Abstract
Purpose:
We introduce a methodology to calculate the microdosimetric quantity dose-mean lineal energy for input into the microdosimetric kinetic model (MKM) to model the relative biological effectiveness (RBE) of proton irradiation experiments.
Methods and Materials:
The data from seven individual proton RBE experiments were included in this study. In each experiment, the RBE at several points along the Bragg curve was measured. Monte Carlo (MC) simulations to calculate the lineal energy probability density function of 172 different proton energies were carried out with use of Geant4 DNA. We calculated the fluence-weighted lineal energy probability density function (fw(y)), based on the proton energy spectra calculated through MC at each experimental depth, calculated for input into the MKM, and then computed the RBE. The radius of the domain (rd) was varied to reach the best agreement between the MKM-predicted RBE and experimental RBE. A generic RBE model as a function of dose averaged linear energy transfer (LETD) with one fitting parameter was presented and fit to the experimental RBE data as well, to facilitate a comparison to the MKM.
Results:
Both the MKM and LETD based models modeled the RBE from experiments well. Values for rd were similar to those of other cell lines under proton irradiation that were modeled with the MKM. Analysis of the performance of each model revealed neither model was clearly superior to the other.
Conclusions:
Our three key accomplishments include the following: (1) Developed a method that uses the proton energy spectra and lineal energy distributions of those protons to calculate dose-mean lineal energy. (2) Demonstrated that our application of the MKM provides theoretical validation of proton irradiation experiments that show that RBE is significantly greater than 1.1. (3) Showed that there is no clear evidence that the MKM is better than LETD based RBE models.
Summary
Using Monte Carlo simulations, the microdosimetric kinetic model (MKM) was applied to three previously published proton relative biological effectiveness (RBE) studies. The MKM fitting was compared to a generic RBE model as a function of dose averaged linear energy transfer (LETD). Analysis of the fittings reveal no clear advantage to using one model over the other and we conclude that both the MKM and LETD based proton RBE models are appropriate.
Introduction
With increased adoption of proton therapy in clinics around the world, researchers are increasingly focusing on the issue of relative biological effectiveness (RBE). In proton therapy, RBE is defined as the ratio of x-ray dose to proton dose needed to reach a specified biological end point, most often clonogenic cell survival. Based on early experiments, and summarized by Paganetti et al. [1], RBE in proton therapy was set to a constant value of 1.1 at all points along the beam and for all end points. Experiments, however, have shown a wide range of RBE values and demonstrate RBE’s dependency on factors such as dose, cell type, point in the cell cycle, biological end point, etc. An updated, comprehensive study of proton RBE performed by Paganetti [2], using the available experimental data from published literature, revealed that due to a lack of standardization across experiments and large uncertainties, RBE values on average are ~1.1 but increase at the end of the proton beam’s range. This increased RBE at the end of the range may have clinical consequences, as demonstrated by Peeler et al. and Underwood et al. [3, 4]. Thus, it is important to take into account variable RBE during the treatment planning process.
Several phenomenological RBE models have been developed [5-11], based on in vitro experiments, in order to model the observed variable RBE. Dose-averaged linear energy transfer (LETD) is often used as input for these models. However, as summarized by Mohan et al. [12], for a broad spectrum of proton energies, protons with higher LET should, in theory, contribute more to cell killing than do the lower LET protons in the spectrum for the same dose deposited if RBE is not linear with LET. At the Bragg peak and distal edge of a proton beam, the proton energy spectrum is indeed broad, meaning there is a correspondingly large spread in LET. This large variability in LET could make use of LETD and the reference radiation α and β values alone an inappropriate measure of biological effect. One purpose of this study was to compare an RBE model as a function of LETD to that of the MKM. This allows the comparison of RBE models with different underlying expressions for radiation quality: LETD and .
The microdosimetric kinetic model (MKM) proposed by Hawkins [13, 14], in contrast, uses lineal energy probability density function f(y) as an input. In this work, we present a method with which to calculate f(y) for a monoenergetic proton beam with use of Monte Carlo simulation. Then, we calculated a fluence-weighted lineal energy probability density function (fw(y)) for a polyenergetic proton beam, calculated the dose mean lineal energy and used this as input into the MKM to predict RBE. Finally, we varied a fitting parameter in the MKM to achieve a best-fit between the MKM and RBE experiments. Our work showed that the MKM can be applied to explain the increase in RBE as a function of in recent proton irradiation experiments. To compare the MKM fitting of RBE as a function of to an RBE model that is a function of LETD, we propose a generic RBE model with one fitting parameter. This parameter was varied to achieve the best fit to experimental data, and then the generic RBE model and the MKM fits were compared using Akaike Information Criterion (AIC) [15] to determine which of the two models best described the data.
Methods and Materials
RBE data
The RBE data and proton energy spectra corresponding to the RBE measurements were obtained from the three studies by Chaudhary and Marshall et al. [16], Guan and Bronk et al. [17], and Patel and Bronk et al. [18]. In all three studies, the authors measured RBE at various depths along a proton beam. In addition, they simulated their beam geometry with Geant4 [19], calculating LETD and the proton energy spectrum at each measurement point. Together, these three studies represent seven total experiments across three different beamlines and four cell lines. Pristine Bragg peaks were used for all three studies, and Chaudhary and Marshall et al. also included spread-out Bragg peak investigations in their study.
Lineal energy (y):
Lineal energy y is defined as the quotient of the energy deposited into a volume of interest ε and the mean chord length through that volume by a single energy deposition event [20], and is expressed as
| (1) |
In this work the volume of interest is called the domain and is defined as a sphere with radius rd. In microdosimetry, y is a stochastic quantity that follows a distribution, called the lineal energy probability density function (f(y)) [21]. The second moment of f(y) is the dose-mean lineal energy and is expressed as
| (2) |
Calculating y
Geant4 DNA version 10.2.0 was used to calculate the f(y) for 172 proton energies. The basis for the selection was to ensure adequate sampling of y values at all proton energies used in the RBE experiments. The Geant4 DNA extension [22, 23] was used to simulate 100 tracks at each energy in a liquid water cube geometry of 5-μm side lengths. 5-μm was chosen to match the voxel size that LETD was scored in. To calculate f(y) at a particular energy, a Matlab script was developed to score the lineal energy y at randomly generated locations around the proton track, inside spherical domains with a radius (rd) of 500 nm for each track. The scored y values were then binned into a histogram with a bin width of 0.1 keV/μm, with a range of [0.01, 300] keV/μm.
Calculating
For a proton of a given energy, the lineal energy follows a distribution rather than holding a single, fixed value. Therefore, to get an accurate account of the energy loss across a spectrum of proton energies, the lineal energy probability density function of a polyenergetic proton beam (fw(y)) has to take into account the various constituent f(y) values and their respective weights in the proton energy spectrum. We calculated fw(y) with use of a fluence-weighting technique. At each point along the Bragg curve where RBE was measured, the proton energy fluence (Φ(E)) was calculated via Monte Carlo simulation. Using the fluence at energy i and f(y)i, fw(y) was calculated as follows:
| (3) |
The calculation of for a polyenergetic beam is then straightforward
| (4) |
Uncertainty analysis of f(y)
We assumed that the number of scored y values in each bin follows a Poissonian distribution, and the fractional error per bin can be calculated as . Since calculating is essentially a sum over all scored y values, as shown in Eqs. 2 and 4, the total fractional error of each value is the quadrature sum of the fractional errors from each bin, divided by the sum of all y samples scored. The number of scores per track was increased until the maximum fractional uncertainty for any f(y) was approximately 1.5×10−4. Table S2 contains the number of scores per track that was performed.
Modeling RBE with the MKM
MKM builds upon the theory of dual radiation action, which was proposed by Kellerer and Rossi [24]. It uses the linear-quadratic (LQ) framework and predicts the α parameter as a function of . We calculated αp by using the method proposed by Kase et al. [25]:
| (5) |
where αx and βx are the linear and quadratic fitting parameters from the reference radiation survival curve, ρ is the mass density of water and rd is the radius of the domain. Eqs. 3 and 4 show how we calculated at each point along the Bragg curve in each experiment. Since RBE is defined as the ratio of doses of two radiation types to reach a specified biological endpoint – in this case, a given surviving fraction (SF) - we calculate RBE by solving for both doses from the LQ model. To calculate the experimental RBE, we use the LQ parameters from each clonogenic survival experiment from Refs. 16-18. The values used for αx and βx are given in S1 and the reader is referred to Refs. 16-18 for the LQ parameters for the proton experiments. The MKM assumes βp = βp, and αp is calculated from Eq. 5. Thus RBE can be calculated by solving the LQ model SF = e−α−βD2 for the corresponding dose in each irradiation modality, giving
| (6) |
Other works have derived similar expressions for RBE as a function of dose or survival fraction [7]. In this work, all RBE values were calculated for a survival fraction (SF) of 10%. To find the best rd for each experiment, rd was varied in Eq. 5 through the least squares method. We assume that is independent of rd for sufficiently small domains so that even changing the value of rd on the order of a 200-300 nanometers will not change the underlying microdosimetric distributions.
Comparing the MKM to a generic RBE model
In order to compare the MKM with a model based on LETD we propose an RBE model whose β is constant (βp = βx) and whose α parameter varies linearly with LETD:
| (7) |
This approach is similar to the one taken by Chaudhary and Marshall et al. [16]. This model and the MKM are only approximately correct because the LETD of an x-ray beam is nonzero, around 0.2 keV/μm. However as LETD increases, Eq. 5 and Eq. 7 become a better model of biological response. We applied this model to each RBE experiment and varied the k term to reach a best fit for the data, through the least squares method. The sum of squares from each model and experiment combination were input into the Akaike Information Criterion [15] to determine which model best describes the data.
Results
Calculating
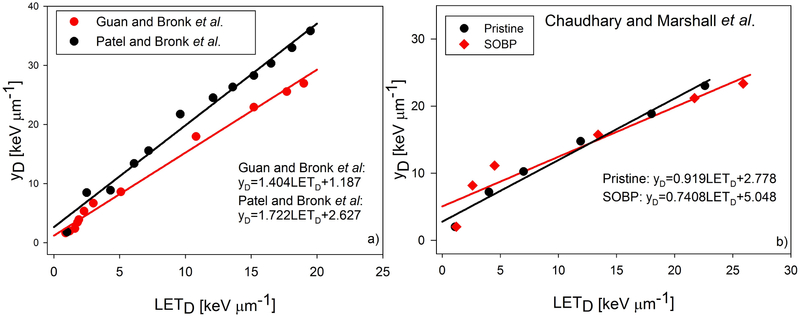
The proton energy spectrum at each point along the Bragg curve for each experiment was used to calculate both the LETD and , with being calculated according to Eq. 3 and 4, and the resulting relationship between the two quantities is noted in Fig. 2. For each experiment, appears to be an approximately linear function of LETD.
Fig. 2.
Plots of as a function of LETD for each of the three experiments. Our results indicate an approximately linear relationship between and LETD. The equation of as a linear function of LETD for each of the four beam configurations are shown in the panels.
Uncertainty analysis of f(y)
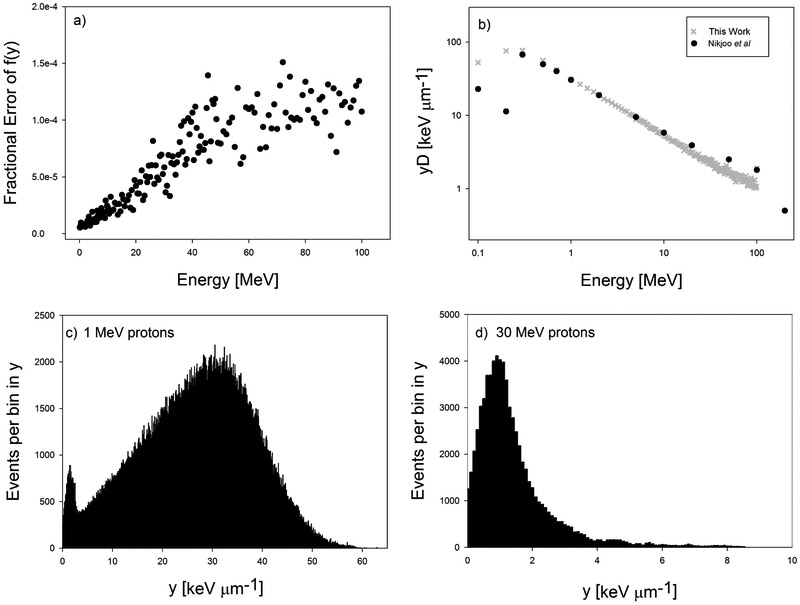
In Fig. 1a, the fractional errors for each f(y) scored is plotted. We input each of our scored f(y) functions into Eq. 2, calculated , and compared our to those reported by Nikjoo et al. [26] in Fig. 1b.
Fig. 1.
(a) Total fractional error from each f(y) scored using Geant4 DNA. (b) Comparison of our values of , using our scored probability density functions (f(y)), to those from Nikjoo et al [26]. (c, d) Histograms for the distribution of y values for 1 MeV and 30 MeV protons, respectively. These histograms were normalized and thus became the f(y) functions.
Modeling RBE with MKM
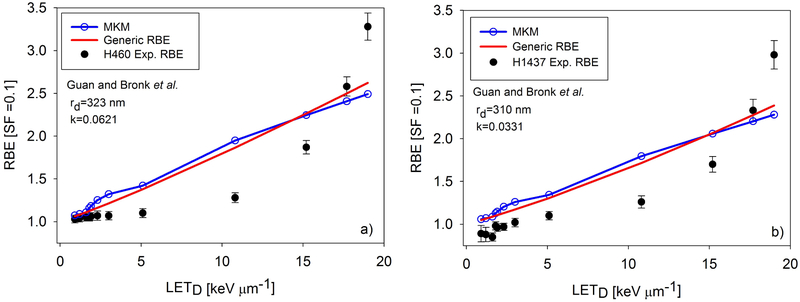
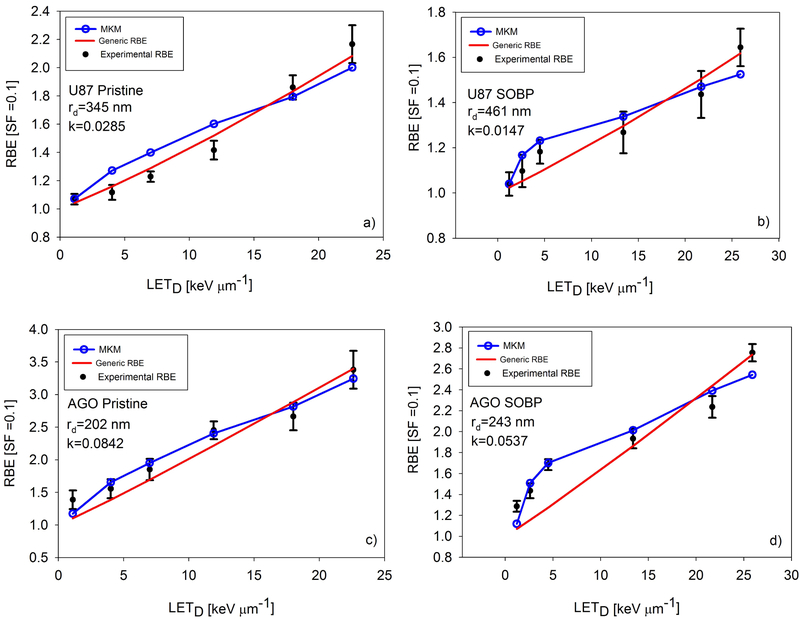
By varying rd, we were able to fit the experimental RBE data well by using the MKM, as shown in Figs. 3-5. Values for the best-fit rd are shown on the plots and in Table 1. Note that in Figs. 3-5, the RBE modeled by the MKM is plotted as a function of LETD, even though the MKM is a function of . This was done to be consistent with past practice and to enable plotting both the MKM and generic RBE model on one plot. While Fig. 2 shows the linear fit for as a function of LETD for each beamline, values were not directly transformed via these equations. Rather, the points were plotted directly against the calculated LETD values.
Fig. 3.
Results of fitting the generic RBE model and the MKM, with fitting parameters. Panel a) contains the experimental data and fits for the H460 data from Guan and Bronk et al, while panel b) contains the data and fits for the H1437 data. The experimental data was performed at MDACC.
Fig. 5.
The results of fitting the generic RBE model and the MKM for the data from Chaudhary and Marshall et al. Panels a) and c) show the pristine Bragg peak irradiations for cell lines U87 and AGO1522, respectively. Panels b) and d) show the spread out Bragg peak irradiations experiments for the U87 and AGO1522 cell lines, respectively. In the low LETD and low region, MKM was better able to model RBE than the generic RBE model. Experiments performed at CATANA, Italy.
Table 1.
Results of AIC analysis between the MKM and generic RBE model. rd: radius of the domain; CI: confidence interval; SS: sum of squares; RL: relative likelihood.
| Guan and Bronk etal | |
|---|---|
| H460 | |
| MKM | Generic |
| rd=323 nm | k=0.0621 |
| CI=[286.7-379.3] | CI=[0.04798-0.07578] |
| SS1=1.473 | SS2=1.061 |
| RL = 0.140 | |
| H1437 | |
| MKM | Generic |
| rd=310 nm | k=0.0330 |
| CI=[274.8-368.8] | CI=[0.02511-0.04073] |
| SS1=1.271 | SS2=0.9396 |
| RL = 0.1632 | |
| Patel and Bronk et al | |
| H460 | |
| MKM | Generic |
| rd=426 nm | k=0.045 |
| CI=[397.5-461.9] | CI=[0.03987-0.05014] |
| SS1=0.3606 | SS2=0.2169 |
| RL = 0.0474 | |
| Chaudhary and Marshall et al | |
| Pristine Bragg Peak, AGO1522 | |
| MKM | Generic |
| rd=202 nm | k=0.08423 |
| CI=[192.4-213.9] | CI=[0.07092-0.09746] |
| SS2=0.1096 | SS1=0.2404 |
| RL = 0.0948 | |
| Pristine Bragg Peak, U87 | |
| MKM | Generic |
| rd =345 nm | k=0.0285 |
| CI=[311.3-394.7] | CI=[0.02554-0.0313] |
| SS1=0.1189 | SS2=0.02386 |
| RL = 0.0081 | |
| SOBP, AGO1522 | |
| MKM | Generic |
| rd=243 nm | k=0.0537 |
| CI=[227.2-262.8] | CI=[0.03957-0.06765] |
| SS2=0.1086 | SS1=0.3402 |
| RL = 0.0325 | |
| SOBP, U87 | |
| MKM | Generic |
| rd = 461 nm | k=0.0147 |
| CI=[420.0-518.0] | CI=[0.01318-0.0194] |
| SS2=0.02759 | SS1=0.01614 |
| RL = 0.2002 | |
Comparing the MKM to a generic RBE model
Table 1 contains the values of each of the k parameters for the generic RBE model given in Eq. 7. The output of the AIC calculation is the relative likelihood that a given model is a better fit than another model and are given in Table 1. In two experiments, the MKM was found to fit the data better than the generic RBE model, while five experiments were better modeled by the generic RBE model. However, in the majority of the experiments the AIC analysis also suggests that there is little evidence to choose between the models as the differences in AICs were typically small.
Discussion
In the present study we calculated at various depths along the Bragg curve, based on the proton energy spectra at that depth. By inputting at various depths along the Bragg curve into the MKM and varying rd we were able to find a best-fit rd that modeled the experimental RBE data well. We then compared this fit of the MKM to a generic RBE model with one fitting parameter to determine if RBE was better modeled by the MKM or a generic RBE model that includes LETD as a measure of radiation quality. AIC analysis indicated that for two experiments, MKM was the superior model and for five experiments, the generic RBE model was better. However in most experiments there was not strong evidence favoring either model.
The values published by Nikjoo et al. [26] provide a useful reference for our values, since both our method and Nikjoo et al.’s published data are modeled with use of a domain radius of 500 nm. When comparing our values to those of Nikjoo et al. in Fig. 1b), we see excellent agreement except the lowest two proton energies, 0.1 and 0.2 MeV. However these energies contribute so little dose in a clinical proton beam spectrum that they do not have a significant impact on our microdosimetric calculations. Our calculations also agree well with those reported by Lindborg et al [27]. The low fractional error and agreement with the values published by Nikjoo et al. and Lindborg et al. give us confidence in the accuracy of the values of our calculations for . Figure 2 shows that and LETD increase in a linear relationship. This relationship has been derived by Kellerer and is where δ2 is the weighted energy loss per collision and d is the diameter of the domain [28].Confidence intervals for the linear relationships between and LETD plotted in Fig. 2 are reported in the Supplementary Data. Recently, a series of microdosimetric measurements were conducted by Anderson et al. [29] where they measured at various depths along a 71.3 MeV and 159.9 MeV proton beam and compared those measurements to calculations of LETD. They found a similar, linear relationship between LETD and , and our values of at similar LETD are similar to theirs. It is important to highlight the differences between LETD and . LETD is calculated by sampling the stopping power, known as the unrestricted LET, of each proton that passes through the scoring volume and averaging the LET values by weighing them according to how much energy was deposited by those protons in the scoring volume. is a measure of the energy deposited into a volume, called a domain. The energy deposited into the domain can come from the primary proton and delta rays. When calculating , we include instances when the proton passes near the domain, but not through it, and delta rays deposit energy into the domain.
In this study, rd was varied for each experiment to achieve a best fit between the MKM and experiments. The values for rd for these experiments range between 0.202 μm and 0.461 μm. Work by Kase et al. [30] showed that when fitting the MKM to other proton RBE data, rd values were 0.26 μm, 0.34 μm, and 0.35 μm for V79, HSG, and T1 cell lines, respectively. Mairani et al. [31] conducted a similar fitting utilizing proton and helium ion irradiation data and found their best fitting rd value was 0.300 μm . We concluded that our rd values were similar to previous studies that applied the MKM to proton RBE experiments. Patel and Bronk and Guan and Bronk used the same cell line, H460, in their experiments. We were able to fit both of them by using the MKM. However, the value of rd to get a best fit was different for each experiment. Based on Guan et al.’s data, rd was 0.323 μm and for Patel et al.’s data, rd was 0.426 μm. The Guan and Bronk et al. data show RBE as a much more nonlinear function of LETD than the similar studies performed by Patel and Bronk et al. and Chaudhary and Marshall et al., and it remains unclear why. Both the generic RBE model and the MKM fit this data set poorly when compared to the fits shown in Figs. 4 and 5. Three RBE models were applied to the Guan and Bronk et al. data by McNamara et al. [9], and Mohan et al. [12] and their models were unable to account for the highest experimental RBE, though their models were better able to fit the RBE at 15 keV/μm and below. The study in Ref. 18 highlights the importance and sensitivity of the settings used in Monte Carlo simulations, particularly when the LETD and dose values from simulations are used for calculating RBE. We suggest that the difference in Monte Carlo settings in the different studies is the reason for the large difference in rd values for the same cell line in two different experiments, and why both the MKM and generic RBE model fit the Patel and Bronk et al. data, and Chaudhary and Marshall et al. data better. In the high gradient region distal to the Bragg peak, there may be significant uncertainties in delivered dose, which may be quite different among the different experiments. To address these issues, we are further investigating the relationship between RBE, and LETD by performing cell irradiation experiments using ramp fields and SOBPs with different dose and LETD combinations [32, 33]. What is also striking about the experimental data in Figs. 3 and 4 is the RBE at LETD > ~15 keV/μm seems to be increasing nonlinearly. This is in contrast to heavy ion studies that show RBE as a linear function of LETD up to around 100 keV/ μm [34]. This nonlinearity at high proton LETD may also point to modifications of the MKM and other RBE models, such as including a variable β term or higher-order terms in the relationship between LET and α.
Fig. 4.
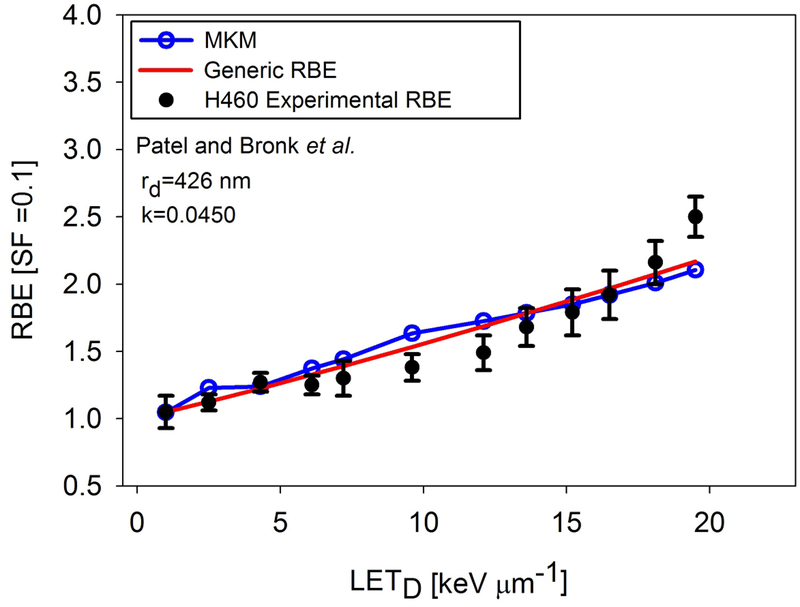
Results of fitting the generic RBE model and the MKM to the Patel and Bronk et al. data. Experimental data was performed at HIT, Germany.
The results of fitting the MKM and the generic RBE model to the experimental data are shown in Figs. 3-5. Overall, both models gave very similar fits to the data. Particularly in the U87 and AGO1522 pristine Bragg peak experiments in the Chaudhary and Marshall et al. data, and the Patel and Bronk et al. data, RBE appears to be a linear function of LETD. For the SOBP experiments from Chaudhary and Marshall et al., the MKM better models the RBE at the lowest three LETD points. The difference in the two models can be significant, as seen in Fig. 5b) and 5d). Others have suggested that LETD may not be the best surrogate for biological response [12]. The superior fit of the MKM shown in Fig. 5b) and 5d) support the hypothesis that better encapsulates biological effect than LETD, particularly in an SOBP, supporting the soundness of its underlying assumptions. However, more studies similar to the one presented here are needed to fully determine the relative merits of MKM or LETD based models in a proton SOBP. To determine which RBE model fits the data best, the AIC analysis in Table 1 was used. Overall, the generic RBE model was preferred in five experiments and the MKM in two. A minor caveat is the relatively weak preference for the generic RBE model in the Guan and Bronk et al. data, where the relative likelihood of MKM being the better model was 0.140 and 0.163 for the H460 and H1437 experiments, respectively.
Based on the AIC analysis between the generic RBE model and the MKM, there is no clear evidence that the MKM models RBE better than a generic LETD based model. We have demonstrated that the MKM can be used to model proton RBE experiments whose RBE values are much higher than the clinically accepted RBE=1.1 model, however we have not demonstrated a clear superiority by using the MKM. Thus we conclude that there is not enough evidence to move away from LETD based RBE models. There may be some end of range proton RBE nonlinearity but at present we conclude that both the MKM and LETD based models provide an accurate model of RBE, even if the underlying RBE model is more complex.
An advantage of this MKM fitting procedure is that its use is not restricted to protons. Any radiation type, such as helium or carbon, whose fw(y) can be calculated, can apply this method to modeling RBE. A modified version of the MKM was incorporated into a treatment planning system for carbon ions by Inaniwa et al. [35] and our group is developing this MKM algorithm to be implemented in a treatment planning system for proton therapy. Future studies will examine the differences in treatment plans optimized by using the traditional RBE=1.1 paradigm and those optimizing on biological effect by using the MKM.
Conclusions
Using data from three independent proton RBE studies, we have presented a method to calculate the dose mean lineal energy of a polyenergetic proton beam and apply the microdosimetric kinetic model (MKM) to the RBE experiments. To our knowledge, this fluence-based method of calculating the probability density function fw(y) based on the proton energy distribution and f(y) for monoenergetic proton beams is unique. This study provides a theoretical basis for modeling the increase in RBE along the distal edge of a proton beam. We also applied a generic LETD based RBE model to the same data and compared the MKM and generic RBE model fittings with AIC analysis. Based on this analysis there is no clear evidence that the MKM, using radiation quality represented by , is superior to a generic model using LETD as a measure of the radiation quality. Given the wide ranges of LET values in a spread out Bragg peak and the concern about the quantity LETD, measuring f(y) distributions and with a tissue equivalent proportional counter may be the preferred way to characterize proton beam quality in the clinic, at least as a calibration of Monte Carlo simulation settings.
Supplementary Material
Acknowledgements:
The authors thank Oleg Vassiliev for his contributions to this work and for Tamara Locke in the Scientific Publications Department at MD Anderson for editing the manuscript. This study was supported in part by the NIH/NCI grant 2U19CA021239.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None.
References
- 1.Paganetti H, Niemierko A, Ancukiewicz M, et al. , Relative biological effectiveness (RBE) values for proton beam therapy. Int J Radiat Oncol Biol Phys, 2002. 53(2): p. 407–421. [DOI] [PubMed] [Google Scholar]
- 2.Paganetti H, Relative biological effectiveness (RBE) values for proton beam therapy. Variations as a function of biological endpoint, dose, and linear energy transfer. Phys Med Biol, 2014. 419(59: p. R419–R472. [DOI] [PubMed] [Google Scholar]
- 3.Peeler CR, Mirkovic D, Titt U, et al. , Clinical evidence of variable proton biological effectiveness in pediatric patients treated for ependymoma. Radiother Oncol, 2016. 121(3: p. 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Underwood T, Grassberger C, Bass R, et al. , OC-0245: Clinical evidence that end-of-range proton RBE exceeds 1.1: lung density changes following chest RT. Radiother Oncol, 2017. 123: p. S123–S124. [Google Scholar]
- 5.Abolfath R, Peeler CR, Newpower M, et al. , A model for relative biological effectiveness of therapeutic proton beams based on a global fit of cell survival data. Sci Rep, 2017. 7(1: p. 8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carabe-Fernandez A, Dale RG, and Jones B, The incorporation of the concept of minimum RBE (RBEmin) into the linear-quadratic model and the potential for improved radiobiological analysis of high-LET treatments. Int J of Radiat Biol, 2007. 83(1: p. 27–39. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y and Ahmad S, Empirical model estimation of relative biological effectiveness for proton beam therapy. Radiat Prot Dosimetry, 2012. 149(2: p. 116–23. [DOI] [PubMed] [Google Scholar]
- 8.Jones B, Towards Achieving the Full Clinical Potential of Proton Therapy by Inclusion of LET and RBE Models. Cancers, 2015. 7(1: p. 460–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNamara AL, Schuemann J, and Paganetti H, A phenomenological relative biological effectiveness (RBE) model for proton therapy based on all published in vitro cell survival data. Phys Med Biol, 2015. 60(21: p. 8399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rorvik E, Thornqvist S, Stokkevag CH, et al. , A phenomenological biological dose model for proton therapy based on linear energy transfer spectra. Med Phys, 2017. 44(6: p. 2586–2594. [DOI] [PubMed] [Google Scholar]
- 11.Wedenberg M, Lind BK, and Hardemark B, A model for the relative biological effectiveness of protons: the tissue specific parameter alpha/beta of photons is a predictor for the sensitivity to LET changes. Acta Oncol, 2013. 52(3: p. 580–8. [DOI] [PubMed] [Google Scholar]
- 12.Mohan R, Peeler CR, Guan F, et al. , Radiobiological issues in proton therapy. Acta Oncol., 2017: p. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins RB, A microdosimetric-kinetic theory of the dependence of the RBE for cell death on LET. Med Phys, 1998. 25(7 Pt 1: p. 1157–70. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins RB, A Microdosimetric-Kinetic Model for the Effect of Non-Poisson Distribution of Lethal Lesions on the Variation of RBE with LET. Journal of radiation research, 2003. 160(1: p. 61–69. [DOI] [PubMed] [Google Scholar]
- 15.Akaike H, A new look at the statistical model identification. IEEE Trans Automat Contr, 1974. 19(6): p. 716–723. [Google Scholar]
- 16.Chaudhary P, Marshall TI, Perozziello FM, et al. , Relative biological effectiveness variation along monoenergetic and modulated Bragg peaks of a 62-MeV therapeutic proton beam: a preclinical assessment. Int J Radiat Oncol Biol Phys, 2014. 90(1: p. 27–35. [DOI] [PubMed] [Google Scholar]
- 17.Guan F, Bronk L, Titt U, et al. , Spatial mapping of the biologic effectiveness of scanned particle beams: towards biologically optimized particle therapy. Sci Rep, 2015. 5: p. 9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel D, Bronk L, Guan F, et al. , Optimization of Monte Carlo particle transport parameters and validation of a novel high throughput experimental setup to measure the biological effects of particle beams. Med Phys., 2017. 44(11: p. 6061–6073. [DOI] [PubMed] [Google Scholar]
- 19.Agostinelli S, Allison J, Amako K, et al. , Geant4—a simulation toolkit. Nucl Instrum Methods Phys Res A, 2003. 506(3): p. 250–303. [Google Scholar]
- 20.ICRU, ICRU Report 51: Quantities and Units in Radiation Protection Dosimetry. 1993. [Google Scholar]
- 21.ICRU, ICRU Report 36: Microdosimetry. 1983. [Google Scholar]
- 22.Bernal MA, Bordage MC, Brown JMC, et al. , Track structure modeling in liquid water: A review of the Geant4-DNA very low energy extension of the Geant4 Monte Carlo simulation toolkit. Physica Medica, 2015. 31(8: p. 861–874. [DOI] [PubMed] [Google Scholar]
- 23.Incerti S, Ivanchenko A, Karamitros M, et al. , Comparison of GEANT4 very low energy cross section models with experimental data in water. Med Phys, 2010. 37(9): p. 4692–4708. [DOI] [PubMed] [Google Scholar]
- 24.Kellerer AM and Rossi HH, The theory of dual radiation action. Curr. Top. Radiat. Res. Q, 1972. 8: p. 85–158. [Google Scholar]
- 25.Kase Y, Kanai T, Matsumoto Y, et al. , Microdosimetric measurements and estimation of human cell survival for heavy-ion beams. Radiat Res, 2006. 166(4: p. 629–38. [DOI] [PubMed] [Google Scholar]
- 26.Nikjoo H, Emfietzoglou D, Liamsuwan T, et al. , Radiation track, DNA damage and response-a review. Rep Prog Phys, 2016. 79(11): p. 116601. [DOI] [PubMed] [Google Scholar]
- 27.Lindborg L, Hultqvist M, Å Carlsson T, et al. , Lineal energy and radiation quality in radiation therapy: model calculations and comparison with experiment. Physics in Medicine & Biology, 2013. 58(10): p. 3089. [DOI] [PubMed] [Google Scholar]
- 28.Kellerer AM Fundamentals of Microdosimetry The Dosimetry of Ionizing Radiation, ed. Kase KR, Bjarngard BE, and Attix FH. Vol. 1 1985, Academic Press: New York. [Google Scholar]
- 29.Anderson SE, Furutani KM, Tran LT, et al. , Microdosimetric measurements of a clinical proton beam with micrometer-sized solid-state detector. Med Phys, 2017. 44(11: p. 6029–6037. [DOI] [PubMed] [Google Scholar]
- 30.Kase Y, Kanai T, Matsufuji N, et al. , Biophysical calculation of cell survival probabilities using amorphous track structure models for heavy-ion irradiation. Phys Med Biol, 2008. 53(1: p. 37–59. [DOI] [PubMed] [Google Scholar]
- 31.Mairani A, Magro G, Tessonnier T, et al. , Optimizing the modified microdosimetric kinetic model input parameters for proton and (4)He ion beam therapy application. Phys Med Biol, 2017. 62(11: p. N244–N256. [DOI] [PubMed] [Google Scholar]
- 32.Geng C, Gates D, Bronk L, et al. Physical parameter optimization scheme for radiobiological studies of charged particle therapy Physica Medica, 51 (2018), pp. 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan F, Geng C, Ma D, et al. RBE Model-Based Biological Dose Optimization for Proton Radiobiology Studies Int J Part Ther Studies, 5 (1) (2018), pp. 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sørensen BS, Overgaard J, and Bassler N, In vitro RBE-LET dependence for multiple particle types. Acta Oncol, 2011. 50(6): p. 757–762. [DOI] [PubMed] [Google Scholar]
- 35.Inaniwa T, Furukawa T, Kase Y, et al. , Treatment planning for a scanned carbon beam with a modified microdosimetric kinetic model. Phys Med Biol, 2010. 55(22: p. 6721–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.