressed with multiciliated ependymal cell markers, such as S100β, while it is absent from B1 cells (Jacquetetal., 2009). FoxJ1 is also essential for E1 cell differentiation (Jacquet et al., 2009) and is implicated in centriole docking (Stubbs et al., 2008; Yu et al., 2008). Although the molecular link between transcription factors controlling the differentiation of E1 cells and the expansion of the apical domain remains unknown, data suggest that molecular programs that guide the initial differentiation of E1 cells are well underway by the time FoxJ1 is expressed. GemC1 and Mcidas are upstream of E1 cell fate (Kyrousi et al., 2015), and c-Myb directly regulates FoxJ1 expression (Tan et al., 2013). c-Myb is also co-expressed with FoxJ1 in the wall of the lateral ventricles in the postnatal brain until P5, suggesting that FoxJ1 activation is mediated by c-Myb (Tan et al., 2013). Deletion of c-Myb and/or FoxJ1 disrupts the generation of multiple cilia and causes ependymal cell layer alteration, associated with enlarged lateral ventricles or hydrocephalus and reduced NSC proliferation (Malaterre et al., 2008; Jacquet et al., 2009).
In contrast to E1 cells, differentiating B1 cells maintain a smaller apical domain size, similar to RG cells (Figure 2A). This difference in size results in the formation of pinwheels that can contain multiple B1 cells at their cores (Mirzadeh et al., 2008). Our results show that pinwheels develop around P5 in a ventral-to-dorsal and posterior-to-anterior gradient, similar to the direction of E1 cell differentiation and maturation (Figure 3; Spassky et al., 2005). However, for pinwheels to form, B1 cells must also maintain adhesion among themselves so that their small apical domains can coalesce into the cores of pinwheels. E1 cells also adhere to B1 cells in pinwheel cores. VCAM1 has been shown to be expressed on the apical domain of quiescent B1 cells (Kokovay et al., 2012; Codega et al., 2014; Hu et al., 2017), and blocking antibodies against VCAM1 destabilize junctional complexes and disrupt epithelial cytoarchitecture, resulting in a loss of pinwheels (Kokovay et al., 2012). However, the time at which VCAM1 function is necessary for the development of pinwheels, and the potential involvement of other adhesion molecules expressed by E1 cells, B1 cells, or both are not well understood. We found that expression of VCAM1, which is expressed by most RG cells by E17.5, preceded the onset of RG apical domain expansion that occurred between P0 and P5 (Figures 2A and 2D–2F). Recent work has also demonstrated that widespread VCAM1 expression in RG cells at birth is progressively lost by epithelial cells with large apical domains, likely corresponding to differentiating and maturing E1 cells (Huetal.,2017). Interestingly, VCAM1 expression persists during E1 cell differentiation for a short period (Figures 2B and 2C). This subpopulation of epithelial cells had increased apical domain size but expressed both VCAM1 and FoxJ1 (Figure 2B). Some of these VCAM1+ cells also had deuterosomes (Figure 2C). We do not know whether VCAM1 expression detected in differentiating E1 cells represents the remnants of slowly degrading protein, or whether it continues to be expressed and play a role during E1 cell differentiation. Given that epithelial layer integrity is essential for V-SVZ organization and function, and the maintenance of adult neurogenesis (Kuo et al., 2006; Rasin et al., 2007; Jacquet et al., 2009; Paez-Gonzalez et al., 2011; Kokovay et al., 2012), it is possible that this adhesion protein is essential to stabilize the apical contacts between differentiating RG cells to maintain a continuous epithelium during lateral wall expansion.
Our data not only reveal some of the cellular mechanisms by which the V-SVZ becomes organized by demonstrating the co-ordinated changes in the apical domains of E1 and B1 cells (Figures 6A and 6B), but also show that the origins of E1 and B1 cells are closely linked (Figure 6C). Lineage-tracing experiments using four alternative methods showed that embryonic RG cells divide to give rise to clones in the V-SVZ that contain either E1 or B1 cells, or a combination of both. It is possible that some clonally related sibling cells were not identified here due to cell death, migration away from the ventricle, or consuming neurogenic divisions (Obernier et al., 2018). The retrovirus library showed clonal relationships between E1 cells and neurons in the OB, which suggested that E1 and B1 cells could be derived from the same progenitor cell in the embryo (Fuentealba et al., 2015; data not shown). Clonal analysis using Ubc-StarTrack, Nestin::CreER;Ai14, and Nestin::CreER;Confetti mice confirmed this hypothesis by directly revealing pairs of clonally related B1 and E1 cells in the adult V-SVZ (Figures 4 and 5). This indicates that at least some RG are bipotent. While the majority of bipotent clones (E1-B1) appeared around E14.5, a peak time for E1 and B1 cell generation (Spassky et al., 2005; Fuentealba et al., 2015), mixed clones were also observed at other time points of clonal labeling. This suggests that some bipotent progenitors are present throughout embryonic development.
Figure 6. Summary and Proposed Model of the Developmental Origins of E1 and B1 Cells.
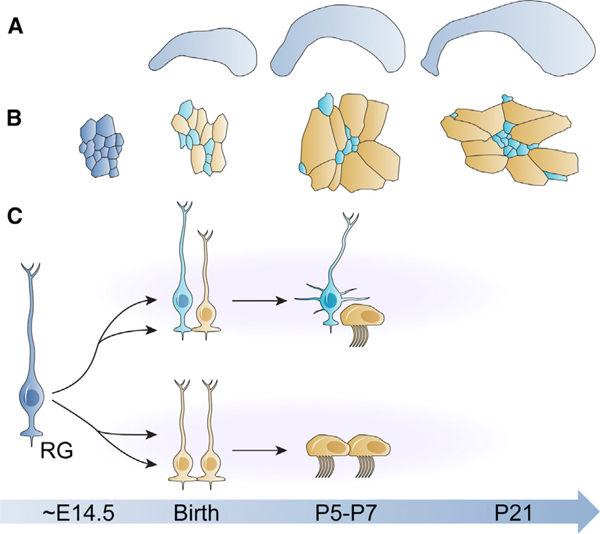
(A) The lateral wall of the lateral ventricle nearly doubles In size after birth, despite decreasing numbers of ventricular epithelial cells.
(B) The apical surface sizes of ventricular epithelial cells increase in late fetal development and postnatally. As E1 (gold) and B1 cells (teal) differentiate from RG cells (dark blue), they start to express cell type-specific markers, and E1 cells begin to dramatically expand their apical surfaces. By P5, pinwheels start to form in the P-V region of the whole mount and are found in all regions of the lateral wall after P10.
(C) An embryonic RG cell (left) divides around E14.5 to give rise to two types of clones: mixed E1-B1 clones (top row) or E1-E1 clones (bottom row). Morphological and molecular maturation of E1 and B1 cells begins around birth and is largely completed by P7. The age associated with each cartoon panel in (A)-(C) is located on the blue timeline directly below the image.
Taken together, these results demonstrate that a subpopulation of RG cells gives rise to E1 and B1 cells, in addition to other RG cell populations that generate clones containing only E1 cells (Figure 6C). The intrinsic or extrinsic signals that determine the fate of RG cells remain elusive. The proportion of B1 to E1 cells varies along both dorsal-ventral and anterior-posterior axes of the lateral wall (Mirzadeh et al., 2008), and B1 cells give rise to different types of OB interneurons based on their location in the lateral wall (Kelsch et al., 2007; Kohwi et al., 2007; Merkle et al., 2007; Ventura and Goldman, 2007; Young et al., 2007). The mechanisms that drive this apparent spatially encoded heterogeneity remain unknown, but its origins are likely present in embryonic RG cells (Fuentealba et al., 2015).
Here, we have charted the transformation of the embryonic VZ into the adult V-SVZ. This process is key for the generation of an epithelium that combines two different functions important to the postnatal brain: CSF homeostasis and postnatal generation of neurons. We show how closely interrelated the postnatal development of E1 and B1 cells’ apical domain is, and how this process is key to the formation of pinwheels. The emerging V-SVZ expands, not by cell addition, but by a dramatic expansion and reorganization of E1 cells. In addition, we provide direct evidence that E1 and B1 cells, which together form key structural components of the V-SVZ niche and are functionally linked in their regulation of adult neurogenesis, also have a common cell of origin in embryonic development.
Together, these findings reveal the timing and cellular changes that underlie the transformation of the embryonic VZ into an adult V-SVZ. Our findings raise specific questions, including how the relative numbers of E1 and B1 cells are controlled, and how their lineages diverge. The common origin of cells so different in structure and function also highlights the divergent nature of the last stages of differentiation of the embryonic neuroepithelial cells. The large postnatal apical expansion of E1 cells and the retraction of B1 cells are carefully orchestrated to form a properly sized ventricle despite decreasing numbers of ventricle-contacting cells. Our study should help identify molecular determinants for this dramatic epithelial remodeling that follows a precise spatiotemporal pattern. This study defines the cellular origins and key stages of cell maturation and tissue remodeling necessary to understand the formation and, ultimately, the function of the V-SVZ.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-acetylated tubulin - 1:500 | Sigma | Cat# T6793; RRID:AB_477585 |
| mouse anti-β-catenin - 1:500 | BD Biosciences | Cat# 610153; RRID:AB_397554 |
| rabbit anti-β-catenin -1:1000 | Sigma-Aldrich | Cat# C2206; RRID:AB_476831 |
| rat anti-BrdU - 1:250 | Abcam | Cat# ab6326; RRID:AB_305426 |
| mouse anti-FoxJ1 -1:300 | Thermo Fisher Scientific | Cat# 14-9965-80; RRID:AB_1548836 |
| mouse anti-γ-tubulin - 1:500 | Abcam | Cat# ab11316; RRID:AB_297920 |
| rabbit anti-γ-tubulin -1:1000 | Sigma-Aldrich | Cat# T5192; RRID:AB_261690 |
| chicken anti-GFAP - 1:1000 | Abcam | Cat# ab4674; RRID:AB_304558 |
| rabbit anti-GFAP - 1:1000 | Dako | Cat# Z0334; RRID:AB_10013382 |
| chicken anti-GFP (for CFP, YFP) - 1:700 | Aves Labs | Cat# GFP-1020; RRID:AB_10000240 |
| rabbit anti-RFP (for tdTomato) - 1:500 | Rockland | Cat# 600-401-379; RRID:AB_2209751 |
| rabbit anti-DsRed (for tdTomato) -1:500 | Clontech Laboratories, Inc. | Cat# 632496; RRID:AB_10013483 |
| rat anti-VCAM1 - 1:100 | BD Biosciences | Cat# 550547; RRID:AB_393741 |
| Secondary antibodies: conjugated to AlexaFluor dyes (donkey or goat polyclonal) |
Thermo Fisher Scientific | N/A |
| Bacterial and Virus Strains | ||
| QmGFP-OL | Fuentealba et al., 2015 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tmx | Sigma-Aldrich | Cat# T5648 |
| Bromodeoxyuridine (BrdU) | Sigma-Aldrich | Cat# B5002 |
| Aqua Poly/Mount medium | Polysciences | Cat# 18606-5 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Nestin::CreERT2: B6.Cg-Tg(Nes-cre/ERT2)KEisc | A. J. Eisch lab (Lagace et al., 2007) | N/A |
| Mouse: Ai14: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory (Madisen etal., 2010) | Cat# 007914 |
| Mouse: Confetti: Gt(ROSA)26Sortm1(CAG- Brainbow2.1)Cle/J | The Jackson Laboratory (Snippert etal., 2010) | Cat# 013731 |
| Mouse: CD-1 | Charles River Laboratories | Strain Code: 482 |
| Mouse: C57BL/6 | Cajal Institute | N/A |
| Oligonucleotides | ||
| primer S161 (5’-GACAACCACTACCTGAGCACC CAGT-3’) | Fuentealba et al., 2015 | N/A |
| primer S126 (5’-GGCTCGTACTCTATAGGCTTCAG CTGGTGA-3’ ) | Fuentealba et al., 2015 | N/A |
| primer S160 (5’- ATCACATGGTCCTGCTGGAGTT CGTGA-3’) | Fuentealba et al., 2015 | N/A |
| primer S128 (5’- ATTGTTGAGTCAAAACTAGAGC CTGGACCA-3’) | Fuentealba et al., 2015 | N/A |
| Recombinant DNA | ||
| UbC-StarTrack plasmids | Figueres-Oñate et al., 2016 | N/A |
| Software and Algorithms | ||
| R version 3.4.3 (2017-11-30) | The R Foundation | N/A |
| ImageJ | NIH | N/A |
| Stereo Investigator | MBF Bioscience | N/A |
| Neurolucida | MBF Bioscience | N/A |
| MAFFT version 7 | CBRC, Japan | N/A |
| MultiAlin | GenoToul bioinformatics | N/A |
| Zeiss PALM MicroBeam LCM (version 4.3.2.13) | Carl Zeiss MicroImaging | N/A |
| Other | ||
| PEN-membrane slides-2 μm | Leica Microsystems | Cat# 11505158 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Arturo Alvarez-Buylla (ABuylla@stemcell.ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
For UbC-StarTrack experiments, wild-type C57BL/6 mice from the Cajal Institute animal facility were treated according to the European Union Council Guidelines on the use and welfare of experimental animals (2010/63/EU) and those of the Spanish Ministry of Agriculture (R.D. 120½005 and L. 32/2007). The CSIC Bioethical Committee approved all procedures. Males and females were used for all experiments.
All other animal experiments were approved by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center at UCSF, and maintained following the NIH, American Veterinary Medical Association, and UCSF guidelines. Nestin::CreER (Lagace et al., 2007), Ai14 (Madisen et al., 2010), and R26R-Confetti (Snippert et al., 2010) mice have been previously generated and were genotyped as described. Wild-type mice were CD1 (Charles River Laboratories) unless otherwise noted. Males and females were used for all experiments.
METHOD DETAILS
Whole-mount Dissections
Whole-mount dissections were done as described in Mirzadeh et al. (2008, 2010a). Briefly, mouse brains at different ages were extracted and the walls of the lateral ventricles dissected out from the caudal aspect of the telencephalon, the hippocampus, and septum. Whole mounts were fixed in 4% paraformaldehyde with 0.1% Triton X-100 at 4°C overnight and then stored in PBS/ 0.1% sodium azide at 4°C until immunohistochemical processing. After immunostaining, a ~200–300 mm thick section of the lateral wall was carefully dissected away from the remaining brain and mounted in Aqua Poly/Mount medium (Polysciences) on microscope slides. Whole mounts were left to settle for at least twelve hours prior to imaging.
Immunostaining
Whole mounts were incubated in blocking solution, 0.5% Triton X-100/10% normal goat or donkey serum/PBS, for 1–2 hr at room temperature. Primary antibodies were incubated for 24–48 hr at 4°C, followed by appropriate secondary antibodies for 2–48 hr at 4°C, depending on target antigens. For bromodeoxyuridine (BrdU) immunostaining, samples were incubated for 40 min at 37°C with 2N HCl followed by 10 min at 25° C with 0.1 M boric acid (pH 8.5) prior to immunostaining procedures. Whole mounts were embedded in Aqua Poly/Mount medium (Polysciences) on microscope slides for imaging.
BrdU and Tmx Administration
For embryonic Tmx administration, two to three month-old, timed-pregnant Nestin::CreER;Ai14 or Nestin::CreER;Confetti females received a single dose of Tmx, 1.67mg/kg and 100mg/kg respectively, via oral gavage (Ihrie et al., 2011). Nestin::CreER;Ai14 mice received two repeated intraperitoneal injections of BrdU (50mg/kg, six hours apart) at the indicated ages in the text. The morning of the plug was considered E0.5. Mice were sacrificed and brains were processed for microscopy at the ages indicated in the text. UbC-StarTrack animals were injected with a single dose of 75 mg/kg Tmx at P1.
In Utero Surgeries
In utero electroporation was performed as described previously (Figueres-Oñate et al., 2015). Briefly, E14.5 timed-pregnant mice were anesthetized with 1.5% isofluorane/O2 inhalation and their uterine horns were exposed by midline laparotomy. Intraventricular lateral injections of the UbC-StarTrack mixture into E14.5 embryo brains were guided by trans-illumination. Five consecutive electric square wave pulses (37V, E15; 50 ms duration) were applied to each embryo, after which the uterine horns were replaced into the abdominal cavity. Dams were placed in clean cage to recover and they were monitored closely.
QUANTIFICATION AND STATISTICAL ANALYSIS
For all experiments, male and female mice were used, and sample sizes were not determined beforehand. Statistical analyses were performed using R version 3.4.3 (2017–11-30) (Figure 1). One-way ANOVA and Tukey Honest Significant Difference multiple comparison correction (95% confidence interval) were used to determine statistical significance. p values < 0.05 were considered significant. All applied statistical tests and outputs, including p values, are listed in Table 1.
Imaging and Quantification
Immunofluorescence images, except for UbC-StarTrack experiments in Figure 4 (see below), were acquired using Leica SP5 and SP8 confocal microscopes.
Data in Figure 1 were generated using Stereo Investigator and Neurolucida software (MBF Bioscience) on a Zeiss Axiovert 200M epifluorescence microscope. Whole-mount areas were traced based on p-catenin staining (Figures 1B and 1C). The dorsal-ventral length of the lateral wall was traced in serial coronal brain sections and reconstructed in 3-D (Figures 1D and 1E) with Neurolucida software (MBF Bioscience). Average apical surface sizes and numbers (Figures 1F and 1G) were estimated using systematic random sampling OpticalFractionator and Nucleator (vertical section aligned to dorsal-ventral whole-mount axis, four measuring rays) stereological probes and β-catenin staining (Figures 1F and 1G, respectively). Sampling parameters were as follows: 200 sites per whole mount were sampled using square 64 μm2 (ages < P15) or 144 μm2 (ages = > P15) counting frames. Only estimates with Schmitz-Hof coefficients of error < 0.10 were included in analyses.
For data in Figure 2, images were acquired using Leica SP5 and SP8 confocal microscopes. Apical domain sizes of epithelial cells were measured using ImageJ software (Schindelin et al., 2012; Schneider et al., 2012) from one field per each of the four regions of one lateral wall per age (92.35×92.35 μm2 fields). Apical surfaces were delimited by beta-catenin staining, and surface areas were segmented using the Squassh algorithm in the Mosaic plugin (Paul et al., 2013; Rizk et al., 2014). Segmented image masks were manually checked for accuracy against the original beta-catenin staining, and corrected with the pen tool if necessary. Apical surfaces sizes were measured using the Fiji Analyze Particles algorithm, with a minimum cell size of two μm2. Cell-type specific markers were annotated by hand using the Multi-point tool. Marker points, corresponding to VCAM1, FoxJ1, or none, were assigned to each apical surface ROI using an ImageJ macro.
UbC-StarTrack fluorescent labeling (Figure 4) was visualized under a confocal microscope using acquisition parameters previously described (Figueres-Oñate et al., 2016). Briefly, color codes were determined using an ImageJ macro. Each cell was separately evaluated for fluorescence intensity in six independent channels for different emission wavelengths. This was done for the nucleus and for the cytoplasm. Fluorophore intensity measurements above background levels were considered as positive for that reporter; this was done for each channel. Cells containing the same pattern of reporter expression were considered clonally related. One additional detection channel was available after StarTrack fluorophore analysis, and so we immunostained whole mounts for FoxJ1 to identify E1 cells. B1 cells were identified by morphology. Clones containing FoxJ1+ cell(s) are reported in the figure.
Tamoxifen-based clonal analysis (Fig. 5) was conducted as follows: Whole mounts were prepared at ages indicated in the text (P10 or P21), and immunostained. Entire whole-mount surfaces were imaged at 10x or 20x magnification to identify locations of potential clones. Clonal pairs of cells containing at least one E1 cell which were then imaged at 63x magnification with 0.5 μm z-plane intervals from the surface of the ventricle wall through the cell bodies of reporter-positive cells. Image stacks were then analyzed to confirm clonality and cell type composition. Pairs of cells containing at least one E1 cell are reported in the figure.
Highlights.
Numbers of cells in the lateral ventricle wall decrease despite ventricle growth
Ependymal cells expand apical surface area to accommodate larger ventricle size
Apical ependymal expansion and clustering of stem cells results in pinwheel formation
Ependymal and adult neural stem cells share common embryonic progenitors
ACKNOWLEDGMENTS
The authors would like to thank members of the Alvarez-Buylla laboratory for helpful discussions, William Walantus for technical expertise, and Kenneth Xavier Probst ofXavierStudioforcell illustration designs. This studywassup-ported by funding from the Howard Hughes Medical Institute/Helen HayWhitney Foundation (to L.C.F.); Becas Chile, Ministerio de Educacion (to J.I.P.); Deutsche Forschungsgemeinschaft (to K.O.); NIH grant F32 NS103221 (to S.A.R.); MINECO (BFU2016–80360-R; to M.F.-O.); and NIH grants R37 HD32116 and R01 NS28478, the Heather and Melanie Muss Endowed Chair, and a generous gift from the John G. Bowes Research Fund (to A.A.-B.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Al Jord A, Lemaître A-II, Delgehyr N, Faucourt M, Spassky N, and Meunier A (2014). Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature 516, 104–107. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, and Song H (2011). In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari F, Michel J, Baumgart EV, Theis F, Götz M, and Ninkovic J (2015). Fast clonal expansion and limited neural stem cell self-renewal in the adult subependymal zone. Nat. Neurosci. 18, 490–492. [DOI] [PubMed] [Google Scholar]
- Cepko C, Ryder EF, Austin CP, Walsh C, and Fekete DM (1995). Lineage analysis using retrovirus vectors. Methods Enzymol. 254, 387–419. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Ryder E, Austin C, Golden J, Fields-Berry S, and Lin J(1998). Lineage analysis using retroviral vectors. Methods 14, 393–406. [DOI] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, and Doetsch F (2014). Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 82, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarolis NA, Mechanic M, Petrik D, Carlton A, Ables JL, Malhotra S, Bachoo R, Götz M, Lagace DC, and Eisch AJ (2013). In vivo contribution of nestin-and GLAST-lineage cells to adult hippocampal neurogenesis. Hippocampus 23, 708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bigio MR (2010). Ependymal cells: biologyand pathology. Acta Neuropathol. 119, 55–73. [DOI] [PubMed] [Google Scholar]
- Doetsch F, Caillé I, Lim DA, García-Verdugo JM, and Alvarez-Buylla A(1999). Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716. [DOI] [PubMed] [Google Scholar]
- Figueres-Oñate M, García-Marqués J, Pedraza M, De Carlos JA, and Lopez-Mascaraque L (2015). Spatiotemporal analyses of neural lineages after embryonic and postnatal progenitor targeting combining different reporters. Front. Neurosci. 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueres-Oñate M, García-Marqués J, and Lopez-Mascaraque L (2016). UbC-StarTrack, a clonal method to target the entire progeny of individual progenitors. Sci. Rep. 6, 33896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster P, Daclin M, Asm S, Faucourt M, Boletta A, Genovesio A, and Spassky N (2017). mTORC1 signaling and primary cilia are required for brain ventricle morphogenesis. Development 144, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, and Alvarez-Buylla A (2015). Embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Miya H, Watanabe T, Kawai H, Yamasaki N, Harada Y, Imayoshi I, Nelson M, Nakayama KI, Hirabayashi Y, and Gotoh Y (2015). Slowlydividing neural progenitors are an embryonicorigin ofadult neural stem cells. Nat. Neurosci. 18, 657–665. [DOI] [PubMed] [Google Scholar]
- Golden JA, Fields-Berry SC, and Cepko CL (1995). Construction and characterization of a highly complex retroviral library for lineage analysis. Proc. Natl. Acad. Sci. USA 92, 5704–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao B, Meunier A, Mortaud S, Aguilar A, Corsi J-MM, Strehl L, Hirota Y, Desoeuvre A, Boutin C, Han Y-GG, et al. (2010). Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat. Cell Biol. 12, 341–350. [DOI] [PubMed] [Google Scholar]
- Hu X-L, Chen G, Zhang S, Zheng J, Wu J, Bai Q-R, Wang Y, Li J, Wang H, Feng H, et al. (2017). Persistentexpression ofVCAM1 in radial glial cells is required forthe embryonic origin ofpostnatal neural stem cells. Neuron 95, 309–325.e6. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Shah JK, Harwell CC, Levine JH, Guinto CD, Lezameta M, Kriegstein AR, and Alvarez-Buylla A (2011). Persistent sonic hedgehog signaling in adult brain determines neural stem cell positional identity. Neuron 71, 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, and Ghashghaei HT (2009). FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 136, 4021–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsch W, Mosley CP, Lin C-WW, and Lois C (2007). Distinct mammalian precursors are committed to generate neurons with defined dendritic projection patterns. PLoS Biol. 5, e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, and Mitchell BJ (2013). Deuterosome-mediated centriole biogenesis. Dev. Cell 27, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Petryniak MA, Long JE, Ekker M, Obata K, Yanagawa Y, Rubenstein JL, and Alvarez-Buylla A (2007). A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1-and Dlx5/6-expressing progenitors. J. Neurosci. 27, 6878–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokovay E, Wang Y, Kusek G, Wurster R, Lederman P, Lowry N, Shen Q, and Temple S (2012). VCAM1 is essential to maintain the structure ofthe SVZ niche and acts as an environmental sensor to regulate SVZ lineage progression. Cell Stem Cell 11, 220–230. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Mirzadeh Z, Soriano-Navarro M, Rasin M,Wang D, Shen J, Sestan N, Garcia-Verdugo J, Alvarez-Buylla A, Jan LY, and Jan Y-NN (2006). Postnatal deletion ofNumb/Numblike reveals repairand remodeling capacity in the subventricular neurogenic niche. Cell 127,1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrousi C, Arbi M, Pilz G-AA, Pefani D-EE, Lalioti M-EE, Ninkovic J, Götz M, Lygerou Z, and Taraviras S (2015). Mcidas and GemC1 are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development 142, 3661–3674. [DOI] [PubMed] [Google Scholar]
- Lagace DC, Whitman MC, Noonan MA, Ables JL, DeCarolis NA, Arguello AA, Donovan MH, Fischer SJ, Farnbauch LA, Beech RD, et al. (2007). Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 27, 12623–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaterre J, Mantamadiotis T, Dworkin S, Lightowler S, Yang Q, Ransome MI, Turnley AM, Nichols NR, Emambokus NR, Frampton J, and Ramsay RG (2008). c-Myb is required for neural progenitor cell proliferation and maintenance ofthe neural stem cell niche in adult brain. Stem Cells 26, 173–181. [DOI] [PubMed] [Google Scholar]
- Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, and Alvarez-Buylla A (2006). Origin ofoligodendrocytes in the subventricular zone ofthe adult brain. J. Neurosci. 26, 7907–7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Tramontin AD, García-Verdugo JMM, and Alvarez-Buylla A (2004). Radial gliagive riseto adult neural stem cells in thesubventricularzone. Proc. Natl. Acad. Sci. USA 101, 17528–17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, and Alvarez-Buylla A (2007). Mosaic organization of neural stem cells in the adult brain. Science 317, 381–384. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, and Alvarez-Buylla A (2008). Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell 3, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Doetsch F, Sawamoto K, Wichterle H, and Alvarez-Buylla A (2010a). The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 2010, 1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Han Y-GG, Soriano-Navarro M, García-Verdugo JM, and Alvarez-Buylla A (2010b). Cilia organize ependymal planar polarity. J. Neurosci. 30, 2600–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernier K, Cebrian-Silla A, Thomson M, Parraguez JII, Anderson R, Guinto C, Rodas Rodriguez J, Garcia-Verdugo J-MM, and Alvarez- Buylla A (2018). Adult neurogenesis is sustained by symmetric self-renewal and differentiation. Cell Stem Cell 22, 221–234.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata S, Nakatani J, Herranz-Pérez V, Cheng J, Belinson H, Inubushi T, Snider WD, García-Verdugo JM, Wynshaw-Boris A, and Alvarez-Buylla A (2014). Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron 83, 558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata S, Herranz-Pérez V, Nakatani J, Boletta A, García-Verdugo, and Álvarez-Buylla A (2015). Mechanosensory genes Pkd1 and Pkd2 contribute to the planar polarization of brain ventricular epithelium. J. Neurosci. 35,11153–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega F, Gascón S, Masserdotti G, Deshpande A, Simon C, Fischer J, Dimou L, Chichung Lie D, Schroeder T, and Berninger B (2013). Oligoden-drogliogenic and neurogenic adult subependymal zone neural stem cells constitute distinct lineages and exhibit differential responsiveness to Wnt signalling. Nat. Cell Biol. 15, 602–613. [DOI] [PubMed] [Google Scholar]
- Paul G, Cardinale J, and Sbalzarini IF (2013). Coupling image restoration and segmentation: A generalized linear model/Bregman perspective. Int. J. Comput. Vis. 104, 69–93. [Google Scholar]
- Paez-Gonzalez P, Abdi K, Luciano D, Liu Y, Soriano-Navarro M, Rawlins E, Bennett V, Garcia-Verdugo JM, and Kuo CT (2011). Ank3-dependent SVZ niche assembly is required for the continued production of new neurons. Neuron 71, 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin M-RR, Gazula V-RR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li H-SS, Jan LY, Jan Y-NN, Rakic P, and Sestan N (2007). Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat. Neurosci. 10, 819–827. [DOI] [PubMed] [Google Scholar]
- Rizk A, Paul G, Incardona P, Bugarski M, Mansouri M, Niemann A, Ziegler U, Berger P, and Sbalzarini IF (2014). Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat. Protoc. 9, 586–596. [DOI] [PubMed] [Google Scholar]
- Sawamoto K, Wichterle H, Gonzalez-Perez O, Cholfin JA, Yamada M, Spassky N, Murcia NS, Garcia-Verdugo JM, Marin O, Rubenstein JL, et al. (2006). Newneuronsfollowtheflowofcerebrospinal fluid intheadult brain. Science 311, 629–632. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9,676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, and Clevers H (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. [DOI] [PubMed] [Google Scholar]
- Spassky N, Merkle FT, Flames N, Tramontin AD, García-Verdugo JMM, and Alvarez-Buylla A (2005). Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 25, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisuía Belmonte JC, and Kintner C (2008). The forkhead protein Foxj1 specifies node-like cilia in Xenopusand zebrafish embryos. Nat. Genet. 40, 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FE, Vladar EK, Ma L, Fuentealba LC, Hoh R, Espinoza FH, Axelrod JD, Alvarez-Buylla A, Stearns T, Kintner C, and Krasnow MA (2013). Myb promotes centriole amplification and later steps of the multicilio-genesis program. Development 140, 4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong CK, Fuentealba LC, Shah JK, Lindquist RA, Ihrie RA, Guinto CD, Rodas-Rodriguez JL, and Alvarez-Buylla A (2015). A dorsal SHH-dependent domain in the V-SVZ produces large numbers of oligodendroglial lineage cells in the postnatal brain. Stem Cell Reports 5, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontin AD, García-Verdugo JMM, Lim DA, and Alvarez-Buylla A (2003). Postnatal development of radial glia and the ventricular zone (VZ): a continuum of the neural stem cell compartment. Cereb. Cortex 13, 580–587. [DOI] [PubMed] [Google Scholar]
- Ventura RE, and Goldman JE (2007). Dorsal radial glia generate olfactory bulb interneurons in the postnatal murine brain. J. Neurosci. 27, 4297–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KM, Fogarty M, Kessaris N, and Richardson WD (2007). Subven-tricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci. 27, 8286–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, and Roy S (2008). Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 40, 1445–1453. [DOI] [PubMed] [Google Scholar]
- Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, et al. (2015). In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]


