SUMMARY
The diversity of cardiac lineages contributes to the heterogeneity of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). Here, we report the generation of a hiPSC TBX5Clover2/NKX2-5TagRFP double reporter to delineate cardiac lineages and isolate lineage-specific subpopulations. Molecular analyses reveal that four different subpopulations can be isolated based on the differential expression of TBX5 and NKX2-5: TBX5+NKX2-5+ TBX5+NKX2-5−, TBX5−NKX2-5+, and TBX5−NKX2-5− subpopulation, mimicking the first heart field, epicardial, the second heart field, and endothelial lineage, respectively. Genetic and functional characterization indicates that each subpopulation differentiates into specific cardiac cells. We further identify CORIN as a cell surface marker for isolating the TBX5+NKX2-5+ subpopulation and demonstrate the use of lineage-specific CMs for precise drug testing. We anticipate that this tool will facilitate the investigation of cardiac lineage specification and isolation of specific cardiac subpopulation for drug screening, tissue engineering, and disease modeling.
Graphical Abstract

In Brief
Wu and colleagues create a double knock-in human TBX5Clover2/NKX2-5TagRFP reporter to delineate cardiac lineages during differentiation. Molecular and functional analyses uncover that differential expression of TBX5 and NKX2-5 specifies different cardiac fate. This reporter is applicable for studying cardiac lineage determination in vitro and isolating cardiac subpopulations.
INTRODUCTION
Cardiogenesis is orchestrated by temporally and spatially regulated expression of cardiac transcription factors, leading to specification of multiple lineages (Buckingham et al., 2005). During heart development, cardiac progenitor cells (CPCs) of the “first heart field” (FHF) marked by TBX5, NKX2-5, and HCN4 contribute to the left ventricle (Buckingham et al., 2005; Spater et al., 2013), whereas CPCs of the “second heart field” (SHF) marked by NKX2-5, ISL1, and MEF2C develop into the right ventricle and atria (Domian et al., 2009). Moreover, the epicardial lineage marked by TBX5, TBX18, and WT1 further differentiates into cardiomyocytes (CMs), smooth muscle cells, endothelial cells (ECs), and fibroblasts (Cai et al., 2008). It should be noted that NKX2-5+ atrial and ventricular subtypes are distinct from the NKX2-5− nodal subtype that is specified by TBX18 (Wiese et al., 2009), TBX5 (Puskaric et al., 2010), and SHOX2 (Espinoza-Lewis et al., 2009). These findings suggest that combinatorial expression of cardiac transcription factors is essential for determining cardiac lineages and differentiated cell types.
TBX5 and NKX2-5 are two important cardiac transcription factors that control many aspects of heart development. They can interact physically to cooperatively activate cardiac gene expression in atrial and ventricular CMs (Bruneau et al., 2001), whereas they seem to be mutually exclusive in nodal CMs. The expression of NKX2-5 needs to be repressed by coordinated interactions between TBX5 and SHOX2 to determine the nodal cell fate (Espinoza-Lewis et al., 2009; Puskaric et al., 2010). These studies further corroborate that fine-tuned regulation of cardiac transcription factor expression is essential for cell fate determination.
The ability of human induced pluripotent stem cells (hiPSCs) to differentiate into CMs has reshaped our approaches to studying heart development. In vitro cardiac differentiation of hiPSCs can recapitulate many cellular aspects of cardiac lineage specification and commitment (Birket et al., 2015). Moreover, hiPSC-CMs represent a heterogeneous pool of ventricular, atrial, and nodal CMs, indicating multiple co-existing cardiac lineages. The nature of heterogeneity allows us to delineate different cardiac lineages and isolate lineage-specific cardiovascular cells.
In this study, we identified four discrete subpopulations using a TBX5Clover2/NKX2-5TagRFP hiPSC double reporter. Our findings show that TBX5+NKX2-5+ (G+R+) subpopulation is close to the FHF lineage and mainly differentiates into ventricular CMs, whereas TBX5+NKX2-5− (G+R−) subpopulation exhibits epicardial lineage characteristics and contributes to nodal CMs. TBX5−NKX2-5+ (G−R+) subpopulation mimics the SHF lineage and primarily differentiates into atrial CMs. Lastly, the progeny of TBX5−NKX2-5− (G−R−) subpopulation has EC properties. Transcriptome analysis of each subpopulation identifies CORIN as a cell surface marker for enriching the G+R+ subpopulation. Furthermore, we demonstrate that lineage-specific CMs provide a useful model for precise drug testing. Overall, this study provides a unique tool for isolation and application of human lineage-specific cardiovascular cells.
RESULTS
Generation of hiPSC TBX5Clover2/NKX2-5TagRFP dual reporter
To delineate cardiac lineages, we developed a hiPSC double reporter expressing Clover2 and TagRFP from the loci of TBX5 and NKX2-5, respectively, using CRISPR/Cas9 (Figure 1A, Figure S1A-H, and Table S1). The Clover2 or TagRFP element was placed upstream of TBX5 or NKX2-5 start codon and translation was mediated via an IRES. The protein level of TBX5 and NKX2-5 in hiPSC-CMs was comparable between the reporter and parental line (Figure S1I). The expression kinetics of Clover2 and TagRFP mirrored the expression of TBX5 and NKX2-5, respectively (Figure S1J). On day 6 of differentiation, ~55% of cells were TBX5+ (G+), and the remaining 45% was TBX5− (G−) (Figure 1B-C). From day 7 onward, four discrete subpopulations were identified: G+R+, G+R−, G−R+, and G−R− (Figure 1B and 1D).
Figure 1. Isolation of cardiac subpopulations from TBX5Clover2/NKX2-5TagRFP hiPSC-derived cells.
(A) Representative images show G+R+ (yellow color), G+R− (asterisk), and G−R+ (arrow) subpopulations. Scale bars, 100 μm.
(B) Fluorescence-activated cell sorting (FACS) plots show separation of subpopulations. Numbers in the quadrants represent the respective percentage of subpopulations.
(C-D) Percentage of subpopulations from 5 to 16 independent experiments.
(E) Gene expression in individual subpopulation isolated on day 10 (n=3).
(F) Gene expression in G+R−, G−R+, and G+R+ derivatives on day 35 (n=3). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (mean±SEM). See also Figure S1, Tables S1, S2, and S4.
TBX5Clover2/NKX2-5TagRFP hiPSCs reveal distinct cardiac lineages
We first analyzed the expression of lineage markers to identify each subpopulation. TBX5 was upregulated over 60-fold in both G+R+ and G+R− subpopulations (Figure 1E and Table S2). NKX2-5 expression was upregulated over 5- and 10-fold in G+R+ and G−R+ subpopulations, respectively. The significant enrichment of TBX5 and NKX2-5 in designated subpopulations confirmed the fidelity of the reporter. All three colored subpopulations (i.e., G+R+, G+R−, and G−R+) underwent cardiogenic lineage path with upregulation of GATA4 and TNNT2. Interestingly, HCN4, a marker for FHF and nodal CMs (Stieber et al., 2003), was over 5-fold higher in G+R+ and G+R− subpopulations, whereas SHF markers, MEF2C and ISL1, were upregulated in the G−R+ subpopulation. WT1 and TBX18, the epicardial lineage markers, were highly expressed in the G+R− subpopulation. Additionally, the expression of PECAM1 and KDR was higher in G+R− and G−R− subpopulations. These results reveal that each subpopulation is defined by a set of lineage marker genes, suggesting that our hiPSC double reporter can isolate four different lineage subpopulations.
We then cultured G+R+, G+R− and G−R+ subpopulations to day 35 to investigate their derivatives (Figure 1F). Both TBX5 and NKX2-5 were still upregulated in the designated subpopulations, further confirming the fidelity of the reporter. HAND1 was significantly upregulated in the G+R+ subpopulation, although the expression of HAND2 was not different among the three subpopulations. Interestingly, ventricular CM markers (MYL2 and IRX4) were significantly upregulated in the G+R+ subpopulation compared with G+R− and G−R+ subpopulations.
Epicardial cells can form tight junctions identified by the presence of ZO1 along the cell membrane (Austin et al., 2008). Immunostaining of G+R− cells exhibited that ZO1+ cells were also WT1+, validating their epicardial phenotype (Figure S2A). Flow cytometry analyses showed that 72% of G+R− cells was WT1+, whereas hiPSC-derived epicardial cells (hiPSC-Epi) contained 92% WT1+ cells (Figure S2B-C). The WT1 expression was lower in G+R− cells than hiPSC-Epi, but the TBX18 expression was not different (Figure S2D). Interestingly, nodal cell ion channel genes (HCN1, HCN4, and KCNJ3) were highly expressed in the G+R− subpopulation (Figure 1F). Cardiac transcription factors, including SHOX2 and TBX3, which play a significant role in specifying nodal cells (Espinoza-Lewis et al., 2009; Hoogaars et al., 2007), were also enriched in the G+R− subpopulation. However, the expression of TBX2, a marker for the atrioventricular node (AVN) (Aanhaanen et al., 2009), was not different among subpopulations. The atrial markers including NR2F2, MYL7, and PITX2 (Devalla et al., 2015) were significantly upregulated in the G−R+ subpopulation, although ~78% of G−R+ cells was cardiac troponin+ (cTnT+) (Figure S2E). It should be noted that the expression of TBX1, a marker for anterior SHF (Paige et al., 2015), was low in the G−R+ subpopulation and TBX1+ hiPSC-CMs were scarce (Churko et al., 2018) (Figure S2F-G). Moreover, the upregulation of TCF21, ACTA2, MYH11, and PECAM1 in the G+R− subpopulation suggested a mixture of fibroblasts, smooth muscle cells, and ECs in its derivatives, but the G−R+ subpopulation was only enriched for smooth muscle cell and EC markers (Figure 1F and Figure S2H).
During heart development, gene expression shows dynamic changes that define distinct cardiac progenitors (Kokkinopoulos et al., 2015). We also found that some cells in G+R+, G+R−, and G−R+ subpopulations showed a dynamic change in the expression of TBX5 or NKX2-5 (Figure S2I). For example, around 10% of day 8 G+R− cells initiated the expression of NKX2-5 to become G+R+ cells on day 12, whereas approximately 40% G+R− cells turned off the expression of TBX5 to become G−R− cells. However, from days 8 to 12, almost all G−R− cells retained their identity.
G+R+ subpopulation differentiates into ventricular CMs
To further reveal the influence of TBX5 and/or NKX2-5 on cell fate, we next characterized functional properties of each subpopulation. We utilized a fluorescent voltage sensor, ASAP2 (Chamberland et al., 2017), to record action potentials (APs). The combination of brightness and voltage sensitivity of ASAP2 enabled us to distinguish the AP morphology of all three CM subtypes: ventricular, atrial, and nodal subtypes (Figure 2A). Interestingly, only ventricular and atrial AP morphology were captured in the G+R+ subpopulation, with 90% being ventricular. However, three CM subtypes were distinguished in the non-sorted hiPSC-CMs (Figure 2B). To confirm the fidelity of ASAP2, we performed the whole cell patch-clamp recordings. Consistent with the optical imaging, the major subtype of G+R+ CMs was ventricular (93%) and no nodal cells were identified (Figure 2C-D). These results provide strong evidence that ASAP2 faithfully recapitulates the AP morphology of hiPSC-CMs and is suitable for subtype phenotyping. Additionally, immunostaining also demonstrated that the majority of G+R+ CMs were ventricular subtype (Figure S3A).
Figure 2. Functional characterization of individual subpopulation.
(A) Optical imaging of action potentials (APs) using ASAP2 shows “ventricular-like” (top), “atrial-like” (middle), and “nodal-like” (bottom) hiPSC-CMs between days 35 and 40.
(B) Percentage of subtypes in G+R+ and non-sorted hiPSC-CMs. Numbers indicate the recorded cell number.
(C) Representative whole cell patch-clamp recordings of G+R+ and non-sorted hiPSC-CMs.
(D) Percentage of subtypes in G+R+ and non-sorted hiPSC-CMs by patch-clamp. Numbers indicate the recorded cell number.
(E) Maximum diastolic potential (MDP).
(F) Expression of ion channel genes (n=3).
(G-H) Western blots and quantification of the expression of Kir2.1 (n=3).
(I-J) Quantification of contraction velocity and contraction deformation distance (G+R+, n=14; non-sorted, n=33).
(K) Seahorse extracellular-flux assays measuring oxygen consumption rate under palmitate condition.
(L) Analyses of Seahorse extracellular-flux assay results (n=8).
(M) Optical imaging of G+R− and G−R+ hiPSC-CM APs using ASAP2.
(N) Percentage of subtypes in G+R− and G−R+ hiPSC-CMs. Numbers indicate the recorded cell number.
(O-Q) Quantification of beating rate, AP amplitude, and AP duration (APD) in G+R−, G−R+, G+R+, and non-sorted hiPSC-CMs (G+R+, n=92; G+R−, n=58; G−R+, 68; Non-sorted, n=99).
(R) Gene expression of endothelial cell (EC) markers in G−R− cells on day 35 (n=3).
(S-T) G−R− cells formed “vasculature-like” structure (S) in vitro and (T) in vivo. Scale bars, 30 μm. *p<0.01, **p<0.01, ***p<0.001, ****p<0.0001 (mean±SEM). See also Figures S2 and S3 and Table S4.
We also found that G+R+ ventricular CMs displayed a more negative maximum diastolic potential (MDP) (Figure 2E). To further probe the molecular contribution to this phenotype, we analyzed the expression of key ion channel genes and found a higher expression of KCNJ2 encoding the Kir2.1 inward-rectifier potassium (Ik1) channel in G+R+ CMs (Figure 2F-H). Moreover, G+R+ CMs showed a higher contraction velocity and contraction deformation distance than non-sorted hiPSC-CMs (Figure 2I-J), suggesting that purified G+R+ CMs possess functional advantages over the heterogeneous hiPSC-CMs.
We next examined the metabolic status of CMs by measuring mitochondrial oxygen consumption. We observed a significant increase in mitochondrial respiration in G+R+ CMs under basal and pyruvate conditions (Figure S3B-E), but not under the glucose condition (data not shown). More importantly, when palmitate, a type of fatty acid, was used as the substrate, G+R+ CMs showed a significantly higher oxygen consumption rate than non-sorted hiPSC-CMs (Figure 2K-L). These results indicate that G+R+ CMs favor mitochondrial respiration and use fatty acid more efficiently than non-sorted hiPSC-CMs.
Nodal and atrial CMs are the major subtypes derived from G+R− and G−R+ subpopulations, respectively
Lineage analysis identified that the sinoatrial node is derived from TBX18+SHOX2+NKX2-5− progenitors (Mohan et al., 2017). As the G+R− subpopulation is defined as NKX2-5− with an enriched expression of TBX18 and SHOX2, we hypothesized that G+R− CMs would be nodal subtype. Indeed, approximately 80% of G+R− CMs had a typical nodal CM AP morphology. However, 90% of G−R+ CMs were atrial subtype (Figure 2M-N). Furthermore, G+R− and G−R+ CMs exhibited distinct AP parameters from G+R+ CMs (Figure 2O-Q). Importantly, the percentage of CM subtypes and AP properties of day 60 CMs remained similar to that at the earlier stage (~day 35) in each subpopulation (Figure S3F-I and Figure 2M-Q). In sum, these results provide strong evidence that different lineage subpopulations lead to differentiation of specific subtypes that are determined by differential expression level of TBX5 or NKX2-5.
G−R− subpopulation differentiates into ECs
EC markers (VWF, PECAM1, CD144, KDR, and NOS3) were upregulated in the G−R− subpopulation compared with the G+R+ subpopulation, although the purity and expression level of those markers were lower than bona fide ECs (Figure 2R and Figure S3J-L). When placed on Matrigel in the presence of VEGF, G−R− cells formed a “tube-like” structure (Figure 2S). To further validate their in vivo angiogenesis potential, G−R− cells were implanted subcutaneously (s.c.) in NOD/SCID mice for 4 weeks and immunostaining of tissue sections showed vessel formation (Figure 2T). These data suggest that the G−R− subpopulation contains functional ECs.
Transcriptome profiling shows inter-subpopulation differences
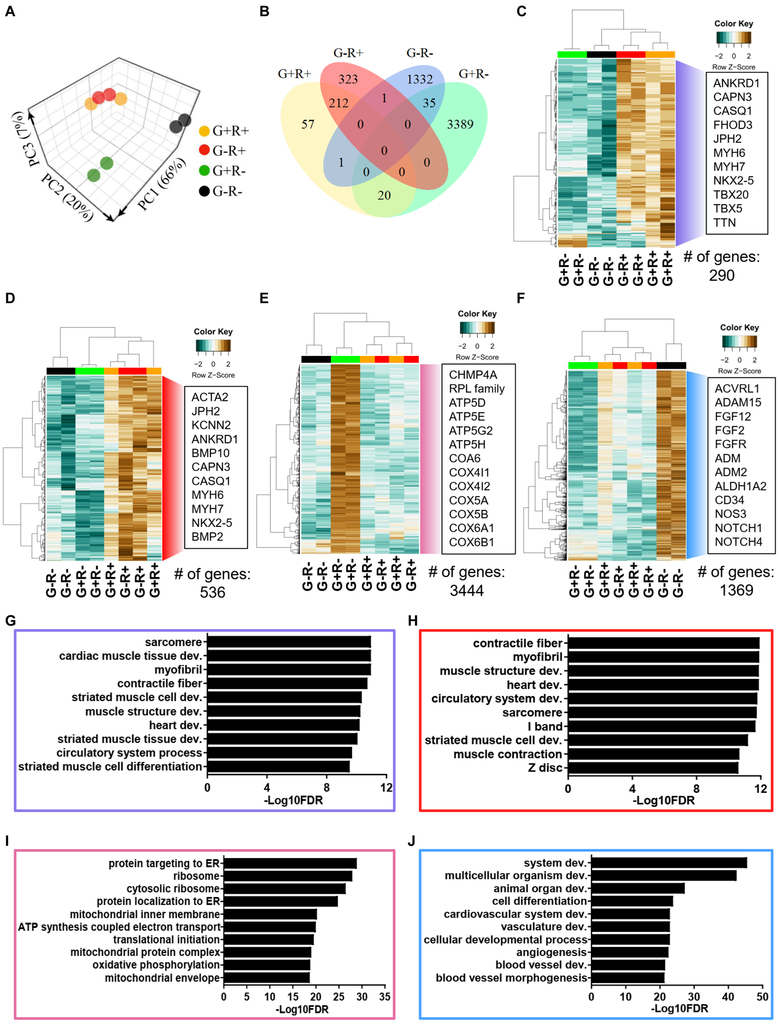
To achieve a detailed understanding of the genetic programs modulated by TBX5 and/or NKX2-5 during lineage divergence, we next performed RNA sequencing (RNA-seq) analysis on purified subpopulations. Principal component analysis (PCA; Figure 3A) and unsupervised hierarchical clustering (Figure S4A) revealed a closer correlation in G+R+ and G−R+ subpopulations with more overlapped genes (Figure 3B and Table S3). Analyses of changes in gene ontology (GO) terms between subpopulations allowed us to understand the TBX5/NKX2-5−dependent regulation of gene expression (Figure 2C-J and Figure S4A). A unique gene group associated with protein synthesis (cluster 1) was upregulated in the G+R− subpopulation (Figure S4A, Figure 3E and 3I). Concordant with the high proliferation capacity of epicardial cells (G+R−) and ECs (G−R−), a group of genes associated with cell cycle and division were upregulated (cluster 2) (Figure S4A), suggesting that NKX2-5 plays a repressive role in proliferation. The transcriptomic profiles of G+R+ and G−R+ representing the FHF and SHF lineage, respectively, were not distinct, indicating the dominant role of NKX2-5 in regulating cardiac differentiation and contraction at the early stage (Figure 3C-D, 3G-H and clusters 3 and 4 in Figure S4A). Motif enrichment analyses predicted that multiple cardiac transcription factors, including ISL1, MEF2 family, and GATA family, bound to genes enriched in clusters 3 and 4, confirming the fate of cardiac lineages of G+R+ and G−R+ subpopulations (Figure S4B). We also identified a group of genes (cluster 6) that were upregulated in G−R− (Figure 3F, 3J, and Figure S4A). GO term analyses indicated that the TBX5- and NKX2-5−independent expression of genes was associated with organ development and vasculature development. Additionally, motif enrichment analyses indicated that the GATA family bound to the promoters of these genes in cluster 6, which is in line with previous findings that GATA transcription factors promoted endothelial differentiation in avian (Kamei et al., 2011) and that ECs were differentiated from NKX2-5− cardiac progenitors (Giacomelli et al., 2017). Taken together, these data indicate that individual subpopulations not only have differential gene expression profiles but also exhibit distinct transcriptional regulatory networks.
Figure 3. Transcriptomic analyses of subpopulations.
(A) Principal component analysis (PCA) plots with the computation of the closest neighboring subpopulations.
(B) Venn diagram shows the overlap between differentially expressed genes in subpopulations.
(C-F) Heatmaps show hierarchical clustering of differentially expressed genes that are enriched in (C) G+R+, (D) G−R+, (E) G+R−, and (F) G−R− with at least 2-fold higher than at least two other subpopulations. Representative upregulated genes are listed in the box.
(G-J) GO analyses of upregulated genes in (G) G+R+, (H) G−R+, (I) G+R−, and (J) G−R−. The significance is shown as –Log10FDR. See also Figure S4 and Table S3.
Identification of a cell surface marker for the G+R+ subpopulation
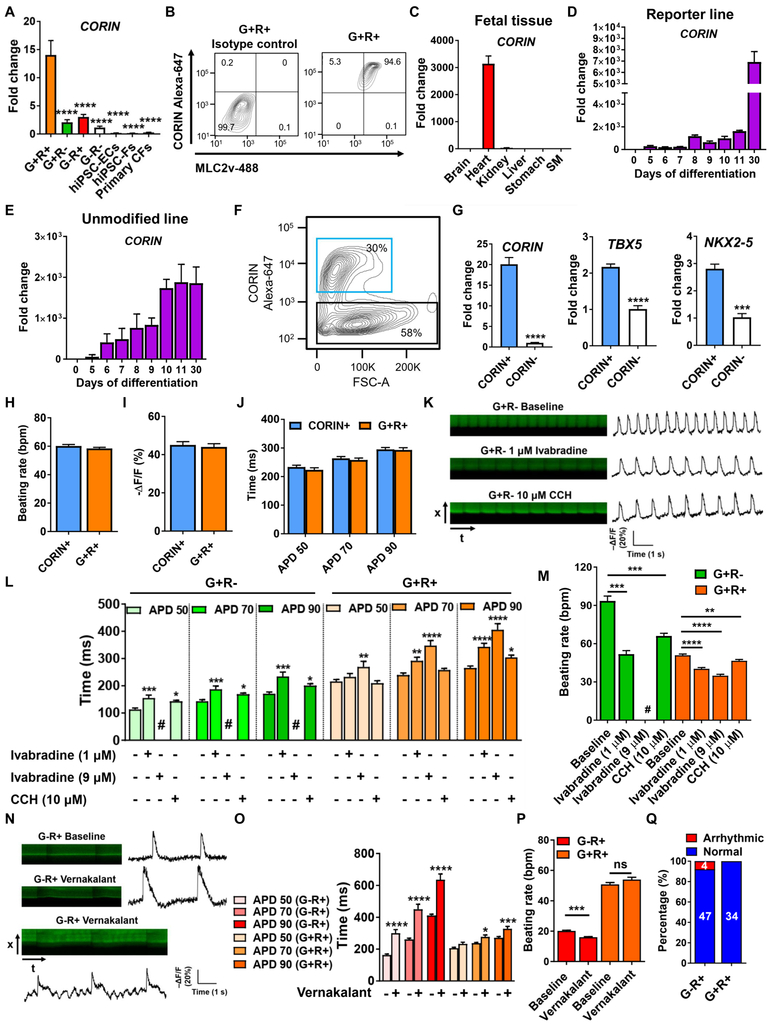
As functional analyses show advantages of G+R+ CMs, we next sought to identify a cell surface marker for isolating the G+R+ subpopulation. We discovered one gene called CORIN encoding a type II transmembrane serine protease (Figure 3B and Table S3), which can enrich the G+R+ subpopulation. Quantitative PCR (qPCR) analysis confirmed the significant upregulation of CORIN in the G+R+ subpopulation (Figure 4A). Moreover, the expression of CORIN in primary cardiac fibroblasts (CFs), induced fibroblasts (hiPSC-Fs), and hiPSC-ECs was negligible (Figure 4A). Flow cytometry analyses showed that the majority of day 30 CORIN+ subpopulation were MLC2v+ (Figure 4B). We conducted additional analyses of human fetal tissues to confirm the dominant expression of CORIN in the heart (Figure 4C). The dynamic expression of CORIN showed that its expression was first detectable at the cardiac progenitor stage and continued to increase until day 30 post-differentiation (Figure 4D-E). We then used CORIN to sort hiPSC-CMs derived from unmodified hiPSCs on day 10 (Figure 4F). Indeed, CORIN+ cells showed upregulation of TBX5 and NKX2-5 compared to CORIN− cells (Figure 4G). Furthermore, electrophysiological properties of day 30 CORIN+CMs, including beating rate, AP amplitude, and APD, were comparable to those of G+R+ CMs (Figure 4H-J). Overall, these results demonstrate the feasibility of using CORIN to isolate the TBX5+NKX2-5+ subpopulation.
Figure 4. Application of TBX5Clover2/NKX2-5TagRFP hiPSC reporter.
(A) Expression of CORIN in isolated subpopulations, hiPSC-derived endothelial cells (hiPSC-ECs), hiPSC-derived fibroblasts (hiPSC-Fs), and primary cardiac fibroblasts (CFs) (n=3).
(B) Flow cytometry of CORIN and MLC-2v in day 30 G+R+ CMs.
(C) Expression of CORIN in human fetal tissue.
(D-E) Dynamic expression of CORIN during cardiac differentiation in (D) the reporter and (E) unmodified hiPSC-CMs from three independent differentiation.
(F) Representative flow cytometry plot of day 10 live cells shows CORIN+ (blue box) and CORIN− (black box) cells.
(G) Expression of CORIN, TBX5, and NKX2-5 (n=3).
(H-J) Comparable AP properties including beating rate, AP amplitude, and APD between day 30 CORIN+ and G+R+ CMs (CORIN+, n=38; G+R+, n=55).
(K) Optical imaging of APs in G+R− hiPSC-CMs with and without treatment of ivabradine or carbamylcholine chloride (CCH).
(L-M) The effect of ivabradine and CCH on (L) APD and (M) beating rate (G+R− baseline, n=36; G+R− 1 μM ivabradine, n=21; G+R− CCH, n=19; G+R+ baseline, n=31; G+R+ 1 μM ivabradine, n=20; G+R+ 9 μM ivabradine, n=16; G+R+ CCH, n=22). #, stop beating.
(N) Optical imaging of APs in G−R+ hiPSC-CMs with and without treatment of vernakalant.
(O-P) The effect of vernakalant on (O) APD and (P) beating rate (G−R+ baseline, n=45; G−R+ vernakalant, n=47; G+R+ baseline, n=25; G+R+ vernakalant, n=34).
(Q) Vernakalant induces arrhythmias in G−R+ CMs. Numbers indicate the cell number with and without arrhythmias. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (mean±SEM). See also Table S4.
Application of lineage-specific CMs in precise drug testing
We next investigated whether lineage-specific CMs were applicable to assess subtype-specific drug effects. Ivabradine, a “funny” current (If) inhibitor, has been used as a heart rate reducing drug. Ivabradine (1 μM) significantly prolonged APD50 by 38% in G+R− CMs, but not in G+R+ CMs (Figure 4K-L). Notably, despite ivabradine increased APD70 and APD90 in G+R+ CMs, its effect on G+R− CMs was greater than G+R+ CMs (31% vs 21% in APD70, and 37% vs 29% in APD90) (Figure 4L). This greater effect on APD in G+R− CMs led to a concurrent reduction in beating rate to a greater extent (45%) in G+R− CMs than G+R+ CMs (20%) (Figure 4M). Strikingly, a higher dose of ivabradine (9 μM) abolished G+R− CM beating, although it prolonged APD and reduced beating rate in G+R+ CMs (Figure 4L-M). Carbachol (CCH) is a cholinergic agonist that enhances IKACh through interaction with acetylcholine (ACh) receptors, leading to a negative chronotropic effect. Although ACh receptors are expressed in all three CM subtypes, nodal cells tend to show higher densities of IKACh than ventricular CMs (Protze et al., 2017). CCH had a more pronounced effect on beating rate and APD in G+R− than G+R+ CMs (Figure 3K-M).
To test whether G−R+ CMs also act as a preclinical model for precise drug testing, we examined the effect of vernakalant (a drug used for cardioversion of recent-onset atrial fibrillation) on G−R+ CMs (Savelieva et al., 2014). Vernakalant significantly prolonged APD to a greater extent in G−R+ CMs than G+R+ CMs, leading to a significant decrease in beating rate in G−R+ but not G+R+ CMs (Figure 4N-P). Vernakalant also caused arrhythmias in 9% of G−R+ CMs but not in G+R+ CMs, suggesting that G−R+ CMs are more susceptible to vernakalant-induced AP effects (Figure 4N and 4Q).
In summary, these findings not only corroborate that G+R− and G−R+ CMs belong to nodal and atrial subtypes, respectively, but also bear the potential for precise testing of the potency and safety window of new drugs targeting specific CM subtypes.
DISCUSSION
Recent decades have seen significant progress in understanding control of cardiogenesis that establishes the foundation for generating cardiac lineage cells from hiPSCs. In this study, we delineated human cardiac lineages using a hiPSC TBX5Clover2/NKX2-5TagRFP double reporter. We isolate four different cardiac lineage subpopulations during cardiac differentiation with distinct molecular and functional features.
We identified a TBX5+NKX2-5+ subpopulation marked by FHF markers (TBX5, NKX2-5, and HCN4) and predominantly differentiated into ventricular CMs. Overexpression of Tbx5 in mouse embryonic stem cells favored development of the FHF (Herrmann et al., 2011). Single cell RNA-seq analysis of mouse E11.5 CMs showed a higher expression of Tbx5 in the left ventricular CMs compared with the right counterparts, confirming that Tbx5 is an important regulator of the FHF lineage (DeLaughter et al., 2016). These lines of evidence suggest that the TBX5+NKX2-5+ CMs most likely represent left “ventricular-like” CMs.
The high expression of KCNJ2 in TBX5+NKX2-5+ ventricular CMs underscored the low MDP. Animal studies found that the expression of Kir2.1 and corresponding IK1 current density was higher in the left than right ventricular CMs (Samie et al., 2001; Warren et al., 2003), further indicating that TBX5+NKX2-5+ ventricular CMs are functionally closer to the left ventricular CMs. Given the high purity and functional properties of TBX5+NKX2-5+ ventricular CMs, this subpopulation holds great potential for further applications. For example, by subjecting engineered heart tissues (EHTs) to electrical stimulation with increased intensity, the maturity of EHTs has been significantly improved (Ronaldson-Bouchard et al., 2018). Despite of this feature, the EHTs still represent a heterogeneous pool of CMs including all three CM subtypes, which may significantly limit the therapeutic application and drug testing. However, the purified TBX5+NKX2-5+ ventricular CMs may further advance the tissue engineering field by generating pure ventricular EHTs.
Our study indicates that CORIN is enriched in the TBX5+NKX2-5+ subpopulation and enables easy and routine access to the “FHF-like” subpopulation. CORIN was first identified as a mosaic transmembrane serine protease in the heart by in situ hybridization (Yan et al., 1999). In mouse embryo (E9.5), CORIN is detected in both atrium and ventricle and has been suggested to play a role in heart development (Yan et al., 1999). It remains unclear why CORIN is dominantly expressed in the TBX5+NKX2-5+ subpopulation during cardiac differentiation, further in vivo lineage trace study may reveal the lineage origin of CORIN expressing CMs. The identification of CORIN as a cell surface marker for the TBX5+NKX2-5+ subpopulation allows to isolate lineage-specific ventricular CMs used for preclinical studies. One recent study also identified a cell surface marker, CD235a, to monitor the mesoderm that differentiates into ventricular CMs (Lee et al., 2017). As noted in their study, it is unknown whether CD235a+ mesoderm generates left or right ventricular CMs or a mixture of both. However, here we demonstrate that the TBX5+NKX2-5+ CMs appear to have a genetic and functional similarity to the left ventricular CMs.
We also identified two other cardiogenic lineage subpopulations: TBX5+NKX2-5− and TBX5−NKX2-5+ subpopulations. The dominant CM subtype in TBX5−NKX2-5+ subpopulation was atrial subtype, but these cells displayed a slower beating rate compared with atrial CMs in the other subpopulations, as well as atrial CMs generated by modulating the retinoic acid pathway (Lee et al., 2017). This distinct phenotype may be attributed to the relatively high expression ratio of NKX2-5 to TBX5, which is known to regulate ion channel expression associated with AP and beating rate (Arnolds et al., 2012). Despite these differences, consistent with a previous study (Devalla et al., 2015), we found that TBX5−NKX2-5+ atrial CMs exhibited greater sensitivity to vernakalant-induced changes in APs. We were surprised to detect only a small number of ventricular CMs in the G−R+ subpopulation, given the fact that the SHF lineage contributes to the right ventricle and atria in vivo. It is worth noting that within the SHF, progenitors giving rise to the atria and inflow tract are considered posterior SHF that is marked by only ISL1, whereas TBX1+ISL1+ precursor cells develop into the right ventricle and outflow tract (Paige et al., 2015). The low expression of TBX1 in the G−R+ subpopulation is in line with the dominant differentiation of atrial CMs. It remains unclear why only posterior SHF is induced under our standard differentiation condition, but the absence of 3D microenvironmental cues could explain the discrepancy between in vitro differentiation and in vivo heart development.
We found that the TBX5+NKX2-5− subpopulation was highly enriched with epicardial lineage markers and primarily differentiated into nodal CMs. These findings conform with studies showing that activation of TBX5 and repression of NKX2-5 are required for epicardium and nodal cell fate determination (Witty et al., 2014). Our results are also consistent with recent findings that pacemaker-like CMs are derived from the NKX2-5− population (Protze et al., 2017). The TBX5+NKX2-5− nodal CMs have similar molecular features such as the upregulated expression of SHOX2, TBX5, and KCNJ3 and electrophysiological properties compared with nodal CMs generated using the direct differentiation protocol (Protze et al., 2017).
In summary, we present here a approach for deconvolution of human cardiac lineages and prospective isolation of lineage-specific cardiovascular cells. This is the first demonstration of the ability to identify four distinct cardiovascular lineage subpopulations, providing insights into the genetic regulation of human cardiac lineage specification and paving the way toward preclinical applications.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Joseph Wu (joewu@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary human samples
The protocols used in this study were approved by the Institutional Review Board (IRB) at Stanford University. The written consent was obtained from all the donors involved in this study. Fetal tissues including brain, heart, kidney, liver, stomach, skeletal muscle were acquired from a male aborted fetus at 16.4 weeks of age.
Human iPSCs and culture condition.
hiPSCs from healthy donors were obtained from Stanford Cardiovascular Institute Biobank. hiPSCs were routinely maintained in the E8 medium (GIBCO, Life Technologies) and grown on Matrigel-coated (Fisher Scientific) 6-well plates. Confluent cells were expanded into 6-well plates at a 1:12 ratio after a 6-min incubation with gentle cell dissociation reagent (STEMCELL Technologies) at 37 °C. Cells were maintained in the E8 medium supplemented with 10 μM Y-27632, ROCK inhibitor (Fisher Scientific) for 24 hr after re-plating, and the medium was then switched to E8 medium. A medium change was performed every 24 hr. Cultures were maintained at 37 °C in a humidified incubator with 5% CO2 and 5% O2.
CRISPR/Cas9-mediated genome editing.
To establish the TBX5Clover2/NKX2-5TagRFP hiPSC double reporter line, two gRNAs were designed and cloned into the PX458 plasmid (Addgene). Clover2 or TagRFP was placed upstream of TBX5 and NKX2-5 start codon, respectively, followed by IRES to allow expression of target transcription factor genes. The fragments of 5’ homology arm of TBX5 or NKX2-5 targeting vectors and adjunct half Clover2 or TagRFP sequences were synthesized as GeneArt® Strings™ DNA fragments (Life Technologies). Oligonucleotides were cloned into the PB-MV-PGK-Puro-TK (or PB-MV-PGK-Neo-TK) vector (Transposagen) facilitated by NotI and BsiWI restriction sites. The fragments of 3’ homology arm of targeting vectors and adjunct half Clover2 or TagRFP sequences were synthesized as described above, and cloned to vectors using Nsil and AscI restriction sites. Both CRISPR/Cas9 plasmids and targeting vectors were delivered into hiPSCs by Lipofectamine® 3000 according to the manufacturer’s instructions (Life Technologies). Subsequently, cells with targeting vector integration were selected by puromycin (0.25 μg/mL) (Life Technologies) and Geneticin (400 μg/mL) (Life Technologies). Targeted clones were identified using a PCR screening strategy. To excise the selection cassette, transient expression of piggyBac transposase was performed by transfection of excision-only piggyBac™ transposase mRNA (Transposagen Biopharmaceuticals) using Lipofectamine® MessengerMAX™ (Life Technologies), followed by negative selection using gancyclovir (2 μM) (Sigma Aldrich). The established clones were genotyped by PCR and sequencing.
METHOD DETAILS
Differentiation of hiPSCs to CMs.
For monolayer-based CM differentiation, cells were passaged at a 1:6 ratio, and grown for three days to 85% confluence. The medium was replaced with RPMI supplemented with B27 without insulin (Life Technologies) and 8 μM CHIR-99021 (Selleck Chemicals) and cells were transferred to a humidified incubator with 5% CO2 and 21% O2 at 37 °C. Forty eight hours later, the medium was changed to RPMI-B27 without insulin for 24 hr, and then to RPMI-B27 without insulin supplemented with 5 μM IWR-1 (Selleck Chemicals) for 48 hr. On day 5, the medium was changed back to RPMI-B27 without insulin for 48 hr, and then switched to RPMI-B27 for 72 hr. On day 11, the medium was transiently changed to RPMI-B27 without D-glucose (Life Technologies) for 72 hr to allow metabolic purification of CMs. Cells were then dissociated after a 6-min incubation with Accutase (Sigma Aldrich) at 37 °C followed by seeding into 6-well plates. A second metabolic purification was performed 48 hr after re-plating as described above. After the second purification, CMs were continuously cultured in RPMI-B27 until further use for experiments. The CMs that underwent metabolic purification were composed of the non-sorted population.
Differentiation of hiPSCs to endothelial cells (ECs).
hiPSCs were split at a 1:14 ratio, and grown for 4 days to 75% confluence. Media was changed to RPMI-B27 without insulin as described above, supplemented with 6 μM CHIR99021 for two days. Media was then replaced by RPMI-B27 without insulin, supplemented with 2 μM CHIR99021 for two more days, followed by 24 hr of RPMI-B27 without insulin. On day 5 of endothelial differentiation, the medium was changed to EGM2 (Lonza), supplemented with 50 ng/mL VEGF, 20 ng/ml BMP4, and 20 ng/ml FGFb for 7 days. On day 12, cells were dispersed using TrypLE (5 min, at 37 °C) and the CD144-positive population was sorted using MACS columns and CD144-conjugated magnetic microbeads (Miltenyi Biotec), as per the manufacturer’s instructions. After selection, cells were seeded on 0.2% gelatin-coated plates, and maintained in EGM2 media. ECs were used for further experiments.
Differentiation of hiPSCs to epicardial cells and fibroblasts.
The differentiation of hiPSCs to epicardial cells was followed using the previously published protocol with modifications (Bao et al., 2017). Briefly, cells were passaged at a 1:6 ratio, and grown for three days to 85% confluence. The medium was replaced with RPMI supplemented with B27 without insulin and 6 μM CHIR-99021. Forty eight hours later, the medium was changed to RPMI-B27 without insulin for 24 hr, and then to RPMI-B27 without insulin supplemented with 5 μM IWR-1 (Selleck Chemicals) for 48 hr. On day 5, the medium was changed back to RPMI-B27 without insulin for 24 hr. On day 6, cells were dissociated with Accutase and re-plated at a 1:12 split in an epicardial differentiation medium containing Advanced DMEM/F12 (ThermoFisher Scientific) with 2.5 mM GlutaMAX (ThermoFisher Scientific) and 100 μg/ml ascorbic acid (Sigma-Aldrich), supplemented with 10 μM Y-27632 for 24 hr. On days 7 and 8, the medium was replaced with the epicardial differentiation medium with 6 μM CHIR-99021. On day 9, the medium was changed to the epicardial differentiation medium supplemented with 2 μM SB431542 (Selleck Chemicals) every other day. Flow cytometry and gene expression analysis of WT1+ cells were conducted on day 30. The differentiation of epicardial cells into fibroblasts was followed by the previous protocol (Bao et al., 2017). Briefly, hiPSC-epicardial cells were treated with 10 ng/ml bFGF for 6 days.
Immunofluorescence staining and confocal microscopy imaging.
hiPSC-CMs grown on Matrigel-coated glass coverslips (Warner Instruments) were fixed in 4% paraformaldehyde (PFA) for 10 min and permeabilized with 0.2% Triton X-100 in phosphate buffer saline (PBS; pH 7.4) for 1 hr at room temperature. Cells were then blocked with 5% bovine serum albumin (BSA) in 0.2% Triton X-100, followed by an overnight incubation with primary antibodies to Cardiac troponin (cTnT) (Abcam; ab45932), MLC-2v (Synaptic Systems; 310111), ZO1 (ThermoFisher Scientific; 33-9100), WT1 (Abcam; ab89901), and α-smooth muscle actin (α-SMA) (Abcam; ab7817) at a 1:100 dilution in PBS with 0.1% Triton X-100 and 1% BSA at 4 °C. Cells were washed three times with 0.2% Tween-20 PBS solution for 5 min each, and then incubated for 1 hr at room temperature in the dark with goat anti-mouse or anti-rabbit immunoglobulin G conjugated to Alexa Fluor 488 and 594 secondary antibodies (Life Technologies) (1:200, diluted in PBS with 0.1% Triton X-100 and 1% BSA). Cells were washed again three times as described above, mounted with ProLong® Gold Antifade Mountant with DAPI (Life Technologies; P36935) on Superfrost Plus slides (Thermo Scientific). Fluorescent images were captured on a Zeiss LSM 710 confocal microscope (20×, NA: 0.8 and 40× oil-immersions, NA: 1.3 objectives). Image processing and data analysis were performed using ImageJ.
Flow cytometry and cell sorting.
hiPSC-derived cells (from day 5 to day 10 post-differentiation) were dissociated with Accutase as described above, filtered through a 35-μm cell strainer snap cap, and collected in a 5 ml FACS tube (Corning). Live cells were sorted on a BD Biosciences FACS Aria II instrument fitted with a 100 μm nozzle using FACSDiva software. Live cells were gated on the basis of forward scatter and side scatter. Cells were sorted into RPMI B27 with 10 μM Y-27632 and 5% KnockOut serum replacement (ThermoFisher Scientific). Flow cytometry gates were set to minimize cross-contamination by neighboring subpopulations. For experiments of genetic and functional characterization, cells without metabolic purification were sorted on day 10, followed by re-plating on 12-well plates. Colored (G+R+, G+R−, and G−R+) cells were cultured in the RPMI-B27 medium, and G−R− cells were cultured in EGM-2 (Lonza) supplemented with 50 ng/mL VEGF, 20 ng/ml BMP4, and 20 ng/ml FGFb for later characterization.
Flow cytometry analyses of fixed cells were conducted following BD staining protocol. Briefly, cells were fixed in BD Cytofix/Cytoperm (Fisher Scientific) for 20 min at 4 °C. Primary antibodies were labeled in BD Perm/Wash buffer with 1: 200 dilution for 30 min at room temperature. Secondary antibodies were labeled BD Perm/Wash buffer with 1: 250 dilution on ice for 30 min, unless conjugated primary antibodies [CORIN Alexa-647 (Bioss Antibodies, bs-7685R-A647) and CD31 Alexa-488 (ThermoFisher Scientific, BD558068)] were used. Flow cytometry analyses were conducted using the BD Biosciences FACS Aria II instrument fitted with a 100 μm nozzle using FACSDiva software. For live cell sorting experiment, hiPSC-CMs were dissociated into single cell suspension and incubated with CORIN Alexa-647 in HBSS (Fisher Scientific) with 5% FBS and 10 μM Y-27632 on ice for 30 min. Cells were sorted as described above.
Lentiviral vector infection and optical imaging of ASAP2.
hiPSC-CMs were seeded on Matrigel-coated 35 mm glass bottom dish with 20 mm micro-well (Cellvis) and infected with ASAP2 lentivirus with the multiplicity of infection (MOI) of three in a RPMI-B27 medium. The ASAP2 lentiviral vector was a gift from Michael Lin’s laboratory and lentivirus package was conducted by VectorBuilder. The medium containing lentivirus was replaced with fresh RPMI-B27 24 hr after infection. Optical imaging of ASAP2 was performed five days after infection. Cells were maintained in Tyrode’s solution at 37 °C during recording. ASAP2 was excited at 488 nm and emission was collected over 510 nm. Line scan images were acquired on a Zeiss LSM710 confocal microscope (Zeiss) equipped with a 20× objective (NA: 0.8). Line scan images were processed to enhance brightness for presentation purpose using ImageJ. Raw imaging data were analyzed using a custom-written MATLAB program. ASAP2 data were presented as -ΔF/F.
Whole cell patch-clamp recordings.
hiPSC-CMs from days 35 to 40 were seeded on Matrigel-coated glass coverslips (Warner Instruments) for patch-clamp recordings. These recordings were conducted using an EPC-10 patch-clamp amplifier (HEKA). Glass pipettes with 3-4 MΩ resistance were prepared using thin-wall borosilicate glass (A-M System) with a micropipette puller (Sutter Instrument). For recording, coverslips containing CMs were transferred to a RC-26C recording chamber (Warner Instruments) mounted on the stage of an inverted microscope (Nikon). Action potentials (APs) were recorded from CMs superfused with Tyrode solution at 37 °C (TC-324B heating system, Warner, USA). The Tyrode solution consisted of NaCl (140 mM), KCl (5.4 mM), CaCl2 (1.8 mM), MgCl2 (1 mM), HEPES (10 mM), and glucose (10 mM); pH was adjusted to 7.4 with NaOH. The pipette solution consisted of KCl (120 mM), MgCl2 (1 mM), Mg-ATP (3 mM), HEPES (10 mM), and EGTA (10 mM); pH was adjusted to 7.2 with KOH. Data were acquired using PatchMaster software (HEKA) and digitized at 1.0 kHz. Data were analyzed using a custom-written MATLAB program. Ventricular and atrial subtypes were classified using a ratio of (APD30-40/APD70-80). Ventricular-like APs had a ratio (APD30–40/APD70–80) of ~2.5 compared with the atrial-like ratio of ~1.1; thus APs with a ratio >1.5 were categorized as ventricular like (Ma et al., 2011).
Seahorse extracellular metabolic-flux assay.
Day 35 hiPSC-CMs were plated at 30,000 per well in Synthemax II-coated Seahorse plates in RPMI-B27. The bioenergetic responses of cells were measured with the Seahorse Bioscience XF96 flux Analyzer following instruction in the XF cell Mito Stress Test Kit User Guider. Baseline oxygen consumption rate (OCR) measurements were performed first, followed by injection of oligomycin to a final concentration of 5 μM. After three measurements, FCCP was injected to a final concentration of 5 μM and three measurements were conducted. Lastly, rotenone and antimycin A were injected to reach a final concentration of 2 μM followed by three measurements. CMs were subjected to four conditions: basal assay medium (DMEM only), D-glucose (5 μM), palmitate (167 μM), or pyruvate (1 mM). OCR was normalized to live cell count using a PrestoBlue dye according to manufacturer’s protocol and quantified on a TECAN Pro1000 machine (Stanford High-Throughput Bioscience Center).
Phase-contrast video image-based contraction assays.
Phase-contrast cell motion movies of CM contraction were captured using a high-speed digital CMOS camera (KP-FM400WCL, Hitachi Kokusai Denki Engineering) mounted on an inverted microscope (Eclipse Ti, Nikon). Video images of day 35 hiPSC-CMs in 12-well plates (37 °C and 5% CO2) were recorded using SI8000C View Software (Sony) as sequential phase-contrast images with a 10× objective (NA: 0.3) at a frame rate of 75 frame per second (fps) and a resolution of 1024 × 1024 pixels. Data analysis was performed using SI8000C Analyzer software (Sony).
Immunoblotting.
Protein samples were prepared using RIPA lysis and an extraction buffer (ThermoFisher Scientific; 89900) supplemented with a protease and phosphatase inhibitor cocktail (ThermoFisher Scientific; 78440). Each sample was subjected to electrophoresis on 4–12% NuPAGE Bis–Tris gradient gels (Life Technology; NP0335), and proteins were transferred to mini nitrocellulose membranes (Biorad; 1704158) using a semi-dry-based transfer system (Bio-Rad). Membranes were incubated overnight with rabbit Kir2.1 antibody (Abcam; ab109750, 1:1000), rabbit TBX5 antibody (Abcam; ab137833, 1: 500), or mouse NKX2-5 antibody (Santa Cruz Biotechnology; sc-376565, 1:1000), followed by incubation for 1 hr with anti-rabbit (Abcam; ab6721, 1:1000) or anti-mouse (Abcam; ab6789, 1 :1000) horseradish peroxidase (HRP)-conjugated secondary antibody. The housekeeping protein GAPDH was blotted by incubating 1 hr with GAPDH-loading control antibody conjugated with HRP (ThermoFisher Scientic; MA5-15738-HRP, 1:10,000). Signals were detected by chemiluminescence (ThermoFisher Scientic; 34580). Blot intensity was quantified using Image Lab 5.2.1 (Biorad).
In vitro tube formation assay.
To assess the angiogenesis ability of TBX5−NKX2-5− cells in vitro, day 35 1×104 TBX5−NKX2-5− cells were seeded in wells pre-coated with Matrigel using an EGM-2 medium containing 50 ng/ml VEGF. Tube-like structures were imaged after 24 hr incubation.
In vivo angiogenesis assay.
To investigate the angiogenesis in vivo, day 35 1 × 106 TBX5−NKX2-5− cells were mixed in 200 μl growth factor-reduced Matrigel with 20 ng/ml VEGF (PeproTech; 100-20). The cell mixture was subcutaneously injected into NOD/SCID mice. Matrigel plugs were removed from mice after 4 weeks, then mounted in an OCT-embedding compound and kept in −80 °C. Tissue sections were cut using a cryostat and stained with human CD31 (Cell Signaling; 3528S) and VE-Cadherin antibody (Cell Signaling; 2500S) for imaging analysis.
RNA extraction and quantitative real-time polymerase chain reaction (qPCR).
Total RNA was extracted and purified using an RNeasy Mini kit (QIAGEN; 74134). The quantity and quality of RNA were determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific), and then RNA was reverse transcribed using iScript™ cDNA synthesis kit (Biorad; 1708890) according to the manufacturer’s instructions. The qPCR was performed using TaqMan® Universal PCR Master Mix (Fisher Scientific; 4304437) and Taqman probes (Life Technologies) (see Table S4) on a StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific) in 20 μL reaction. Relative mRNA levels were normalized to those of 18S mRNA in each reaction. Three biological replicates per group were used for qPCR.
RNA-sequencing (RNA-seq).
Each subpopulation was isolated on day 7.5, and RNA was extracted as described above. Library preparations were conducted using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs) and subjected to sequencing on a Hiseq 4000 platform (Novogene).
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed and graphed using Prism (GraphPad). Data were presented as mean±SEM Comparisons were conducted via one-way ANOVA test followed by multiple comparisons (Tukey post hoc analysis), or via an unpaired two-tailed Student’s t-test with significance defined by p<0.05 (*), p<0.01 (**), p<0.001 (***), and p<0.0001 (****).
RNA-seq Analysis.
RNA-seq reads were aligned to human reference genome hg19 by Tophat2 (Kim et al., 2013). FeatureCounts was applied to assign the reads to genes based on GENCODE genome annotation (version 19) (Liao et al., 2014). The differentially expressed gene (DEG) analysis (adjusted p<0.01) was implemented by DESeq in R. Hierarchical clustering was applied on DEGs by R. Functional enrichment analysis was implemented using Bioconductor GeneAnswers (https://www.bioconductor.org/packages/release/bioc/html/GeneAnswers.html).
Motif enrichment analysis was implemented by Homer (Heinz et al., 2010). GO pathway enrichment analysis was implemented using a previously described method (Xue et al., 2014). The sequences around the promoters of genes in each cluster were extracted (upstream: 400 bp, downstream: 100 bp), and only transcription factors that were significantly enriched (FDR < 0.00001) in any cluster were included.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data gene rated in this paper is GSE102202.
Supplementary Material
Table S3 (Related to Figure 3). Gene lists of Venn diagram.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-cTnT | Abcam | ab45932 RRID: AB_956386 |
| Anti-αSMA | Abcam | ab7817 RRID: AB_262054 |
| Anti-MLC-2V | Synaptic systems | 310111 RRID: AB_887738 |
| Anti-Kir2.1 | Abcam | ab109750 RRID:AB_10858446 |
| Anti-ZO1 | ThermoFisher Scientific | 33-9100 RRID:AB_2533147 |
| Anti-CD31 | Cell Signaling | 3528S RRID:AB_2160882 |
| Anti-WT1 | Abcam | ab89901 RRID:AB_2043201 |
| Anti-VE-Cadherin | Cell Signaling | 2500S RRID:AB_2077969 |
| CORIN-Alexa 647 | Bioss Antibodies | bs-7685R-A647 |
| Anti-GAPDH | ThermoFisher Scientific | MA5-15738-HRP RRID:AB_2537659 |
| CD31-Alexa 488 | BD Biosciences | BD558068 |
| Bacterial and Virus Strains | ||
| ASAP2 lentiviral vector | Gift from Michael Lin’s lab | NA |
| Biological Samples | ||
| Human fetal tissue | Wu lab tissue bank | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Essential 8™ medium | Life Technologies | A1517001 |
| RPMI no glucose | Life Technologies | 11875-020 |
| RPMI | Life Technologies | 11875-119 |
| B27 supplement minus insulin | Life Technologies | A1895601 |
| B27 supplement | Life Technologies | 17504-044 |
| EGM-2 BulleKit | Lonza | CC-3162 |
| CHIR 99021 | Selleck Chemicals | S2924 |
| IWR-1 | Selleck Chemicals | S7086 |
| Matrigel matrix | Corning | 354277 |
| Advanced DMEM/F12 | ThermoFisher Scientific | 12634028 |
| GlutaMAX | ThermoFisher Scientific | 35050061 |
| Ascorbic acid | Sigma-Aldrich | A8960 |
| Gentle cell dissociation reagent | STEMCELL Technologies | 07174 |
| Accutase | Sigma-Aldrich | A6964 |
| BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit | Fisher Scientific | BD 554714 |
| VEGF | PeproTech | 100-20 |
| bFGF | PeproTech | 100-18B |
| Ivabradine | Sigma-Aldrich | SML0281 |
| Vernakalant | AdooQ BioScience | A15275-5 |
| Carbachol | Sigma-Aldrich | C4382 |
| Y-27632 (ROCK inhibitor) | Selleck Chemicals | S1049 |
| SB431542 | Selleck Chemicals | S1067 |
| Puromycin | Life Technologies | A11138-03 |
| Geneticin | Life Technologies | 10131035 |
| piggyBac™ transposase mRNA | Transposagen Biopharmaceuticals | PBx-m |
| Lipofectamine® MessengerMAX™ | Life Technologies | LMRNA008 |
| Lipofectamine 3000 | Life Technologies | L3000015 |
| HBSS with Ca2+ and Mg2+ | Fisher Scientific | 14025092 |
| KnockOut serum replacement | ThermoFisher Scientific | 10828028 |
| ProLong® Gold Antifade Mountant with DAPI | Life Technologies | P36935 |
| TaqMan® Universal PCR Master Mix | Fisher Scientific | 4304437 |
| Critical Commercial Assays | ||
| RNeasy plus Mini Kit | QIAGEN | 74136 |
| iScript™ cDNA synthesis kit | BioRad | 1708890 |
| NEBNext® Ultra™ II RNA Library Prep Kit for Illumina® | NEB | E7630L |
| Deposited Data | ||
| RNA-seq | This paper | GSE102202 |
| Experimental Models: Cell Lines | ||
| Human iPSCs | Stanford CVI Biobank | N/A |
| TBX5Clover2/NKX2-5TagRFP human iPSCs | This paper | N/A |
| Oligonucleotides | ||
| See Table S4 for qPCR primers | ThermoFisher Scientific | N/A |
| Software and Algorithms | ||
| GraphPad Prism version 7.0 | GraphPad Softward | N/A |
| FlowJo v10 | FLOWJO, LLC | N/A |
| ImageJ | NIH | N/A |
| MATLAB | Mathworks | N/A |
| Tophat2 | Johns Hopkins University | https://ccb.jhu.edu/software/tophat/index.shtml |
| FeatureCounts | Walter Eliza Hall Institute of Medical Research | http://bioinf.wehi.edu.au/featureCounts/ |
| DESeq | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq.html |
| GeneAnswers | Bioconductor | https://www.bioconductor.org/packages/release/bioc/html/GeneAnswers.html |
| Homer | University of California San Diego | http://homer.ucsd.edu/homer/ |
Highlights:
TBX5Clover2/NKX2-5TagRFP reporter enables purification of 4 cardiac subpopulations
Different cardiac lineages differentiate into specific cardiac cell types
CORIN is a cell surface marker for TBX5+NKX2-5+ subpopulation
Lineage-specific cardiomyocyte subtypes can be used for precise drug testing
ACKNOWLEDGEMENTS
We thank Professor Michael Z Lin (Department of Bioengineering, Stanford University) for providing ASAP2 lentiviral vector. This work was supported by American Heart Association Merit Award, NIH R01 HL130020, NIH R01 HL133272, NIH R01 HL128170, (J.C.W.), NIH R01 NS089533 (H.M.B.), NIH K01 HL135455 (N.S), NIH K99 HL133473 (H.W), American Heart Association Postdoctoral Fellowship 16POST31150011 (H.W), and TRDRP Fellowship 497329 (J.Z.Z).
Footnotes
DECLARATION OF INTERESTS
J.C.W is a co-founder of Khloris Biosciences, but the work presented was performed independently and has no competing interests.
H.M.B. is co-founder of Myoforte Therapeutics, but the work presented was performed independently and has no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, et al. (2009). The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ. Res 104, 1267–1274. [DOI] [PubMed] [Google Scholar]
- Arnolds DE, Liu F, Fahrenbach JP, Kim GH, Schillinger KJ, Smemo S, McNally EM, Nobrega MA, Patel VV, and Moskowitz IP (2012). TBX5 drives Scn5a expression to regulate cardiac conduction system function. J. Clin. Investig 122, 2509–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin AF, Compton LA, Love JD, Brown CB, and Barnett JV (2008). Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFbeta. Dev Dyn 237, 366–376. [DOI] [PubMed] [Google Scholar]
- Birket MJ, Ribeiro MC, Verkerk AO, Ward D, Leitoguinho AR, den Hartogh SC, Orlova VV, Devalla HD, Schwach V, Bellin M, et al. (2015). Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol 33, 970–979. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. (2001). A murine model of Holt-Oram syndrome defines roles of the T-Box transcription factor Tbx5 in cardiogenesis and disease. Cell 106, 709–721. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, and Zaffran S (2005). Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet 6, 826–837. [DOI] [PubMed] [Google Scholar]
- Cai C-L, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. (2008). A myocardial lineage derives from Tbx18 epicardial cells. Nature 454, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberland S, Yang HH, Pan MM, Evans SW, Guan S, Chavarha M, Yang Y, Salesse C, Wu H, Wu JC, et al. (2017). Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife 6, e25690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churko JM, Garg P, Treutlein B, Venkatasubramanian M, Wu H, Lee J, Wessells QN, Chen SY, Chen WY, Chetal K, et al. (2018). Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat. Commun 9, 4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaughter Daniel M., Bick Alexander G., Wakimoto H, McKean D, Gorham Joshua M., Kathiriya Irfan S., Hinson John T., Homsy J, Gray J, Pu W, et al. (2016). Single-cell resolution of temporal gene expression during heart development. Dev. Cell 39, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, Jackson C, Gkatzis K, Elliott DA, Chuva de Sousa Lopes SM, Mummery CL, et al. (2015). Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med 7, 394–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Chiravuri M, van der Meer P, Feinberg AW, Shi X, Shao Y, Wu SM, Parker KK, and Chien KR (2009). Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science 326, 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, et al. (2009). Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol 327, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E, Bellin M, Sala L, van Meer BJ, Tertoolen LG, Orlova VV, and Mummery CL (2017). Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann F, Bundschu K, Kuhl SJ, and Kuhl M (2011). Tbx5 overexpression favors a first heart field lineage in murine embryonic stem cells and in Xenopus laevis embryos. Dev Dyn 240, 2634–2645. [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, et al. (2007). Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 21, 1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei CN, Kempf H, Yelin R, Daoud G, James RG, Lassar AB, Tabin CJ, and Schultheiss TM (2011). Promotion of avian endothelial cell differentiation by GATA transcription factors. Dev. Biol 353, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinopoulos I, Ishida H, Saba R, Ruchaya P, Cabrera C, Struebig M, Barnes M, Terry A, Kaneko M, Shintani Y, et al. (2015). Single-Cell Expression Profiling Reveals a Dynamic State of Cardiac Precursor Cells in the Early Mouse Embryo. PLoS One 10, e0140831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Protze SI, Laksman Z, Backx PH, and Keller GM (2017). Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21, 179–194.e174. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, Kamp TJ, Kolaja KL, Swanson BJ, and January CT (2011). High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol 301, H2006–H2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan R, Boukens BJ, and Christoffels VM (2017). Lineages of the Cardiac Conduction System. J Cardiovasc Dev Dis 4, pii: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Plonowska K, Xu A, and Wu SM (2015). Molecular regulation of cardiomyocyte differentiation. Circ. Res 116, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, and Keller GM (2017). Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol 35, 56–68. [DOI] [PubMed] [Google Scholar]
- Puskaric S, Schmitteckert S, Mori AD, Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H, and Rappold G (2010). Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum. Mol. Genet 19, 4625–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, and Vunjak-Novakovic G (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie FH, Berenfeld O, Anumonwo J, Mironov SF, Udassi S, Beaumont J, Taffet S, Pertsov AM, and Jalife J (2001). Rectification of the background potassium current: a determinant of rotor dynamics in ventricular fibrillation. Circ. Res 89, 1216–1223. [DOI] [PubMed] [Google Scholar]
- Savelieva I, Graydon R, and Camm AJ (2014). Pharmacological cardioversion of atrial fibrillation with vernakalant: evidence in support of the ESC Guidelines. Europace 16, 162–173. [DOI] [PubMed] [Google Scholar]
- Spater D, Abramczuk MK, Buac K, Zangi L, Stachel MW, Clarke J, Sahara M, Ludwig A, and Chien KR (2013). A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat. Cell Biol 15, 1098–1106. [DOI] [PubMed] [Google Scholar]
- Stieber J, Herrmann S, Feil S, Löster J, Feil R, Biel M, Hofmann F, and Ludwig A (2003). The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc. Natl. Acad. Sci. USA 100, 15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M, Guha PK, Berenfeld O, Zaitsev A, Anumonwo JM, Dhamoon AS, Bagwe S, Taffet SM, and Jalife J (2003). Blockade of the inward rectifying potassium current terminates ventricular fibrillation in the guinea pig heart. J. Cardiovasc. Electrophysiol 14, 621–631. [DOI] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, and Christoffels VM (2009). Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res 104, 388–397. [DOI] [PubMed] [Google Scholar]
- Witty AD, Mihic A, Tam RY, Fisher SA, Mikryukov A, Shoichet MS, Li R-K, Kattman SJ, and Keller G (2014). Generation of the epicardial lineage from human pluripotent stem cells. Nat. Biotechnol 32, 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, et al. (2014). Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Sheng N, Seto M, Morser J, and Wu Q (1999). Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J. Biol. Chem 274, 14926–14935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3 (Related to Figure 3). Gene lists of Venn diagram.
Data Availability Statement
The accession number for the RNA-seq data gene rated in this paper is GSE102202.