Abstract
DNA alkylguanine DNA alkyltransferases (AGTs) are evolutionary conserved proteins that repair alkylation damage in DNA, counteracting the effects of agents inducing such lesions. Over the last years AGTs have raised considerable interest for both the peculiarity of their molecular mechanism and their relevance in cancer biology. AGT knock out mice show increased tumour incidence in response to alkylating agents, and over-expression of the human AGT protein in cancer cells is frequently associated with resistance to alkylating chemotherapy. While all data available point to a function of AGT proteins in the cell response to alkylation lesions, we report for the first time that one of the two AGT paralogs of the model organism C. elegans, called AGT-2, also plays unexpected roles in meiosis and early development under physiological conditions. Our data suggest a role for AGT-2 in conversion of homologous recombination intermediates into post-strand exchange products in meiosis, and show that agt-2 gene down-regulation, or treatment of animals with an AGT inhibitor results in increased number of germ cells that are incompatible with producing viable offspring and are eliminated by apoptosis. These results suggest possible functions for AGTs in cell processes distinct from repair of alkylating damage.
Subject terms: RNAi, Genetic models
Introduction
DNA alkylguanine DNA alkyltransferases (AGTs) are evolutionary conserved, peculiar proteins that repair alkylation damage in DNA, mainly at the O6-position of guanines, thus counteracting the effects of alkylating agents that form such adducts1–3. AGTs are widely present in organisms of the three living kingdoms (bacteria, archaea, eukaryotes) and typically comprise two domains: a less conserved N-terminal domain, which might play regulatory functions, and a highly conserved C-terminal domain, which houses the functional elements required for DNA binding and repair4–10. This latter occurs in a single-step SN2-type reaction, in which the alkyl moiety is covalently bound to a conserved cysteine residue in the protein active site, leading to release of an intact double strand and a protein molecule which carries the alkyl adduct1,7,11. This alkylated protein form is irreversibly inactivated and directed to intracellular degradation pathways12. AGT inactivation and degradation can also be triggered by competitive inhibitors of AGTs, such as O6-benzylguanine (O6-BG)13. Several lines of evidence suggest a link between AGT and cancer. First, knock out of the AGT-encoding gene in mouse results in increased tumour incidence in response to alkylating agents14,15. Moreover, over-expression of the human AGT protein (hAGT) in cancer cells is frequently associated with resistance to alkylating chemotherapy; for this reason, chemotherapy regimens of O6-BG in combination with alkylating agents are in clinical development13,16.
Over the last years, the nematode C. elegans has emerged as a model for genetic, molecular, and cellular analysis of DNA repair pathways. In particular, the C. elegans gonad is a useful toolkit for the study of germ line DNA repair as well as apoptosis, which occur both physiologically and in response to exogenous DNA damage, and their progress can be easily followed thanks to the gonad precise spatiotemporal organization17. Importantly, most pathways and key factors in these processes are conserved from worms to humans. These include homologous recombination (HR), non-homologous end-joining (NHEJ), mismatch repair, nucleotide excision repair, interstrand crosslinking repair18; orthologs of several human disease-linked genes belonging to these pathways are conserved in the nematode, including the DNA damage checkpoint gene ATR (atl-1 in C. elegans)19, the breast cancer predisposition BRCA1 and BRCA2 genes (brc-1 and brc-2, respectively)20–23, the Fanconi Anemia FANCJ, FANCD-2, FANC-I and FANCM genes (dog-1, fcd-2, fnci-1, fncm-1, respectively)24–28, making C. elegans a useful model to study disease-related genes.
Whereas most prokaryotic and eukaryotic species encode a single AGT ortholog, the C. elegans genome comprises two ORFs potentially coding for two distinct AGT orthologs, known as AGT-1 and AGT-229. A truncated form of AGT-2 purified in recombinant form was shown to be endowed with DNA alkyltransferase activity in vitro and to confer resistance to alkylating agents when expressed in Escherichia coli29, thus suggesting that the protein is a bona fide AGT. No data about the in vivo function of either protein have been reported.
In this paper we used genetic tools combined with high-resolution microscopy to investigate the function of AGT-2 in C. elegans. Our data reveal that the agt-2 gene plays unexpected roles in the nematode meiosis and early development under physiological conditions.
Methods
C. elegans strains and culture
All strains (Supplementary Table S1) were cultured at 20 °C under standard conditions as described by Wood30. The N2 Bristol strain was used as the wild type background. The agt-2(tm6462) strain was provided by the Mitani Laboratory31, and backcrossed three times into the N2 wild type strain. The sequence of agt-2 wild type and agt-2(tm6462) alleles were checked by PCR amplification and sequencing using the oligonucleotides listed in Supplementary Table S2. Double and triple mutants (agt-2; spo-11, agt-2; syp-2, agt-2; cep-1, agt-2; syp-2; lig-4) were generated by genetic crosses and genotyped by PCR analysis using primers flanking each deletion. All primers are listed in Supplementary Table S2.
RNA isolation and real-time RT-PCR
Wild type and agt-2 young adult nematodes were broken by snap freezing in liquid nitrogen and then ground to a powder with a mortar and pestle. Total RNA was extracted using the RNeasy Plant Mini Kit (Quiagen) according to the manufacturer’s instructions. Residual DNA in RNA preparations was removed by using the DNA-free DNA Removal kit (Ambion), the absence of DNA contamination was checked by PCR analysis. Purified RNA was quantified by a Nanodrop instrument (Thermofisher) and RNA integrity was checked by 1.5% agarose gel electrophoresis. cDNA was generated using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem) according to the manufacturer’s instructions. Reactions were performed in 50 μl and contained 5 μg of RNA, 1X enzyme mix (including Mulv and RNase inhibitor protein), and 1X RT Buffer mix (including dNTPs, random octamers, and oligo dT-16). Reactions were incubated for 60′ at 37 °C (step1) and then for 5′ at 95 °C (step 2). Real-time quantitative PCR reactions were performed using the Power SYBR Green Master Mix (Applied Biosystem), according to the manufacturer’s instructions. Each reaction was prepared in a total volume of 20 μl containing 10 ng of cDNA and 0.25 μM of primers (HPLC purified by Eurofins; sequences are listed in Supplementary Table S3). For each biological replicate, three technical replicates were run using a CFX96 Real Time System (Bio‐Rad). The RT parameters were: 36 cycles of amplification; T = 56 °C for annealing. The specificity of amplified products was checked by 1.5% agarose gel electrophoresis. The qPCR parameters were validated by the CFX Maestro software. Results were recorded as relative gene expression changes after normalizing for the housekeeping pmp-3 gene expression and computed using the comparative CT method (2–CT) as previously described32. The 2–CT value was >1 for gene more highly expressed in the mutant strain; 2–CT value was <1 for gene more highly expressed in the wild-type strain. Shown are the means ± SD from three independent experiments.
RNA interference
RNAi was performed by feeding as described previously33, using clones from the Ahringer library (Gurdon Institute, Cambridge, UK)34. The procedure is described in Supplementary Fig. S1A,B. Briefly, HT115 bacteria were transformed with a vector (L4440) for IPTG-inducible expression of double-stranded RNA (dsRNA). Animals were synchronized via standard hypochlorite treatment and grown on OP50 seeded NGM plates. L4 worms were washed with M9 buffer and transferred to fresh plates seeded with RNAi bacteria immediately and consecutively after 1 hr, 12 hrs and then every 24 hrs. Laid eggs, dead embryos and developmental defects were scored after 72 hrs.
Screening of phenotypes
The procedure is described in the Supplementary Fig. S1B. Young adult worms were picked and individually cloned onto freshly seeded plates. Each worm was transferred to a fresh plate every 12 hrs, and laid eggs, embryonic lethality, males, developmental defects and larval arrests were scored after 72 hrs. Embryonic lethality was calculated as the ratio of the unviable eggs on laid eggs. The percentage of males, aberrant phenotypes and larval arrests was calculated as the ratio of males/aberrant/larval phenotype on hatched eggs. Data are from at least four independent experiments for each genotype.
O6-BG treatment
The procedure is described in the Supplementary Fig. S1C. Synchronized animals were grown on OP50-seeded NGM plates in the presence of O6-BG (SIGMA) at different concentrations (0.05–2 mM). Young adult worms were picked and transferred onto fresh plates (containing the same concentration of O6-BG than the original plate) every 24 hrs for 72 hrs and embryonic lethality was calculated. The phenotypes were scored on progenies during the development. For each O6-BG concentration at least three independent experiments were performed.
Immunolocalization
Gonads of synchronous young adult hermaphrodites were dissected, fixed, and processed for immunostaining as described35. The primary rabbit antibody against RAD-5136 was used at 1:200, The secondary antibody was the Texas Red anti-rabbit (Invitrogen) used at 1:400. The number of RAD-51 foci in each germ line zone (gonadal tip, mitotic zone, transition zone, early pachytene, mid-pachytene, late pachytene) was recorded.
Image acquisition and analysis
Images were collected using a Leica DM6 fluorescence microscope and Hamamatsu camera under the control of Leica LAS AF 6000 software. Images were deconvolved and analyzed using the Leica LAS AF 6000 software and Image J program. Quantitative analysis of RAD-51 foci and DAPI-stained bodies along the germ line were performed on z series. Optical sections were collected at 0.18 µm and 0.50 µm increments respectively. The t-Student test for independent samples was used for the analysis of apoptosis levels and RAD-51 foci. At least five gonads were quantified for each genotype or condition.
DNA damage sensitivity
For methylation DNA damage, synchronized L4 worms grown on standard plates were transferred to 10 ml of OP50-containing M9 Buffer (3 g/L KH2PO4, 6 g/L Na2HPO4, 5 g/L NaCl, 1 mM MgSO4) with or without N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) or methyl-methane-sulphonate (MMS) and incubated at 20 °C with shaking. After 16 hrs worms were washed 3 times with M9 buffer and transferred to OP50 standard plates for 24 hrs. For each treatment, 9 worms were transferred to three fresh plates (three worms for each plate) and laid eggs and dead embryos were scored for 24 hrs.
For methylation treatment in combination with O6-BG, synchronized animals were grown on OP50-seeded NGM plates in the presence of O6-BG as described before. Young adult worms were picked and transferred on OP50-NGM plates containing MMS (0.5 mM) and incubated at 20 °C. After 16 hrs worms were transferred to OP50 standard plates (three worms for each plate) and laid eggs and dead embryos were scored for 72 hrs.
For camptothecin (CPT) treatment synchronized L4 worms were grown on NGM plates containing 5 μM CPT (Sigma-Aldrich) at 20 °C. The gonads were dissected immediately after CPT-treatment (T0) or incubated in standard conditions for 8 h (T 8) and 24 h (T 24) and analyzed by immunolocalization as described before.
Apoptosis assay
Apoptotic germ corps were measured by Syto-12 vital staining as previously described35. To measure apoptosis levels after DSB induction, synchronized young adult worms were exposed to 20 Gy of γ-rays using a Caesium-137 source. Following treatment, worms were transferred to OP50-seeded plates, incubated at 20 °C and analysed after 2 hrs. To measure apoptosis levels after DNA alkylation damages, worms were treated with MMS and analysed at young adult stage. Each condition was replicated at least three times in independent experiments. Apoptotic corps from at least 80 gonads were quantified for each genotype or condition.
Quantitative analysis of DAPI-stained bodies in diakinesis nuclei
Synchronized adult worms were transferred to glass slides in 15 μl of M9 solution. Samples were permeabilized, fixed and washed with absolute ethanol, then 15 μl of a 2 ng/μl solution of 4′, 6′-diamidino-2-phenylindole hydrochloride (DAPI) diluted in M9 was added. Quantitative analysis was performed on z series of images acquired using a Leica DM6000 fluorescence microscope, Leica DC 350 FX camera under the control of Leica LAS AF 6000 software. Optical sections were collected at 0.50 μm increments.
Results
The agt-2 gene is involved embryonic and post-embryonic development
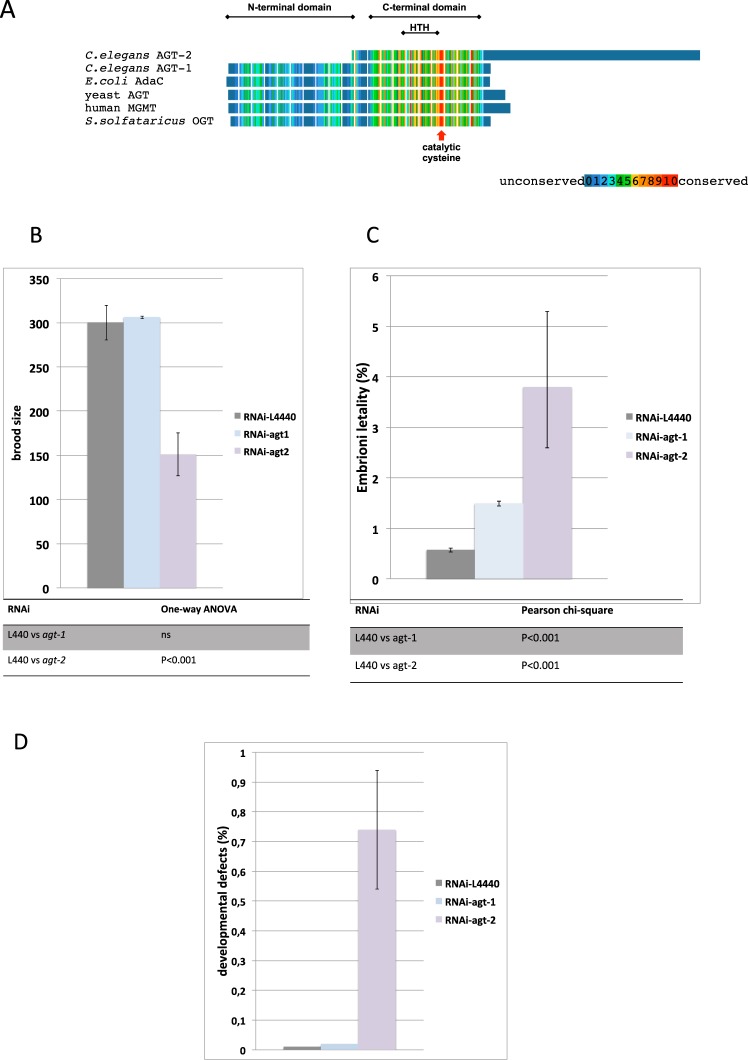
Of the two C. elegans AGT paralogs, the putative AGT-1 protein (ORF Y62E10A.5) is structurally similar to other members of this protein family, whereas AGT-2 (ORF F09E5.13) predicted primary sequence is highly divergent29. This 274 amino acid protein displays significant similarities to canonical AGTs only in the region from residues 62 to 96, which contains the cysteine acceptor site and the DNA helix-turn-helix (HTH) binding motif; even in this region, the active site motif is –PCHP– instead of –PCHR– found in hAGT, and few of the residues in the presumptive HTH motif are identical to the corresponding residues in hAGT (Fig. 1A and Supplementary Fig. 2A). More strikingly, the N-terminal domain found in all other AGTs is totally absent in AGT-2. In contrast, AGT-2 has a much longer C-terminal sequence, not found in any known AGTs, which is rich in proline and basic amino acids and shows weak (about 25%) sequence similarity with human histone 1 C (Fig. 1A and Fig. S2A). Whereas the N-terminal portion did show alkyl-transferase activity in vitro, the whole protein could not be obtained in recombinant form (29 and our unpublished results); thus, the function of the AGT-2 C-terminal domain has not been established.
Figure 1.
(A) Schematic representation of sequence alignment of selected AGTs. The N- and C- terminal AGT domains, the highly conserved catalytic cysteine and the HTH motif are indicated. The long blue C-terminal extension in AGT-2 is the domain with weak similarity with histone H129. The alignment was obtained by the PRALINE program (http://www.ibi.vu.nl/programs/pralinewww/) and adjusted manually to fit the highly divergent AGT-2 sequence. (B–D) Silencing of agt-1 and agt-2 by RNAi. See Supplementary Fig. 1B,C for the scheme of the experiment. Histograms show: brood size (B); embryonic lethality, plotted as a percentage of the hatching of each genotype (C); developmental defects, plotted as a percentage of the hatching of each genotype (D). Statistical analysis is reported below each histogram (ns = not significant). Number of scored P0 worms: N2 (L440) = 27; N2 (RNAi-agt-1) = 27; N2 (RNAi-agt-2) = 36.
Given these puzzling observations, we aimed at investigating the function of the two C. elegans AGT-encoding genes. We thus down-regulated the agt-1 and agt-2 genes by RNAi and subjected interfered animals to phenotypic screening: whereas RNAi of agt-1 had no apparent effect on fertility and embryonic lethality, down regulation of agt-2 resulted in significant reduction of the brood size, and increase in the frequency of dead embryos as well as post-embryonic developmental defects (Fig. 1B–D).
This result suggests that AGT-2 plays a role in C. elegans physiology under normal growth conditions and was unexpected, since all available data point to a function of AGT proteins in repair of alkylating agents. We thus decided to investigate the role of agt-2 in the nematode biology.
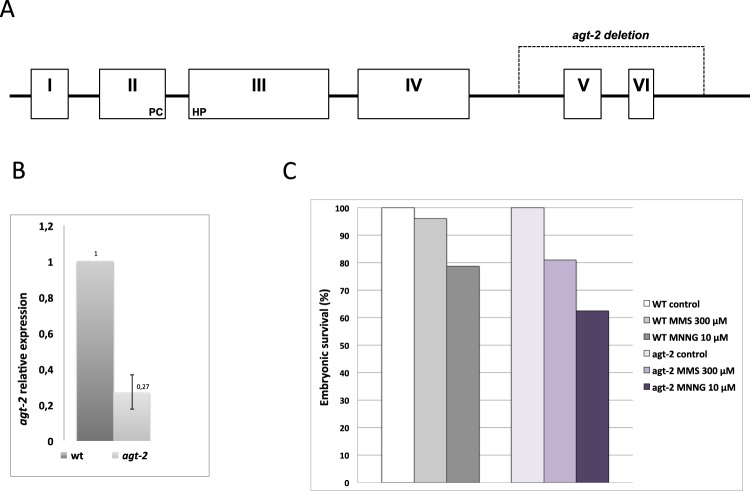
In order to confirm the RNAi results we looked for stable agt-2 mutants in C. elegans genetic banks and retrieved the tm6462 strain, we will refer to as the agt-2 mutant. This strain holds a 369 bp deletion spanning the last two agt-2 exons, the last intron and part of the 3′ UTR (Fig. 2A). If translated, a full-length transcript covering the whole agt-2 (tm6462) allele would eventually result in a protein identical to the wild type AGT-2 protein up to residue 234. This putative deleted protein would contain the predicted catalytic motif and, based on the results obtained in vitro on a shorter truncated protein29, could in principle catalyse the trans-alkylation reaction; however, we cannot currently make any prediction on the effect of the truncation of the C-terminal portion on the protein activity/stability. Quantitative RT-PCR analysis showed that the level of agt-2 transcripts is reduced by about five fold in the agt-2 mutant as compared to the wild type (Fig. 2B), thus suggesting that the tm6462 deletion induces down-regulation or destabilization of the agt-2 mRNA. We conclude that, even if the mutant AGT-2 version encoded by the agt-2 (tm6462) allele was as functional as the wild type protein, its level would be only 20% of that found in the N2 strain. Thus, agt-2 (tm6462) can be considered at least a hypomorphic mutant. A parallel analysis detected no agt-1 transcripts in either wild type or agt-2 strains (data not shown) suggesting that, under the conditions used, this gene is not expressed or its transcript level is below the detection threshold of the experiment.
Figure 2.
(A) Schematic representation of the agt-2 gene structure. Exons are indicated by numbered boxes and introns by black lines. Residues PHCP of the catalytic site (exons II and III) are indicated. The dashed line span exons V and VI represents the portion deleted in the mutant agt-2(tm6462) allele. (B) Analysis of agt-2 gene expression in the N2 background by real-time RT–PCR. Data were normalized to the expression level of the pmp-3 housekeeping gene as previously described35 and expressed as the relative mRNA level compared with the average expression level in wild type animals. The mean of three independent experiments from two biological replicates is shown. (C) Effect of alkylation damage on wild type and agt-2 mutant worms. L4 worms were treated with either MMS or MNNG at the indicated concentrations. Embryonic viability 48 hrs after DNA damage treatment was plotted as a percentage of the hatching normalized by that in untreated worms of each genotype. Number of scored P0 worms: untreated wild type = 9; MMS-treated wild type = 18; MNNG-treated wild type = 18; untreated agt-2 = 18; MMS-treated agt-2 = 18; MNNG-treated agt-2 = 18. (chi-squared test: WT-MMS vs agt-2 MMS P < 0.001; WT-MNNG vs agt-2 MNNG P < 0.001).
Since the agt-1 expression could be subjected to a trigger, we analysed the transcript after DNA damage induction, however no transcript was found after treatment of wild type or agt-2 strains with alkylating agent MMS (data not showed).
We tested the sensitivity of the agt-2 mutant to two monofunctional alkylating agents, namely MNNG and MMS (Fig. 2C). The agt-2 mutant resulted reproducibly, although not dramatically, more sensitive to both drugs, as compared to the wild type (p < 0.001 for both treatment). The moderate sensitivity of the agt-2 mutant to alkylating damage is compatible with the notion that these lesions can be repaired by multiple repair systems37. Thus, these results confirmed that the AGT-2 protein is involved in efficient repair of DNA alkylation lesions and that the tm6462 deletion impairs, at least in part, this activity.
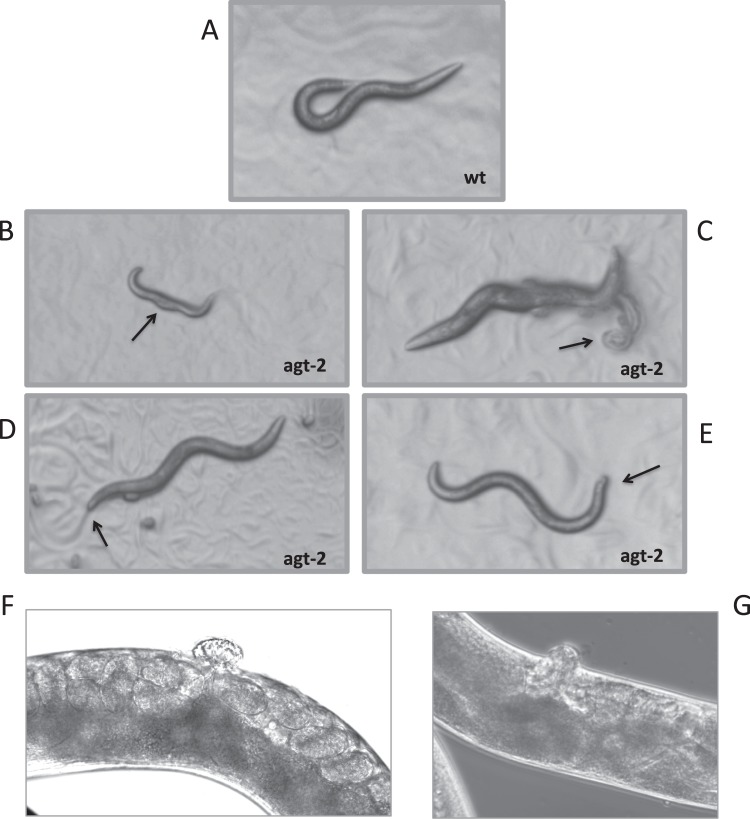
In the above experiment we noticed a higher number of dead progeny in untreated agt-2 as compared to wild type worms (not shown), as observed in the agt-2 RNAi. To confirm this observation, we performed systematic phenotypic screening of agt-2 mutant worms (Table 1). Interestingly, this strain showed significant reduction of brood size and increase in embryonic lethality as compared to the wild type; in addition, we observed elevated rate of larval arrests in the agt-2 population and, among the live progeny, of developmental defects (Table 1 and Fig. 3). In contrast, the incidence of males, reflecting the frequency of X-chromosome non-disjunction in C. elegans meiosis, was similar in the agt-2 and wild type strains (Table 1).
Table 1.
Screening of phenotypes in agt-2 and wild type populations.
| WT | agt-2 | |
|---|---|---|
| Parental | 20 | 28 |
| Laid eggs | 5341 | 5769 |
| Hatched eggs | 5314 | 5699 |
| Brood size | 267 | 206 |
| Dead embryos (%) | 0.5 | 1.2 |
| Males (%) | 0.06 | 0.04 |
| Larval arrest (%) | 0 | 0.9 |
| Developmental defects (%) | 0.04 | 0.8 |
Embryonic lethality was calculated as the percentage of unviable eggs on the total laid eggs. Developmental defects were calculated as the percentage of aberrant larval phenotypes on hatched eggs.
Figure 3.
Developmental defects found in agt-2 mutant populations. (A) A typical N2 worm at L4 stage. (B) larval arrest with a bulging of the body. (C) A dead worm with burst vulva. (D,E) Animals with misshapen tail. (F,G) Magnification of protruding/ectopic vulva.
Collectively, these data suggest that, as expected, agt-2 contributes to C. elegans response to alkylating agents; surprisingly, it is also involved in embryonic and post-embryonic development under normal growth conditions.
AGT-2 protein inhibition induces embryonic lethality and developmental defects
O6-BG is a competitive and highly specific inhibitor of most, although not all AGTs, since it reacts with the active site cysteine forming a covalent adduct, which inactivates irreversibly the protein. In human cells, O6-BG induced AGT inactivation triggers the protein degradation; thus, the inhibitor is being tested to improve the efficacy of chemotherapeutic regimens based on alkylating drugs16. A truncated form of AGT-2 was reported to be sensitive to the inhibitor in vitro29, thus suggesting that the protein might be O6-BG target in vivo as well; however, the effects of O6-BG on live worms have never been tested.
We thus exposed wild type animals to different concentrations of the inhibitor and performed phenotypic screenings (Table 2). Worms treated with the drug at 0.05 mM showed brood size reduction and increased embryonic lethality as well as developmental defects, as compared to control worms; interestingly, these phenotypes were very similar to those observed in agt-2 and agt-2 (RNAi) animals. Higher concentrations of O6-BG (2 mM) exacerbated all phenotypes and in particular induced high rate of embryonic lethality and developmental defects (Table 2).
Table 2.
Screening of phenotypes in wild type worms after treatment with O6-BG at the indicated concentrations.
| Control | O6-BG (0.05 mM) | O6-BG (2 mM) | |
|---|---|---|---|
| Parental | 18 | 18 | 18 |
| Laid eggs | 4046 | 3250 | 3145 |
| Brood size | 225 | 181 | 175 |
| Dead embryos (%) | 0.5 | 2.9 | 15 |
| Developmental defects (%) | 0 | 0.8 | 7 |
The percentage of embryonic lethality was calculated as the ratio of the total unviable eggs on the total laid eggs. The percentage of developmental defects was calculated as the ratio of aberrant larval phenotypes on hatched eggs.
We also tested the sensitivity of O6-BG treated worms to alkylation agent MMS showing an increased embryonic lethality as compared to O6-BG untreated worms.
(wt-MMS = 2.5%; wt-O6-BG-MMS = 4.2%; WT-MMS-O6-BG = 2.4%; Supplementary Fig. 2B).
Taken together the results are consistent with the prediction that the observed phenotypes are due to inactivation and/or degradation of the AGT-2 protein, reinforcing the hypothesis that the protein plays a role in DNA damage repair and also in the important step(s) of C. elegans embryonic and post-embryonic development in the absence of exogenous DNA damage.
AGT-2 is involved in meiotic DSB repair
The above experiments clearly showed that embryonic lethality is increased when the agt-2 gene is mutated or down-regulated, and when wild type animals are treated with an inhibitor of AGT proteins. In general, elevated levels of embryonic lethality can be associated to problems arising during C. elegans meiosis. In order to identify the nature of the agt-2 mutant defect, we analysed the key meiotic steps in these animals. Earlier in meiotic prophase -after premeiotic DNA replication- homologous chromosomes recognize each other and pair. Chromosome pairing is coupled with the assembly of the synaptonemal complex (SC), and is followed by formation of multiple physiological double strand breaks (DSBs) induced by the SPO-11 protein, which trigger meiotic recombination (for recent review see18). Homologous pairing and recombination are tightly coordinated to ensure the proper chromosome inheritance.
To establish whether agt-2 animals have defects in chromosome pairing or synapsis, we performed cytological analysis of mutant worms from the leptotene to diplotene stages of meiosis I, and we found no morphological defects that might suggest homolog alignment or SC problems (data not shown). In addition, we stained mutant and wild type gonads with an antibody against the SYP-1 protein, an SC component38. The SC complex appeared normal in agt-2 mutants (Supplementary Fig. 3), thus suggesting that these latter do not have major problems in homologous chromosome pairing and synapsis.
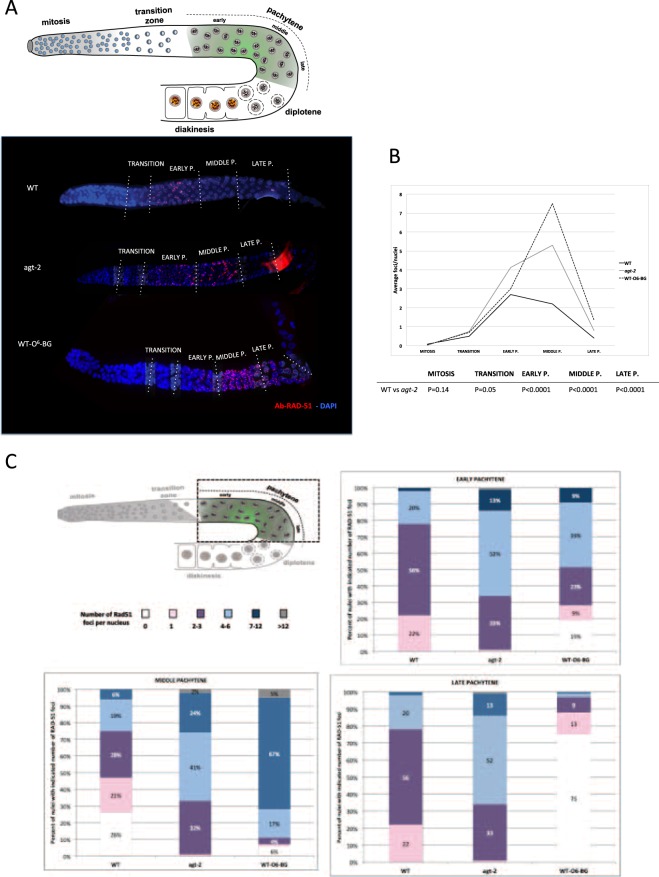
We then wondered whether agt-2 mutants initiate correctly meiotic recombination. To answer this question, we used antibodies against the DNA strand-exchange protein RAD-5136. RAD-51 binds single-stranded regions adjacent to resected DSBs, forming distinct foci, which follow precise kinetics in meiotic prophase and are widely used as a cytological marker of recombination intermediates and DBSs after DNA damages. Increase and/or persistence of such foci reflect a DNA repair defect36,39. We observed that the number of RAD-51 foci was significantly higher in agt-2 as compared to wild type oocytes (Fig. 4A,B). In addition, quantitative analysis of RAD-51 foci in the different zones along the gonad showed that the foci persistence followed a different kinetics in the two strains: in the wild type, the number of RAD-51 foci typically increased from zygotene to early pachytene, declined in mid pachytene and disappeared by late pachytene, indicating that meiotic DSBs had been successfully repaired (Fig. 4B,C). In the mutant, the peak of RAD-51 foci was shifted to mid pachytene, and a significant number of foci persisted until late pachytene. In both strains RAD-51 foci were absent in the mitotic zone (Fig. 4B,C).
Figure 4.
Analysis of RAD-51 foci in C. elegans germline. (A) Upper panel: schematic depiction of a single gonad arm. Germ cells are arranged in a “conveyor belt-like” fashion with mitotic germ cells at the distal end of the gonad arm (mitosis). As cells move along the distal arm they enter meiosis in the transition zone and progress through the different stages of pachytene. Starting from transition zone, the DBSs are formed and then repaired along the pachitene. In diplotene through diakinesis, the paired chromosomes condense to form bivalents that will be packaged into each oocyte. In the lower panel, representative images of DAPI stained DNA (blue) and RAD-51 foci (red) detected with the anti-RAD-51 antibody in the indicated zones of the gonad (P.: pachytene). (B,C) Quantitative analysis of RAD-51 foci distribution in WT, WT-O6-BG and agt-2 animals. (B) The y-axis shows the average of RAD-51 foci per nucleus; the x-axis represents the position (zone) along the gonad. At least five gonads were scored for each genotype or treatment. Statistical analysis was carried out using the two-tailed paired Student’s t tests27,35. A significance value of p < 0.05 was used. (C) Upper panel: schematic drawing of pachytene indicating the position of the zones scored for RAD-51 foci. In the lower panel, stacked bar graph depicting quantitation of RAD-51 foci in early pachytene nuclei (EARLY P.), middle pachytene nuclei (MIDDLE P.) and late pachytene nuclei (LATE P.) of indicated genotypes. Different colored segments in the histograms represent the percentage of nuclei scored with the indicated number of RAD-51 foci (see the color code on the top of the graph).
Interestingly, wild type animals treated with 2 mM O6-BG also showed increased number and persistence of RAD-51 foci as compared to controls; in this case, the number of RAD-51 foci per nucleus was even higher than in the agt-2 mutant (Fig. 4A–C).
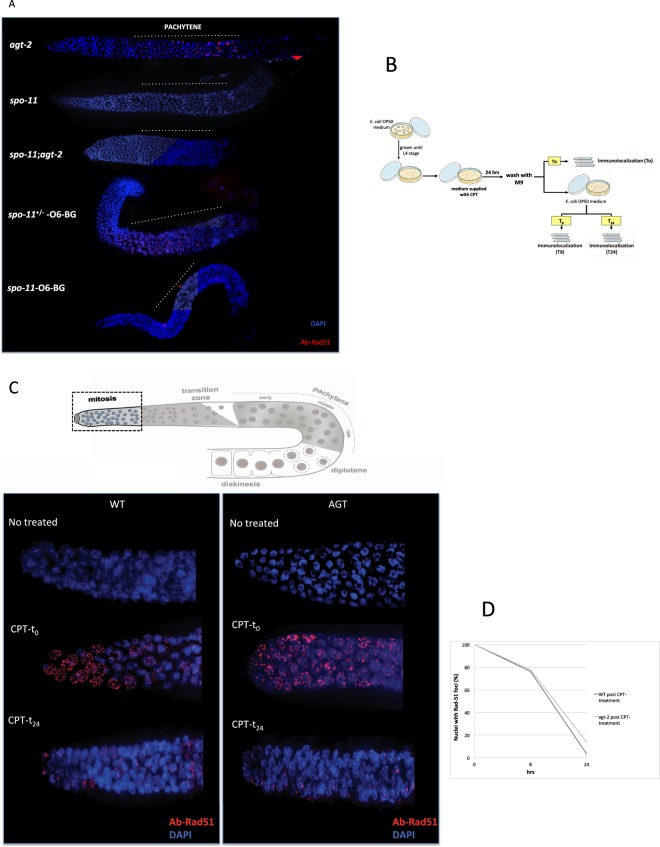
The fact that RAD-51 foci were not observed in mitotically dividing oocytes in the agt-2 mutant or in the presence of O6-BG suggests that the defect we are observing is meiotic and not a consequence of problems occurring during premeiotic replication. In order to confirm this hypothesis, we generated an agt-2; spo-11 double mutant and analysed the presence of RAD-51 foci in this strain: as shown in Fig. 5A, no RAD-51 foci were observed in this mutant, as in the spo-11 strain. In addition, when the spo-11 mutant was treated with O6-BG, virtually no RAD-51 foci were observed, whereas, as expected, a high number of RAD-51 foci were present in meiotic prophase in the spo-11/+ heterozygous treated with O6-BG (Fig. 5A).
Figure 5.
(A) Elevation of RAD-51 foci in the absence of a fully functional AGT-2 is dependent on meiotic DSBs. Gonads from young adult hermaphrodites were dissected and processed for immunofluorescence using the anti-RAD-51 antibody (red). DNA was visualised by DAPI staining (blue). All gonads are orientated with the distal end to the left and the proximal end to the right. Dashed lines shows the pachytene zone. Representative images of the indicated genotype or treatment are shown. At least three gonads were analyzed for each genotype or treatment. The spo-11+/− heterozygote was used as a positive control for the O6-BG treatment. (B–D) Formation of RAD-51 foci in germline nuclei in response to CPT. (B) Schematic representation of CPT treatment: L4-stage WT and agt-2 mutant were treated with 5 μM CPT for 24 h. (C) Upper panel: Schematic depiction of C. elegans germline highlights the position of mitotic nuclei scored for RAD-51 foci. Lower panel: representative images of fixed mitotic nuclei at the distal end of the germline immunostained with RAD-51 antibody and counterstained with DAPI. The WT and agt-2 mutant gonads were processed immediately post CPT treatment (t0) or after 24 h (t24). (D) The percentage of germline nuclei with RAD-51-positive foci in the gonad tip were determined at indicated times post CPT treatment.
In conclusion, the increased number of SPO-11-dependent RAD-51 foci in meiotic prophase and their persistence indicates that a defect in repairing meiotic DSBs occurs when agt-2 is mutated or wild type animals are treated with the AGT inhibitor. These results suggest a role for AGT-2 in the efficient repair of SPO-11-dependent meiotic DSBs.
The defects observed in agt-2 mutant may also reflect a role of AGT-2 in sensing the presence of DNA damage. To test this possibility we checked the RAD-51 foci formation after DNA damage (DBSs) in mitotic germline nuclei. The DBSs were inducted by the topoisomerase I inhibitor camptothecin (CPT), which causes replication fork stalling and collapse in actively cycling cells (Fig. 5B-D). After treatment with CPT (CPT-t0) the RAD-51 foci are largely diffuses in mitotic nuclei both in wild type and agt-2 mutant, suggesting that the accumulation of RAD-51 at damage-inducted DBSs doesn’t depends from AGT-2 (Fig. 5C,D). We also examined the kinetic of DBSs repair from 24 hrs post CPT treatment (CPT-t24) demonstrating that more then 80% of the nuclei did not show RAD-51 foci after 24 hrs, both in WT and agt-2 mutant (Fig. 5C,D). Taken together these results imply that AGT acts upon spo-11 dependent DNA breaks and is dispensable for sensing and repair the DNA damage.
AGT-2 involvement in crossover formation and inter-sister recombination
By late pachytene, homologous recombination leads to the formation of one and only one inter-homolog crossing over (CO) event per each chromosome. The rest of SPO-11 induced DSBs are repaired by non-CO mechanisms. During late prophase, SC disassembles and homologous chromosomes remain linked by chiasmata (physical attachments provided by COs in combination with sister chromatid cohesion). At diakinesis each condensed chromosome pair forms a compact body, which can easily visualized by DNA staining (e. g. DAPI); diakinetic nuclei normally contain six such bodies18. In contrast, if crossing over is prevented either through loss of the synaptonemal complex (SC; e. g., in syp mutants) or a block to meiotic DSB formation (e. g., in spo-11 mutants), then 12 unsynapsed chromosomes (univalents) are observed at diakinesis. Thus, the morphology and number of diakinesis chromosomes are a useful readout of meiotic recombination defects.
Cytological analysis revealed that most of the diakinetic nuclei in agt-2 mutant contain the wild-type complement of six bivalents at diakinesis; however, a significant number of agt-2 nuclei had seven and, occasionally, eight DAPI stained bodies (Fig. 6A,B). The presence of more than six DAPI-stained bodies is usually observed in older wild type animals or in meiotic mutants in which proper segregation is inefficient due to lack of CO between two homologs. For instance, brc-1 mutants are specifically defective in establishing a correct CO between the X chromosomes, which ultimately results in increased frequency of males20,22. Opposite to brc-1, agt-2 has a normal frequency of males, while showing increased embryonic lethality and larval arrests (Table 1), which might result from missegregation of autosomes. This result might suggest that agt-2 is largely dispensable for CO on the X chromosome, although it seems to contribute in establishing COs on autosomes.
Figure 6.
Analysis of DAPI stained bodies in diakinesis nuclei of the indicated genotypes. (A) Upper panel: Schematic depiction highlights the formation of bivalents (pairs of homologs) during dikinesis. Lower panel: Representative images of single DAPI stained oocyte nucleus at diakinesis from WT and agt-2 mutant. The number of DAPI-stained bodies in each nucleus is shown. Scale bar: 2 μm. (B,C) Quantitation of DAPI-stained bodies. The y-axis in the histograms represents the percentage of nuclei in each class and the x-axis indicates the number of DAPI-stained bodies. Statistical analysis was carried out using one-way ANOVA multiple comparison test. A significance value of P < 0.05 was used; ns = not significant. (wt vs agt-2: p = 0.0015; syp-2 vs agt-2; syp-2: p < 0.001; agt-2; syp-2 vs agt-2; syp-2; lig.4: ns). Data are from five independent experiments. (D) Representative images of DAPI stained oocyte nuclei at diakinesis from indicated genotypes Scale bar: 2 μm. Nuclei for each genotype: N2 = 285, agt-2 = 250, syp-2 = 140, agt-2; syp-2 = 240, agt-2; syp-2; lig-4 = 100, spo-11: 100, agt-2; spo-11: 100.
In syp-2 mutants meiotic DSBs occur normally; however, because of the absence of SC, which prevent homologous chromosome pairing, meiotic DSBs persist until late prophase, and can eventually be repaired using sister chromatids as template, maintaining chromosome integrity. Thus, syp-2 meiocytes that reach diakinesis show 12 almost intact univalents17. Mutants with problems in the use of the sister chromatid as a template for meiotic DSB repair, such as brc-1, show higher number of univalent at diakinesis20,22.
We found that only about half of agt-2; syp-2 diakinetic nuclei contain the expected 12 univalents, whereas a significant number of oocytes contained less then 12 DAPI stained bodies or unstructured, misshapen chromatin (Fig. 6C,D). These defects were not observed if meiotic DSBs were prevented: the expected complement of 12 univalents was found in the agt-2, spo11 double mutant (Supplementary Fig. S4A,B), thus suggesting that the anomalies found in agt-2, syp-2 diakinetic nuclei depend on meiotic DSBs and are due to inefficient repair of these lesions by using the sister chromatid as a template.
The reduced number of diakinetic chromosomes observed in the agt-2; syp-2 mutants contrasts with chromosome fragmentation observed in brc-1; syp-2 mutants, and is rather reminiscent of fcd-2; syp-2 diakinetic oocytes27. These cells possess between 6 and 12 DAPI-stained bodies presumably due to inappropriate engagements between nonhomologous chromosomes during DNA repair, which were shown to be dependent on the alternative pathway for repair of DSBs, namely nonhomologous end joining (NHEJ). In order to establish whether NHEJ might play a role in the diakinetic abnormalities found in agt-2, we obtained the triple mutant agt-2; syp-2; lig-4, in which the NHEJ lig-4 gene was mutated. No substantial rescue of the agt-2 diakinetic defects was observed in the triple mutant (Fig. 6C). Thus, although agt-2; syp-2 diakinetic oocytes do show problems in efficient use of sister chromatids to repair meiotic DSBs, this defect results neither in fragmentation, as observed in the absence of BRC-1, nor in NHEJ-dependent illegitimate repair between nonhomologous chromosomes, as in the absence of FCD-2. One possibility is that promiscuous non covalent chromosome aggregations occur in a fraction of agt-2; syp-2 oocytes which are unable to perform correct sister-chromatid dependent homologues recombination (HR) to repair meiotc DSBs; these aggregations may result in reduction of the number of diakinetic chromosomes or, at most, massive chromosome destructuring (Fig. 6C,D).
In conclusion, the results reported in this paragraph indicate that agt-2 is involved in both efficient CO on the autosomes and efficient inter-sister repair of meiotic DSBs.
AGT-2 deficiency induces high levels of germ line apoptosis
Under normal growth conditions, about half of the oocytes in the C. elegans germ line undergo physiological cell death by the end of pachytene40. In addition, failure to faithfully execute meiosis triggers two checkpoints, called DNA damage and synapsis checkpoint, respectively41–43. Accumulation of DNA damage in the germ line, either in response to genotoxic stress, or upon failure to execute faithfully meiotic recombination triggers the DNA damage checkpoint, which result in cell cycle arrest at the G2 phase in order to allow for DNA repair. However, when level of DNA damage is excessive, cells might undergo apoptosis. Checkpoint-induced germ line apoptosis depends on the C. elegans p53 orthologue, CEP-1, removes defective meiotic nuclei to prevent aneuploidy and defective gametes and is restricted to late pachytene stage cells44.
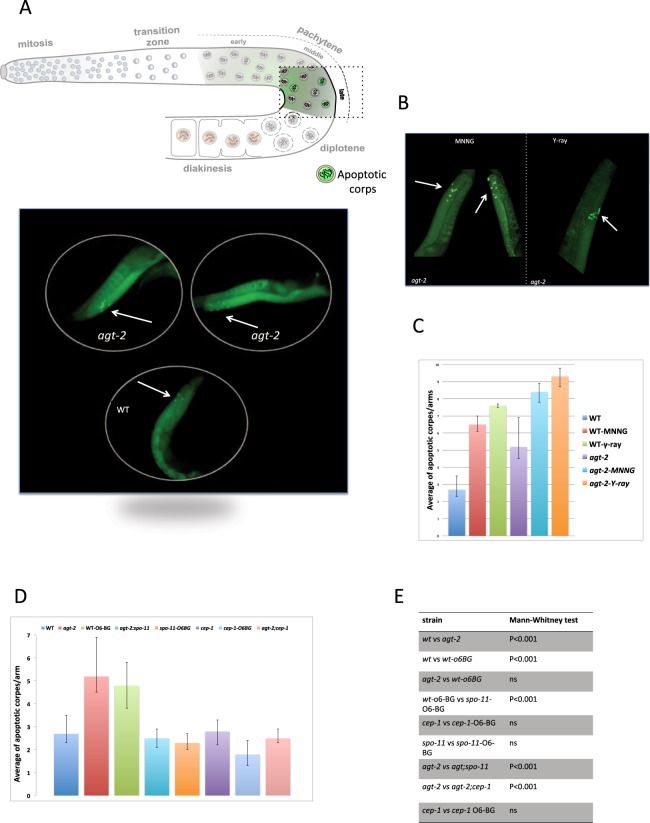
Interestingly, we found that the average number of apoptotic pachytene nuclei was significantly higher in agt-2 mutants than in the wild type during unperturbed growth (Fig. 7A,C and Supplementary Fig. 5). The number of apoptotic nuclei in agt-2 animals was further increased by exposure to ionizing radiation or MNNG (Fig. 7B,C), indicating that the mutant strain is able to respond normally to DNA damage activating the DNA damage checkpoint (Fig. 7D,E).
Figure 7.
Analysis of apoptosis in the indicated genotypes or treatment. (A) Upper panel: germ cell apoptosis occurs mainly within the gonad loop region nearly as developing oocytes exit the pachytene stage of meiotic prophase (late pachytene). Schematic depiction shows the position of apoptotic corps highlighted in green. Lower panel: representative images showing apoptotic corps stained with SYTO-12, in the late pachytene zone of wild type and agt-2 gonads. The arrows indicate the location of apoptotic nuclei. (B) Representative images showing apoptotic corps in the late pachytene zone of agt-2 gonads treated with the indicated damaging agent; images were taken 16 hrs and 2 hrs after MNNG (10 μM) or γ-ray, respectively. (C,D) Quantification of apoptotic corps. Histograms show the average of apoptotic corps per gonad arm in the indicated genotypes or treatment. Error bars represent the standard error of the means (S.E.M.) calculated from at least three independent experiments. (E) Statistical comparisons between data sets were conducted using a two-tailed Mann-Whitney test. A significance value of P < 0.05 was used; ns = not significant. At least 80 gonads were scored for each condition.
We were therefore interested in determining whether the increased apoptosis observed in agt-2 mutant was mediated by the cep-1 pathway. For this, we generated double mutant strain agt-2; cep-1 and then analyzed its phenotypes. We found that the embryonic lethality and brood size of the double mutant were similar to those observed in the agt-2 mutant (brood size = 207 ± 6; embryonic lethality = 1.3% ± 0.2). In contrast to agt-2 mutant, the increase in the number of apoptotic corps was not observed in agt-2; cep-1 either under physiological conditions or after DNA damages induction. In addition, wild type worms treated with 2 mM O6-BG showed the same increase in the number of apoptotic nuclei found in untreated agt-2 mutants (Fig. 7D,E and Supplementary Fig. 5). However, O6-BG did not induce apoptosis increase when cep-1 was mutated (Fig. 7D,E). In order to clarify the nature of the damage triggering apoptosis elevation we analysed the apoptosis level in the absence of SPO-11: in agt-2; spo-11 mutants and in spo-11 mutants treated with O6-BG the level of apoptosis was similar to that found in untreated wild type animals (Fig. 7D,E).
Taken together, these results suggest the elevated apoptosis levels observed in the agt-2 mutant or when the wild type was treated with the AGT inhibitor depend on inefficient repair of meiotic DSBs, which trigger the DNA damage checkpoint.
Discussion
The predicted C. elegans AGT-2 protein has a peculiar primary structure, as compared with canonical AGT proteins; we here demonstrated that it also plays unexpected roles in the nematode biology. We have used different strategies to reduce or hamper AGT-2 activity: we analyzed an agt-2 deletion strain, performed RNAi to down-regulate agt-2 expression and used the AGT inhibitor O6-BG. In all circumstances we found the same range of relevant phenotypes: (i) brood size reduction; (ii) elevation of embryonic lethality and rate of developmental defects; (iii) accumulation and persistence of SPO-11 dependent RAD-51 foci; (iv) increased levels of SPO-11 and CEP-1 dependent germ line apoptosis. The increase in embryonic lethality and larval arrests, combined with the presence of more than six DAPI-stained bodies at diakinesis, suggests inefficient segregation of autosomes. In addition, we also evidenced that mutation of agt-2 causes inefficient repair of meiotic DSBs using sister chromatids as template.
Collectively, these results suggest that AGT-2 is involved in efficient processing of meiotic DSBs, and that down-regulation or inactivation of the protein results in increased number of meiocytes that are incompatible with producing viable offspring and are eliminated by apoptosis.
Our results point to a role for AGT-2 in efficient conversion of HR intermediates into post-strand exchange products in meiosis. This result was unexpected, since the body of evidence available from experiments in other organisms suggests a specific role for AGT proteins in repair of alkylation damage. Experiments in vitro29 and evidence in vivo (this paper) suggest that AGT-2 is endowed with DNA-alkyl transferase activity. Indeed, agt-2 mutant animals show increased sensitivity to alkylating agents and, most important, treatment of wild type animals with the AGT inhibitor O6-BG elicits the same phenotypes observed when the agt-2 gene is mutated or down-regulated.
It is currently not clear whether the meiotic phenotypes observed are the consequence of the inactivation or impairment of the AGT-2 biochemical activity, and is difficult to envisage a role for such a specific activity in meiotic DSBs processing. One possibility is that AGT-2 plays a double role, i. e., it might catalyse direct repair of alkylating damage, thanks to its biochemical activity, and participate in the processing of meiotic DSBs by a still unclear mechanism. In this view, it is interesting to recall that several other C. elegans DNA repair genes have been implicated in meiotic recombination. These include, among others, brc-1, fcd-2, msh-4 and msh-522,27,36,45. The exact role of these genes in meiosis is not completely understood and is likely distinct. For instance, brc-1 mutants show problems in X-chromosome segregation and inter-sister recombination20,22; fcd-2 mutants compensate defects in HR by up-regulation of the NHEJ activity27; msh-4 and msh-5 mutants do not perform interhomolog COs and do not activate DNA-damage induced apoptotic response45. Notably, the range of the agt-2 mutant phenotypes is not completely overlapping to that of any of the above mutants.
Data on the role of AGT proteins besides alkylation repair in other organisms are scarce. Knock out mice are viable and show no apparent defects, apart from higher sensitivity to alkylating agents-induced tumours14,15, although careful inspection of meiotic details in these animals has not been reported. On the other hand, several lines of evidence suggest that AGT proteins might participate in a number of important cellular functions. Proteomic analysis showed interaction of hAGT with a variety of cellular proteins46. Of particular interest is the interaction with proteins involved in DNA replication, recombination and repair (such as MCM2, PCNA, ORC1, DNA polymerase d, MSH-2, and DNA-dependent protein kinase), as well as histones. Notably, purified histone H1 and a combined histone fraction stimulate the AGT-catalyzed reaction in vitro29, an intriguing result in light of the peculiar presence, in AGT-2, of a histone-like domain, which raises the possibility that this domain has regulatory functions.
Intriguingly, in mouse embryonic fibroblasts and human cell lines, interaction of AGT with BRCA2 induced degradation of both proteins after treatment with alkylating agents or O6-BG47. Like BRCA2, the C. elegans hortholog BRC-2 facilitates nuclear localization of RAD-51 and its loading onto DNA21. This observation suggests a link between AGT-2 and the BRC-2-RAD-51 complex, which needs to be tested in further experiments.
Finally, AGT-2 might play a signaling role. This role is reminiscent of ATL proteins, that are very similar to AGTs, but catalytically inactive due a substitution of the active site cysteine. These proteins are believed to act as signal molecules retrieving other factors to damaged sites48. Thus, AGT-2 might function to integrate different DNA damage and repair signals, to ultimately ensure and maintain genomic stability.
Supplementary information
Acknowledgements
This paper is dedicated to the memory of our dear colleague Maria Ciaramella who recently passed away: she inspired, mentored and supported all of as. This study was partially supported by MIUR FIRB-Futuro in Ricerca RBFR12OO1G_002 “Nematic” Project. M.S. worked at this project as part of his PhD thesis in Biology at University of Naples “Federico II”. We thank the Mitani Laboratory (National Bioresource Project for the Experimental Animal Nematode C. elegans) for the agt-2 strain and Elia Di Schiavi for agt-1 and agt-2 RNAi bacterial strains from the Ahringer library. We thank Giovanni del Monaco for technical support.
Author Contributions
M.S. and C.F. designed and conducted the experiments, analyzed the results; A.A. and G.P. designed the experiments and analyzed the results N.R. conducted the experiments; M.C. and A.V. designed the experiments, analyzed the results and wrote the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mario Serpe and Chiara Forenza contributed equally.
Maria Ciaramella is deceased.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-43394-1.
References
- 1.Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat. Res. 2000;462:83–100. doi: 10.1016/S1383-5742(00)00017-X. [DOI] [PubMed] [Google Scholar]
- 2.Pegg AE. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem. Res. Toxicol. 2011;24:618–639. doi: 10.1021/tx200031q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soll JM, Sobol RW, Mosammaparast N. Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities. Trends Biochem. Sci. 2017;3:206–218. doi: 10.1016/j.tibs.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Q, Kanugula S, Pegg AE. Function of domains of human O6-alkyl-guanine-DNA alkyltransferase. Biochemistry. 2005;44:15396–15405. doi: 10.1021/bi051460d. [DOI] [PubMed] [Google Scholar]
- 5.Miggiano R, et al. Biochemical and structural studies of the Mycobacterium tuberculosis O6-methylguanine methyltransferase and mutated variants. J. Bacteriol. 2013;195:2728–2736. doi: 10.1128/JB.02298-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miggiano R, et al. Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA. Biochem. J. 2016;473:123–133. doi: 10.1042/BJ20150833. [DOI] [PubMed] [Google Scholar]
- 7.Miggiano R, et al. Every OGT Is Illuminated … by Fluorescent and Synchrotron Lights. Int. J. Mol. Sci. 2017;5(12):18. doi: 10.3390/ijms18122613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perugino G, et al. Activity and regulation of archaeal DNA alkyltransferase: Conserved protein involved in repair of DNA alkylation damage. J. Biol. Chem. 2012;287:4222–4231. doi: 10.1074/jbc.M111.308320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perugino G, et al. Structure-function relationships governing activity and stability of a DNA alkylation damage repair thermostable protein. Nucleic Acids Res. 2015;43:8801–8816. doi: 10.1093/nar/gkv774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrone C, et al. Interdomain interactions rearrangements control the reaction steps of a thermostable DNA alkyltransferase. Biochim. Biophys. Acta. 2017;1861:86–96. doi: 10.1016/j.bbagen.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Drabløs F, et al. Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair. 2004;11:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23:823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 13.Morales-Ramírez P, Vallarino-Kelly T, Cruz-Vallejo VL. Effect of O6-chloroethylguanine DNA lesions on the kinetics and mechanism of micronucleus induction in vivo. Environ Mol Mutagen. 2010;51:236–42. doi: 10.1002/em.20538. [DOI] [PubMed] [Google Scholar]
- 14.Blumenthal DT, et al. A Phase III study of radiation therapy (RT) and O6-benzylguanine + BCNU versus RT and BCNU alone and methylation status in newly diagnosed glioblastoma and gliosarcoma: Southwest Oncology Group (SWOG) study S0001. Int J Clin Oncol. 2015;20:650–8. doi: 10.1007/s10147-014-0769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bugni JM, Meira LB, Samson LD. Alkylation-induced colon tumorigenesis in mice deficient in the Mgmt and Msh6 proteins. Oncogene. 2009;28:734–41. doi: 10.1038/onc.2008.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabik CA, Njoku MC, Dolan ME. Inactivation of O6-alkylguanine DNA alkyltransferase as a means to enhance chemotherapy. Cancer Treat Rev. 2006;32:261–276. doi: 10.1016/j.ctrv.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Hillers KJ, Jantsch V, Martinez-Perez E, Yanowitz JL. Meiosis. WormBook. 2017;4(2017):1–43. doi: 10.1895/wormbook.1.178.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose A. Replication and repair. WormBook. 2014;4:1. doi: 10.1895/wormbook.1.54.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Muse T, Boulton SJ. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 2005;24:4345–55. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulton SJ, et al. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 2004;14:33–39. doi: 10.1016/j.cub.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 21.Martin JS, et al. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 2005;25:3127–39. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamo A, et al. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008;9:287–292. doi: 10.1038/sj.embor.7401167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko E, Lee J, Lee H. Essential role of brc-2 in chromosome integrity of germ cells in C. elegans. Mol. Cells. 2008;26:590–4. [PubMed] [Google Scholar]
- 24.Lee KY, et al. Developmental stage- and DNA damage-specific functions of C. elegans FANCD2. Biochem. Biophys Res. Commun. 2007;352:479–85. doi: 10.1016/j.bbrc.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 25.Youds JL, et al. Rose AM. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol. Cell. Biol. 2008;28:1470–9. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youds JL, Barber LJ, Boulton SJ. C. elegans: a model of Fanconi anemia and ICL repair. Mutat. Res. 2009;668:103–116. doi: 10.1016/j.mrfmmm.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Adamo A, et al. Preventing non homologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 28.Lee KY, Chung KY, Koo HS. The involvement of FANCM, FANCI, and checkpoint proteins in the interstrand DNA crosslink repair pathway is conserved in C. elegans. DNA Repair (Amst). 2010;9:374–82. doi: 10.1016/j.dnarep.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 29.Kanugula S, Pegg AE. Novel DNA repair alkyltransferase from Caenorhabditis elegans. Environ. Mol. Mutagen. 2001;38:235–43. doi: 10.1002/em.1077. [DOI] [PubMed] [Google Scholar]
- 30.Wood, W. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory (1988).
- 31.Gengyo-Ando K, Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys Res. Commun. 2000;269:64–9. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- 32.Livak KG, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Timmons A. Fire Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 34.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 35.Germoglio Marcello, Adamo Adele. A Role in Apoptosis Regulation for the rad-51 Gene of Caenorhabditis elegans. Genetics. 2018;209(4):1017–1028. doi: 10.1534/genetics.118.301152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colaiácovo MP, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–74. doi: 10.1016/S1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 37.Klapacz J, et al. Contributions of DNA repair and damage response pathways to the non-linear genotoxic responses of alkylating agents. Mutat. Res. Rev. Mutat. Res. 2016;767:77–91. doi: 10.1016/j.mrrev.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–42. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, La Volpe A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–9. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 41.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/S1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 42.Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310:1683–6. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- 43.Bailly A, Gartner A. Germ cell apoptosis and DNA damage responses. Adv. Exp. Med. Biol. 2013;757:249–76. doi: 10.1007/978-1-4614-4015-4_9. [DOI] [PubMed] [Google Scholar]
- 44.Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 2001;11:1722–1727. doi: 10.1016/S0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 45.Silva N, Adamo A, Santonicola P, Martinez-Perez E, La Volpe A. Pro-crossover factors regulate damage-dependent apoptosis in the Caenorhabditis elegans germ line. Cell. Death Differ. 2013;20:1209–18. doi: 10.1038/cdd.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niture SK, Doneanu CE, Velu CS, Bailey NI, Srivenugopal KS. Proteomic analysis of human O6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry. Biochem. Biophys Res. Commun. 2005;337:1176–84. doi: 10.1016/j.bbrc.2005.09.177. [DOI] [PubMed] [Google Scholar]
- 47.Philip S, et al. Degradation of BRCA2 in alkyltransferase-mediated DNA repair and its clinical implications. Cancer Res. 2008;68:9973–9981. doi: 10.1158/0008-5472.CAN-08-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reissner T, Schorr S, Carell T. Once overlooked, now made visible: ATL proteins and DNA repair. Angew. Chem. Int. Ed. Engl. 2009;48:7293–5. doi: 10.1002/anie.200904042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.