Abstract
Radiomics is a fast-growing research area based on converting standard-of-care imaging into quantitative minable data and building subsequent predictive models to personalize treatment. It has been proposed as a study objective in clinical trial concepts and a potential biomarker for stratifying patients across interventional treatment arms. In recognizing the growing importance of radiomics in oncology a group of medical physicists and clinicians from NRG Oncology reviewed the current status of the field and identified critical issues, providing general assessment and early recommendations for incorporation in oncology studies.
Keywords: Radiomics, NRG Oncology
Radiomics and Applications
Radiomics introduction
The concept of relating imaging information to predicting prognosis and therapeutic response traces its root to the early days of robotics and computer vision in the 1960s, but its systemic application to quantitative imaging analysis dates to the beginning of the 1980s in areas such as computer-aided detection or diagnosis (CAD) 1. The application of this approach to biological markers and therapeutic endpoints only started in the past decade, when the concept of personalized medicine arose following the increasing use of genomics. Some early examples include the investigation of correlations between hepatocellular carcinoma imaging phenotypes with gene expression 2, and between PET-based features and radiotherapy response 3.
When initial studies to investigate whether MRI-based measurements of breast cancer volume could accurately assess the response to neoadjuvant chemotherapy proved promising 4, they were used as a springboard for a series of American College of Radiology Imaging Network (ACRIN) trials. The ACRIN trials investigated using serial MRI studies to predict therapeutic response to chemotherapy; this was called the ‘I-SPY TRIAL’ for ‘Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging And moLecular Analysis’ 5.
Since 2010, this field has been formalized with the term “radiomics”6. The term originates from the word “radio”, which refers to radiology, the science of acquiring medical images through the use of radiation (e.g., CT, PET, MRI). The suffix “omics” follows from the wholesome notion, which was firstly used in the term genomics to indicate the whole mapping of human genetics7. Currently, this process of extraction of massive quantitative information from anatomical/molecular images and relating them to corresponding biological information and clinical endpoints is an emerging field referred to as ‘radiomics’8.
Radiomics applications
With the recent advances of imaging techniques, imaging has extended its role to the whole spectrum of cancer management, from detection and diagnosis, to treatment response monitoring, and further risk surveillance. The clinical application of radiomics is expected to play important roles in every aspect of cancer management.
Tumor detection and diagnosis
One of the earliest applications for radiomics-driven method is in tumor detection, with the greatest success in lung and breast imaging. US Food and Drug Administration (FDA)-approved systems are currently being used in the clinic. A recent study utilizing commercially-available clinical tools identified many lung cancers that were initially missed by radiologists in the International Early Lung Cancer Action Program (I-ELCAP) trial 9.
Currently the trend moves to a direction called discovery radiomics 10,11. Instead of using pre-defined radiomics feature set, the image data are directly fed into the discovery engine, where a customized radiomics sequencer is constructed using deep learning architecture such as convolutional neural network (CNN), and then the descriptive radiomics sequencer can be applied to identify normal or abnormal tissues. Besides tumor detection, radiomics features have also been demonstrated helpful to identify various types of lesions. For example, Li et al. suggested that mammographic images contain computer-extractable information, which may distinguish between BRCA1/2 mutation carriers and non-carriers 12. Grimm et al. also observed that imaging features from DCE-MRI were strongly associated with Luminal A and Luminal B hormonal receptor positive molecular subtypes 13. Overall, early detection and identification of tumors could be useful for better stratification of patients and identification of subsequent treatment options.
Treatment outcome prediction for decision making support
Recently, significant interest in utilizing radiomics for early prediction of treatment response has emerged. In predicting pathological complete response (pCR) following neoadjuvant chemoradiation for locally advanced rectal cancer, Nie et al. showed improved prognostic values could be achieved using a voxelized radiomics analysis approach over conventional imaging metrics 14. Zhang et al., identified MRI-based radiomics as prognostic factor for progression-free survival in patients with nasopharyngeal carcinoma (NPC). The prediction power significantly outweighed than traditional TNM (tumor-node-metastasis) staging 15. Although the TNM staging is the cornerstone for treatment decision making. It is typically assessed based on gross anatomy information, not reflecting the intra-tumor heterogeneity. While the radiomics approach can characterize the intra-tumor heterogeneity noninvasively thus can add incremental value to the clinical information in assessing the treatment outcome.
Radiomics features from CT, PET/CT and CBCT have also shown predictive value for response to treatment 16–24. Investigators analyzed the daily non-contrast CTs, acquired during routine IGRT using an in-room CT, from patients with head and neck 25, lung 26 and pancreatic cancers 27,40. They reported that radiation can induce patient specific changes in CT texture features, and that these changes can be detected in the early phase of radiation therapy. Ohri et al., showed pre-treatment metabolic tumor volume (MTV) and heterogeneity textural metrics on PET/CT can be good prognostic factors for locally advanced non-small cell lung cancer (NSCLC) patients treated with chemo-radiotherapy based on data from ACRIN 6668/RTOG 0235 28. Buizza et al., in addition, showed the longitudinal temporal and spatial changes from PET/CT image could improve the early survival prediction for chemoradiation treatment29. Radiomics has also been used to predict radiation induced normal tissue toxicities, such as radiation pneumonitis 21 or xerostomia 30,31. These results suggest that a radiomics-based signature may emerge as an accepted imaging biomarker for predicting therapeutic outcome and for improving decision support in cancer treatment.
Risk assessment
Radiomics has also been extended to risk surveillance in several cancers. Liu et al., investigated the association between imaging features and low-grade gliomas (LGG) related epilepsy, and proposed a radiomics-based model for the prediction of associated risk32. Similarly, it is well known that mammographic density is an independent risk factor, and radiomics may provide much more information than breast density. 33–35. Li et al. investigated breast parenchymal patterns in mammographic images in 456 patients (53 with BRCA1/2 gene carrier, 75 with unilateral cancer and 328 with low-risk) 12. They demonstrated that women at high risk tend to have dense breasts with coarse and low-contrast texture patterns. Haberle et al. performed a case-control study with 864 cases vs. 418 controls 36. Of the 470 radiomics features explored, 46 remained in the final risk model; the radiomics model outperformed than the conventional risk model with mammographic density. These studies may promote future breast cancer prevention trials to investigate the role of radiomics to measure breast tissue composition in individual woman for personalized risk management.
Radiogenomics
In radiogenomics, the radiomics phenotype is correlated with a genomic profile. The hypothesis is that imaging may provide insight into tumor phenotypes which are driven by the heterogeneity of the genetic evolution. Radiogenomics is a very young field due to the lack of data consisting of both imaging and genomic measurements on the same set of tumors 37,38.
Recent studies in brain tumors 37,39,40, lung cancer 40–42 and breast cancer 13,43 suggest value for radiogenomics. National shared databases, such as the Cancer Imaging Archive (TCIA) and the Cancer Genome Atlas (TCGA), provide researchers opportunities to explore this field. Using MRI images from TCIA and clinical, histopathologic, and genomic data from TCGA, Li et al. investigated the relationship between MRI imaging phenotypes and multi-gene assays including Oncotype Dx, MammoPrint and PAM50 43. Multiple linear regression analyses demonstrated significant associations between radiomics signatures and multigene assay recurrence scores. Zinn et al. identified an association between high T2 FLAIR volumes, upregulation of periostin (POSTN), and downregulation of miR-219 using data from TCGA39. They noted high levels of POSTN were associated with mesenchymal tumors and shorter survival and further concluded that this approach may be valuable for identifying new targets for molecular inhibition or future therapies.
Although numerous radiomics features can be extracted from medical images, the method by which tumor pathophysiological processes give rise to imaging phenotypes remain unclear. More studies are required to confirm these associations to further elucidate the biological meaning of the radiomics features.
Radiomics Processes and Components
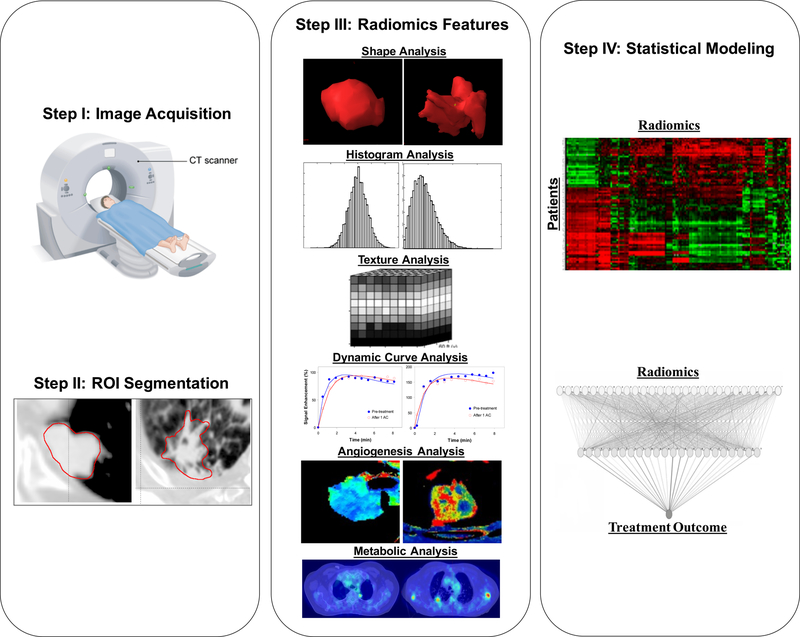
One goal of radiomics is to convert images into data that can be mined using high throughput computing. The process follows several general steps, as shown in Figure 1: (a) image acquisition, (b) region of interest (ROI) identification and segmentation, (c) quantitative image feature extraction, (d) data mining and informatics analysis.
Figure 1.
The general radiomics study workflow. Step I: Image acquisition; step II: region of interest identification and segmentation; step III: quantitative image feature extraction and Step IV: data mining and informatics analysis.
Image acquisition
The first step of image-based radiomics phenotyping involves image acquisition. Different image acquisitions will provide different, and often complementary information. For example, size or volume-based analysis can be obtained using anatomical MRI or CT. Measurements of perfusion can be determined from a series of dynamic-contrast-enhanced (DCE) MRI or CT acquisitions. Functional MRI, such as diffusion-weighted-imaging (DWI) can be used for tissue microcirculation and cellularity evaluation 14. Metabolic changes, such as rate of glucose metabolism, can be measured using fluorodeoxyglucose (FDG)-PET 44. Emerging functional and molecular imaging methods that are increasingly used in clinical trials may offer additional biomarkers. Human experts can help guide the choice of the imaging modalities tailored to the disease site of interest, clinical endpoints and potential treatment options.
Historically, imaging devices are designed for subjective interpretation of images, allowing clinicians to identify, for example, the presence and the location of a lesion. Subsequent technical innovation has largely focused on improving imaging quality, shortening scanning time, or integrating with treatment machines. Conversely, they are not primarily to provide quantitative measurements in a reproducible manner. The standardization of imaging acquisition protocols is typically lacking, and wide variations in reconstruction or acquisition parameters can exist. Zhao et al. studied repeat computed tomography (CT) data set from lung cancer patients and concluded that smooth and sharp reconstruction algorithms should not be used interchangeably 45. Galavis et al. assessed the variability of radiomics features extracted from PET due to different acquisition modes, reconstruction algorithms, post-filtering, and iteration numbers. 40 out of 50 features were shown to have substantial variability, up to 30%46. Due to the gradient strength of the scanner, pulse sequence used, method of contrast agent administration, k-space trajectory sampling, and other factors, results from MRI can vary more significantly 47–49. Because the quality of the radiomics data depends on the reliability of the acquisition protocols used in clinical centers, the impact of these variations on the stability of radiomics features needs to be thoroughly investigated and understood in future studies.
Region of interest (ROI) identification and segmentation
Defining regions of interest is a fundamental task within the practice of oncology. In radiology, human experts identify the presence, location and size of the suspicious areas, for diagnosis, staging or response assessment. In radiation oncology, the human experts must identify the tumor extent for treatment and organ-at-risks (OARs) for radiation sparing. Manual outlining by experienced radiologists or radiation oncologists is often treated as the gold standard. Yet it is labor intensive and suffers from high inter-operator or even intra-operator variabilities. ROIs may be contoured more consistently using semi- or fully automated methods such as thresholding, region growing, classifiers, clustering, Markov random field models, artificial neural networks, deformable models and atlas-guided approaches 50,51. Although full automation may present a new opportunity for standardized segmentation method, challenges related to complex anatomy or areas with low soft tissue contrast persist, and manual correction of contours by an experienced physician is often required. A rather new idea to avoid segmentation pitfalls is the use of a “digital biopsy”, which samples rather than segmenting the region of interest. This approach has been recently applied to sampling CT lung nodules 52. More recently, advanced machine learning based algorithms has been applied for image segmentation or sampling 53,54. There are also several large initiatives aimed at developing automatic segmentation solutions using deep learning. These include Google’s DeepMind 55, Microsoft’s InnerEye 56, Mirada’s DLCExpert 57, and the Grand Challenges in Biomedical Image Analysis 58. These automated segmentation tools have been shown to improve efficiency of structure set generation, particularly for organs-at-risk (OARs). In the near future, deep-learning-based segmentation tools may be robust enough for routine radiomics applications.
Radiomics features
The core of radiomics is the extraction of high-dimensional feature sets to quantify images. The features extracted from images can be divided into static (i.e., a snapshot of enhancement at one point in time) or dynamic (i.e., time variant) features according to the acquisition protocol used at the time of scanning 59.
Static image features
Several types of static image features can be applied to radiomics studies, including: (a) morphology-based features which are used to capture three-dimensional (3D) shape characteristics, such as volume and surface area, and sphericity which quantifies how close a 3D volume is to a sphere 60. A higher sphericity indicates a round shape while a lower value indicates an irregular or elongated shape, (b) intensity-based features which are used to quantify the gray-level distribution inside the ROI. Examples of first-order intensity-based features include mean, standard deviation, percentiles, kurtosis, and skewness. They are used to characterize the overall intensity variability. While second-order intensity-based features, also referred to as texture features, look into the local distribution. Example metrics include the gray-level co-occurrence matrices (GLCM) 61, gray-level run length matrices (GLRLM) 62, and gray-level size zone matrices (GLSZM) 63. GLCM gives the probability of observing a pair of values in voxels at a given distance in a given direction 61. GLRLM measures the number of consecutive voxels with the same value aligned in a given direction 62 and GLSZM reflects the number of neighboring voxels with the same value 63. Higher-order intensity values may be achieved using image transformations (e.g., Laplacian or Gaussian) 64,65 with different filter grids (e.g. Laws’ filters) 66, which highlight edge structures, or by using wavelet composition 67, which characterize sharp transitions in the intensity frequency spectrum.
Dynamic image features
In order to quantify the dynamic behavior of a contrast agent or other tracer within a region (which can be one or more voxels), pharmacokinetic modeling is typically used. In general, pharmacokinetic model considers tissue concentration as a convolution between the arterial input function (AIF) and the residual function for the decay of contrast agent inside the region of interest. The intravascular and interstitial space can be modeled under different assumptions. The most widely used kinetic model, Toft’s model, for example, assumes instantaneous mixing of contrast upon the arrival in the intravascular and interstitial space, while the extended Toft’s model considers a delay effect of the tissue concentration transferred from the artery 68. The adiabatic tissue homogeneity model (AATH) is motivated by the fact that the concentration of contrast agent in the extravascular distribution volume changes slowly relative to that in the intravascular space 69. Thus, it assumes that there exists a finite transit time for contrast solutes to travel from the arterial to the venous phase. The Patlak model is a linearization of irreversible compartment models in an equilibrium state where the tracers flow into the tissue without leaving 70. The Logan model is also a linearization of reversible compartment model in the equilibrium states where the tracers can move freely back into the plasma 70. Another approach is to directly fit the residual function using deconvolution without making any additional model assumptions 71. Typically-derived dynamic image features include regional blood flow (rBF), regional blood volume (rBV), mean transit time (MTT), extraction fraction, permeability surface area product, and most frequently volume transfer constant (Ktrans), and extravascular extracellular volume (ve). With an increasing emphasis on imaging of the tumor microenvironment, DCE-CT/MRI and FDG-PET have evolved as important functional techniques in this setting.
Overall, the current radiomics pipeline typically incorporates thousands of extracted radiomics features and these are still expected to further widen as the field continues to evolve.
Analytical tools
As in many other “-omics” fields, the number of input variables often far exceeds the number of patients. To reduce the probability of false positives, feature selection as dimension reduction is often needed, and filter-based score-ranking approaches, such as Wilcoxon, Chi-square, principle-component-analysis (PCA) are typically used 72. This can be carried out either using univariate methods, as the scoring criterion only depends on the feature relevancy, or multivariate methods using a weighted sum to maximize relevancy and minimize redundancy 72,73. Feature selection can also be combined with feature classification into a single model; examples include least-absolute-shrinkage-and-selection-operator (LASSO) 74 and Elastic Net 75.
Once a feature set is obtained, a data-driven model can be constructed. These include supervised and non-supervised approaches 76. Unsupervised analysis does not provide an outcome variable, but summary information of the data. The most frequently used graphical display is a heat map, which simultaneously reveals cluster structures in a data matrix 77. Supervised analysis, in contrast, creates models that attempt to separate the data with respect to a treatment outcome, such as responders vs. non-responders. Typical classification methods include conventional logistic regression or more advanced machine learning techniques 78,79.
Outcome modeling by logistic regression
Logistic regression is a common tool for multi-metric modeling. A logit transformation is used:
| (a) |
where n is the number of cases (patients), xi is a vector of the input variable values (i.e.,image features) used to predict f (xi ) for outcome yi (e.g., tumor control or toxicity) of the ith patient,
| (b) |
where d is the number of model variables and the β’s are the set of model coefficients determined by maximizing the probability that the data gave rise to the observations. This gives a linear combination of selected features with coefficients of respective weightings to the outcome, providing an intuitive tool for clinicians to interpret the associations between selected variables and the outcome.
Outcome modeling by machine learning
There are a wide class of AI techniques (e.g., neural networks, decision trees, support vector machine), which are able to provide a non-linear association of input variables to the outcome 80,81. Indeed, prognostic biomarkers developed using these machine-learning methods have increased performance when compared with conventional statistical methods 78,82,83. Recently, deep learning algorithms such as convolutional neural networks (CNN) have achieved breakthrough prediction power in a variety of medical studies, including detection of lung nodules on CT 84,85,86, and detection of breast cancer on mammogram 87,88. A comparison in mortality prediction from chest CT between a deep-learning framework and a standard framework with radiomics features, showed increased accuracy with CNN based classification 89. It is anticipated that multi-task learning will help to provide a degree of interpretation for deep learning approaches 76,81–90. Given enough high-quality data (text and images), it is expected that the role of CNNs will continue to expand in medicine and quantitative imaging. Despite these advances, however, concerted efforts are needed to promote detailed understanding of these approaches, including the relationship between dataset sizes, possible confounders and performance of outcome prediction.
Quality Assurance of the Images and Methodologies
Despite the promise that radiomics may hold for precision medicine, there are significant concerns regarding the lack of reproducibility in results within and across modalities and among multiple institutions. In this section, the challenges associated with the clinical translation of radiomics are highlighted, and recommendations are provided for application in NCTN clinical trials.
Standardization of image acquisition parameters
In recent years, the field has strived to improve standardization by defining standard acquisition protocols. National efforts have been led by the Quantitative Imaging Network (QIN) initiated by the National Cancer Institute (NCI) 91,92, by Radiological Society of North of America (RSNA) Quantitative Imaging Biomarker Alliance (QIBA) 93,94, and others 95–96. The NCI-Quantitative Image Excellence (CQIE) project was initiated in 2010 and the NCTN is a key focus for this effort 96,97. The CQIE provides PET/CT and MRI phantoms and protocols for site qualification, while QIBA provides consensus ‘profiles’ on the measurement accuracy of quantitative imaging biomarkers and the requirement/procedures needed to achieve this level of accuracy 98.
Despite the progress made by these groups, there still no universal acquisition protocols for any imaging modality in clinical practice. For studies involved with radiomics in NTCN trials, therefore, we recommend:
a comprehensive description of the image acquisition parameters should be documented, including manufacture, model, types of images (e.g., CT, MRI, contrast-enhanced), contrast agents, image acquisition parameters (e.g., slice-by-slice or three-dimensional acquisition, MRI magnetic field/TR/TE/flip angle, CT tube current/voltage, axial or helical mode), reconstruction package, software version, image resolution, signal-to-noise ratio, management of motion artifact;
If clinical trial is being conducted in institutes with the same scanners, the same scanning protocols should be strictly followed. Comparison across institutions with different scanners may be difficult. So it is suggested, when possible, to use each patient as his/her own control and the delta changes instead of the absolute value, or other corrections means for such variability including accounting for contrast;
direct measurement from scans with contrast should be used cautiously since uptake and the time from injection to imaging will differ and cause large variability. A practical strategy is to control the normal tissue ROIs as a baseline. For instance, on an individual basis, the average background of parenchyma or muscle can be used to normalize breast scans 99,100;
using radiomics feature(s) that are less dependent to variations in image acquisition protocol and/or platform into the final model.
Standardization of image pre-/post-processing
Following image acquisition, there can still be a large range of voxel intensities and image noise, therefore, filtering procedures may be needed to enhance the signal and reduce the unwanted noise 101. Fave et al. tested the effect of different image pre-processing filters, such as bit-depth resampling and smoothing filters, on radiomics features, concluding that the correlation of extracted radiomics features with clinical outcome changes with different filters 102. The impact of noise, which directly impacts intensity and GLCM features, has also been studied 103. In addition, images obtained on two different scanners may result in different pixel values due to different detector materials, image resolution or acquisition techniques 76. A very recent study by Reuze et al., showed GLCM-entropy values were higher on a scanner equipped with time-of-flight capabilities, leading to very different cutoff values for predicting recurrence 76. Therefore, some post-image acquisition processing as filtering and normalization is necessary.
To reduce the impact of noise, the SNR from all acquired images should be well-controlled. Some smoothing filtering procedure maybe used so that all images come close to a target spatial resolution value 104–106. A consequence of this approach is that images with the highest initial resolution will be degraded, which is especially adverse for analysis of image texture. CT images without contrast may be used directly if geometry distortions are calibrated properly and consistency in terms of HU can be obtained. MR images in general are susceptible to different distortions. A typical correction procedure would follow normalization prior to radiomics analysis 107. PET images tend to have more noise than other imaging modalities used in radiomics analysis, a typical pre-processing procedure to correction procedure has been to use Poisson-to-Gaussian conversion (a root-squared transform) 108.
Additional procedures include the discretization signal intensities into finite intervals for intensity-based feature analysis, using either absolute (fixed bin size) or relative (fixed number of bins) discretization 109. The choice of method is important, as the extracted features will vary. It has demonstrated that absolute discretization shows better repeatability, lower sensitivities to changes, and is not volume dependent 109,76. Thus, absolute discretization with fixed bin size should be adopted for intensity-based radiomics analysis. Each discretization method, however, has advantages and drawbacks and can lead to substantially different results 76.
Overall, effects of these pre-/post-processing techniques on variations in radiomics analysis remain as an open area. Although specific recommendations are being worked out by the IBSI group 110 and other, more general pitfalls to avoid including:
to include a detailed description of filtering technique, noise reduction technique, intensity-correction, intensity discretization and bit-depth resampling, should be provided; and
to apply radiomics feature(s) that are less dependent to variations in image pre-/post-processing techniques into the final model.
Reproducibility of radiomics features
Differences in segmentation methods are likely to bias the stability of shape metrics and intensity-based features. Parmar et al., performed a stability analysis based on an inter-observer study 111. Fifty-six radiomics features, quantifying shape, intensity and texture were extracted from lung CT images. They showed for manual delineation among 5 experienced operators, only 52% of these features had high reproducibility compared to 88% based on semi-automatic segmentation.
Moreover, contours of regions-of-interest (ROIs) are typically stored into two image formats: directly as voxels (e.g., NIfTI), or as (x,y,z) coordinates (e.g., DICOM-RT STRUCT). We need to determine which voxel centers lie within the space enclosed by the contour polygon. Thus different interpolation and partial-volume fraction threshold would also affect the contour-based feature calculation. Researchers showed contour data-rendered volumes exhibited large variations across the commercialized stereotactic radiosurgery (SRS) platforms using both the phantom and patient cases by up to 20% 112.
The methodologies to calculate radiomics feature can vary as well. Radiomics features can be obtained from either a 2D image slice 113,13 or from a reconstructed 3D volume 114. As another example, GLCM texture analysis can be calculated either by averaging the values of the matrices computed for 13 distinct directions or a single matrix that accounts for tumor co-occurrence information in all 13 directions 102,115. In addition, features computed from different matrices can have the same name 116. For example, entropy can be computed either from a histogram of intensities or from the GLCM matrix accounting for spatial similarity 117. Thus the impact of different feature identification methods used in radiomics also needs to be carefully studied.
Furthermore, the variability of radiomics indices has been found to be highly feature dependent. Recent studies on mammography datasets reported that robust features were those that described spatial patterns rather than directionality or image intensity118. Deformable registration of CT lung data was shown to alter underlying texture but certain features could still be identified that were robust to registration effects 21. Consensus has been reached that first-order and shape features in both CT and CBCT are generally more repeatable than texture or higher-order features 119–120,121. First-order statistically-derived features from a standard uptake value (SUV) histogram are generally robust with respect to segmentation, while texture features consistently showed greater sensitivity to segmentation differences 111,119,122.
Regarding the computation of radiomics features, we recommend:
unambiguous definitions of each radiomics feature should be provided and evaluated;
if contouring is involved, describe how ROIs are delineated in the image. Specify if segmentation is performed manually, semi-automatically or automatically, by how many users/experts and how consensus has been formed. The reproducibility of the radiomics features based on segmentations by multiple observers needs to be assessed;
the stability and the accuracy of features should be confirmed in terms of calculation algorithms, such as interpolation, criteria to include or exclude voxels from an ROI mask, 2D/3D calculations; or use features that do not require accurate segmentation;
additional suggestions include through the use of test-retest scan during the developmental phase (i.e., scanned on the same scanner but repositioned in between scans) with physical phantom and/or patient studies, to compare the means and standard deviations of these results as well as their correlations. Within the radiation oncology work flow, there are instances that could mimic the test-retest scenario. One example is a 4DCT scan if each phase is treated as a separate CT scan. A good practice policy is to eliminate features that prove to be unreliable in the test-retest. To this end, several datasets are public available. Of note, the RIDER dataset allows validation of results in the same set of patients with two scans taken 15mins apart 123. Investigators are encouraged to develop and share additional shared test-retest datasets;
again, using radiomics feature(s) that are less dependent to above variabilities to build the final model.
Radiomics phantoms
To help in addressing some of the challenges described above, the design of radiomics specific phantoms is now an active area of research 124–126. One example is the Credence Cartridge Radiomics (CCR) phantom, which was designed for use in studies of texture feature robustness 125. Using the CCR phantom, Mackin et al. investigated the inter-scanner variability of the calculated texture features, looking for clustering effects due to the scanner manufacturer and CT acquisitions parameters 127. They subsequently developed a correction technique to reduce or eliminate the variability in radiomics features due to differences in image pixel size based on CT images using the physical CCR phantom 128.
Digital phantoms such as the the Zobel and NCAT phantoms have long been used in image processing applications 129. It is unclear, however, whether these phantoms are suited for radiomics, and particularly for texture analysis. An alternative approach is to use standardized patterns such as the Brodatz Textures, 130 though the accompanying signatures are not necessarily clinically relevant. Alternately, simulated images can be used to determine optimal parameters for feature analysis. McGurk et al. augmented PET images from 30 soft tissue sarcoma patients by varying the extent of axial data combined per slice (‘span’) 131. Simulated T1-weighted and T2-weighted MR images were acquired by varying the repetition time and echo time in a spin-echo pulse sequence, respectively. The impact of PET and MR image acquisition parameter variation on individual textures was investigated to assess the global response and the predictive properties of a texture-based model. The results suggested that such a process is feasible for identifying an optimal set of image acquisition parameters to improve prediction performance 131.
Overall, those radiomics phantoms may be helpful in assessing the inter-/intra-scanner variabilities, and thus protocols for regular phantom QA may be worth of being developed, similar to dosimetric study in current radiotherapy QA program, to monitor inter-scan and inter-vendor variability of image-derived features. In addition, phantom studies will also be useful in optimizing imaging protocols and image pre-/post-processing techniques that allow for reliable radiomics characterization. Ultimately, specific acquisition protocols optimized to generate superior radiomics measurements for a given clinical problem should be developed and standardized and thereafter validated using clinical scanners.
Robustness of modeling with radiomics
In the current iteration of radiomics, image features have to be extracted with high throughput, putting a premium on statistical modeling and machine learning algorithm development. Currently there is no consensus on which feature selection or learning methods should be used. Parmar et al. carried out a comparative study using 12 machine-learning algorithms combined with 14 feature selection methods, for radiomics-based prediction of 2-year survival of lung cancer patients treated with radical radiotherapy alone or with chemotherapy 78. They concluded the 30% variation was observed for different classification method but not the feature selection algorithm. Given the state of the art, an analysis of the impact of different learning approaches on the stability and robustness of the proposed models is needed.
The general understanding is that simpler models involving few radiomics features are more robust. To avoid over-fitting, a reasonable rule of thumb with feature selection or dimension reduction is that 10 positive samples (patients) are needed for each feature selected for binary classification according to the Harrell’s guidelines 132,133. For example, radiomics analysis can be performed for 100 positive events, which will result no more than 10 features selected for the final prediction model.
Recently, deep learning has emerged as a productive force across many healthcare disciplines, especially in diagnostic radiology and pathology. However, there is a significant mismatch between the perceived capabilities of AI compared to their actual capabilities in present imaging studies 134,135. The scarce availability of high quality data, as well as the lack of standardized processes among institutions are the key-barriers. In addition, interpretation of non-linear relationship between input variables vs. outcome, as given by many current “black box” machine learning models, requires high-level expertise in this field. Additional complementary information such as patient demographics, genomics, histology, and biomarkers may be helpful in interpreting the results.
Nevertheless, robust models:
need to assess the over-fitting risk by using cross-validation methods during the classification step and controlled by dimension reduction methods.
need to be built upon rigorous training, testing and validation. Estimation of predictive performance in single-institution cohorts should include multiple-folded repeated cross validation to minimize the risk of overfitting, while validation with external dataset is highly recommended; and
may need to consider to accommodate clinical information, and with covariates of genomic profiles, histology, biomarkers, patient histories etc., to generate clinically understandable and acceptable decisions.
Overview of Commercial and Open-Source Radiomics Systems
Commercial systems
Several commercial applications providing radiomics capabilities applicable to cancer clinical trials have been created, including HealthMyne.com 136, Texrad.com 137, and Oncoradiomics.com 138. The commercial entities offer platforms that automate image analysis and clinical interpretation for radiologists and oncologists. Radiomic feature extraction is included as part of image analysis functionality, and intensity, shape and texture metrics, along with conventional size measurements used in routine image interpretation as RECIST (Response Evaluation Criteria In Solid Tumors), therapy response monitoring, and cancer screening, are generated on approval of structure segmentation. Along with imaging data (CT, MR and PET), these systems can import and display electronic health record (EHR) information via an HL7 interface, and imported and generated data is stored in a minable patient database that can be examined.
Open source systems
There are several open source software packages capable of performing radiomics analysis. Examples include: IBEX 139, MaZda 140, CGITA 141, pyradiomics 142,143, CERR144. Because there is no official index of the various open source radiomics packages, this list is not necessarily exhaustive. These open-source packages are typically capable of calculating first-order texture, GLCM, and GLRLM features, but not necessarily more sophisticated ones such as fractal features. Certain packages can calculate texture features in 2D and 3D (IBEX, CGITA, pyradiomics) while others are limited to 2D datasets (MaZda). Furthermore, the ability to alter calculation parameters, such as the number of gray-levels and directions for GLCMs, is not available in all open-source packages (e.g., MaZda). Some packages were tailored for certain imaging modalities. For example, IBEX was initially developed for CT data whereas MaZda was initially developed for MRI studies. As a result, both IBEX and MaZda rescale pixel values in an image to remove any non-negative pixel values 145. Algorithmic implementation of features in each package could also vary. Some efforts have been made, such as the image biomarker standardization initiative (IBSI) 110, to standardize feature definitions. Some packages such as pyradiomics attempt to adhere to IBSI definitions. Beyond open-source software tools, a number of groups have also developed in-house tools for radiomics analyses. One such example is an open source Matlab that calculates different texture matrices in addition to global metrics. It also supports multivariate model building using a logistic regression model with bootstrapping for order selection 146. Overall, when selecting an open-source package, the user should verify that it meets their task-specific need and provides expected results.
It is recommended that:
investigators provide access to code and datasets to ensure results can be reproduced. The datasets should include images, segmentations and clinical information if available and compliant with HIPAA regulations.
investigators promote an open source approach to radiomics software as an important step toward independent validation and to general dissemination of the approach.
Implementation in Clinical Trials and Recommendations
In order for radiomics to gain broad acceptance in clinical medicine, the value that the technology brings to clinical assessment and decision making must be affirmed in the context of clinical trials. Ideally, clinical trials that are conducted across diverse institutional environments and that are agnostic to vendor or technological platform will provide the most robust validation of the significance of radiomics. High quality clinical research is resource intensive and consumes time, energy and resources of physicians, research staff, and patients alike. The recently published roadmap for imaging biomarkers is a notable advancement, showcasing key recommendations for clinical translation of radiomics 147. These considerations being applied to the role of genomics in clinical trials may also apply to radiomics. Due to the expense of clinical trials, it is crucial that any prospective trials involving radiomics are carefully designed.
An important clinical question is what type of studies will be best suitable to test the value of radiomics. Strategies must be defined, mitigate and/or quantify uncertainties, risk and cost associated with any potential biomarker in making research or clinical.
Trials need to be carefully designed considering both the initial and any secondary analyses to ensure that the proper data has been collected to support the study questions. Whether there is and how much value of radiomics would add into the current gold standard clinical measurement needs to be verified. It is also prudent to incorporate radiomics studies as exploratory aims in the initial phase of the implementation into clinical trials. It should be clearly specified whether radiomics input will be used to stratify or determine patient eligibility or integrated within the trial to assess the quality of the test.
For clinical trials, a standard radiomics data format, similar to the needs defined by the Advanced Technology Consortium to support clinical trials, one of the main proponents for the DICOM-RT standard, may be necessary. Such a standard would need to be developed by researchers, clinicians, and manufacturers together to address the competing needs for robust quantitative evaluation of data. In addition, a guideline for reporting results from radiomics studies should established. A concept of radiomics quality score (RQS) has been proposed as possible evaluation criteria for radiomics studies 148. The RQS contains sixteen key components that intend to minimize bias and enhance the usefulness of the radiomics models. These recommendations may establish a reporting guidelines for future radiomics studies.
For a given trial, being able to share a standard acquisition protocol in support of the trial across participating institutions would establish a strong foundation for meaningful radiomics data and analysis.
Summary
Radiomics is an evolving field that is growing at a rapid pace with great potential to impact the design of future clinical trials in oncology. As was noted in a 2009 report, however, many genomics-based studies have been published that contained significant analytical errors 110, and scientists reported that they failed to replicate 47 out of 53 landmark studies 149. These findings raise similar concerns regarding the accountability of the statistical analysis, the transparency of the raw data accessibility and validation of results in radiomics studies. As the ultimate goal of radiomics is to build reliable imaging biomarkers to assist clinical decision-making, a prospective investigation must be trained, tested and validated against a completely independent data set, with a systematic study design with uniform treatment delivery, complete reporting of results, and robust statistical analysis. Protocols for standardizing image acquisition, feature extraction, and analysis will help to streamline the process for clinical trials. This can build on already existing expertise in the community in credentialing and evaluation of other metrics (e.g., treatment plan delivery, imaging QA,) to accelerate the process.
Nevertheless, the promise of radiomics is still quite positive. Medical imaging is redefining its role as a valuable data source for precision medicine in the guise of image-based phenotyping. Further, it could characterize tumor heterogeneity at a macroscopic level, a critical limitation of biopsy-based or current genomic approaches, which could potentially provide important complementary information for precision medicine. The successful introduction of these methods into clinical care will require much additional research to determine how underlying driving biologic phenomena are related to the tumor imaging phenotypes. In the foreseeable future, we expect that the data gleaned from oncology examinations will be converted into quantitative data which will be interfaced with knowledge databases to improve the diagnostic and prognostic power for clinical decision support.
Acknowledgement
This project was supported by NCI grants U24CA180803(IROC), U10CA180868(NRG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Publisher's Disclaimer: Disclaimer
The materials presented represent the expert consensus among a group of NRG Oncology investigators developed specifically for use in NCTN or NCTN endorsed clinical trials. Authors are not responsible for use in any other context.
References:
- 1.Doi K Computer-Aided Diagnosis in Medical Imaging: Historical Review, Current Status and Future Potential. Computerized medical imaging and graphics : the official journal of the Computerized Medical Imaging Society 2007;31(4–5):198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. Journal of vascular and interventional radiology : JVIR 2007;18(7):821–831. [DOI] [PubMed] [Google Scholar]
- 3.El Naqa I, Grigsby P, Apte A, et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit 2009;42(6):1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partridge SC, Gibbs JE, Lu Y, et al. MRI measurements of breast tumor volume predict response to neoadjuvant chemotherapy and recurrence-free survival. AJR American journal of roentgenology 2005;184(6):1774–1781. [DOI] [PubMed] [Google Scholar]
- 5.Hylton NM, Gatsonis CA, Rosen MA, et al. Neoadjuvant Chemotherapy for Breast Cancer: Functional Tumor Volume by MR Imaging Predicts Recurrence-free Survival-Results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL (1527-1315 (Electronic)). [DOI] [PMC free article] [PubMed]
- 6.Gillies RJ, Anderson AR, Gatenby RA, Morse DL. The Biology Underlying Molecular Imaging in Oncology: From Genome to Anatome and Back Again. Clinical radiology 2010;65(7):517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avanzo M, Stancanello J, El Naqa I. Beyond imaging: The promise of radiomics. Phys Med 2017;38:122–139. [DOI] [PubMed] [Google Scholar]
- 8.Lambin P, Rios-Velazquez E Fau - Leijenaar R, Leijenaar R Fau - Carvalho S, et al. Radiomics: extracting more information from medical images using advanced feature analysis (1879-0852 (Electronic)). [DOI] [PMC free article] [PubMed]
- 9.Liang M, Tang W, Xu DM, et al. Low-Dose CT Screening for Lung Cancer: Computer-aided Detection of Missed Lung Cancers. Radiology 2016;281(1):279–288. [DOI] [PubMed] [Google Scholar]
- 10.Kumar V, Gu Y Fau - Basu S, Basu S Fau - Berglund A, et al. Radiomics: the process and the challenges (1873-5894 (Electronic)).
- 11.Shafiee MJ, Chung AG, Khalvati F, Haider MA, Wong A. Discovery radiomics via evolutionary deep radiomic sequencer discovery for pathologically proven lung cancer detection (2329-4302 (Print)). [DOI] [PMC free article] [PubMed]
- 12.Li H, Giger ML, Lan L, Janardanan J, Sennett CA. Comparative analysis of image-based phenotypes of mammographic density and parenchymal patterns in distinguishing between BRCA1/2 cases, unilateral cancer cases, and controls. J Med Imaging (Bellingham) 2014;1(3):031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: Luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. Journal of magnetic resonance imaging : JMRI 2015;42(4):902–907. [DOI] [PubMed] [Google Scholar]
- 14.Nie K, Shi L, Chen Q, et al. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22(21):5256–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, Tian J, Dong D, et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma (1078-0432 (Print)). [DOI] [PubMed]
- 16.Goh V, Ganeshan B, Nathan P, Juttla JK, Vinayan A, Miles KA. Assessment of response to tyrosine kinase inhibitors in metastatic renal cell cancer: CT texture as a predictive biomarker. Radiology 2011;261(1):165–171. [DOI] [PubMed] [Google Scholar]
- 17.Yip C, Davnall F, Kozarski R, et al. Assessment of changes in tumor heterogeneity following neoadjuvant chemotherapy in primary esophageal cancer. Diseases of the esophagus : official journal of the International Society for Diseases of the Esophagus 2015;28(2):172–179. [DOI] [PubMed] [Google Scholar]
- 18.Aerts HJ, Grossmann P, Tan Y, et al. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci Rep 2016;6:33860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015;114(3):345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez CJ, Nagornaya N, Parra NA, et al. Association of Radiomics and Metabolic Tumor Volumes in Radiation Treatment of Glioblastoma Multiforme. International journal of radiation oncology, biology, physics 2017;97(3):586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunliffe AR, Armato SG 3rd, Fei XM, Tuohy RE, Al-Hallaq HA. Lung texture in serial thoracic CT scans: registration-based methods to compare anatomically matched regions. Med Phys 2013;40(6):061906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palma DA, van Sornsen de Koste J, Verbakel WF, Vincent A, Senan S. Lung density changes after stereotactic radiotherapy: a quantitative analysis in 50 patients. International journal of radiation oncology, biology, physics 2011;81(4):974–978. [DOI] [PubMed] [Google Scholar]
- 23.Diot Q, Kavanagh B, Schefter T, Gaspar L, Stuhr K, Miften M. Regional normal lung tissue density changes in patients treated with stereotactic body radiation therapy for lung tumors. International journal of radiation oncology, biology, physics 2012;84(4):1024–1030. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Galperin-Aizenberg M, Pryma D, Simone CB, Fan Y. Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy. Radiotherapy and Oncology 2018;129(2):218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng M, Yang C, Chen X, et al. Computed tomography number changes observed during computed tomography-guided radiation therapy for head and neck cancer. International journal of radiation oncology, biology, physics 2015;91(5):1041–1047. [DOI] [PubMed] [Google Scholar]
- 26.Paul J, Yang C, Wu H, et al. Early Assessment of Treatment Responses During Radiation Therapy for Lung Cancer Using Quantitative Analysis of Daily Computed Tomography. International journal of radiation oncology, biology, physics 2017;98(2):463–472. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Oshima K, Schott D, et al. Assessment of treatment response during chemoradiation therapy for pancreatic cancer based on quantitative radiomic analysis of daily CTs: An exploratory study. PLoS One 2017;12(6):e0178961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohri N, Duan F, Snyder BS, et al. Pretreatment 18F-FDG PET Textural Features in Locally Advanced Non-Small Cell Lung Cancer: Secondary Analysis of ACRIN 6668/RTOG 0235 (1535-5667 (Electronic)). [DOI] [PMC free article] [PubMed]
- 29.Buizza G, Toma-Dasu I, Lazzeroni M, et al. Early tumor response prediction for lung cancer patients using novel longitudinal pattern features from sequential PET/CT image scans (1724-191X (Electronic)). [DOI] [PubMed]
- 30.Nakatsugawa M, Cheng Z, Goatman KA, et al. Radiomic Analysis of Salivary Glands and Its Role for Predicting Xerostomia in Irradiated Head and Neck Cancer Patients. International Journal of Radiation Oncology • Biology • Physics 2016;96(2):S217. [Google Scholar]
- 31.Nardone VA-Ohoo, Tini P, Nioche C, et al. Texture analysis as a predictor of radiation-induced xerostomia in head and neck patients undergoing IMRT (1826-6983 (Electronic)). [DOI] [PubMed]
- 32.Liu Z, Wang Y, Liu X, et al. Radiomics analysis allows for precise prediction of epilepsy in patients with low-grade gliomas. NeuroImage : Clinical 2018;19:271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Titus-Ernstoff L, Tosteson AN, Kasales C, et al. Breast cancer risk factors in relation to breast density (United States). Cancer causes & control : CCC 2006;17(10):1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd NF, Guo H, Martin LJ, et al. Mammographic density and the risk and detection of breast cancer. The New England journal of medicine 2007;356(3):227–236. [DOI] [PubMed] [Google Scholar]
- 35.Vachon CM, Brandt KR, Ghosh K, et al. Mammographic breast density as a general marker of breast cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2007;16(1):43–49. [DOI] [PubMed] [Google Scholar]
- 36.Haberle L, Wagner F, Fasching PA, et al. Characterizing mammographic images by using generic texture features. Breast cancer research : BCR 2012;14(2):R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013;267(2):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zinn PO, Mahajan B, Sathyan P, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One 2011;6(10):e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong SJ, Kim TJ, Choi YW, Park JS, Chung JH, Lee KW. Radiogenomic correlation in lung adenocarcinoma with epidermal growth factor receptor mutations: Imaging features and histological subtypes. European radiology 2016;26(10):3660–3668. [DOI] [PubMed] [Google Scholar]
- 41.Hsu JS, Huang MS, Chen CY, et al. Correlation between EGFR mutation status and computed tomography features in patients with advanced pulmonary adenocarcinoma. Journal of thoracic imaging 2014;29(6):357–363. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Kim J, Balagurunathan Y, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clinical lung cancer 2016;17(5):441–448.e446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Zhu Y, Burnside ES, et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology 2016;281(2):382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi NC, Chun TT, Niemierko A, et al. Potential of (18)F-FDG PET toward personalized radiotherapy or chemoradiotherapy in lung cancer. European Journal of Nuclear Medicine and Molecular Imaging 2013;40(6):832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao B, Tan Y, Tsai W-Y, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Scientific Reports 2016;6:23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta oncologica (Stockholm, Sweden) 2010;49(7):1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bologna M, Corino VDA, Montin E, et al. Assessment of Stability and Discrimination Capacity of Radiomic Features on Apparent Diffusion Coefficient Images. J Digit Imaging 2018. [DOI] [PMC free article] [PubMed]
- 48.Ford J, Dogan N, Young L, Yang F. Quantitative Radiomics: Impact of Pulse Sequence Parameter Selection on MRI-Based Textural Features of the Brain. Contrast Media Mol Imaging 2018;2018:1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molina D, Perez-Beteta J, Martinez-Gonzalez A, et al. Lack of robustness of textural measures obtained from 3D brain tumor MRIs impose a need for standardization. PLoS One 2017;12(6):e0178843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annual review of biomedical engineering 2000;2:315–337. [DOI] [PubMed] [Google Scholar]
- 51.El Naqa I, Yang D, Apte A, et al. Concurrent multimodality image segmentation by active contours for radiotherapy treatment planning. Med Phys 2007;34(12):4738–4749. [DOI] [PubMed] [Google Scholar]
- 52.Echegaray S, Nair V, Kadoch M, et al. A Rapid Segmentation-Insensitive “Digital Biopsy” Method for Radiomic Feature Extraction: Method and Pilot Study Using CT Images of Non-Small Cell Lung Cancer. Tomography 2016;2(4):283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation (1361-8423 (Electronic)). [DOI] [PubMed]
- 54.Birenbaum A, Greenspan H. Multi-view longitudinal CNN for multiple sclerosis lesion segmentation. Engineering Applications of Artificial Intelligence 2017;65:111–118. [Google Scholar]
- 55. https://www.deepmind.com/health.
- 56.Clinicians PIMIAtE. https://www.microsoft.com/en-us/research/project/medical-image-analysis/.
- 57.Aljabar PGM. The cutting edge: delineating contours with Deep Learning 2017. [Google Scholar]
- 58.challenges G-c-a. https://grand-challenge.org/.
- 59.El Naqa I The role of quantitative PET in predicting cancer treatment outcomes. Clinical and Translational Imaging 2014;2(4):305–320. [Google Scholar]
- 60.Volume Wadell H., Shape, and Roundness of Quartz Particles. The Journal of Geology 1935;43(3):250–280. [Google Scholar]
- 61.Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics 1973;SMC-3(6):610–621. [Google Scholar]
- 62.Dong-Hui Xu ASK, Jacob D. Furst, Daniela S. Raicu. Run-length encoding for volumetric texture. Paper presented at: 4th IASTED Int Conf Vis Imaging Image Process VIP2004. [Google Scholar]
- 63.Thibault G, Fertil B, Navarro C,L, et al. Texture indexes and gray level size zone matrix. Application to cell nuclei classification. Paper presented at: 10th International Conference on Pattern Recognition and Information Processing, PRIP 2009; 2009, 2009; Minsk, Belarus. [Google Scholar]
- 64.Wu X, Wang K, Zhang D. Palmprint Texture Analysis Using Derivative of Gaussian Filters. Paper presented at: 2006 International Conference on Computational Intelligence and Security; 3–6 Nov. 2006, 2006. [Google Scholar]
- 65.Du H, Jin X, Willis PJ. Two-level joint local laplacian texture filtering %J Vis. Comput 2016;32(12):1537–1548. [Google Scholar]
- 66.Laws KI. Rapid Texture Identification. Paper presented at: 24th Annual Technical Symposium1980. [Google Scholar]
- 67.Livens S Wavelets for texture analysis, an overview. 6th International Conference on Image Processing and its Applications; 1997. [Google Scholar]
- 68.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. Journal of magnetic resonance imaging : JMRI 1999;10(3):223–232. [DOI] [PubMed] [Google Scholar]
- 69.St Lawrence KS, Lee TY. An adiabatic approximation to the tissue homogeneity model for water exchange in the brain: I. Theoretical derivation. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism 1998;18(12):1365–1377. [DOI] [PubMed] [Google Scholar]
- 70.Niu TYP; Sun X; Mao T; Xu L; Yue N; Kuang Y; Shi L; Nie K;. Variations of Quantitative Perfusion Measurement on Dynamic Contrast Enhanced CT for Colorectal Cancer: Implication of Standardized Image Protocol. Phys Med Biol 2018(Accepted). [DOI] [PubMed]
- 71.Jerosch-Herold M, Swingen C, Seethamraju RT. Myocardial blood flow quantification with MRI by model-independent deconvolution. Med Phys 2002;29(5):886–897. [DOI] [PubMed] [Google Scholar]
- 72.Chandrashekar G, Sahin F. A survey on feature selection methods. Computers & Electrical Engineering 2014;40(1):16–28. [Google Scholar]
- 73.Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence 1997;97(1):273–324. [Google Scholar]
- 74.Guyon I, Andr, #233, Elisseeff. An introduction to variable and feature selection. J Mach Learn Res 2003;3:1157–1182. [Google Scholar]
- 75.Zou H, Hastie T. Regularization and Variable Selection via the Elastic Net. Journal of the Royal Statistical Society Series B (Statistical Methodology) 2005;67(2):301–320. [Google Scholar]
- 76.Reuze S, Schernberg A, Orlhac F, et al. Radiomics in Nuclear Medicine Applied to Radiation Therapy: Methods, Pitfalls, and Challenges. International journal of radiation oncology, biology, physics 2018. [DOI] [PubMed]
- 77.Wilkinson L, Friendly M. The History of the Cluster Heat Map. The American Statistician 2009;63(2):179–184. [Google Scholar]
- 78.Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ. Radiomic Machine-Learning Classifiers for Prognostic Biomarkers of Head and Neck Cancer. Frontiers in oncology 2015;5:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao X, Chu C, Li Y, et al. The method and efficacy of support vector machine classifiers based on texture features and multi-resolution histogram from (18)F-FDG PET-CT images for the evaluation of mediastinal lymph nodes in patients with lung cancer (1872-7727 (Electronic)). [DOI] [PubMed]
- 80.Lao J, Chen Y, Li Z-C, et al. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Scientific Reports 2017;7(1):10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Scientific Reports 2017;7(1):5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu W, Parmar C, Grossmann P, et al. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Frontiers in oncology 2016;6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLaren CE, Chen WP, Nie K, Su MY. Prediction of malignant breast lesions from MRI features: a comparison of artificial neural network and logistic regression techniques. Acad Radiol 2009;16(7):842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang X, Shan J, Vaidya V. Lung nodule detection in CT using 3D convolutional neural networks. Paper presented at: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017); 18–21 April 2017, 2017. [Google Scholar]
- 85.da Silva GLF, Valente TLA, Silva AC, de Paiva AC, Gattass M. Convolutional neural network-based PSO for lung nodule false positive reduction on CT images (1872-7565 (Electronic)). [DOI] [PubMed]
- 86.Jin H, Li Z, Tong R, Lin L. A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection (2473-4209 (Electronic)). [DOI] [PubMed]
- 87.Borges Sampaio W, Moraes Diniz E, Corrêa Silva A, Cardoso de Paiva A, Gattass M. Detection of masses in mammogram images using CNN, geostatistic functions and SVM. Computers in Biology and Medicine 2011;41(8):653–664. [DOI] [PubMed] [Google Scholar]
- 88.Charan S, Khan MJ, Khurshid K. Breast cancer detection in mammograms using convolutional neural network. Paper presented at: 2018 International Conference on Computing, Mathematics and Engineering Technologies (iCoMET); 3–4 March 2018, 2018. [Google Scholar]
- 89.Carneiro G, Oakden-Rayner L, Bradley AP, Nascimento J, Palmer L. Automated 5-year mortality prediction using deep learning and radiomics features from chest computed tomography. Paper presented at: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017); 18–21 April 2017, 2017. [Google Scholar]
- 90.Thompson RF, Valdes G, Fuller CD, et al. Artificial intelligence in radiation oncology: A specialty-wide disruptive transformation? Radiother Oncol 2018. [DOI] [PMC free article] [PubMed]
- 91.Clarke LP, Nordstrom RJ, Zhang H, et al. The Quantitative Imaging Network: NCI’s Historical Perspective and Planned Goals. Translational oncology 2014;7(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shankar LK. The clinical evaluation of novel imaging methods for cancer management (1759-4782 (Electronic)). [DOI] [PubMed]
- 93.Huang EP, Wang XF, Choudhury KR, et al. Meta-analysis of the technical performance of an imaging procedure: guidelines and statistical methodology (1477-0334 (Electronic)). [DOI] [PMC free article] [PubMed]
- 94.Sullivan DC, Obuchowski NA, Kessler LG, et al. Metrology Standards for Quantitative Imaging Biomarkers (1527-1315 (Electronic)). [DOI] [PMC free article] [PubMed]
- 95.Program. NCICI. National Cancer Institute. Cancer Imaging Program https://imaging.cancer.gov/.
- 96.Rosen M, Kinahan PE, Gimpel JF, et al. Performance Observations of Scanner Qualification of NCI-Designated Cancer Centers: Results From the Centers of Quantitative Imaging Excellence (CQIE) Program. Academic radiology 2017;24(2):232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kurland BF, Gerstner ER, Mountz JM, et al. QIN. Promise and pitfalls of quantitative imaging in oncology clinical trials. Magnetic resonance imaging 2012;30(9):1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. https://www.rsna.org/QIBA/.
- 99.Wang J, Kato F, Oyama-Manabe N, et al. Identifying Triple-Negative Breast Cancer Using Background Parenchymal Enhancement Heterogeneity on Dynamic Contrast-Enhanced MRI: A Pilot Radiomics Study (1932-6203 (Electronic)). [DOI] [PMC free article] [PubMed]
- 100.Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Annals of oncology : official journal of the European Society for Medical Oncology 2017;28(6):1191–1206. [DOI] [PubMed] [Google Scholar]
- 101.Wells WM. Efficient Synthesis of Gaussian Filters by Cascaded Uniform Filters. IEEE Transactions on Pattern Analysis and Machine Intelligence 1986;PAMI-8(2):234–239. [DOI] [PubMed] [Google Scholar]
- 102.Fave X, Cook M, Frederick A, et al. Preliminary investigation into sources of uncertainty in quantitative imaging features. Computerized medical imaging and graphics : the official journal of the Computerized Medical Imaging Society 2015;44:54–61. [DOI] [PubMed] [Google Scholar]
- 103.Fave X, Mackin D, Yang J, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med Phys 2015;42(12):6784–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lasnon C, Majdoub M, Lavigne B, et al. (18)F-FDG PET/CT heterogeneity quantification through textural features in the era of harmonisation programs: a focus on lung cancer (1619-7089 (Electronic)). [DOI] [PubMed]
- 105.Boellaard R Standards for PET image acquisition and quantitative data analysis. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2009;50 Suppl 1:11S–20S. [DOI] [PubMed] [Google Scholar]
- 106.Quak E, Le Roux P-Y, Hofman MS, et al. Harmonizing FDG PET quantification while maintaining optimal lesion detection: prospective multicentre validation in 517 oncology patients. European Journal of Nuclear Medicine and Molecular Imaging 2015;42(13):2072–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magnetic resonance imaging 2004;22(1):81–91. [DOI] [PubMed] [Google Scholar]
- 108.Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. Journal of Medical Imaging 2015;2(4):041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leijenaar RT, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015;5:11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.A Zwanenburg SL Vallières M, Löck S. Image biomarker standardisation initiative. arXiv preprint arXiv:161207003 2016.
- 111.Parmar C, Rios Velazquez E, Leijenaar R, et al. Robust Radiomics Feature Quantification Using Semiautomatic Volumetric Segmentation. PLOS ONE 2014;9(7):e102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ma L, Sahgal A, Nie K, et al. - Reliability of contour-based volume calculation for radiosurgery. J Neurosurg 2012;117:203–210. [DOI] [PubMed] [Google Scholar]
- 113.Nie K, Chen JH, Chan S, et al. Development of a quantitative method for analysis of breast density based on three-dimensional breast MRI. Med Phys 2008;35(12):5253–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baek HM, Chen JH, Nie K, et al. Predicting pathologic response to neoadjuvant chemotherapy in breast cancer by using MR imaging and quantitative 1H MR spectroscopy. Radiology 2009;251(3):653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hatt M, Majdoub M, Vallieres M, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2015;56(1):38–44. [DOI] [PubMed] [Google Scholar]
- 116.Buvat I, Orlhac F, Soussan M. Tumor Texture Analysis in PET: Where Do We Stand? (1535-5667 (Electronic)). [DOI] [PubMed]
- 117.Dercle L, Ammari S, Bateson M, et al. Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Scientific Reports 2017;7(1):7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mendel KR, Li H, Lan L, et al. Quantitative texture analysis: robustness of radiomics across two digital mammography manufacturers’ systems. J Med Imaging (Bellingham) 2018;5(1):011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. International journal of radiation oncology, biology, physics 2018. [DOI] [PMC free article] [PubMed]
- 120.van Timmeren JE, Leijenaar RTH, van Elmpt W, et al. Test-Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? (2379-139X (Electronic)). [DOI] [PMC free article] [PubMed]
- 121.Bagher-Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ. On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers (2473-4209 (Electronic)). [DOI] [PubMed]
- 122.van Velden FH, Kramer GM, Frings V, et al. Repeatability of Radiomic Features in Non-Small-Cell Lung Cancer [(18)F]FDG-PET/CT Studies: Impact of Reconstruction and Delineation (1860-2002 (Electronic)). [DOI] [PMC free article] [PubMed]
- 123.NIH. https://wiki.nci.nih.gov/display/CIP/RIDER.
- 124.Nordstrom RJ. The Quantitative Imaging Network in Precision Medicine. Tomography 2016;2(4):239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mackin D, Fave X, Zhang L, et al. Measuring Computed Tomography Scanner Variability of Radiomics Features. Investigative radiology 2015;50(11):757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shafiq-Ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep 2018;8(1):10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mackin D, Fave X, Zhang L, et al. Measuring CT scanner variability of radiomics features. Investigative radiology 2015;50(11):757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mackin D, Fave X, Zhang L, et al. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLOS ONE 2017;12(9):e0178524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McGurk R, Seco J, Riboldi M, Wolfgang J, Segars P, Paganetti H. Extension of the NCAT phantom for the investigation of intra-fraction respiratory motion in IMRT using 4D Monte Carlo. Physics in medicine and biology 2010;55(5):1475–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. http://www.ux.uis.no/~tranden/brodatz.html.
- 131.Vallieres M, Laberge S, Diamant A, El Naqa I. Enhancement of multimodality texture-based prediction models via optimization of PET and MR image acquisition protocols: a proof of concept. Phys Med Biol 2017;62(22):8536–8565. [DOI] [PubMed] [Google Scholar]
- 132.Steyerberg EW, Eijkemans Mj Fau - Harrell FE Jr., Harrell Fe Jr Fau - Habbema JD, Habbema JD. Prognostic modeling with logistic regression analysis: in search of a sensible strategy in small data sets (0272-989X (Print)). [DOI] [PubMed]
- 133.Harrell FE Jr, Lee Kl Fau - Mark DB, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors (0277-6715 (Print)). [DOI] [PubMed]
- 134.Obermeyer Z, Lee TH. Lost in Thought - The Limits of the Human Mind and the Future of Medicine (1533-4406 (Electronic)). [DOI] [PMC free article] [PubMed]
- 135.Chen JH, Asch SM. Machine Learning and Prediction in Medicine — Beyond the Peak of Inflated Expectations 2017;376(26):2507–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. https://www.healthmyne.com/.
- 137. http://texrad.com/.
- 138. https://www.oncoradiomics.com/.
- 139.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 2015;42(3):1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. http://www.eletel.p.lodz.pl/programy/mazda/.
- 141.Fang YH, Lin CY, Shih MJ, et al. Development and evaluation of an open-source software package “CGITA” for quantifying tumor heterogeneity with molecular images. Biomed Res Int 2014;2014:248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arimura H, Soufi M, Kamezawa X, Ninomiya K, Yamada M Radiomics with artificial intelligence for precision medicine in radiation therapy. J Radiat Res 2018. [DOI] [PMC free article] [PubMed]
- 144.Apte AP, Iyer A, Crispin-Ortuzar M, et al. Technical Note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research 2018;45(8):3713–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Foy JJ, Robinson KR, Li H, Giger ML, Al-Hallaq H, Armato SG. Variation in algorithm implementation across radiomics software. J Med Imag 5(4) 044505. 2018;5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vallieres M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60(14):5471–5496. [DOI] [PubMed] [Google Scholar]
- 147.O’Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14(3):169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sanduleanu S, Woodruff HC, de Jong EEC, et al. Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score. Radiother Oncol 2018;127(3):349–360. [DOI] [PubMed] [Google Scholar]
- 149.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature 2012;483(7391):531–533. [DOI] [PubMed] [Google Scholar]