SUMMARY
Developing and adult tissues use different cis-regulatory elements. Although DNA at some decommissioned embryonic enhancers is hypomethylated in adult cells, it is unknown whether this putative epigenetic memory is complete and recoverable. We find that in adult mouse cells, hypomethylated CpG dinucleotides preserve a nearly complete archive of tissue-specific developmental enhancers. Sites that carry the active histone mark H3K4me1, and are therefore considered ‘primed’, are mainly cis-elements that act late in organogenesis. In contrast, sites decommissioned early in development retain hypomethylated DNA as a singular property. In adult intestinal and blood cells, sustained absence of Polycomb Repressive Complex 2 indirectly reactivates most –and only– hypomethylated developmental enhancers. Embryonic and fetal transcriptional programs re-emerge as a result, in reverse chronology to cis-element inactivation during development. Thus, hypomethylated DNA in adult cells preserves a ‘fossil record’ of tissue-specific developmental enhancers, stably marking decommissioned sites and enabling recovery of this epigenetic memory.
Graphical abstract

eTOC blurb
Although cells decommission embryonic enhancers by erasing histone marks, these regions retain hypomethylated DNA into adulthood, preserving an accurate archive of tissue-specific developmental cis-elements. This epigenetic memory is recovered after prolonged absence of Polycomb Repressive Complex 2, when transcription factors selectively reactivate hypomethylated developmental enhancers and their tissue-restricted target genes.
INTRODUCTION
Adult tissues arise through sequential activation of selected enhancers and target genes in embryonic primordial (Bonn et al., 2012; Long et al., 2016). cis-elements used during development are subsequently inactivated (Long et al., 2016), yielding to regulatory and transcriptional programs unique to each adult tissue, but it is unclear if adult cells preserve a recoverable memory of silenced embryonic enhancers. This process is important to understand because induced pluripotency in somatic cells represents reversal of ontogeny and transcription factor (TF)-based reprogramming can leave vestiges of the original cell lineage (Kim et al., 2010; Polo et al., 2010). Moreover, cancers reactivate selected fetal genes, usually from the same or a closely related lineage (Hu and Shivdasani, 2005; Kho et al., 2004); pancreas cancer metastases, for example, reactivate embryonic foregut-specific enhancers (Roe et al., 2017).
Active enhancers display accessible chromatin, transcription factor binding, and the histone marks H3K4me1 and H3K27ac, which together associate best with expressed genes (Barski et al., 2007; Heintzman et al., 2007). The histone mark H3K27me3 is associated in ESC and adult tissues with repressed promoters, especially those of morphogenetic and TF genes (Bernstein et al., 2006; Boyer et al., 2006; Jadhav et al., 2016), but H3K27me3 is typically absent at adult enhancers (Saxena et al., 2017; Zentner et al., 2011). Instead, many candidate enhancers carry H3K4me1 but not H3K27ac; enhancers similarly marked in ESC appear to be ‘primed’ (Rada-Iglesias et al., 2010) or ‘poised’ (Creyghton et al., 2010; Zentner et al., 2011) for activation upon acquiring H3K27ac (Figure S1A). However, H3K4me1 is not necessary for placement of H3K27ac (Dorighi et al., 2017), and it is unclear why large numbers of enhancers are ‘primed’ in terminally differentiated cells.
Beyond modified histones, enhancers show reduced DNA methylation. Whereas most CpG dinucleotides in mammalian DNA are fully methylated (me), long stretches of high CG density (islands, CGIs) near transcription start sites (TSSs) are unmethylated in those tissues where the gene is active (Deaton and Bird, 2011). In contrast, enhancers have short stretches of CG-poor DNA with 20% to 50% meCpG (Hon et al., 2013; Stadler et al., 2011; Ziller et al., 2013). This intermediate state reflects oxidation of meCpG (Bogdanovic et al., 2016; Stadler et al., 2011) and likely the net effect of methylation, mediated by DNA methyltransferases (DNMTs), and demethylation mediated by TET enzymes (Hon et al., 2014; Stroud et al., 2011). Reduced meCpG at selected adult enhancers is attributed to their prior activity in embryos (Hon et al., 2013; Lee et al., 2015). However, most cis-elements used in embryos remain unidentified and >50% of sites with hypomethylated DNA in the fetal brain were fully methylated in adult brains (Hon et al., 2013). Moreover, the multitude of brain cell types precludes knowing which fetal enhancers lose or preserve this feature in given cell populations. Most importantly, it is unclear if decommissioned developmental enhancers retain memory that adult cells can recover.
In contrast to the brain and other adult tissues, the intestinal epithelium descends lineally from a well-defined region of gut endoderm, without cellular admixture. Proliferating stem and progenitor cells in adult intestinal crypts continually produce terminally differentiated, short-lived villus cells of one predominant type: enterocytes (Clevers, 2013) (Figure 1A). Villus epithelial cells are readily separated from crypts and underlying mesenchyme (Weiser, 1973) and their developmental forebears can be purified from embryos (Sherwood et al., 2009). We took advantage of these features to study recruitment and decommissioning of cis-elements during mouse gut development. We found that adult intestinal cells retain a record of ~90% of tissue-specific developmental enhancers in the form of hypomethylated DNA. Enhancers used early in development retain no other discernible mark, whereas thousands of sites used late in gestation retain some H3K4me1 but not H3K27ac. These H3K4me1+H3K27ac− regions are therefore not necessarily ‘poised’ or ‘primed’ for activation, but are sites that were decommissioned late in development. Remarkably, prolonged absence of Polycomb Repressor Complex 2 (PRC2) results in indirect and delayed reactivation of silenced developmental enhancers, in roughly reverse order to their use in embryos. In diverse PRC2-null cells, only tissue-restricted hypomethylated enhancers –and nearly all such enhancers– are reactivated, with attendant expression of developmental genes.
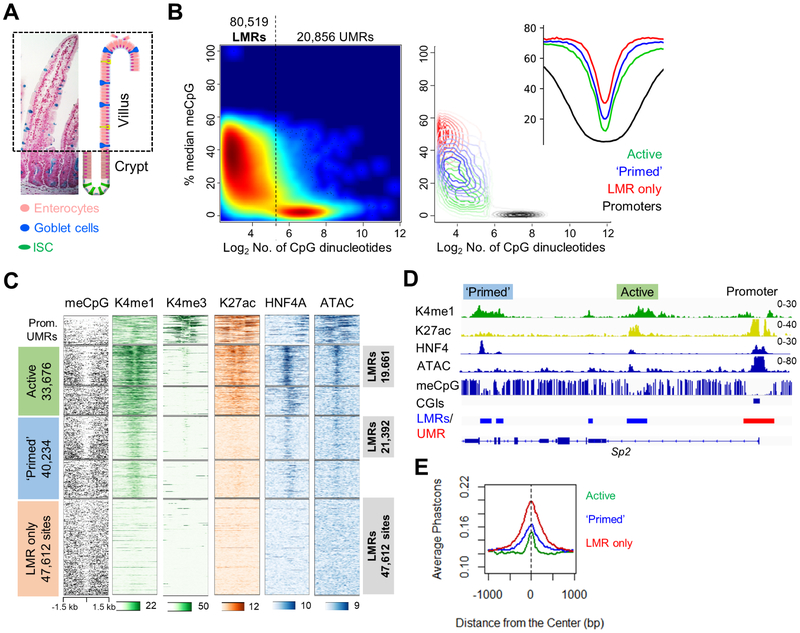
Figure 1. Low-methylated regions (LMRs) in adult mouse intestinal villus epithelium.
(A) Micrograph and drawing of mouse duodenal epithelium stained with Alcian blue and eosin, showing predominance of enterocytes in post-mitotic intestinal villi. (B) Left: distribution of all un- (UMRs) and low- (LMRs) methylated regions in purified villus epithelium. Right: contour map and aggregate meCpG density plots show the distribution and meCpG profiles of promoters, active and ‘primed’ enhancers, and sites recognized only by reduced meCpG. (C) Signals for methylated DNA (meCpG), histone marks H3K4me1/3 and H3K27ac, occupancy of the TF HNF4A, and open chromatin (ATAC) at 12,710 promoter UMRs, active and ‘primed’ enhancers, and LMR-only regions. H3K4me1+ enhancers are partitioned into groups with (LMRs) or without hypomethylated DNA. (D) Representative Integrative Genome Viewer (IGV) tracks showing key cis-element features. Numbers refer to the scales for relative ChIP or ATAC signals. (E) High evolutionary conservation of enhancers in each group, including LMR-only sites.
See also Figures S1 and S2.
meCpG is confidently implicated in X chromosome inactivation, gene imprinting, and silencing of endogenous viruses, but its role in other forms of epigenetic memory is uncertain (Bestor et al., 2015; Schubeler, 2015). Our findings reveal meCpG as a key determinant of bona fide and recoverable epigenetic memory dating to the period of organogenesis.
RESULTS
Short stretches of hypomethylated DNA vastly outnumber marked, active enhancers
In adult mouse duodenal villus epithelium, ChIP-seq (Table S1 and Figure S1B) identified 33,676 distant (>−2 and >+1 kb from TSSs) H3K4me1+ regions with, and 40,234 sites lacking, H3K27ac; genes near H3K27ac+ enhancers were expressed at high levels, whereas those near H3K27ac− sites were not (Figure S1C). Because the proportion of primed enhancers seemed high for short-lived, terminally differentiated cells and the extent of meCpG at these sites is unknown, we assessed genome-wide DNA methylation at base resolution. Our whole-genome bisulfite sequencing (WGBS) data showed high concordance with published results (Sheaffer et al., 2014) and comparably low meCpG fractions in both groups of H3K4me1+ sites (Figure S1D). Defined by established criteria (Stadler et al., 2011), 53% to 58% of sites in each group showed reduced meCpG, revealing hypomethylated DNA as a common but not universal enhancer property. Notably, ~32,000 regions far from TSSs showed <50% meCpG, identified at a false discovery rate (FDR) of 0.05, but lacked H3K4me1 or H3K27ac, indicating that sites with hypomethylated DNA outnumber those with active histone marks. Applying an FDR of 0.1, we identified ~12,000 additional hypomethylated sites (17.5% increase, Figure S2A) in purified duodenal villus epithelium and hence mapped a total of 20,856 unmethylated (UMRs) and 80,519 low-methylated regions (LMRs, corresponding to FDR 0.1 and meCpG <59%, Figure 1B). UMRs overlapped extensively with promoter CGIs, but included 8,146 non-promoter sites (Figure S2B), whereas LMRs encompassed active regions, ‘primed’ enhancers, and 47,612 sites that lack H3K4me1 or H3K27ac (Figure 1B-C). Going forward, we consider all non-promoter regions with either active mark or reduced meCpG as potential enhancers (Figure 1C and Figure S2C). The conclusions that follow apply whether we consider 68,517 LMRs (FDR 0.05) or 80,519 LMRs (FDR 0.1, Figure S2A); the latter, more inclusive set gives a fuller picture of embryonic enhancer usage and recrudescence. Of note, the LMR profile, including LMR-only regions, was similar in villus and Lgr5+ intestinal stem cells (Figure S2C), indicating that it characterizes the tissue and not just enterocytes.
Active enhancers showed accessible chromatin and binding of HNF4A, a TF that binds most enterocyte enhancers (San Roman et al., 2015); these features were absent or much reduced among ‘primed’ enhancers, and totally lacking in the regions showing only low meCpG (Figure 1C-D). Of note, although levels of hypomethylated DNA were comparable at active and ‘primed’ enhancers, H3K4me1 was generally weaker at the latter sites and histone marks were more robust at active enhancers with reduced meCpG than in heavily methylated regions (Figure 1C). Moreover, the marked nucleosomes at active enhancers flanked a central area of DNA hypomethylation, accessible chromatin, and TF occupancy, whereas nucleosomes at ‘primed’ enhancers coincided with areas of low meCpG (Figure 1C), in agreement with their diminished chromatin access and infrequent HNF4A binding. These data identify distinct patterns of histone marking and DNA hypomethylation at active and ‘primed’ enhancers, and reveal thousands of sites with reduced meCpG as a solitary feature.
To examine the bona fides of the latter group, first we identified LMRs at FDR 0.1 in public WGBS data from mouse blood (An et al., 2015). At least 45% of these LMRs lacked H3K4me1 (Figure S2D), revealing LMR-only sites in comparable abundance to the intestine. Moreover, intestinal LMRs lacking H3K4me1 showed even higher enrichment of TF sequence motifs (Figure S2E), and evolutionary conservation (Figure 1E) than did H3K4me1+ enhancers. Whereas active and ‘primed’ enhancers were enriched for DNA sequence motifs associated with known intestinal TFs and nearby genes are enriched for enterocyte functions, both ‘primed’ and LMR-only sites were enriched for the motifs of developmental TFs, such as FOX factors, and nearby genes are enriched for developmental functions (Figure S2E-F). Thus, intestinal LMRs encompass both active and seemingly inactive cis-elements of two types, H3K4me1+ and H3K4me1−, and features of the inactive sites suggest prior activity during development.
Adult cells retain a comprehensive archive of developmental enhancers
A fraction of hypomethylated areas of DNA in adult mouse (Hon et al., 2013) and zebrafish (Lee et al., 2015) cells show cis-element activity in embryos, but >50% of sites with reduced meCpG in the fetal brain were fully methylated in adult brain cells (Hon et al., 2013). Because the brain contains numerous cell types, it is unclear which fetal enhancers lose or preserve hypomethylated DNA in a given population. In contrast, the intestinal epithelium descends directly from region-specific endoderm, and after embryonic day (E) 11, the surface protein EPCAM selectively marks prospective epithelial cells (Sherwood et al., 2009), allowing their isolation by flow cytometry (Figure S3A). To trace the possible origins of hypomethylated regions in the adult intestine, we assessed gene activity and enhancer dynamics in endodermal cells purified from different stages in intestine development (Table S1 and Figure S3B-C).
Between E11.5 and adult intestinal epithelium, EPCAM+ cells showed 12,266 differences in mRNA levels (>2-fold, q <0.05) between any two stages; these alterations occurred in waves that coincide with early, mid, and late gestation (Figure S3D). Over the same period, ATAC-seq (Buenrostro et al., 2015) on EPCAM+ cells identified 68,510 unique areas of open chromatin >1 kb from promoters. Unsupervised k-means clustering placed these sites in 10 clusters (Figure S3E), which formed 4 distinct groups (Figure 2A): (1) accessible at all stages, (2) open only before E14.5, (3) accessible mainly in mid- (E14.5) to late (E16.5) gestation, and (4) open at and after E16.5. Developmental stage-specific expression of nearby genes correlated with these waves of chromatin access (Figure S3E), indicating that regions identified by ATAC represent active enhancers. Indeed, intestinal endoderm purified at E16.5 (this study) or E12.5 (Kazakevych et al., 2017) showed H3K4me1 or H3K27ac, respectively, in many areas marked by ATAC before E14.5 (Figure S3F). At >96% of sites with open chromatin before E16.5, the chromatin was closed in adult cells (Figure 2B). These ~26,000 ATAC+ sites function during intestine development and are subsequently decommissioned; because ChIP- and ATAC-seq have some limitations in sensitivity, the full complement of inactivated developmental enhancers is likely even larger.
Figure 2. Chromatin and meCpG dynamics during mouse intestine development.
(A) Profiles of open chromatin in purified EPCAM+ intestinal cells across 4 gestational (E) ages and in adult villus cells at regions >−2 or >1 kb from TSSs. Among the 68,510 sites identified by ATAC-seq at any stage, successive waves of accessible chromatin identify candidate embryonic, fetal, and adult enhancers. (B) Limited overlap of accessible chromatin (ATAC) sites in developing and adult intestinal epithelium. Most of the 38,376 regions open in fetal or embryonic intestinal endoderm were closed in adults. (C) Profiles of hypomethylated DNA at ATAC+ sites, extracted from WGBS data on undifferentiated E6.5 epiblast (Seisenberger et al., 2012), developing endoderm, and adult villus epithelium. Representative IGV tracks show open chromatin, H3K4me1, meCpG, and mRNA dynamics at representative embryonic (Hapln1) and fetal (Myl1) loci. (D) meCpG states in adult, fetal, and embryonic intestinal epithelium. All but 8,914 of the 53,350 LMRs (16.7%) identified objectively in the developing intestine met stringent LMR criteria in adults. Most enhancers that became inaccessible after development (B) thus retained hypomethylated DNA.
See also Figure S3.
To determine enhancer DNA methylation dynamics during intestine development, we performed WGBS on purified E12.5 and E16.5 intestinal epithelium. ATAC-identified enhancers were fully methylated in the epiblast and meCpG was reduced sequentially: first at sites showing open chromatin at E11.5 and E12.5, later in regions that opened in mid-gestation, and last at active adult enhancers (Figure 2C). Moreover, we objectively identified 53,350 unique non-promoter regions hypomethylated (FDR <0.1) at E12.5 and E16.5; these sites correspond to the areas we had identified in adult villus cells as the ‘LMR-only’ and ‘primed’ groups, respectively (Figure S3G). Notably, 83% of sites hypomethylated at E12.5 and 92% of sites hypomethylated at E16.5 remained so in adult villus cells (Figure 2C-D); only 8,914 enhancers became fully methylated. Thus, after decommissioned enhancers relinquish chromatin access, active histone marks and regulatory functions, they retain reduced meCpG as a singularly stable feature, preserved over the hundreds of cell divisions that separate embryonic from adult cells. Thousands of sites hypomethylated in the epiblast, however, were almost fully methylated in adult intestinal cells (Figure S3H), indicating that tissue-restricted enhancers are archived only after cells are specified. Within this archive, sites active late in gestation retain H3K4me1 and occasional weak TF binding and open chromatin in adult cells. Accordingly, these are not ‘primed’ cis-elements per se, but transient and late-acting developmental enhancers. In contrast, sites active early in organogenesis retain none of the features we examined, other than hypomethylated DNA.
Prolonged PRC2 inactivity selectively reactivates developmental enhancers
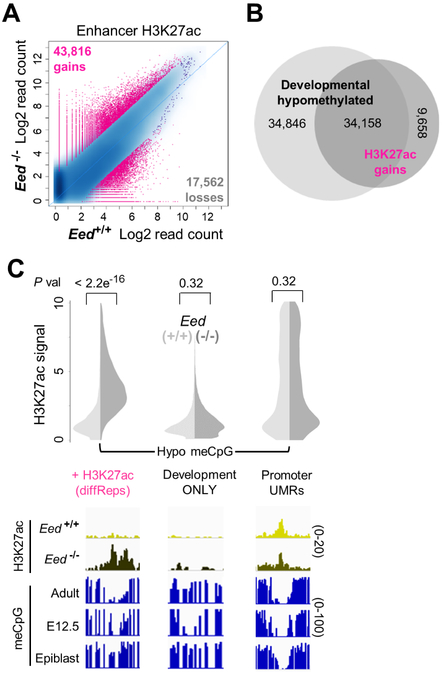
Inactivation of developmental genes is a cardinal function of PRC2, which places the histone mark H3K27me3. However, up to 9 days after loss of PRC2 from intestinal epithelium in Villin-CreER-T2;EedFl/Fl mice, only the few genes with tissue-specific bivalent (H3K4me3+ H3K27me3+) promoters are derepressed (Jadhav et al., 2016). All methylated H3K27 forms were abrogated within 9 days of Cre recombinase activation, and among 10 other covalent histone marks we examined by mass spectrometry, only H3K27ac was modestly increased (Figure 3A). Because these findings hinted that absence of PRC2 might expose unmethylated H3K27 residues to indiscriminate acetylation, we examined the H3K27ac distribution in Eed−/− villus epithelium. To preclude any contribution from Eed-proficient ‘escaper’ crypts, we administered tamoxifen over a prolonged period; when mice became moribund, at 14 days, the epithelium lacked H3K27me3 or dividing cells (Figure S4A). Even within 9 days of PRC2 depletion, ChIP revealed H3K27ac at silenced fetal enhancers, and by day 14 the marking was stronger and now also evident at embryonic enhancers (Figure 3B). We verified these gains by meticulous normalization using Drosophila chromatin ‘spike-in’ controls (Methods and Table S1). The same sites also acquired H3K4me1 (Figure 3B), indicating increased enhancer activity rather than passive acetylation of unmethylated H3K27.
Figure 3. Reactivation of developmental enhancers after prolonged PRC2 deficiency.
(A) Epithelium-restricted loss of all methylated H3K27 forms in Villin-CreER-T2;EedFl/Fl mice 9 days after the 1st of 5 daily injections of tamoxifen; residual fluorescence signals are confined to sub-epithelial cells, separated from the overlying epithelium by dashed lines. Pie charts depict the fractions of each covalently modified form of H3K27 and the table lists the proportions of other histone modifications measured by mass spectrometry in purified WT and Eed−/− villus epithelium (N=3 each). All H3K27me was markedly reduced and H3K27ac was slightly increased, while 10 other histone marks were unperturbed. (B) Accumulation of H3K27ac and H3K4me1 at fetal and embryonic enhancers in Eed−/− intestinal villus epithelium and preferential loss of these histone marks from non-hypomethylated active enhancers. Sites are arranged in the same order as Figure 1C, with adult (active) and fetal (‘primed’) enhancers partitioned into those with and without LMRs. IGV tracks below illustrate the H3K27ac and H3K4me1 gains (numbers refer to the scales for ChIP-seq signals).
See also Figure S4A.
Among millions of nucleosomes depleted of H3K27me in Eed−/− cells, however, diffReps (Shen et al., 2013) identified only 43,816 sites of H3K27ac gain and 17,562 sites of reduced H3K27ac located >1 kb from promoters (Figure 4A). Thus, sustained absence of PRC2 in adult intestinal cells results in highly selective histone modulation. Nearly 80% of sites that acquired H3K27ac by these objective measures coincided with hypomethylated developmental enhancers (P=0, Fisher exact test), and conversely, nearly 1/2 of all hypomethylated enhancers inactivated during development gained H3K27ac (Figure 4B). In addition, many embryonic and fetal enhancers with discernible H3K27ac and H3K4me1 gains (Fig. 3B) did not meet stringent diffReps criteria. To assess site specificity, we considered the 8,914 sites that had low meCpG in embryonic or fetal intestine, but became fully methylated in adults (Figure 2D), and the ~13,000 promoter UMRs (Figure S2B). Unlike developmental enhancers with persistent hypomethylation, neither group acquired significant H3K27ac in Eed−/− intestines (Figure 4C). Thus, histone activation was almost entirely restricted to decommissioned developmental enhancers.
Figure 4. Reactivation is confined to hypomethylated decommissioned enhancers.
(A) Strict quantitation of H3K27ac gains and losses (>1.5-fold, q<0.01) in mutant cells by diffReps (Shen et al., 2013). (B) Areas of objective H3K27ac gain overlap significantly with embryonic and fetal enhancers that are hypomethylated in adult intestinal epithelium. (C) In contrast, H3K27ac did not accumulate at the 8,914 enhancers that are hypomethylated in development but methylated in adults, or at the 12,710 promoter UMRs. P values were calculated using the Wilcoxon signed rank test. IGV tracks below each split-violin plot (WT, light; Eed−/− dark) illustrate H2K27ac changes at cis-elements with different levels of basal (WT) meCpG.
Basis of enhancer reactivation in the absence of PRC2
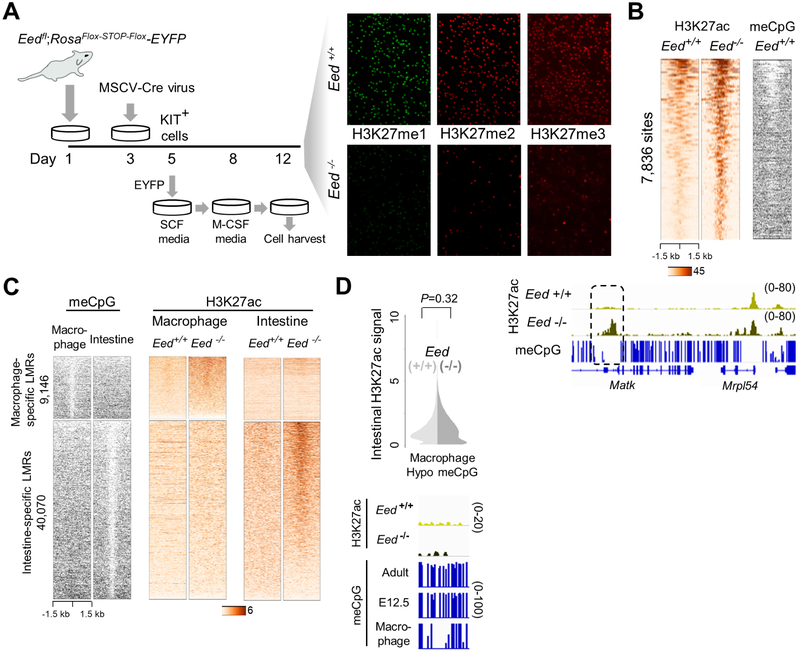
To examine the specificity of this response to PRC2 loss, first we identified LMRs in public WGBS data from mouse skin and brain (Figure S4B), just as we had done earlier for blood cells (Figure S2D). In keeping with the tissue specificity attributed to enhancers, LMRs identified in villus epithelium or any other tissue were usually fully methylated in the others, whereas UMRs (mostly promoters) usually lacked meCpG in all tissues (Figure S4C-D). To determine if H3K27ac gains occur at LMRs after PRC2 loss in other cell types, we cultured bone marrow from EedFl/Fl mice, deleted Eed by viral CRE expression, induced macrophage differentiation in vitro, and confirmed that mutant CD11b+ macrophages lacked H3K27me (Figure 5A). In PRC2-null macrophages, diffReps identified 7,836 sites of quantitative H3K27ac gain, and these occurred selectively at hypomethylated enhancers (Figure 5B). We also identified nearly 50,000 inactive (H3K27ac−) enhancers with unambiguous differential methylation between macrophages and intestinal cells. In each case, H3K27ac accumulated only in the corresponding PRC2-null tissue, as is evident in heatmaps (Figure 5C) and by strict quantitation of macrophage sites in intestinal cells (Figure 5D). In summary, only silenced developmental enhancers with low meCpG acquire active histone marks in the absence of PRC2; this occurs with high tissue specificity and, in the intestine, sooner at fetal than at embryonic enhancers.
Figure 5. Enhancer flux in PRC2-null macrophages.
(A) Erasure of all methyl-H3K27 forms in Eed−/− macrophages, cultured as depicted in the experimental schema and examined by immunofluorescence. (B) Relation of 7,836 sites of objective H3K27ac gain (diffReps >1.5-fold, q<0.01) in Eed−/− macrophages to the meCpG state of each in WT cells. H3K27ac accumulated at sites with basal hypomethylated DNA; IGV tracks are shown at a representative locus. (C) LMRs in macrophages and intestine that lack H3K27ac in the respective WT cells show tissue-specific H3K27ac gains in PRC2-null cells. (D) Macrophage-restricted enhancers (n=21,083) do not acquire H3K27ac in Eed−/− intestine. P values were calculated using the Wilcoxon signed rank test. IGV tracks below split-violin plot (WT, light; Eed−/− dark) illustrate presence or absence of H2K27ac gains at cis-elements with different levels of basal (WT) meCpG in intestine.
See also Figure S4B-D.
To investigate the basis for significant enhancer modulation in PRC2-null cells, we used ChIP to map the distributions in adult wild-type (WT) villus cells of all PRC2-modified forms of H3K27 (mono-, di-, and tri-methyl) and the PRC2 component SUZ12 (Table S1). Mirroring the distributions reported in ESC (Ferrari et al., 2014), H3K27me1 predominated at active loci, H3K27me2 in intergenic regions, and SUZ12 and H3K27me3 at silenced promoters and genes (Figure S5A). Enhancers generally lacked SUZ12 or H3K27me3, and when present, H3K27me3 was dispersed across large regions, at levels considerably lower than those found at repressed bivalent promoters (Figure S5B). Likewise, H3K27me1 and H3K27me2 were distributed over hundreds of kb throughout the genome, with no focal enrichment in areas that acquire H3K27ac in Eed−/− cells (Figure S5B-C). Thus, modulation of these enhancers is likely a secondary effect stemming from the direct, short-term consequence of PRC2 loss: activation of numerous bivalent TF genes that mediate early intestine development (Jadhav et al., 2016).
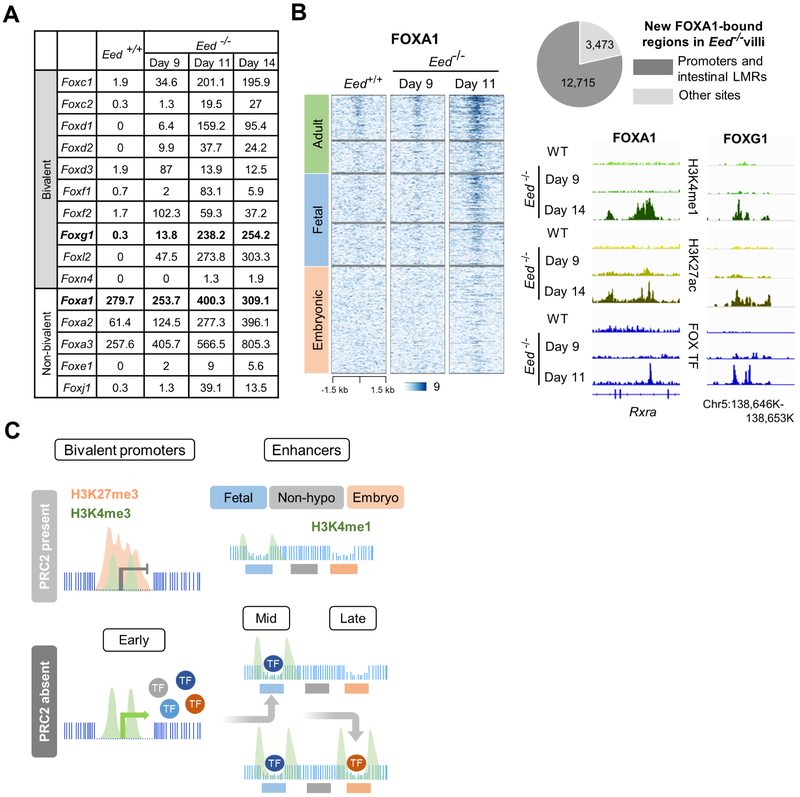
Such mRNAs, including many FOX-family TFs, continued to accumulate after day 9 (Figure 6A) and showed persistent association with basal promoter H3K4me3 levels (Figure S6A); prolonged PRC2 loss also increased levels of certain non-bivalent FOX genes (Figure 6A). To investigate the likely proximate cause of enhancer reactivation, we performed ChIP-seq for the two FOX TFs against which ChIP-grade antibodies are available. FOXA1 binding was robust and >78% of sites newly occupied in Eed−/− villus cells mapped to hypomethylated adult and developmental enhancers (Figure 6B). FOXG1 binding was less strictly quantifiable, but we nevertheless detected unambiguous occupancy at fetal and embryonic enhancers showing H3K27ac gains, some of which also bound FOXA1 (Figure S6B). Thus, following promoter-based early gene reactivation, binding of FOX and other TFs likely accounts in aggregate for cis-element reactivation as an indirect consequence of PRC2 deficiency (Figure 6C).
Figure 6. Enhancer reactivation follows on early consequences of PRC2 deficiency.
(A) Partial list of representative induced and stable FOX-family TF mRNAs (normalized read counts) expressed from bivalent and non-bivalent genes in the mouse intestine. (B) New enhancer FOXA1 binding in Eed−/− cells occurs predominantly at sites defined in this study, including hypomethylated fetal enhancers that acquired H3K4me1 and H3K27ac. Enhancers are arranged in the same order as in Figures 1C and 3B. IGV tracks show FOXA1 and FOXG1 occupancy at enhancers within 11 days of initial Eed deletion. (C) Inferred sequence of events following PRC2 loss. After TF and other genes are activated early from bivalent promoters, some TFs reactivate first hypomethylated fetal and then embryonic enhancers; fully methylated regions avoid reactivation.
See also Figures S5 and S6A-B.
Transcriptional consequences of enhancer recrudescence
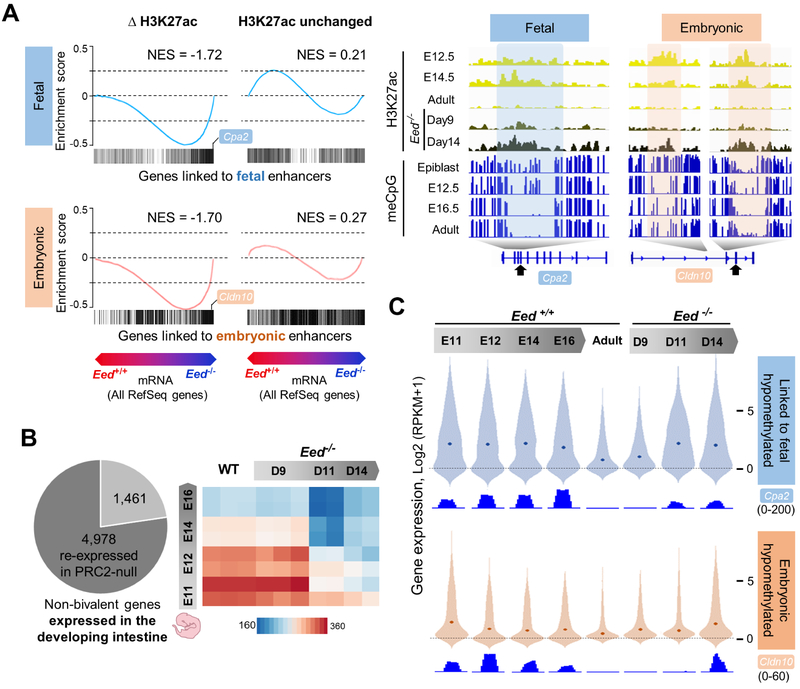
Compared to the few bivalent genes activated within 9 days of initial CreE-T2 activation, 6,127 additional genes increased (>2-fold, q <0.05) by 14 days, the limit to which mice tolerated intestinal PRC2 loss. Most corresponding promoters had high basal levels of H3K4me3 and no H3K27me3 in WT intestines (Figure S6C-D). Thus, promoter H3K4me3 is the limiting factor –or a reliable proxy for one– at genes activated soon after PRC2 loss (Jadhav et al., 2016), but it does not account for genes activated after sustained PRC2 deficiency. Instead, genes activated after day 9 showed striking association with fetal and embryonic enhancers that acquired H3K27ac, but not with enhancers that lacked H3K27ac gains (Figure 7A).
Figure 7. Expression of developmental genes linked to reactivated enhancers.
(A) Gene set enrichment analysis (GSEA) showing the distribution of genes near (<50 kb) fetal and embryonic enhancers among the genes reactivated by day 14 in Eed−/− villus cells. All RefSeq genes are arrayed in order of relative expression in WT and Eed−/− cells. Recommissioned H3K27ac+ fetal and embryonic enhancers (left) were significantly associated with reactivation of genes within 50 kb, while those lacking H3K27ac gains were not (right). NES, normalized enrichment score. Representative IGV tracks (right) show recrudescence of decommissioned enhancers in PRC2-null (Eed−/−) intestines. Arrows point to exons at which mRNA levels are shown in panel C. (B) Left: Fraction of all non-bivalent genes expressed during intestine development that were reactivated in Eed−/− villus cells. Right: Relatedness (Euclidean distances) among RNA-seq profiles of triplicate (WT and 9-day Eed−/−) or duplicate (all others) samples of WT and Eed−/− adult villus epithelium and WT intestinal endoderm. (C) Distributions of mRNA levels during intestine development and after adult PRC2 depletion of all genes located <50 kb from hypomethylated fetal or embryonic enhancers. Dots in each violin plot represent median values. WT levels of both transcript groups were comparably low, fetal genes were reactivated earlier than embryonic genes, and both groups were re-expressed at levels approaching those seen in developing WT endoderm. IGV tracks show representative RNA-seq data for the exons marked with arrows in panel B.
See also Figures S6C-D and S7.
More than 77% of non-bivalent genes decommissioned during intestine development were activated in the prolonged absence of PRC2, and intestinal villus cells accordingly expressed both adult and embryonic transcriptomes (Figure 7B). Moreover, among genes activated after sustained PRC2 deficiency, 70.2% had been expressed in the developing intestine, compared for example to 23.3% and 21.4% that were expressed in developing heart or lungs (P <0.0001), and most of the latter genes were also expressed in the developing intestine (Figure S7). Thus, activation was largely confined to, and encompassed most, intestine-specific developmental genes. Those expressed late in gestation and linked to silenced fetal (H3K4me1+) enhancers were reactivated by day 11 and further increased at day 14, whereas genes expressed early in development and associated with decommissioned embryonic (LMR-only) enhancers were re-expressed later (Figure 7C). Basal expression of both gene groups was equally low in WT adult villi and the timing of re-expression in Eed−/− cells correlated with sequential reactivation of fetal and embryonic enhancers (Figure 3B). Transcript levels of both groups in adult Eed−/− cells approached those present during development (Figure 7C). Thus, histone and mRNA fluxes together reveal enhancer DNA hypomethylation as the crucial property that underlies memory and reactivation of tissue-restricted developmental programs.
DISCUSSION
Methylated DNA inactivates endogenous viruses, sex chromosomes and imprinted genes, but its roles in tissue-specific and developmental gene control draw on correlative findings and remain controversial (Bestor et al., 2015). Separately, it has been unclear if decommissioning of developmental enhancers is irrevocable. This study reveals low meCpG as a persistent adult feature of almost 90% of those decommissioned enhancers that were hypomethylated during development and the signature of sites that reactivate after prolonged PRC2 deficiency. Among the millions of cis-elements in the genome, only developmental enhancers with low meCpG were activated. The significant overlap with sites that gained H3K27ac (Figure 4B) is incomplete because formal definition of LMRs (FDR 0.1) and H3K27ac gains (q <0.01) requires arbitrary cut-offs. Indeed, gains of H3K27ac and H3K4me1 were evident at most developmental LMRs (Figure 3B) and in hypomethylated regions with 60% to 90% meCpG. Thus, hypomethylated DNA determines which enhancers are reactivated or at least signifies the potential for robust, tissue-specific reactivation. These sites re-emerged despite their lack of focal H3K27me or PRC2 binding in wild-type cells. Together with binding of embryonic TFs at activated sites, our findings imply that enhancer recrudescence is unrelated to H3K27me per se.
Hypomethylated DNA, presumably generated during development by TET enzymes (Hon et al., 2014; Stroud et al., 2011), is stably preserved from embryonic tissues to adult organs. The maintenance methyltransferase DNMT1 offers a simple explanation because in each cell cycle it reproduces existing meCpG states on the new DNA strand (Bestor et al., 2015; Li et al., 1992; Schubeler, 2015). Thus, reduced meCpG could be preserved through faithful copying of fetal and embryonic meCpG templates over many cell generations; alternatively, it may reflect continually opposing DNMT and TET activities. Methylated histone H3K4 is thought to repel DNMT3L to help maintain absence of meCpG at promoters and CGIs in ES and germ cells (Ooi et al., 2007) and a recent study implicates DNMT3A in persistent hemimethylation of DNA at cohesin- and CTCF-binding sites during ES cell replication (Xu and Corces, 2018). Most LMRs in our study, however, lack H3K4me or other overt barriers to DNA methylation, and de novo DNMTs decline appreciably after development (Schubeler, 2015). Of note, epiblast LMRs are largely methylated in adult intestinal cells and embryonic LMRs have slightly larger meCpG fractions than fetal enhancers. These observations imply that the copying mechanism may be imperfect over long periods, in agreement with Holliday and Pugh’s ideas about ‘developmental clocks’, wherein DNMTs and demethylases are inherently inefficient over many cell cycles (Holliday and Pugh, 1975).
Both embryonic and fetal enhancers lack H3K27ac and their target genes are comparably silent in adults, but fetal enhancers retain low levels of H3K4me1, the solitary mark associated with enhancer ‘priming’ or ‘poising’ in ESC (Creyghton et al., 2010; Rada-Iglesias et al., 2010; Zentner et al., 2011). In adult intestines, however, enhancers bearing only H3K4me1 are those that were decommissioned late in organogenesis. As vestiges of fetal life, these sites likely have little physiologic function, though they do activate sooner in PRC2-null cells than H3K4me1− enhancers. Enhancers that act only during development often elude detection by assays for accessible chromatin or modified histones. Hypomethylated DNA, which preserves a faithful record of these enhancers into adulthood, can therefore be used to identify embryonic cis-elements in purified adult cells. However, only ~60% of active enhancers are categorically hypomethylated (Figure 1C), owing in part to limited CpG content and because some enhancers act through methyl-insensitive TFs (Schubeler, 2015). Accordingly, whole-genome meCpG profiles (Hon et al., 2013; Ziller et al., 2013) will likely identify a significant but incomplete fraction of all cis-elements.
At least in PRC2-deficient cells, reduced enhancer meCpG is necessary and sufficient for embryonic gene reactivation, which seems constrained by the repertoires of latent enhancers and available TFs. To the extent that these constraints apply generally, our findings may explain certain features of cellular reprogramming and of aberrant gene activity in cancer. TF-driven induction of pluripotency in adult somatic cells can leave vestiges of the starting cell type and limit the differentiation potential of resulting iPS cells (Bar-Nur et al., 2011; Polo et al., 2010). This is attributed in part to retained promoter CGI fingerprints (Kim et al., 2010; Ma et al., 2014), but the restricted potential may reflect residual tissue-specific enhancer hypomethylation. In the examples of TF-mediated modulation of one cell type into another, only selected heterologous cell fates are achieved, e.g., CEBPA-driven conversion of lymphocytes into macrophages (Xie et al., 2004), possibly constrained by the repertoire of tissue-specific developmental enhancers. Three TFs –PDX1, NEUROG3 and MAFA– together convert gut endocrine cells into insulin-producing beta cells in vivo (Chen et al., 2014). This process is markedly more efficient in the gastric antrum, which shares an origin with the pancreas, than in the intestine (Ariyachet et al., 2016), possibly reflecting the pool of available hypomethylated enhancers. Finally, the finite enhancer repertoire in any cell, delineated during development, suggests a basis for oncofetal gene activation, such as tissue-specific fetal genes in colon and brain tumors (Hu and Shivdasani, 2005; Kho et al., 2004) and embryonic foregut-restricted genes in pancreatic cancer metastases (Roe et al., 2017).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests will be fulfilled by the Lead Contact, Ramesh Shivdasani (ramesh_shivdasani@dfci.harvard.edu), after execution of a Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Lgr5EGFP-IRES-CreERT2 mice (Barker et al., 2007) were purchased from Jackson Laboratories and Eedfl/fl;Villin-CreER(T2) mice were described previously (Jadhav et al., 2016). Animals were maintained on the C57Bl/6 background and housed at 23 ±°C, 55 ±15% humidity, and 12 hr light/dark cycles, with food and water available ad libitum. Mice were weaned at 21 days of age and handled according to ethical and procedural guidelines from the Animal Care and Use Committee of the Dana-Farber Cancer Institute. Animals were genotyped by PCR before weaning and again at the time of experiments, which were conducted using animals of both sexes with littermates as controls.
METHOD DETAILS
Mouse treatments
Animals 8 weeks or older were injected intraperitoneally with 2 mg tamoxifen on 5 consecutive days and, in some experiments, with 1 mg tamoxifen on alternate days thereafter (Figure S4A).
Isolation of intestinal villus and stem cells
The proximal 1/3 small intestine (duodenum) was used for histology and to collect cells for RNA and chromatin studies. Intestines harvested immediately after euthanasia were washed with cold phosphate-buffered saline (PBS), followed by rotation for 40 min in 5 mM EDTA in PBS (pH 8) at 4°C, with manual shaking every 10 min. Villus epithelium was recovered by filtering the resulting suspension over 70-μm filters (B-D Falcon). Villi retained on these filters were washed with ice-cold PBS and used to extract RNA, chromatin, or DNA. To purify Lgr5+ ISC, Lgr5EGFP-IRES-CreERT2 mouse (Barker et al., 2007) intestines were washed in PBS and villi were depleted by scraping with glass slides. Crypts were extracted by rotating for 30 min in 5 mM EDTA in PBS (pH 8) at 4°C, with manual shaking every 10 min, fo llowed by discarding the supernatant and adding fresh EDTA solution for 10 additional min. Crypts in the 70-μm filtrate were dissociated into single cells by treatment with 4% TrypLE solution (Invitrogen) at 37°C for 30 min and GFPhi ISC were isolated from the viable (DAPI−) cell fraction by flow cytometry on a BD FACSAria II SORP instrument.
Isolation of embryonic and fetal mouse endoderm
The morning of the copulation plug was designated embryonic day (E) 0.5 and embryos were harvested from pregnant dams on 11.5, E12.5, E14.5, and E16.5 into ice-cold PBS. The small intestine (digestive tract distal to the pylorus and proximal to the cecum) was digested with 0.25% trypsin (Life Technologies) for 30 min at 37°C to release single cells, followed by neutralization with fetal bovine serum (FBS, Life Technologies). Cells were passed over 40-μm filters to remove tissue fragments, centrifuged at 1,200 g for 5 min, washed in cold PBS, suspended in fluorescent-activated cell sorting (FACS) buffer (5 mM EDTA in PBS and 2% FBS), stained with APC-conjugated EpCAM antibody (Biolegend 118214, Lot B217174, 1:100) for 1 h at 4°C. Viable (DAPI−) EpCAM+ cells were isolated by flow cytometry on a FACS Aria II SORP instrument.
Purification and treatment of macrophages
Bone marrow cells from EedFFl/Fl;Rosa26REYFP mice were cultured in media supplemented with SCF, IL-3 and IL-6 (R&D Systems, 10 ng/ml each) for 3 days, followed by infection with MSCV-Cre retrovirus, prepared by cloning Cre cDNA into the BglII and XhoI restriction sites in an MSCV-Hygro vector (Clontech). After 48 h, KIT+ cells were enriched using CD117 microbeads (Miltenyi Biotech). Cre-infected (Eed−/− EYFP+) and uninfected (EedFl/Fl, EYFP−) cell fractions were separated by flow cytometry and cultured for 3 additional days in media with 10 ng/ml SCF and IL-3, followed by 4 days in media with 10 ng/ml IL-3 and M-CSF to obtain populations highly enriched for CD11b+ macrophages.
ChIP-seq.
Intestinal epithelium or FACS-sorted cells were fixed by in 1% formaldehyde for 25 min at room temperature immediately following isolation. ~1×106 to 5×106 cells were lysed in buffer containing 30 mM Tris-HCl (pH 8), 1% SDS, 10 mM EDTA, and protease inhibitors (Roche), and chromatin was sheared by sonication in a Covaris E210 sonicator for 50 min with 5-min on/off cycles at 4°C. After centrifugation to remove debris, chromatin was incubated overnight at 4°0 with Ab against H3K4me1 (Diagenode C15410194, Lot A1862D), H3K4me3 (Diagenode C15410003, Lot A1052D), H3K27ac (Active Motif 39135, Lot F1311), H3K27me1 (Active Motif 61015, Lot 35813006), H3K27me2 (Cell Signaling D18C8, Lot 12), H3K27me3 (Millipore 07-449, Lot 2607758), Suz12 (Cell Signaling, D39F6, Lot 6), FOXG1 (Active Motif 61211, Lot 34711001), HNF4A (Santa Cruz sc-6546 (C-19)X, Lot F1311), or 1:1.5 combination of FOXA1 (Abcam 23738, Lot gr292351-2 and Abcam 5089, Lot gr122110-14) antibodies (Ab). For the samples indicated in Table S1, before Ab incubation we added 10 ng Spike-in chromatin from Drosophila Line 2 (S2, Active Motif 53083, Lot 11316004) and 2 μg Spike-in Ab against the Drosophila-specific histone variant H2Av (Active Motif 61686, Lot 17316003). Ab-bound chromatin was captured with magnetic beads (Dynal) and washed sequentially in low-salt (20 mM Tris-HCl pH 8.1, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% TritonX-100), high-salt (20 mM Tris-HCl pH 8.1, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% TritonX-100), and lithium chloride (10 mM Tris pH 8.1, 0.25 M LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholate) buffers. Crosslinks were reversed using 1% SDS and 0.1 M NaHCO3 for 6 h at 65°C, DNA was purified using columns (Qiagen), and ChIP-seq libraries were prepared using ThruPLEX kit (Rubicon, R400427). DNA size distribution in the libraries was determined using high sensitivity DNA Chip detection on Bioanalyzer 2100 (Agilent Genomics) and 75-bp single-end reads were sequenced on a NextSeq 500 instrument (Illumina).
Whole genome bisulfite sequencing (WGBS)
Genomic DNA was purified from cells using MasterPure DNA purification kit (Epicenter MCD85201) and 50 ng DNA was treated with the EZ DNA Methylation-Gold kit (Zymo Research D5005) for bisulfite conversion. We amplified 10 ng of bisulfite-converted DNA and prepared whole genome bisulfite sequencing (WGBS) libraries using the EpiGenome Methyl-Seq kit (Epicenter EGMK81312). Libraries were purified using AMPure beads (Beckman Coulter) and DNA size range of 200-800bp confirmed using high sensitivity DNA Chip detection (Bioanalyzer 2100, Agilent Genomics). Libraries were sequenced on a NextSeq 500 instrument (Illumina) with up to 50% PhiX phage DNA (Illumina) to obtain 150-bp paired-end reads.
RNA-seq
Tissues or purified cells were lysed in Trizol (Life Technologies) and total RNA was extracted. RNA-seq libraries were prepared using TruSeq RNA Sample Preparation Kit V2 (Illumina RS-122-2001) for adult cells or the SMARTer-Seq v4 Low Input mRNA library kit (Clontech) for embryonic (E11.5, E12.5, E14.5, and E16.5) samples. Libraries were sequenced on a NextSeq 500 instrument (Illumina) to obtain 75-bp single-end reads.
ATAC-seq
ATAC-seq (Buenrostro et al., 2015) was performed on 5,000 to 30,000 FACS-sorted epithelial cells from embryonic or adult intestinal epithelium, as described previously (Banerjee et al., 2018). Freshly isolated cells were washed with cold PBS, resuspended in 50 μl cold ATAC lysis buffer (10 mM Tris·Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% (v/v) Igepal CA-630), and nuclei were isolated by centrifugation at 500 g at 4°. Nuclear pellets were treated with Nextera Tn5 Transposase (Illumina, FC-121-1030) for 30 min at 37° in 50 μl reactions. Transposed DNA was column-purified (Qiagen, 28004) and amplified using high-fidelity 2X PCR Master Mix (New England Biolabs) with a common forward primer and different reverse primers carrying sample-specific barcodes. After 5 cycles of amplification, 5 μl of the reaction-mix was amplified using qPCR for 20 cycles; the remaining 45 μl was then amplified for the number of cycles necessary to achieve 1/3 of the maximum fluorescence intensity in qPCR. Primer dimers (<100 bp) were removed from the amplified ATAC-seq library using AMPure beads (Beckman Coulter, A63880), library size distribution was determined using high sensitivity DNA Chip detection on Bioanalyzer 2100 (Agilent Genomics), and sequencing was done on a NextSeq 500 instrument (Illumina) to obtain 75 bp single-end reads.
Detection of proteins
Tissues were fixed overnight in 4% paraformaldehyde at 4°C, washed in PBS, dehydrated in ethanol, and embedded in paraffin; 5-μm tissue sections were deparaffinized and rehydrated. Sections were stained with hematoxylin and eosin or treated with 10 mM sodium citrate (pH 6) for antigen retrieval and incubated overnight at 4°C with antibodies (Ab) against H3K27me1 (Active Motif 61015, Lot 35813006, 1:1000), H3K27me2 (Cell Signaling D18C8, Lot 12, 1:1000), H3K27me3 (Millipore 07-449, Lot 2607758, 1:1000) or KI67 (Vector VP-K452, 1:500) in PBS. After washing in PBS for 10 min, sections were incubated with anti-rabbit or anti-mouse IgG conjugated to Cy3, FITC or biotin (Jackson Laboratories, 1:1000) and signals were detected by fluorescence or by staining with Vectastain Elite ABC Kit (Vector) and 3,3' diaminobenzidine tetrahydrochloride (Sigma P8375). Cultured macrophages were pelleted onto glass slides using Cytospin3 (Shandon) at 800 rpm for 5 min and fixed with 1% paraformaldehyde for 5 min. Slides were washed with PBS and processed as above for H3K27me1/2/3 immunohistochemistry. Analyses were conducted on tissues from at least 5 mice of each genotype.
Post-translational histone modifications
Mouse intestinal epitelial cells were lysed with an AFA Focused-Ultrasonicator (Covaris). Histones were purified from the lysates (Active Motif, Cat. No. 40026) and desalted by off-line reversed phase chromatography on an Agilent 1200 tower using a Jupiter 5 μm C4 300 Å Column 150 × 2 mm (Phenomenex). The resulting peak area was used to estimate concentration against histone preparations of known concentration. Desalted histones were lyophilized and spiked 1:1 with histones isolated from 5×106 HeLa cells grown in RPMI 1640 SILAC heavy arginine (13C6 15N4) medium (Cambridge Isotope Labs), followed by treatment with NHS-propionyl synthesized at neutral pH and digestion with trypsin (Promega). Peptides were lyophilized and treated again to obtain a homogenous population of derivatized lysines and new N-termini, followed by high resolution – high mass accuracy LC-MS/MS in an Orbitrap Elite instrument (Thermo Scientific) equipped with a nanoACQUITY UPLC tower with a 1×100mm HSS T3 1.8 μm column (Waters). Mass spectrometry data were interpreted using Mascot Distiller (Matrix Science) for identification, followed by manual verification. The peak area was processed using Skyline Software v1.4.0.422 (University of Washington) to quantify histone peptides bearing specific post-translational modifications.
QUANTIFICATION AND STATISTICAL ANALYSIS
Delineation and analysis of enhancers and expressed genes
For ATAC- and ChIP-seq, reads were aligned to the mouse genome (Mm9, NCBI build 37) using Bowtie2 (Langmead and Salzberg, 2012) v2.1.0; PCR duplicates and reads aligned to multiple locations were removed from the raw alignment (bam) files and peaks (P <10−5) were detected using MACS (Zhang et al., 2008) v1.4. For further analysis, peaks were divided into promoters (<2 kb upstream and <1 kb downstream from TSSs) or enhancers (non-promoter). Read distributions were visualized on the Integrated Genomics Viewer (Robinson et al., 2011) v2.3 after conversion into signal files (bigWig) using DeepTools (Ramirez et al., 2016) v2.1.0. Signals across samples were quantile normalized with Haystack (Pinello et al., 2014), using 50-bp windows across the genome. For libraries with Drosophila chromatin spike-in, reads were aligned to the Drosophila genome (dm6). A normalization factor (NF) was derived for each library: NF = Drosophila reads in library with the lowest count / Drosophila reads in that library. Each library was then down-sampled to the read counts proportional to its NF.
RNA-seq reads were aligned to the mouse genome (Mm9, NCBI build 37) using STAR aligner (Dobin et al., 2013) v2.5.3a. Data quality measures, including per-base sequence quality, per-read GC content (~50%), comparable read alignments to +/− strands, exonic vs intronic read distributions and 3’ bias, were determined using RSeQC (Wang et al., 2012) v2.6.2. Gene specific read counts were determined using HTSeq (Anders et al., 2015) v0.6.1, followed by normalization in DESeq2 (Love et al., 2014). Normalized tag counts were converted into reads per kb of transcript length per 1M mapped reads (RPKM) and heatmaps representing this measure were generated using GENE-E software (Broad Institute). Differential gene expression between cell types was determined in DESeq2 using negative binomial GLM fitting and the Wald test to calculate statistical significance (P value). Benjamini-Hochberg correction was used to calculate adjusted P values (FDR); genes with FDR <0.05 and the indicated fold-changes were regarded as differentially expressed. RNA expression in developing tissues (Figure S7B) was determined using data from the ENCODE consortium (www.encodeproject.org). Genes expressed from E12.5 to E16.5 heart and lung and silenced before birth were determined by unsupervised k-means clustering of all differentially expressed genes among the developmental and postnatal stages.
Enhancer ATAC peaks from E11.5, E12.5, E14.5, E16.5, and adult intestinal epithelium were pooled to identify a total of 68,510 unique peaks. Unsupervised k-means clustering was conducted for signal within 1.5 kb from the centres of ATAC peaks, using DeepTools (Ramirez et al., 2016) v2.1.0. Gap statistics were used to determine the optimal number of clusters (Figure S3E) and clusters with visually similar signal patterns were merged to form 4 final groups (Figure 2A). Differential ChIP signals between cell types were determined with diffReps (Shen et al., 2013) v1.55.4 using comparison of read counts over 1-kb windows with step size of 100 bp across the genome; ChIP-seq signals for input DNA samples were used as the background. Negative binomial test was used to calculate P-values, and FDRs were calculated using Benjamini–Hochberg adjustment; differential regions with FDR <0.01 were selected for further processing.
Evolutionary conservation of enhancer groups (Figure 1E) was determined by comparison with all vertebrate genomes using Cistrome (Liu et al., 2011). PhastCons conservation scores were plotted as average profiles centered on enhancer summits. Gene enrichment analysis was conducted using GREAT analysis tool (McLean et al., 2010) v3.0 and default parameters to identify Gene Ontology (GO) terms for biological processes associated with genes within 50 kb of different enhancer groups. Significantly enriched terms were plotted using binomial P-values (Figures S2F and S4D). To identify TF motifs enriched in different groups of enhancers, we used the de novo motif-finding tool HOMER (Heinz et al., 2010) v4.7.2, based on cumulative binomial distributions.
Determination and analysis of unmethylated (UMR) and low-methylated (LMR) regions
WGBS reads were filtered to remove those of poor quality and the error-prone 6 bases from the 5’ end using Trimmomatic (Bolger et al., 2014) v0.32. The mouse genome (mm9, build NCBI37) was bisulfite-converted using Bismark (Krueger and Andrews, 2011) v0.13.1 and trimmed WGBS reads were aligned to this genome using Bowtie2 (Langmead and Salzberg, 2012) within Bismark. Duplicate reads were removed and the number of alignments with C (methylated) or T (unmethylated) were determined for each CpG dinucleotide using Bismark. Percent methylation for individual positions was calculated as the fraction of alignments with C relative to the total number of alignments (C or T). In further analysis of each CpG, counts from the two Cs on complementary strands were combined to determine strand-independent methylation.
UMRs and LMRs were identified using the MethylSeekR package (Burger et al., 2013) with the mouse bisulfite-converted genome (BSgenome.Mmusculus.UCSC.mm9) from Bioconductor (www.bioconductor.org) as the reference. All CpGs with coverage <5 were disregarded and C nucleotides overlapping known SNPs between the reference strains (C57BL/6J and 129/S5) were removed to eliminate spurious effects from polymorphism; methylation levels were smoothed over 3 consecutive CpG dinucleotides. Hypomethylated regions were identified as those with smoothed meCpG levels below various specified cut-offs and containing a minimum numbers of CpGs, to calculate the corresponding false discovery rates (FDRs) (Figure S2A). Regions were divided into UMRs (unmethylated and high number of CpGs) or LMRs (CpG-poor with fractional methylation between 10% and pre-defined upper limits). Average methylation was determined at UMR and LMRs from the intestinal epithelium, skin (He et al., 2014), blood (An et al., 2015), brain (Lister et al., 2013), and E6.5 epiblast (Seisenberger et al., 2012). Tissue specificity was calculated as z-scores by comparing the methylated fraction for each UMR or LMR in one tissue against the average of all other tissues.
Linking cis-regulatory regions and gene expression
To correlate groups of enhancers with gene expression, the Regulatory Potential Score (RPS) for individual genes was calculated from a distance-based metric linking all enhancers within 50 kb of that gene using BETA (Wang et al., 2013). For comparative analysis of gene regulation by different groups of enhancers (Figure S3E), z-scores were calculated using the mean RPS of the designated gene set and embryonic, fetal or adult enhancers. To analyze expression changes for genes linked to different groups of enhancers in PRC2-null intestinal cells (Figure 7A), genes were assigned to adult, embryonic or fetal enhancers based on the highest regulatory potential, as defined by BETA. Enriched expression of genes assigned to each group of enhancers in wild-type or Ee−/− cells was determined using the Gene Set Enrichment Analysis approach (Subramanian et al., 2005). Enrichment scores were determined in relation to 1,000 permutations of random gene sets of similar size.
Additional data analyses and display
Scatter plots with density and contour estimates of DNA methylation levels (Figures. 1B, S1D, S2E and S3G) and violin plots with gene expression, meCpG levels or H3K27ac signal estimates were generated in base R (version 3) or using ggplot2 package (Wickham, 2009) v2.2.1. Aggregate density profiles of ChIP-seq and meCpG read distributions (Figure 1B right, Figures S1D, S5C and S6D) were generated using the SitePro package in Cistrome (Liu et al., 2011). Venn diagrams were created using BioVenn (Hulsen et al., 2008). Heatmaps representing ATAC-seq and ChIP-seq data for the same mark (from the same antibody) were created using signals normalized across all samples. Average signal intensity was calculated over non-overlapping 50-bp bins over the regions indicated in each panel; scales alongside the heatmaps represent the relative signal range.
DATA AND SOFTWARE AVAILABILITY
Accession numbers for the data reported in Table S1 are GEO: GSE111024 and GSE115541. Information on individual files is included in Table S1.
Supplementary Material
Highlights.
Hypomethylated DNA preserves nearly complete catalogs of developmental enhancers
Adult H3K4me1+H3K27ac− enhancers are not poised, but remnants of fetal gene activity
TFs relieved of PRC2 repression selectively reactivate hypomethylated enhancers
Recommissioned enhancers drive tissue-specific fetal and embryonic gene activity
ACKNOWLEDGMENTS
Supported by NIH awards R01DK081113, R01DK082889 and U01DK103152 (R.A.S.), F32DK103453 and K01DK113067 (U.J.), and P50CA127003; DFCI-Novartis Drug Discovery Program; a fellowship from the Italian American Cancer Foundation (A.C.); and gifts from the Lind family. We thank S. Robine for Villin-CreER(T2) mice and M. Brown for valuable discussions. Data from this study (Table S1) are deposited in the Gene Expression Omnibus (accession numbers GSE111024 and GSE115541).
Footnotes
DECLARATION OF INTERESTS
UJ and RAS are co-applicants for a patent related in part to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- An J, Gonzalez-Avalos E, Chawla A, Jeong M, Lopez-Moyado IF, Li W, Goodell MA, Chavez L, Ko M, and Rao A (2015). Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun 6, 10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyachet C, Tovaglieri A, Xiang G, Lu J, Shah MS, Richmond CA, Verbeke C, Melton DA, Stanger BZ, Mooney D, et al. (2016). Reprogrammed Stomach Tissue as a Renewable Source of Functional beta Cells for Blood Glucose Regulation. Cell Stem Cell 18, 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee KK, Saxena M, Kumar N, Chen L, Cavazza A, Toke NH, O'Neill NK, Madha S, Jadhav U, Verzi MP, and Shivdasani RA (2018). Enhancer, transcriptional, and cell fate plasticity precedes intestinal determination during endoderm development. Genes Dev 32, 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, and Benvenisty N (2011). Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 9, 17–23. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, and Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Edwards JR, and Boulard M (2015). Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci USA 112, 6796–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Smits AH, de la Calle Mustienes E, Tena JJ, Ford E, Williams R, Senanayake U, Schultz MD, Hontelez S, van Kruijsbergen I, et al. (2016). Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet 48, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, and Furlong EE (2012). Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44, 148–156. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger L, Gaidatzis D, Schubeler D, and Stadler MB (2013). Identification of active regulatory regions from DNA methylation data. Nucleic Acids Res 41, e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Finkbeiner SR, Weinblatt D, Emmett MJ, Tameire F, Yousefi M, Yang C, Maehr R, Zhou Q, Shemer R, et al. (2014). De novo formation of insulin-producing "neo-beta cell islets" from intestinal crypts. Cell Rep 6, 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H (2013). The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, and Bird A (2011). CpG islands and the regulation of transcription. Genes Dev 25, 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, Still CD 2nd, Garcia BA, Adelman K, and Wysocka J (2017). Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Mol Cell 66, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, and Robine S (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193. [DOI] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, Fischle W, Bonaldi T, and Pasini D (2014). Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53, 49–62. [DOI] [PubMed] [Google Scholar]
- He X, Chatterjee R, Tillo D, Smith A, FitzGerald P, and Vinson C (2014). Nucleosomes are enriched at the boundaries of hypomethylated regions (HMRs) in mouse dermal fibroblasts and keratinocytes. Epigenetics Chromatin 7, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39, 311–318. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, and Pugh JE (1975). DNA modification mechanisms and gene activity during development. Science 187, 226–232. [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, and Ren B (2013). Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet 45, 1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, et al. (2014). 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 56, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, and Shivdasani RA (2005). Overlapping gene expression in fetal mouse intestine development and human colorectal cancer. Cancer Res 65, 8715–8722. [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, and Alkema W (2008). BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav U, Nalapareddy K, Saxena M, O'Neill NK, Pinello L, Yuan GC, Orkin SH, and Shivdasani RA (2016). Acquired Tissue-Specific Promoter Bivalency Is a Basis for PRC2 Necessity in Adult Cells. Cell 165, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakevych J, Sayols S, Messner B, Krienke C, and Soshnikova N (2017). Dynamic changes in chromatin states during specification and differentiation of adult intestinal stem cells. Nucleic Acids Res 45, 5770–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho AT, Zhao Q, Cai Z, Butte AJ, Kim JY, Pomeroy SL, Rowitch DH, and Kohane IS (2004). Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev 18, 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. (2010). Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, and Andrews SR (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Lowdon RF, Maricque B, Zhang B, Stevens M, Li D, Johnson SL, and Wang T (2015). Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat Commun 6, 6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, and Jaenisch R (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. (2013). Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. (2011). Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 12, R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, and Wysocka J (2016). Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 167, 1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K, et al. (2014). Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. (2007). DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L, Xu J, Orkin SH, and Yuan GC (2014). Analysis of chromatin-state plasticity identifies cell-type-specific regulators of H3K27me3 patterns. Proc Natl Acad Sci USA 111, E344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 28, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, and Wysocka J (2010). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, Maiorino L, Tiriac H, Young CM, Miyabayashi K, et al. (2017). Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170, 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Roman AK, Aronson BE, Krasinski SD, Shivdasani RA, and Verzi MP (2015). Transcription factors GATA4 and HNF4A control distinct aspects of intestinal homeostasis in conjunction with transcription factor CDX2. J Biol Chem 290, 1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M, San Roman AK, O'Neill NK, Sulahian R, Jadhav U, and Shivdasani RA (2017). Transcription factor-dependent 'anti-repressive' mammalian enhancers exclude H3K27me3 from extended genomic domains. Genes Dev 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D (2015). Function and information content of DNA methylation. Nature 517, 321–326. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, and Reik W (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48, 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schubeler D, and Kaestner KH (2014). DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev 28, 652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao NY, Liu X, Maze I, Feng J, and Nestler EJ (2013). diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One 8, e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Chen TY, and Melton DA (2009). Transcriptional dynamics of endodermal organ formation. Dev Dyn 238, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. (2011). DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, and Jacobsen SE (2011). 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 12, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, and Liu XS (2013). Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc 8, 2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser MM (1973). Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J Biol Chem 248, 2536–2541. [PubMed] [Google Scholar]
- Wickham H (2009). ggplot2: Elegant graphics for data analysis (New York: Springer-Verlag; ). [Google Scholar]
- Xie H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, and Orkin SH (2014). Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell stem cell 14, 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, and Graf T (2004). Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676. [DOI] [PubMed] [Google Scholar]
- Xu C, and Corces VG (2018). Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. Science 359, 1166–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, and Scacheri PC (2011). Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21, 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. (2013). Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers for the data reported in Table S1 are GEO: GSE111024 and GSE115541. Information on individual files is included in Table S1.