Abstract
Scope:
Small selenium (Se) species play a key role in Se metabolism and act as dietary sources of the essential trace element. However, they are redox active and trigger pro- and antioxidant responses. As health outcomes are strongly species-dependent, species-specific characteristics of Se compounds were tested in vivo.
Methods and results:
In the model organism Caenorhabditis elegans (C. elegans), immediate and sustained effects of selenite, selenomethionine (SeMet) and Se-methylselenocysteine (MeSeCys) were studied regarding their bioavailability, incorporation into proteins, as well as modulation of the cellular redox status. While all tested Se compounds were bioavailable, only SeMet persistently accumulated and was non-specifically incorporated into proteins. However, the protection towards chemically-induced formation of reactive species was independent of the applied Se compound. Increased thioredoxin reductase (TXNRD) activity and changes in mRNA expression levels of antioxidant proteins indicated the activation of cellular defense mechanisms. However, in txnrd-1 deletion mutants, no protective effects of the Se species were observed anymore, which was also reflected by differential gene expression data.
Conclusion:
Se species protect against chemically-induced reactive species formation. The identified immediate and sustained systemic effects of Se species give rise to speculations on possible benefits facing subsequent periods of inadequate Se intake.
Keywords: Antioxidant defense system, Caenorhabditis elegans, Selenium, Oxidative stress, selenoproteins
Graphical Abstract
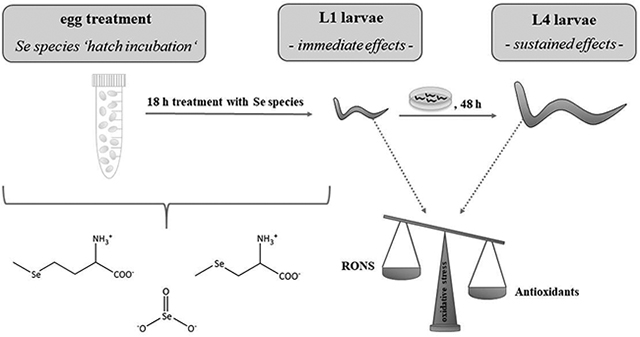
Schematic illustration showing the experimental procedure of an egg treatment (‘hatch incubation’) with different Se species. Immediate effects observed subsequently to treatment in L1 larvae as well as sustained effects in worms further grown to L4 larvae without additional Se species treatment indicated the enhancement of antioxidant defense systems in C. elegans.
1. Introduction
The essential trace element selenium (Se) is implicated in multiple physiological processes fundamentally important to human health, including thyroid and immune function, reproduction and cellular homeostasis.[1, 2] Although Se supplementation has been associated with various health benefits, evidence from epidemiological and in vivo studies is scarce and uncertainty persists in the light of partly contradictory data.[3, 4] A complex interplay of factors defines the biological activity of Se, resulting in either beneficial or adverse health effects, which include baseline status, dosage, speciation and metabolism.[1, 4] Thereby, the either beneficial or detrimental health effects of Se are in a narrow concentration range.[3, 5] In humans, selenoproteins encoded by 25 genes are thought to be responsible for most of the physiological functions of Se.[6] These selenoproteins comprise different enzyme families such as glutathione peroxidases (GPX), iodothyronine deiodinases (DIO) and thioredoxin reductases (TXNRD), with vital functions in regulating thyroid hormone activity (DIO), reduction of hydroperoxides, cellular signaling and redox homeostasis (GPX, TXNRD).[7] Low molecular weight Se species are important food sources that contribute to the Se pool and thereby fuel selenoprotein synthesis. Among the most common dietary Se species are the inorganic form selenite and the seleno-amino acids selenomethionine (SeMet) and Se-methylselenocysteine (MeSeCys), which occur in plant- and animal-based foods or are used in food supplements.[4, 8] Most Se in food is available as selenomethionine (SeMet). Besides being substrates for selenoprotein synthesis, small Se species per se are redox active due to their unique biological activity resulting in the formation of redox active metabolites, such as hydrogen selenide and methylselenol, provoking at the same time pro- and antioxidant mechanisms.[4] Although the significance of the applied Se species has been recognized as a key factor in health outcomes (reviewed in [4]), only few studies have addressed species-specific effects in vivo (e.g.[9–12]). Therefore, the current study aimed for a comparative analysis of three different Se species including selenite, SeMet and MeSeCys in the nematode Caenorhabditis elegans (C. elegans), examining their respective bioavailability, incorporation into proteins, protective role against oxidative stress and impact on the antioxidative system, namely, TXNRD activity as well as the Nrf2/Skn-1 and FoxO/Daf-16 pathways.
The alternative and complementary in vivo model C. elegans is commonly used in mechanistic studies in toxicological, biomedical and aging research, due to its rapid life cycle, genetic manipulability and high degree of evolutionary conserved genes and pathways.[13] Many core metabolic pathways are conserved between humans and C. elegans, including the insulin/IGF-1, AMPK and TOR pathways, as well as transcription factors and nuclear hormone receptors involved in lipid and energy homeostasis.[14] In addition to being a well-established model for neurodegenerative diseases, C. elegans applications to study metabolic disorders, such as diabetes or obesity are available as well (summarized in [15,16]), promoting the implementation of the nematode in nutritional studies. In recent years, a growing body of research on dietary restriction, food choice, nutrient intake and gene-diet interactions has been conducted in C. elegans, demonstrating among other things how diet can have long-lasting effects on animal physiology and even influence future generations (reviewed in [14]). In this context, the current study aimed to address the question if supplementation with Se species in an early life stage has long-term protective effects after supplementation is ceased. The conserved antioxidant signaling pathways and homeostatic regulations of trace element transport processes make C. elegans an invaluable model for studies on the effects of Se in the context of oxidative stress, and have motivated previous work in this field.[17–20] Despite the genetic similarities to higher organisms, the C. elegans genome encodes only one selenoprotein, TXNRD-1.[21, 22] The lack of a more comprehensive selenoproteome clearly entails obvious caveats, but also offers the advantage of obtaining mechanistic indications about this single selenoprotein in the context of a whole, but less complex organism. Although orthologous to the human gene, the function of the nematode TXNRD-1 is poorly understood. Stenvall et al. observed no obvious impairments of growth, reproduction or motility in txnrd-1 deletion mutant worms,[23] which is in contrast to mammals, as systemic TXNRD-1 ablation in mice is embryonically lethal.[24] To better characterize the role of small Se species in C. elegans, especially in the background of oxidative stress, wildtype and txnrd-1 deletion mutant worms were compared.
2. Experimental Section
2.1. C. elegans maintenance and exposure to Se species (‘hatch incubation’)
C. elegans wildtype (N2, Caenorhabditis Genetics Center, Minneapolis, USA) and txnrd-1 deletion mutant worms (VB1414, sv47) were cultivated at 20°C on 8P agar plates seeded with Escherichia coli NA22 as previously described.[25] For each experiment, eggs were isolated from gravid adults using a bleaching solution (1% NaOCl, 0.25 M NaOH) and separated from debris by sucrose floatation. Eggs were hatched overnight in M9 buffer only or M9 buffer containing 100 μM selenite, D/L-SeMet or L-MeSeCys. Stock solutions of selenite (Na2SeO3 ∙ 5 H2O, ≥99%), D/L-SeMet (≥99%, both from Sigma-Aldrich, Steinheim, Germany) and L-MeSeCys (>98%, Abcam, Cambridge, UK) were prepared and diluted in purified water (10 MΩcm, Elix® 15, Merck Millipore) and stored at 4 °C for up to one week (selenite) or two months (SeMet, MeSeCys). After 18 h, hatched L1 larvae were washed four times in M9 buffer and subjected to endpoint analyses. Alternatively, worms were grown to L4 stage on NGM plates seeded with Escherichia coli OP50 without further Se species treatment and utilized for endpoint analyses.
2.2. Dosage information / Dosage regimen
C. elegans were exposed to single doses of 100 μM selenite, D/L-SeMet or L-MeSeCys for 18 h via ‘hatch incubation’ as described in section 2.1. The applied Se species are taken up mainly orally by the hatched larvae, although minor parts may also be taken up via the worms’ cuticles. 100 μM were chosen as a supraphysiological dose for mechanistic studies, resulting in measureable Se concentrations in the worms. The hatch incubation did not cause any toxic effects, as worms hatched normally with hatching rates indistinguishable from untreated control worms.
2.3. Total Se quantification
Total Se quantification was carried out with an ICP-QQQ-MS 8800 mass spectrometer (Agilent, Waldbronn, Germany), applying an isotope dilution method by Marschall et al.[26] 60,000 – 90,000 L1 larvae following ‘hatch incubation’ with the respective Se species were subjected to Se quantification as well as 14,000 L4 stage worms. To each worm pellet purified water was added to a volume of 0.5 mL, followed by sonication and centrifugation. An aliquot of the supernatant was used to determine the protein concentration via the bicinchoninic acid (BCA) assay (Sigma-Aldrich, Steinheim, Germany). The remaining worm suspension was transferred into a 20 mL TFA microwave vessel with 1.5 mL of a digestion mixture containing 30% HNO3 (Merck, Darmstadt, Germany) and 5 μg 77Se per L (Eurisotop SAS, Saarbrücken, Germany). In a closed microwave digestion system (Mars 6, CEM, Kamp-Lintfort, Germany), digestion was carried out applying 650 W to reach 200°C within 15 min and keeping this temperature for additional 20 min. After cooling, samples were filled up to 2.5 mL with water, leading to final concentrations of 20% HNO3 and 3 μg 77Se per L. 3% isopropanol (≥99.999%, Sigma-Aldrich, Steinheim, Germany) were added to each sample before analysis to increase sensitivity, using the carbon enhancement effect.[27] For Se quantification via isotope dilution ICP-QQQ-MS, the instrument was operated with hydrogen (1 mL min−1) and oxygen (0.4 mL min−1) as reaction gases in the collision and reaction cell. The Se isotopes 80Se, 78Se as well as 77Se as isotopic spike were detected on mass and in mass shift mode as oxygen reaction products (+m/z 16). Data analysis was performed by calculating isotope ratios and applying the isotope dilution equation as extensively described elsewhere.[26] The mass transitions m/z were used for quantification, with an LOQ of 0.032 μg Se per L.[26]
2.4. Total Se quantification in worm fractions following TCA precipitation
To determine the amount of Se associated with proteins, fractionation experiments were performed using trichloroacetic acid (TCA). Therefore, 1 mL of ice-cold 20% (w/v) TCA (Roth, Karlsruhe, Germany) was added to the worm pellets, followed by freezing in liquid nitrogen and thawing three times. After sonication and centrifugation, the protein fraction (residue) and the protein-free fraction (supernatant) were obtained. Supernatants were diluted with purified water to a concentration of 10% TCA, and Se quantification was performed by external calibration. The residues were washed with 70% ice-cold EtOH and dried at 37°C under nitrogen flow. Dry residues were transferred into a 20 mL TFA microwave vessel, digested according to the abovementioned protocol and subjected to ICP-QQQ-MS analysis using the isotope dilution method published by Marschall et al.[26]
2.5. Thioredoxin reductase (TXNRD) activity assay
TXNRD activity was estimated by the reduction of 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) to 2-nitro-5-thiobenzoic acid (TNB) by NADPH.[28] Worms were exposed to 100 μM selenite, SeMet or MeSeCys during hatching as described above, grown to L4 stage and washed off the plates using M9 + 0.01% Tween. 10,000 worms were pelletized in 0.1 mL M9 buffer. 30 μL homogenization buffer containing 100 mM Tris, 300 mM KCl and 0.1% Triton X-100 (pH 7.6) and protease inhibitor cocktail (Sigma-Aldrich, Steinheim, Germany) were added followed by three cycles of freezing in liquid nitrogen and thawing. After sonication and centrifugation, an aliquot of the supernatant was taken for protein quantification as described above. 2.5 μL of the worm lysate were mixed with 207.5 μL reaction buffer (100 mM KPO4, 2 mM EDTA, pH 7.4) and 15 μL DTNB (≥ 98%, Sigma-Aldrich, Steinheim, Germany, 50 mM in DMSO) in a 96-well plate. The reaction was started by adding 25 μL of 2 mM NADPH (Sigma-Aldrich, Steinheim, Germany). TNB production was measured in a plate reader (Tecan Infinite M200 Pro) at 412 nm at 25°C after 240 min. TXNRD-independent TNB formation, determined without the addition of NADPH, was subtracted, and data were expressed as mU/mg protein. One unit is defined as the consumption of 1 μmol NADPH, i.e. production of 2 μmol of TNB (extinction coefficient 13.6 mM−1 cm−1 per min).
2.6. Carboxy-DCFH-DA assay
Generation of reactive oxygen and nitrogen species (RONS) was measured in whole worms with a 5(&6)-carboxy-2’,7’-dichloro-dihydrofluorescein-diacetate (Carboxy-DCFH-DA)-based plate reader assay, as previously published with slight modifications.[29] Briefly, a 50 mM stock solution of carboxy-DCFH-DA (Invitrogen) was prepared in DMSO and diluted 1:100 in M9 buffer. Worms were exposed to 500 μM carboxy-DCFH-DA for 1 h (L1 stage) or 2 h (L4 stage) in the dark. After washing worms three times with M9 buffer, 8,000 (L1 stage) or 500 (L4 stage) worms were transferred to each well of a 96-well plate. Worms were incubated with tert-butyl-hydroperoxide (70% (w/w) in H2O, Sigma-Aldrich, Steinheim, Germany) at a final concentration of 350 μM (L1 stage) or 50 μM (L4 stage) to induce RONS. Kinetics of the oxidized DCFH-DA were monitored (excitation 485 nm/emission 535 nm) with a microplate reader (Tecan Infinite M200 Pro) and measured up to 420 min.
2.7. TaqMan gene expression assay
Total RNA was isolated using the Trizol method as published elsewhere.[29] Following isolation, 1 μg total RNA was subjected to cDNA synthesis applying the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions. Quantitative real-time PCR (BioRad) was conducted in duplicate wells for each gene using TaqMan Gene Expression Assay probes (Life Technologies). Data were normalized to the housekeeping gene afd-1 (actin homolog) after calculation of the fold change applying the comparative 2−ΔΔCt method.[30] The following probes were used: skn-1 (assay ID: Ce02407447_g1), daf-16 (assay ID: Ce02422838_m1), sod-3 (assay ID: Ce02404515_g1), gst-4 (assay ID: Ce02458730_g1), gcs-1 (assay ID: Ce02436725_g1), txnrd-1 (Ce02469942_m1) and afd-1 (assay ID: Ce02414573_m1).
2.8. Statistical analysis
Histograms were created using GraphPad Prism (GraphPad Software Inc.). All data presented in the figures and tables are mean values + SEM. Total Se contents and TXNRD activity data were analyzed using unpaired t-tests. Two-way analysis of variance (ANOVA) was performed on the Carboxy-DCFH-DA and Taqman gene expression data, followed by Dunnett’s multiple comparison post-hoc tests. A p-value < 0.05 was considered significant.
3. Results and discussion
3.1. ‘Hatch incubation’ with Se species results in increased Se contents and incorporation into proteins
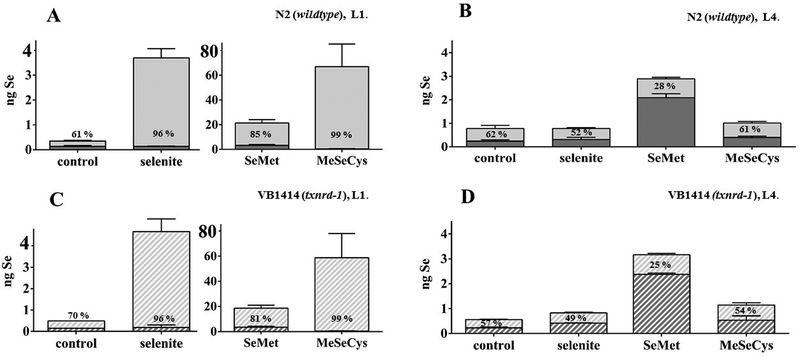
To investigate putative protective effects of the Se species in the developing nematode, C. elegans, a treatment protocol was established to allow for an integration of Se into proteins and to enable adjustments of the antioxidant defense system and metabolism. Additionally, to generate Se-depleted control conditions, agar- and E. coli-free conditions were indispensable. Agar and E. coli contain Se in mixed, unspecified forms and significant total quantities (8P agar: 6.5 ± 0.3 ng Se / g, E. coli NA22: 7.9 ± 0.6 ng Se / mL). Therefore, a ‘hatch incubation’ protocol was carried out, in which synchronized eggs were exposed to the Se species during hatching, which allows for prolonged exposure with the compounds (18 h) in the absence of agar and E. coli at an early life cycle stage. First, bioavailability following hatch incubation was assessed by quantifying total Se contents of the worms (Table 1). In wildtype L1 larvae, treatment with 100 μM of the Se species during hatching resulted in 20- to 80-fold increased Se levels compared to worms hatched in control buffer only. In agreement with observations from an acute high-dose exposure study in C. elegans[31] and studies in other models, e.g.,[32–34] the seleno-amino acids SeMet and MeSeCys accumulated to a greater extent than the inorganic selenite. Similar Se contents were obtained for the txnrd-1 deletion mutant strain VB1414, indicating that the bioavailability of the Se species is independent of the sole C. elegans selenoprotein, which was observed before in the high-dose study.[31]
Table 1.
Total Se contents of N2 (wildtype) and VB1414 (txnrd-1) worms following hatch incubation (18 h) with the Se species, directly after hatching (L1) and 48 h post-treatment (L4). Shown are mean values of at least two independent determinations ± SEM. Statistical analysis via unpaired t-test, **p < 0.01, ***p < 0.001 vs. control of the respective larval stage.
| Incubation | N2 (wildtype) [ng Se/mg protein] |
VB1414 (txnrd-1) [ng Se/mg protein] |
||
|---|---|---|---|---|
| L1 | L4 | L1 | L4 | |
| control | 2.1 ± 0.2 | 4.5 ± 0.4 | 1.3 ± 0.1 | 3.1 ± 0.3 |
| 100 μM selenite | 42.7 ± 0.8*** | 6.2 ± 0.3 | 45.1 ± 12.0** | 6.3 ± 1.1 |
| 100 μM SeMet | 176.0 ± 11.9*** | 19.6 ± 2.1** | 185.8 ± 13.8*** | 17.1 ± 1.9** |
| 100 μM MeSeCys | 152.2 ± 21.6*** | 5.6 ± 0.2 | 165.5 ± 28.1*** | 6.2 ± 0.9 |
Following hatch incubation, L1 larvae were grown to L4 stage without further Se species treatment in order to investigate the persistence of the effects due to Se accumulation as well as a possible incorporation into proteins. Se contents of L4 stage worms hatched in the presence of selenite or MeSeCys were indistinguishable from those of untreated worms (Table 1). Interestingly, hatch incubation with SeMet resulted in considerably higher Se contents in L4 stage in both C. elegans strains. While SeMet and MeSeCys displayed a similar bioavailability in L1 stage, only SeMet was persistently accumulated in L4 stage worms. It has been presumed before that animals cannot distinguish between methionine and SeMet, leading to a non-specific incorporation of SeMet into proteins.[35] However, this question was not addressed before in C. elegans. Therefore, fractionation experiments were carried out to reveal to which extent Se was integrated in worm proteins. As shown in Fig. 1A, total Se in the protein fraction of L1 larvae following SeMet hatch incubation was substantially higher than for the other Se species, indicating that worms incorporate SeMet unspecifically into proteins. This effect could not be observed after incubation with selenite or MeSeCys.
Figure 1.
Total Se contents in protein fractions (dark grey) and non-protein fractions (light grey) of worms following hatch incubation (18 h) with the Se species (100 μM). Stacked columns show absolute values in fractionated worm pellets, whereas percentages give the proportional distribution related to the sum of both fractions, in each case indicated for the non-protein fraction. Shown are mean values of at least two independent experiments with two determinations + SEM. (A) N2 (wildtype), L1 stage. (B) N2 (wildtype), L4 stage (48 h post-treatment). (C) VB1414 (txnrd-1), L1 stage. (D) VB1414 (txnrd-1), L4 stage (48 h post-treatment).
Interestingly, analysis of the Se distribution in L4 stage worms (48 h post-treatment) revealed that the relative percentage of Se associated with proteins was even higher, especially following SeMet hatch incubation (Fig. 1B). The data point out that C. elegans is to a certain extent able to store Se in proteins, while excessive Se measured at L1 in the non-protein fraction is supposed to be excreted until L4. The Se species differ largely in their utilization, with SeMet being incorporated into proteins most efficiently. This corroborates earlier observations (as reviewed in [4, 35]), indicating that SeMet is directly incorporated into proteins while selenite and MeSeCys must first be used to form SeMet or selenocysteine. The Se distribution in txnrd-1 deletion mutants mirrored the distribution in wildtype worms in both developmental stages (Fig. 1C, D). However, in L1 stage txnrd-1 mutants, the amount of Se in the protein fraction was lower than in wildtype worms, which might be attributed to the lack of TXNRD-1. Overall, txnrd-1 worms have a significantly lower Se basal level than wildtype worms.[31] Interestingly, in L4 stage worms, the basal distribution was similar in both strains. Altogether, the data are in line with the assumption that the Se incorporation into proteins occurs in a non-specific rather than a genetically encoded manner.
3.2. Increased TXNRD activity following Se species exposure
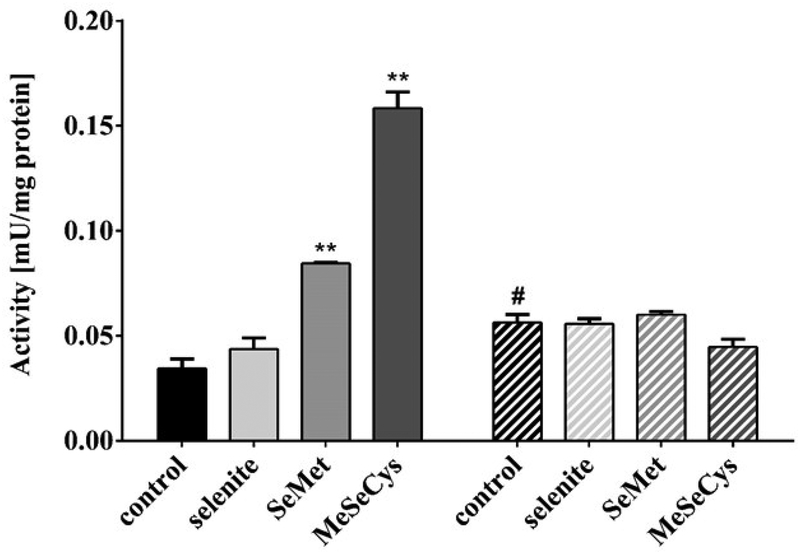
The expression and/or activity of selenoproteins is commonly used and well-recognized as a biomarker of the Se status, as these proteins directly reflect the portion of Se retained in functional forms. In human and animal studies, GPX3 and SELENOP in serum or plasma and GPX1 in blood cells are easily accessible and widely used as functional biomarkers.[36–38] Besides, assays are established for other selenoproteins, including TXNRDs and DIOs, and a dietary intake of 0.1 – 0.2 mg/kg Se has been shown to result in maximal expression of these enzymes in most animal species.[36] With respect to C. elegans, basic understanding of the Se status of these animals under laboratory conditions is lacking. While their actual Se intake in plate culture is difficult to assess and cannot provide information on the worms’ functional Se status, the only C. elegans selenoprotein TXNRD-1 might be a suitable marker. In animals and humans, TXNRD activity is analyzed based on a DTNB reduction assay,[28, 39, 40] which was herein adapted to worm samples. As shown in Fig. 2, TXNRD activity significantly increased 48 h post-treatment following hatch incubation with 100 μM of SeMet or MeSeCys. In contrast, selenite exposure failed to increase TXNRD activity. This indicates that the seleno-amino acids are more efficiently utilized for selenoprotein synthesis than the inorganic form which might be potentiated by the lower bioavailability of selenite. As the TXNRD activity can be further enhanced by increasing the Se status of worms, the basal Se status is supposed to be rather suboptimal under normal conditions. In txnrd-1 deletion mutant worms, treatment with the Se species had no effect on TXNRD activity. Unexpectedly, the basal TXNRD activity is not impaired by genetic ablation of TXNRD-1, but in contrast appears to be even increased. The measured basal activity can be on the one hand attributed to the non-selenoprotein TXNRD-2, as the assay captures only total TXNRD activity due to the lack of specific substrates. On the other hand, other NADPH oxidoreductases that are capable of reducing DTNB, such as glutathione reductase, could contribute to the background activity.[41] The increase in TXNRD activity in mutant worms indicates that one or several of those enzymes are up-regulated to compensate for the loss of TXNRD-1.
Figure 2.
Activity of L4 stage worm lysates (48 h post-treatment) in catalyzing the reduction of 5,5’-dithiobis(2-nitrobenzoic acid) (DTNB) to 5’-thionitrobenzoic acid (TNB). Worms were exposed to the Se species (100 μM) for 18 h during hatching. TNB production was measured 240 min after starting the reaction. Shown is one representative experiment out of four independent experiments with at least three determinations + SEM. Statistical analysis via unpaired t-test, **p < 0.01 vs. control of the respective strain, #p < 0.05 vs. wildtype control. Full bars: N2 (wildtype), striped bars: VB1414 (txnrd-1).
3.3. Protection against t-BOOH-induced RONS formation by Se species pretreatment
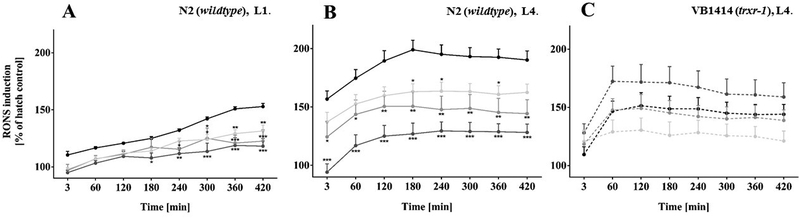
As our results indicated that C. elegans were inadequately supplied with Se, next, we assessed whether Se supplementation has protective effects. As Se is implicated in the antioxidant defense system via selenoproteins,[42] protection from oxidative stress was investigated. RONS levels were measured in worms following hatch incubation with Se species and subsequent exposure to the oxidant tert-butyl-hydroperoxide (t-BOOH). As depicted in Fig. 3A, wildtype L1 larvae showed a time-dependent increase in RONS upon t-BOOH exposure, which was attenuated in larvae hatched in the presence of 100 μM selenite, SeMet or MeSeCys. Protective effects of selenite towards various stressors have been demonstrated before in C. elegans[19, 43] and other models.[44, 45, 48, 49] Likewise, protection by seleno-amino acids against oxidative stress has been reported in various in vitro and rodent studies.[44–49] Although all investigated Se species diminished t-BOOH induced RONS formation, the organic species seemed to be slightly more effective than the inorganic selenite (Fig. 3A, B). Corroborating our observations, previous studies in rats have reported greater antioxidant activity of MeSeCys compared to selenite.[48,49] Interestingly, in the current study, a protective effect was also observed in wildtype worms 48 h post-treatment following hatch incubation (Fig. 3B). As the overall Se content and especially the amount of Se in the protein-free fraction was remarkably lower in L4 stage, the persistent protection can probably not be explained by the ability of the Se species to scavenge free radicals. This hypothesis is supported by data obtained in the txnrd-1 deletion mutant strain. Here, hatch incubation with the Se species did not diminish t-BOOH-induced RONS formation in L4 stage (Fig. 3C). Hence, the only C. elegans selenoprotein TXNRD-1 might be required for the protective effects mediated by the Se species, corroborating previous studies.[19, 50] However, others have reported that TXNRD-1 was not essential for C. elegans for growth, development and reproduction[23, 51] and failed to observe increased sensitivity to acute oxidative stress in txnrd-1 mutants.[19, 23] Surprisingly, in our study, t-BOOH failed to induce RONS in L1 stage txnrd-1 mutants (data not shown). Thus, the antioxidant defense of the VB1414 strain apparently differs strongly from wildtype worms, depending on the developmental stage. Clearly, further efforts are needed to clarify the underlying molecular mechanisms and elucidate TXNRD-1 function in C. elegans. In mice, chronic deletion of TXNRD-1 in hepatocytes resulted in transcriptional upregulation of genes encoding xenobiotic metabolism enzymes, many of which containing Nrf2 binding sites. Thus, TXNRD-1 deficiency in mammalian cells triggers an effective compensatory program involving the activation of the Nrf2 pathway.[52]
Figure 3.
RONS induction by tert-butyl-hydroperoxide (t-BOOH) following exposure to the Se species (100 μM) for 18 h during hatching measured after dye loading and subsequent treatment with 350 μM (L1) or 50 μM (L4) t-BOOH. Data were normalized to the corresponding negative control (w/o t-BOOH) of each hatch treatment group. Shown are representative experiments (n ≥ 2) with mean values of a double determination + SEM. Statistical analysis by two-way ANOVA, followed by Dunnett’s post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001 vs. control. (A) N2 (wildtype), L1 stage. (B) N2 (wildtype), L4 stage (48 h post-treatment). (C) VB1414 (txnrd-1), L4 stage (48 h post-treatment).  selenite,
selenite,  SeMet,
SeMet,  MeSeCys,
MeSeCys,  control.
control.
3.4. Differential effects of Se species on mRNA expression of antioxidant proteins
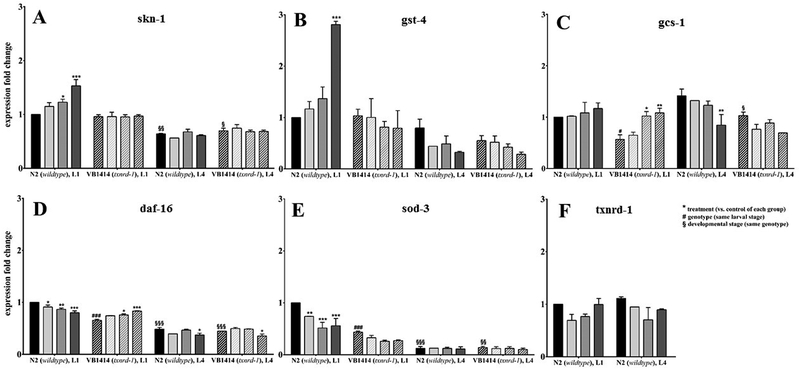
To determine whether the protective effects of Se species in wildtype C. elegans and the differential response of the VB1414 strain might be associated with differences in adaptive stress responses, mRNA expression of proteins implicated in antioxidant defense and stress responses was examined. A selection of genes encoding for antioxidant proteins was investigated in order to obtain first indications of involved pathways. We tested several target genes of the SKN-1 pathway, gst-4 (encoding a glutathione S-transferase), gcs-1 (encoding γ-glutamylcysteine synthetase) and the transcription factor skn-1 itself, which is the orthologue of the vertebrate Nrf2.[53, 54] Another key transcription factor implicated in antioxidant responses is the C. elegans FoxO orthologue DAF-16,[55] regulating the expression of genes such as sod-3 (encoding a manganese superoxide dismutase).[56] Besides DAF-16, also SKN-1 is able to upregulate sod-3 expression.[57] To visualize differences between wildtype and txnrd-1 deletion mutant worms as well as the two different larval stages within the strains, gene expression data were normalized to the respective wildtype L1 stage controls.
In wildtype L1 larvae, hatch incubation with MeSeCys resulted in an upregulation of skn-1 and gst-4 mRNA levels (Fig. 4A, B). A similar trend was observed for selenite and SeMet, with a significant effect for skn-1 following SeMet treatment. Those effects are lost in L1 txnrd-1 deletion mutant worms, which implies that TXNRD-1 is mediating the Se-induced up-regulation of both genes. In mammals, TXNRD-1 is a known regulator of Nrf2.[58] If this happens to be the case in C. elegans as well, it might explain the observed effects. While treatment with MeSeCys induced the expression of skn-1 and its target gene gst-4 in wildtype worms, in txnrd-1 mutants, the crosstalk between TXNRD-1 and SKN-1 seems to be missing. Consequently, MeSeCys failed to induce the expression of those genes. Interestingly, gcs-1 shows the opposite result. In wildtype L1 worms, gcs-1 is unaffected by the Se compounds, but it is up-regulated by SeMet and MeSeCys, when txnrd-1 is missing. This up-regulation, however, just restores basal levels of expression detectable in wildtype L1 worms, because there is a substantial drop in gcs-1 expression in txnrd-1 mutants compared to wildtype worms under basal conditions. Manipulation of the expression of skn-1 as well as a single gst has been described to modulate worm`s responses to oxidative stress.[54, 59] Previous work has shown that skn-1 mutants are vulnerable to oxidative stress,[54] whereas skn-1 overexpression afforded protection against manganese-induced toxicity.[60] Furthermore, selenite exposure of the nematode has been shown to enhance expression of gst-4 as well as gcs-1 under PA14 infection via SKN-1.[61] Recently, Salgueiro et al. (2017) published that sod-3, gcs-1, gst-4 and txnrd-1 mRNA levels are altered following exposure to two 4-phenylchalcogenil-7-chloroquinolines derivatives (PSQ for selenium and PTQ for tellurium-containing compounds), thus suggesting their involvement in the protective effects of these compounds.[62] Overexpression of a single GST increased the resistance to intracellularly induced oxidative stress.[59] Moreover, Tawe et al. (1998) showed that gst-4 (K08F4.7) is inducible by the oxidative agent paraquat.[63] Deletion of gcs-1, which catalyzes the first, rate-limiting step in the biosynthesis of glutathione, results in decreased stress resistance against, for example, arsenic toxicity.[64] Vice versa, it was reported to be inducible by oxidative and thermal stress.[54]
Figure 4.
mRNA expression levels of antioxidant genes following hatch incubation (18 h) with the Se species (100 μM). Relative gene expression was determined by qRT-PCR. Shown are mean values + SEM of two independent experiments in duplicates. Data are normalized to the untreated wildtype at L1 stage and relative to afd-1/β-actin. Statistical analysis using two-way ANOVA: treatment (vs. untreated control of respective group) *p < 0.05, **p < 0.01, ***p < 0.001, genotype (vs. wildtype control of the same larval stage) #p < 0.05, ###p < 0.001, developmental stage (vs. L1 of respective strain) §p < 0.05, §§p < 0.01, §§§p < 0.001.  control,
control,  selenite,
selenite,  SeMet,
SeMet,  MeSeCys.
MeSeCys.
48 h post-treatment (L4 stage), gene expression studies revealed lower mRNA levels of skn-1 compared to L1 stage, while for gst-4 a non-significant downregulation was observed for all Se species treated worms in comparison to L1 (Fig. 4A, B). Independent of the txnrd-1 genotype, no up-regulation by the Se compounds was observed in L4. This is typical for the already described bi-phasic nature of Nrf2/Skn-1 responses, which in the short term are rapidly up-regulated followed by down-regulation at more protracted time points, at times even below basal levels. Gcs-1 expression was also inherently different in the examined strains. Basal gcs-1 levels were increased in L4 in comparison to L1 independent of the txnrd-1 genotype, but in contrast to L1, gcs-1 was down-regulated by MeSeCys in L4 wildtype worms (Fig. 4C). Furthermore, the down-regulation appears to restore basal gcs-1 levels in L1 wildtype worms, indicating that MeSeCys might balance gcs-1 expression, maintaining its levels within certain gates.
In addition to SKN-1, DAF-16 is an important transcription factor for antioxidant stress response.[65] Following hatch incubation with the Se species, mRNA levels of daf-16 and sod-3 were decreased in wildtype L1 stage worms (Fig. 4D, E). Overexpression and/or overactivation of sod-3 are necessary for the induction of pro-longevity response after mild oxidative damage.[66] The lower expression might bear a compensatory effect to counteract oxidative stress in the worm as observed in the RONS formation assay. Interestingly, L4 stage wildtype as well as txnrd-1 worms showed remarkably lower expression levels of daf-16, and to a greater extent, sod-3. Analogous to gcs-1, basal mRNA levels of daf-16 as well as sod-3 were also lower in L1 stage txnrd-1 deletion mutants in comparison to wildtype L1. This might contribute to the enhanced ability of the L1 txnrd-1 deletion mutants to combat oxidative stress. No t-BOOH-induced RONS formation could be observed in the L1 larvae of the txnrd-1 deletion mutant worms up to 1400 μM t-BOOH (LD50: 19.8 mM following 30 min incubation, data not shown). As a known regulator of both, the Nrf2/Skn-1 and FoxO/Daf-16 pathway, TOR signaling might be involved in the observed downregulation of gcs-1, daf-16 and sod-3 in the txnrd-1 mutant. When TORC1 is inhibited genetically in C. elegans, Nrf2/Skn-1 and FoxO/Daf-16 activate protective genes, whereas activation of TORC1 opposes these pathways and inhibits Nrf2/Skn-1 and FoxO/Daf-16 driven transcription.[67] However, whether TOR signaling is activated in the txnrd-1 deletion mutant requires further investigation. 48 h post-treatment, the basal gcs-1 mRNA level was increased in the txnrd-1 deletion mutants as compared to L1 stage worms, while daf-16 as well as sod-3 levels were significantly further reduced. This provides an explanation for the absence of any protective effect of the Se species against oxidant-induced RONS formation in the txnrd-1 deletion mutants (Fig. 3C). Consequences of daf-16 knockdown and downregulation have been discussed before. For example, daf-16 mutants were more sensitive to oxidative stress chemically induced by paraquat[55] as well as juglone.[68] Additionally, DAF-16 mediated higher stress resistance and longevity in nematodes carrying an integrated daf-16::gfp transgene.[69] In another study, transcriptional downregulation of daf-16 was observed in response to the nematicidal Cry6Aa2 toxin, and it has been proven to be closely related to hypersensitivity of the worms towards this pathogen.[70] Interestingly, Li et al. (2013) showed that selenite-mediated protection against oxidative stress was dependent on both, DAF-16 and TXNRD-1.[19] The hatch incubation with the Se species did not affect gene expression of txnrd-1 (Fig. 4F), indicating that in C. elegans the selenoprotein is mainly regulated at the translational or activity level, as discussed before (Fig. 2). However, whether DAF-16 and TXNRD-1 act synchronously along the same pathway or via alternative pathways remains unclear. Consequently, antioxidant defense in both C. elegans strains seems to be differentially regulated and involves different stress response pathways that are also highly dependent upon the developmental stage. Taken together, these results imply altered gene expression levels especially in skn-1 and daf-16 which in conjunction with the protective effect of the Se species in reducing oxidant-induced RONS levels indicate the worms’ ability to counteract oxidative stress.
4. Concluding remarks
Utilizing the tractable model organism C. elegans, we show that exposure of hatching nematodes with Se species had immediate and sustained (48 h post-treatment) systemic effects. For the first time, we demonstrated that SeMet was efficiently and permanently incorporated into worm proteins, in contrast to other Se species. However, protection against t-BOOH-induced RONS formation appeared to be independent of the applied Se species. While this might be partly attributed to direct effects of the Se species on gene expression levels, such as skn-1 induction or daf-16 and sod-3 reduction, distinct effects on stress response pathways have been also identified. Corroborating observations in TXNRD-1-deficient hepatocytes,[46] txnrd-1 mutant worms also show the putative involvement of the Nrf2/SKN-1 pathway. Nevertheless, full understanding of the role of the sole selenoprotein TXNRD-1 in C. elegans requires further investigation. As protective effects were still observed 48 h post-treatment, the question arises if a sufficient Se supply in early life stages may have sustained benefits and could help to endure periods of inadequate Se intake.
Acknowledgements
This work was funded by the German Research Foundation (DFG), grant number SCHW 903/9–1, and the Austrian Science Fund (FWF), project number I 2262-N28, as well as the DFG Research Unit TraceAge (FOR 2558). We thank the German Research Foundation (DFG) further for the financial support of BO 4103/2–1. MA was supported in part by NIH grants NIEHS R01 10563, R01ES07331, and NIEHS R01ES020852. We would also like to thank the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), for providing the N2 strain used in this work. Lastly, we want to thank Elias Arnér for his help with the TXNRD activity assay.
Abbreviations:
- C. elegans
Caenorhabditis elegans
- DIO
iodothyronine deiodinase
- GPX
glutathione peroxidase
- ICP-QQQ-MS
inductively coupled plasma tandem mass spectrometry
- MeSeCys
Se-methylselenocysteine
- RONS
reactive oxygen and nitrogen species
- SeMet
selenomethionine
- t-BOOH
tert-butyl-hydroperoxide
- TXNRD
thioredoxin reductase
Footnotes
Conflict of interest
The authors declare no conflict of interest.
5 References
- [1].Rayman MP, Lancet 2012, 379, 1256. [DOI] [PubMed] [Google Scholar]
- [2].Duntas LH, Benvenga S, Endocrine 2015, 48, 756. [DOI] [PubMed] [Google Scholar]
- [3].Jablonska E, Vinceti M, J. Environ. Sci. Heal 2015, 33, 328. [DOI] [PubMed] [Google Scholar]
- [4].Weekley CM, Harris HH, Chem. Soc. Rev 2013, 42, 8870. [DOI] [PubMed] [Google Scholar]
- [5].Wrobel JK, Power R, Toborek M, IUBMB Life 2016, 68, 97. [DOI] [PubMed] [Google Scholar]
- [6].V Kryukov G, Castellano S, V Novoselov S, V Lobanov A, Zehtab O, Guigó R, Gladyshev VN, Science 2003, 300, 1439. [DOI] [PubMed] [Google Scholar]
- [7].Labunskyy VM, Hatfield DL, Gladyshev VN, Physiol. Rev 2014, 94, 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rayman MP, Goenaga-Infante H, Sargent M, Br. J. Nutr 2008, 100, 238. [DOI] [PubMed] [Google Scholar]
- [9].Davis CD, Feng Y, Hein DW, Finley JW, J. Nutr 1999, 129, 63. [DOI] [PubMed] [Google Scholar]
- [10].Barger JL, Kayo T, Pugh TD, Vann JA, Power R, Dawson K, Weindruch R, Prolla TA, Genes Nutr. 2012, 7, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lennicke C, Rahn J, Kipp AP, Dojčinovićc BP, Müller AS, Wessjohann LA, Lichtenfels R, Seliger B, Biochem. Biophys. Acta 2017, 1861, 3323. [DOI] [PubMed] [Google Scholar]
- [12].Hiller F, Oldorff L, Besselt K, Kipp AP, Nutrients 2015, 7, 2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Leung MCK, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, Meyer JN, Toxicol. Sci 2008, 106, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yen CA, Curran SP, Exp Gerontol. 2016, 86, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Markaki M, Tavernarakis N, Biotechnol. J 2010, 5, 1261. [DOI] [PubMed] [Google Scholar]
- [16].Jones KT, Ashrafi K, Dis Model Mech. 2009, 2, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Morgan KL, Estevez AO, Mueller CL, Cacho-Valadez B, Miranda-Vizuete A, Szewczyk NJ, Estevez M, Toxicol. Sci 2010, 118, 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li W-H, Hsu F-L, Liu J-T, Liao VH-C, Food Chem. Toxicol 2011, 49, 812. [DOI] [PubMed] [Google Scholar]
- [19].Li W-H, Shi Y-C, Chang C-H, Huang C-W, Liao VH-C, Mol. Nutr. Food Res 2013, 863. [DOI] [PubMed] [Google Scholar]
- [20].Stefanello ST, Gubert P, Puntel B, Mizdal CR, de Campos MMA, Salman SM, Dornelles L, Avila DS, Aschner M, Soares FAA, Toxicol. Rep 2015, 2, 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gladyshev VN, Krause M, Xu XM, V Korotkov K, V Kryukov G, Sun QA, Lee BJ, Wootton JC, Hatfield DL, Biochem. Biophys. Res. Commun 1999, 259, 244. [DOI] [PubMed] [Google Scholar]
- [22].Buettner C, Harney JW, Berry MJ, J. Biol. Chem 1999, 274, 21598. [DOI] [PubMed] [Google Scholar]
- [23].Stenvall J, Fierro-González JC, Swoboda P, Saamarthy K, Cheng Q, Cacho-Valadez B, Arnér ESJ, Persson OP, Miranda-Vizuete A, Tuck S, Proc. Natl. Acad. Sci. U. S. A 2010, 108, 1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jakupoglu C, Przemeck GKH, Schneider M, Moreno SG, Mayr N, Hatzopoulos A, Hrabé de Angelis M, Wurst W, Bornkamm GW, Brielmeier M, Conrad M, Mol. Cell. Biol, 2005, 25, 1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Brenner S, Genetics 1974, 77, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Marschall TA, Kroepfl N, Jensen KB, Bornhorst J, Meermann B, Kuehnelt D, Schwerdtle T, Metallomics, 2017, 9, 268. [DOI] [PubMed] [Google Scholar]
- [27].Gammelgaard B, Jøns O, J. Anal. At. Spectrom 1999, 14, 867. [Google Scholar]
- [28].Gromer S, Merkle H, Schirmer RH, Becker K, Methods Enzymol. 2002, 347, 382. [DOI] [PubMed] [Google Scholar]
- [29].Bornhorst J, Chakraborty S, Meyer S, Lohren H, Große Brinkhaus S, Knight AL, Caldwell KA, Caldwell GA, Karst U, Schwerdtle T, Bowman A, Aschner M, Metallomics 2014, 6, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Livak KJ, Schmittgen TD, Methods 2001, 25, 402. [DOI] [PubMed] [Google Scholar]
- [31].Rohn I, Marschall TA, Kroepfl N, Jensen KB, Aschner M, Tuck S, Kuehnelt D, Schwerdtle T, Bornhorst J, Metallomics 2018, 10, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lunøe K, Gabel-Jensen C, Stürup S, Andresen L, Skov S, Gammelgaard B, Metallomics 2011, 3, 162. [DOI] [PubMed] [Google Scholar]
- [33].Marschall TA, Bornhorst J, Kuehnelt D, Schwerdtle T, Mol. Nutr. Food Res 2016, 60, 2622. [DOI] [PubMed] [Google Scholar]
- [34].Dolgova NV, Hackett MJ, Macdonald TC, Nehzati S, James AK, Krone PH, George GN, Pickering IJ, Metallomics 2016, 8, 305. [DOI] [PubMed] [Google Scholar]
- [35].Burk RF, Hill KE, Annu. Rev. Nutr 1993, 13, 65. [DOI] [PubMed] [Google Scholar]
- [36].Combs GF, Nutrients 2015, 7, 2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ashton K, Hooper L, Harvey LJ, Hurst R, Casgrain A, Fairweather-Tait SJ, Am. J. Clin. Nutr 2009, 89, 2025S. [DOI] [PubMed] [Google Scholar]
- [38].Kipp AP, Banning A, Van Schothorst EM, Méplan C, Coort SL, Evelo CT, Keijer J, Hesketh J, Brigelius-Flohé R, J. Nutr. Biochem 2012, 23, 1170. [DOI] [PubMed] [Google Scholar]
- [39].Arnér ESJ, Holmgren A, Curr. Protoc. Toxicol 2005, 24, 7.4.1. [DOI] [PubMed] [Google Scholar]
- [40].Hrdina J, Banning A, Kipp A, Loh G, Blaut M, Brigelius-Flohé R, J. Nutr. Biochem 2009, 20, 638. [DOI] [PubMed] [Google Scholar]
- [41].Brigelius R, Muckel C, Akerboom TPM, Sies H, Biochem. Pharmacol 1983, 32, 2529. [DOI] [PubMed] [Google Scholar]
- [42].Letavayová L, Vlcková V, Brozmanová J, Toxicology 2006, 227, 1. [DOI] [PubMed] [Google Scholar]
- [43].Li W-H, Shi Y-C, Tseng I-L, Liao VH-C, PLoS One 2013, 8, e62387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Khera A, Vanderlelie J, Perkins AV, Placenta 2013, 34, 594. [DOI] [PubMed] [Google Scholar]
- [45].Watson M, van Leer L, Vanderlelie JJ, Perkins AV, Placenta 2012, 33, 1012. [DOI] [PubMed] [Google Scholar]
- [46].Parveen F, Nizamani ZA, Gan F, Chen X, Shi X, Kumbhar S, Zeb A, Huang K, Biol. Trace Elem. Res 2014, 157, 266. [DOI] [PubMed] [Google Scholar]
- [47].Cuello S, Ramos S, Mateos R, Martín MA, Madrid Y, Cámara C, Bravo L, Goya L, Anal. Bioanal. Chem 2007, 389, 2167. [DOI] [PubMed] [Google Scholar]
- [48].Li X, Zhang Y, Yuan Y, Sun Y, Qin Y, Deng Z, Li H, Biol. Trace Elem. Res 2016, 173, 433. [DOI] [PubMed] [Google Scholar]
- [49].Yao Z, Zhang Y, Li H, Deng Z, Zhang X, Trace Elem J. Med. Biol 2015, 29, 182. [DOI] [PubMed] [Google Scholar]
- [50].Li W, Bandyopadhyay J, Hwaang HS, Park B, Cho JH, Lee JI, Ahnn J, Lee S-K, Mol Cells. 2012, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Boehler CJ, Raines AM, Sunde RA, PLoS One, 2013, 8, e71525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Suvorova ES, Lucas O, Weisend CM, Rollins MF, Merrill GF, Capecchi MR, Schmidt EE, PLoS One, 2009, 4, e6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Oliveira RP, Abate JP, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK, Aging Cell 2010, 8, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].An JH, Blackwell TK, Genes Dev. 2003, 17, 1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Essers MAG, de Vries-Smits LMM, Barker N, Polderman PE, Burgering BMT, Korswagen HC, Science, 2005, 308, 1181. [DOI] [PubMed] [Google Scholar]
- [56].McElwee J, Bubb K, Thomas JH, Aging Cell 2003, 2, 111. [DOI] [PubMed] [Google Scholar]
- [57].Zhang L, Jie G, Zhang J, Zhao B, Free Radic. Biol. Med 2009, 46, 414. [DOI] [PubMed] [Google Scholar]
- [58].Cebula M, Schmidt EE, Arnér ESJ, Antioxid. Redox Signal 2015, 23, 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Leiers B, Kampkötter A, Grevelding CG, Link CD, Johnson TE, Henkle-Dührsen K, Free Radic. Biol. Med 2003, 34, 1405. [DOI] [PubMed] [Google Scholar]
- [60].Benedetto A, Au C, Avila DS, Milatovic D, Aschner M, PLoS Genet. 2010, 6, e1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li W-H, Chang C-H, Huang C-W, Wie C-C, Liao VH-C, PLoS One 2014, 9, e105810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Salgueiro WG, Goldani BS, Peres TV, Miranda-Vizuete A, Aschner M, da Rocha JBT, Alves D, Avila DS, Free Radic. Biol. Med 2017, 110, 133. [DOI] [PubMed] [Google Scholar]
- [63].Tawe WN, Eschbach M-L, Walter RD, Henkle-Dührsen K, Nucleic Acids Res. 1998, 26, 1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liao VH-C, Yu C-W, Biometals 2005, 18, 519. [DOI] [PubMed] [Google Scholar]
- [65].Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng H-L, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME, Science, 2004, 303, 2011. [DOI] [PubMed] [Google Scholar]
- [66].Yee C, Yang W, Hekimi S, Cell 2014, 157, 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell KT, Cell Metab. 2012, 15, 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Heidler T, Hartwig K, Daniel H, Wenzel U, Biogerontology 2010, 11, 183. [DOI] [PubMed] [Google Scholar]
- [69].Henderson ST, Johnson TE, Curr. Biol 2001, 11, 1975. [DOI] [PubMed] [Google Scholar]
- [70].Wang B, Wang H, Xiong J, Zhou Q, Wu H, Xia L, Li L, Yu Z, Sci. Rep 2017, 7, 14170. [DOI] [PMC free article] [PubMed] [Google Scholar]