Abstract
The multiplicity of cell types comprising multicellular organisms begs the question as to how cell type identities evolve over time. Cell type phylogenetics informs this question by comparing gene expression of homologous cell types in distantly related taxa. We employ this approach to inform the identity of larval skeletogenic cells of echinoderms, a clade for which there are phylogenetically diverse datasets of spatial gene expression patterns. We determined ancestral spatial expression patterns of alx1, ets1, tbr, erg, and vegfr, key components of the skeletogenic gene regulatory network driving identity of the larval skeletogenic cell. Here we show ancestral state reconstructions of spatial gene expression of extant eleutherozoan echinoderms support homology and common ancestry of echinoderm larval skeletogenic cells. We propose larval skeletogenic cells arose in the stem lineage of eleutherozoans during a cell type duplication event that heterochronically activated adult skeletogenic cells in a topographically distinct tissue in early development.
Subject terms: Phylogenetics, Evolutionary developmental biology
Eric Erkenbrack and Jeffrey Thompson have performed ancestral state reconstructions of echinoderm larval skeletogenic cells to examine their relatedness and provide insights into cell-type evolution. They propose that larval skeletogenic cells originated in the stem lineage of eleutherozoans.
Introduction
Cell types are evolutionary units that have diversified in structure and function since the dawn of multicellularity1. During development, generational iterations of cell types are established by gene regulatory networks (GRNs) comprised of regulatory molecules, e.g., transcription factors and signaling pathways, interacting with cell type-specific DNA regulatory elements2 to control gene expression. Similarly, in evolution, cell types form lineages of common ancestry that are maintained over vast expanses of evolutionary time by networks of coding and noncoding regulatory interactions called cell type identity networks1,3. The diversity of functionally distinct cells in multicellular organisms suggests that their underlying identities are labile enough to generate novelty but also rigid enough to buffer change. Understanding how cell type identity evolves is thus key to explaining cell type diversity. It has been proposed that novel cell types are generated by cell type splitting events that give rise to sister cell types from ancestral cell types4, a model which has been advanced to explain the origins of vertebrate ciliary photoreceptor cells4 and mammalian endometrial stromal fibroblasts5,6. A putative mechanism of cell type splitting is cell type duplication, during which sister cell types arise within different developmental lineages or developmental contexts. Whereas it is clear how these proposed mechanisms would contribute to cell type diversity, detailed examples are still uncommon in the literature.
Cell-type phylogenetics and phylogenetic comparative methods have the potential to resolve questions regarding the evolutionary origin of cell types, and the evolution of the larval skeletogenic cells of echinoderms offers one such opportunity. These cells are specified by an extensively studied GRN7–20 and occur in indirect developing larvae of three of the five classes of echinoderms: ophiuroids (brittle stars), holothurians (sea cucumbers), and echinoids (sea urchins). In echinoids and ophiuroids, larval skeletogenic cells synthesize an elaborate larval skeleton that aids in structural support, feeding, and locomotion21. The evolutionary relatedness of these cells and the homology of larval skeletons has long been a point of contention, with various arguments posited for22–24 or against25–31 homology. As all fossil and extant echinoderms possess adult skeletogenic cells and only a subset of echinoderm lineages are known to possess larval skeletogenic cells32,33, there is agreement that adult skeletogenic cells evolved first. One hypothesis is that heterochronic activation of the adult skeletogenic GRN during early development underlay the origin of the larval skeletogenic cell33. It is still unclear where and how many instances of this heterochronic activation occurred in the echinoderm phylogeny. As the most recent phylogeny of echinoderms places asterozoans (asteroids+ophiuroids) as a sister clade to echinozoans (echinoids+holothurians), it is widely thought that the elaborate larval skeletons in echinoids and ophiuroids are the result of independent evolutionary events13,19,29,34,35. Importantly, only recently have developmental gene expression data for holothurians come to light20. These sea cucumbers also possess larval spicules, however, they do not elaborate into a larval skeleton24, suggesting that their inclusion may help clarify the evolutionary relatedness of larval skeletogenic cells.
Here, we frame spatial gene expression data of regulatory genes driving euechinoid larval skeletogenic cell identity from numerous echinoderms in the context of cell type evolution to inform the relatedness of echinoderm larval skeletogenic cells. We collated spatial gene expression patterns for regulatory genes that are important in specification of euechinoid larval skeletogenic cells. We utilized ancestral state reconstruction to estimate the probability of extant spatial gene expression patterns at each node of the eleutherozoan echinoderm phylogeny. Our analyses are consistent with the hypothesis that larval skeletogenic cells arose once in the stem lineage of eleutherozoan echinoderms. We propose that this event was a cell-type duplication event involving activation of the adult skeletogenic cell during early development. This evolutionary event gave rise to a sister cell type, the larval skeletogenic cell, that was subsequently individuated or lost in different lineages of extant eleutherozoans. Our analysis affords a method to rigorously determine ancestral states of spatial gene expression patterns, thereby revealing how cell-type identity changes over vast expanses of evolutionary time.
Results
Gene selection and ancestral state reconstruction
We conducted ancestral state reconstruction on spatial expression patterns from early larval stages and on regulatory genes for which there exist data in at least five taxa in widely diverged clades (see Methods). Experimental studies have revealed transcription factors, e.g., alx1, erg, ets1, and tbrain, and signaling pathways, e.g., VEGF signaling, that are critical for larval skeletogenic cell specification35. The granular molecular detail of this process in the model euechinoid sea urchin Strongylocentrotus purpuratus has motivated comparative evolutionary developmental research in numerous phylogenetically distant echinoderms36–39. We assembled a database of spatial gene-expression data for similar timepoints in early development for alx1, erg, ets1, tbrain, and vegfr, regulatory genes that underlie specification of these cells based on the published GRN at http://echinobase.org/endomes/, and scored spatial distribution of their expression as character states (Fig. 1 and Supplementary Data 1). We used phylogenetic comparative methods40–44 to reconstruct the ancestral spatial expression patterns of regulatory genes in eleutherozoan echinoderms. To explicitly frame spatial gene expression data in the context of echinoderm phylogeny, we included all species that have both spatial gene-expression data for regulatory genes critical to skeletogenic cell specification as well as gene sequences available for divergence time estimation (see Methods). We used a backbone tree from a composite of published echinoderm molecular phylogenies45,46, and time calibrated our trees using Bayesian fossil-calibrated divergence time estimation in BEAST47,48 (Supplementary Fig. 1). Having obtained time-calibrated estimates of branch lengths for all trees in the posterior distribution of our divergence time estimation analysis, we used Markov models to estimate ancestral states of spatial regulatory gene deployment. We estimated ancestral states at every internal node of our phylogeny on a random sample of 10,000 trees from our posterior in a Bayesian framework42. Analyses were carried out using the Bayestraits wrapper in R (see “Methods”; http://rgriff23.github.io/projects/btw.html). By reconstructing ancestral states in a Bayesian framework, we explicitly accounted for differences in branch lengths between our 10,000 posterior trees, and integrated over uncertainty in these branch lengths42. We performed analyses under a number of different models, as well as with different priors on model parameters, and results were broadly the same regardless of model or prior choice (Supplementary Figs. 2–11; Supplementary Tables 1–26). Each analysis was run for 10,000,000 generations sampling every 1000th generation, allowing us to determine the most probable spatial gene expression pattern present at ancestral nodes. Mean posterior probabilities (PPs) plotted at each node of our phylogeny indicate the probability of a gene expression pattern character state. Throughout the manuscript and in Figs. 2 and 3, we refer to mean PPs derived from analyses resulting from a single-rate model with a Uniform prior on transition rates from 0 to 2.
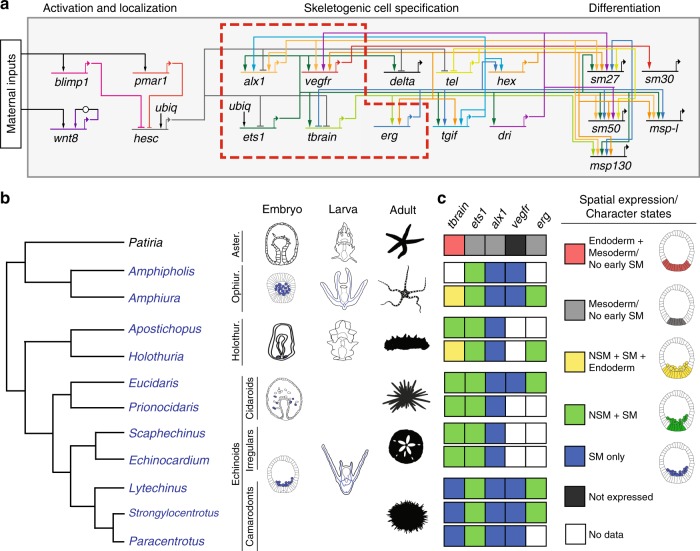
Fig. 1.
Gene regulatory network of the larval skeletogenic cell and spatial distribution of skeletogenic regulatory genes early in development of eleutherozoan echinoderms studied herein. a Gene regulatory network of the S. purpuratus larval skeletogenic cell type showing regulatory genes and their interactions. This study focuses on the evolution of the genes shown in the red dashed box, viz. alx1, erg, ets1, tbrain, and vegfr. The network is based on the GRN found at http://echinobase.org/endomes and various studies11,35. b Phylogeny of eleutherozoan echinoderms showing classes, typical indirect embryonic and larval developmental stages with skeletogenic cells and/or larval skeleton in blue, and typical adult forms. Aster, Asteroidea; Ophiur, Ophiuroidea; Holothur, Holothuroidea. Taxa in blue indicate presence of larval skeletogenic cells. c Spatial distribution of four transcription factors shown to be important for specification of euechinoid larval skeletogenic cells in early development of eleutherozoan echinoderms. Character states used for ancestral state reconstruction are shown at right along with examples of how states were scored. SM skeletogenic mesenchyme, NSM nonskeletogenic mesenchyme. Silhouette images in b were created individually or are in the public domain with the following exceptions: ophiuroid (credit Noah Shlottman, photo from Casey Dunn), clypeasteroid (credit Michelle Site), and camarodont echinoid (credit Frank Förster based on a picture by Jerry Kirkhart; modified by T. Michael Keesey), all of which are used under license http://creativecommons.org/licenses/by-sa/3.0/. No changes were made to the images
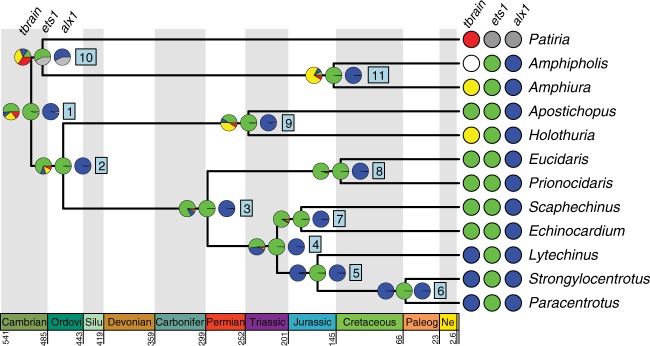
Fig. 2.
Ancestral state reconstruction of spatial gene expression data using a single-rate Markov model42 across eleutherozoan echinoderms. Ancestral states of alx1, ets1, and tbrain, three transcription factors critical to euechinoid larval skeletogenic cell specification. Geological timescale shown at bottom. Nodes are represented by boxed numbers. For each extant character state, the probability that it occurs at the ancestral node was estimated. The pie charts at each node show the mean posterior probability for each spatial expression pattern of the genes shown at right. The colors represent the character states shown in Fig. 1c. Abbreviations in the geological timescale are as follows: Ordovi Ordovician, Silur Silurian, Carbonifer Carboniferous, Paleog Paleogene, Ne Neogene
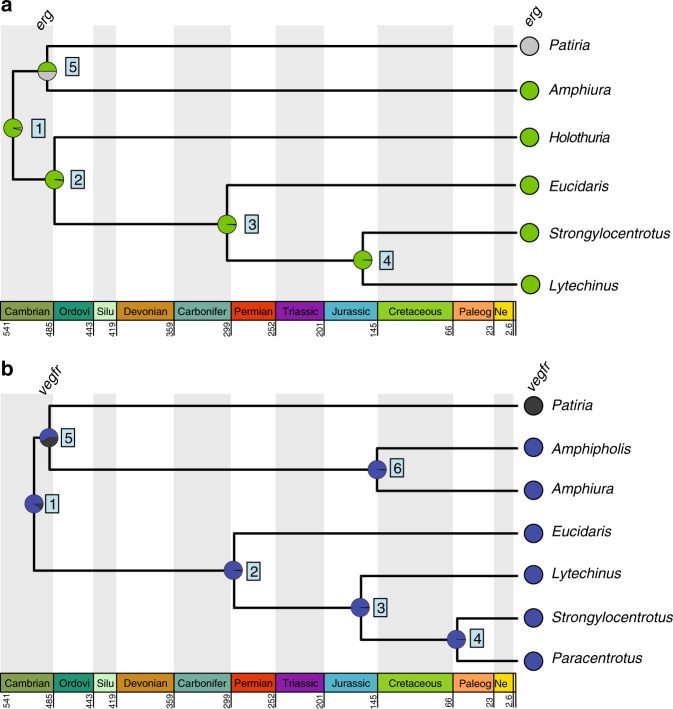
Fig. 3.
Ancestral state reconstruction of spatial gene expression data for erg and vegfr using a single-rate Markov model42 across eleutherozoan echinoderms. a Ancestral state reconstruction of spatial expression patterns of the transcription factor erg. b Ancestral state reconstruction of spatial expression patterns of the signaling gene vegfr. Geological timescale shown at bottom. Nodes are represented by boxed numbers. For each extant character state, the probability that it occurs at the ancestral node was estimated. The pie charts at each node show the mean posterior probability for each spatial expression pattern of the genes shown at right. The colors represent the character states shown in Fig. 1c. Abbreviations in the geological timescale are as follows: Ordovi Ordovician, Silur Silurian, Carbonifer Carboniferous, Paleog Paleogene, Ne Neogene
Considering parsimony alone, plotting the presence of larval skeletogenic cells on the most recent phylogeny of eleutherozoans (Fig. 1b) suggests two evolutionary scenarios for the origin of larval skeletogenic cells: either two gains occurred (one in ophiuroids and one in echinozoans); or one gain occurred at the base of eleutherozoans with one loss in the stem lineage leading to asteroids. To inform these competing evolutionary hypotheses, we performed ancestral state reconstruction on spatial gene expression patterns in early development for four regulatory molecules with detailed functional roles during the specification of larval skeletogenic cells: alx19–11,13,15,18,20,49–54, erg17,20,55,56, ets111,13,15,18,19,50–53,56–59, tbrain7,8,10,11,13,15,18,20,52,59–65, and vegfr17,31,66–68.
Ancestral state reconstruction of skeletogenic cell identity
At all nodes of our phylogeny, we found support for spatial gene expression of alx1 specifically in larval skeletogenic cells, as well as support for broad spatial expression of ets1 in mesoderm (Node 1; PPs = 0.98) (Fig. 2; Supplementary Figs. 3, 4, 7, and 8; Supplementary Tables 2, 3, 7, and 8). These results suggest that the ancestral state of eleutherozoan echinoderms is likely more similar to states seen in extant echinozoans and ophiuroids rather than those observed in asteroids. Similar to results for ets1, though with fewer taxa, ancestral state reconstruction of erg showed support for broad expression in mesodermal cell types in early development since the divergence of eleutherozoans (Nodes 1–4; PPs ≥ 0.96) (Fig. 3a; Supplementary Figs. 5 and 9; Supplementary Table 5). Ancestral state reconstruction for the signaling receptor vegfr, which is critical both for specification and spatial positioning of skeletogenic cells, showed support for skeletogenic specific spatial expression at the most recent common ancestor (MRCA) of eleutherozoans (Node 1, PP = 0.90; Fig. 3b; Supplementary Figs. 2 and 11; Supplementary Tables 6 and 11). However, neither presence nor absence of vegfr in skeletogenic mesoderm in the MRCA of asterozoans is particularly well supported (Node 5; PP for presence = 0.57) (Fig. 3b; Supplementary Fig. 2; Supplementary Table 6). Similarly, support for erg expression in skeletogenic cells or nonskeletogenic mesoderm at this node were equivocal (Node 5; PP = 0.5) (Fig. 3a; Supplementary Fig. 5; Supplementary Table 5) or slightly in favor of echinozoan state if a two rate model is used (Supplementary Fig. 5; Supplementary Table 10). Furthermore, at the ancestral node of asterozoans (Node 10), we find support for alx1 and ets1 states observed in extant ophiuroids rather than in asteroids (PPs = 0.57 and 0.56, respectively). As it could be argued this result may be an artifact of small sampling in asteroids relative to ophiuroids and echinozoans, we conducted a hypothesis test using Bayes Factors and found it offered further support for this result (Supplementary Table 27). We also conducted sensitivity analyses on pruned trees to determine if taxonomic sampling biases were skewing results (Supplementary Fig. 12; Supplementary Tables 28–30). These additional analyses suggested that, in the case of ets1 and alx1, inference in the asterozoan MRCA may be biased by taxon sampling, as results of our pruned analyses were equivocal (Supplementary Fig. 12). In contrast, our inferences at the eleutherozoan MRCA are robust to differential taxon sampling (Supplementary Fig. 12). Nevertheless, our phylogenetically expansive analysis suggest that mesodermal expression of alx1 in early development of asteroids20 is likely an asteroid apomorphy, with one possible explanation being that alx1 participates in other GRN circuits, e.g., basal membrane remodeling and mesenchymal ingression69. Conversely, our sensitivity analysis cannot rule out a reversal back to the ancestral eleutherozoan state in ophiuroids, though we find it unlikely based on the principle of parsimony. We conclude that alx1, erg, ets1, and vegfr have been components of larval skeletogenic cell-type identity since its origin in or before the MRCA of eleutherozoans. Furthermore, given the expression of alx1, ets1, and vegfr in adult skeletogenic cells of asteroids33, ophiuroids70, and echinoids33,71, we conclude that they are likely components of a cell-type identity network possesed by all eleutherozoan skeletogenic cells that likely also drove skeletogenic cell-type identity in ancestral larval skeletogenic cells.
In stark contrast to alx1, erg, ets1, and vegfr, ancestral state reconstruction of the transcription factor tbrain revealed marked lability in its spatial deployment at different ancestral nodes (Fig. 2; Supplementary Figs. 6 and 10; Supplementary Tables 4 and 9). We find support for the presence of tbrain specifically in ancestral larval skeletogenic cells of camarodont euechinoids included in our analysis (Node 5, PP = 0.96), suggesting it functions specifically in the specification of larval skeletogenic cells and is an apomorphy of euechinoids. Moving deeper in evolutionary time to the divergence of the echinoids about 300 million years ago, there is support for tbrain functioning throughout the mesoderm (Node 3, PP = 0.82). At the echinozoan MRCA (Node 2), we also have support for mesodermal tbrain. This results stands in contrast to broader endomesodermal expression patterns observed in asteroids61,63 and hemichordates72, though we note that with inclusion of hemichordate outgroups in the future this result could change. Our results suggest that in the MRCA of echinoids, tbrain functioned more broadly in specifying mesodermal cell-type identities as it does in cidaroid echinoids and holothurians today, and that in the lineage leading to camarodont euechinoids, tbrain lost its functional role in non-skeletogenic mesodermal cell-type identity but maintained its derived function within the skeletogenic cell-type identity network. Thus, in contrast to an ancestral tbrain endomesodermal expression pattern, our analyses support an ancestral mesodermal expression pattern of tbrain with gains of expression in endodermal cells, although it must be noted that the PP for this node is relatively low.
Discussion
To inform the evolution of the echinoderm larval skeletogenic cell, we have presented a framework for cell-type phylogenetic analysis that integrates spatial gene expression data with phylogenetic comparative methods to reconstruct ancestral gene expression. The genes we chose to include in our analyses have been studied in numerous echinoderm taxa and occupy crucial nodes of a well characterized gene regulatory network11,35. As the number of model and nonmodel organisms increases in evolutionary developmental biology, comparative analyses of spatial data will depend more on ancestral state reconstructions than on direct comparisons with an outgroup. However, it should be noted that such analyses are limited by several factors, including knowledge of a detailed GRN, invoking interspecies comparisons of development, and obtaining reliable divergence times and phylogenetic trees. The present study is not exempt from these limitations. Indeed, we chose to analyze five genes with broad phylogenetic sampling over twelve taxa. To support our findings, we ran pruned sensitivity analyses and concluded that decreasing the number of taxa reduced our ability to resolve ancestral states with confidence at certain nodes, especially the Asterozoan and Eleutherozoan MRCAs (Supplementary Fig. 12). Therefore, broad phylogenetic sampling is vitally important to resolve ancestral gene expression patterns. One could argue that increasing the number of genes in the analysis would help resolve the question of interest. However, we suggest that whether or not this is true will depend on the case at hand. For instance, in this study we analyzed genes from a GRN where the functional importance of many regulatory genes is well-known. In most cases, a well characterized GRN will not be available, and it will be equally important to possess a broad sampling of taxa across a phylogeny. For instance, as gene expression in early development becomes available in more asterozoans, which are under-sampled with respect to echinozoans in our analyses, we will gain greater confidence in our inferences at the asterozoan and eleutherozoan MRCAs. Indeed, we also see potential for the approach utilized herein to analyze ancestral states of GRN regulatory architecture. For example, the isolation and characterization of homologous cis-regulatory modules, from ATAC-seq, ChIP-seq, and other omics-level endeavors in phylogenetically distant organisms, could be then incorporated with the method presented here to determine which modules are lineage or cell-type specific.
By framing our results in the context of cell-type evolution, we propose an evolutionary scenario whereby two topographically distinct skeletogenic cell types are the result of a cell-type duplication event that occurred in, or before, the stem lineage of eleutherozoans (Fig. 4a). This event requires heterochronic activation of the adult skeletogenic cell type very early in development, as well as heterotopic activation of the developmental program of adult skeletogenic cells in a distinct developmental context, a similar yet distinct conclusion to a previous analysis33. Importantly, our analyses establish continuity of cell-type homology for the echinoderm larval skeletogenic cell, suggesting the existence of a highly conserved cell-type identity network consisting of, but not limited to, the transcription factors alx1, ets1, erg, and the signaling molecule vegfr that appeared in the stem lineage of eleutherozoan echinoderms, and later was modified when an endomesodermal transcription factor, tbrain, acquired functional importance in the larval skeletogenic GRN.
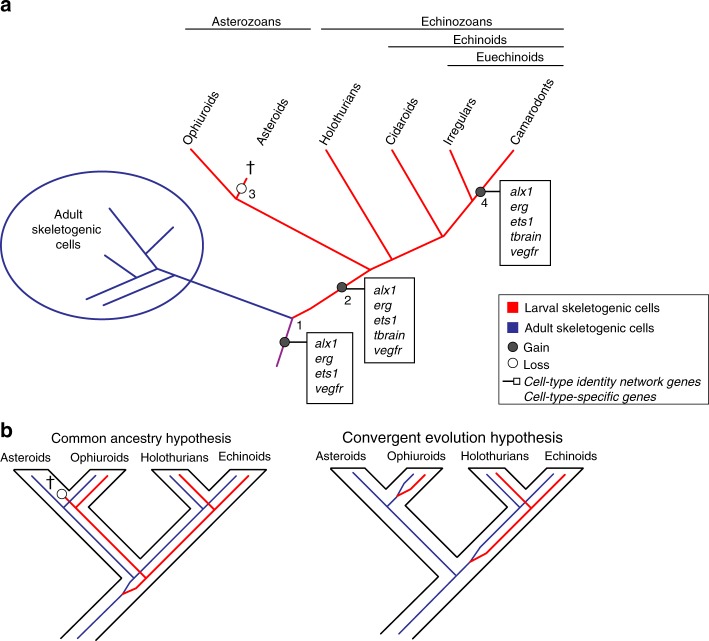
Fig. 4.
Cell-type evolution of echinoderm larval skeletogenic cells. a A cell-type duplication event resulted in activation of the adult skeletogenic cell-type identity network in early development, and thus the presence of skeletogenic cells in ancestral embryonic mesodermal territories (Node 1). Alx1 and vegfr are components of a cell-type specific identity network of larval skeletogenic cells across all extant eleutherozoans. The combination of alx1, erg, ets1, and vegfr comprise the cell-type identity network of skeletogenic cells across this clade. After the duplication event, at least one transcription factor that was ancestrally expressed in embryonic mesoderm domains, tbrain, was subsequently integrated into the larval skeletogenic GRN (Node 2), giving rise to two sister cell types. The larval skeletogenic cell type was lost in asteroids after the divergence with ophiuroids (Node 3). In the lineage leading to extant camarodont euechinoids, tbrain was decoupled from and no longer participated in specification of non-skeletogenic mesodermal cell types through Erg-mediated repression. Different colors represent cell-type lineages. Adult skeletogenic cells, purple. Larval skeletogenic cells, red. b Two competing hypotheses for the origin of larval skeletogenic cells in eleutherozoan echinoderms. The common ancestry hypothesis suggests that all larval skeletogenic cells in eleutherozoans are the result of a cell-type duplication event in the stem lineage of eleutherozoans. These cells would be lost in the lineage leading to extant asteroids. The convergent evolution hypothesis posits that larval skeletogenic cells are the result of at least two evolutionary events, one in the stem lineage of echinozoans and one other in the stem lineage of ophiuroids. Our results support the common ancestry hypothesis
Several observations provide a roadmap as to how the cell-type identity networks driving larval skeletogenic cells evolved. Our results suggest that alx1 and vegfr are part of a cell-type specific suite of regulatory genes that have been specifically expressed in echinoderm larval skeletogenic cells since the MRCA of extant eleutherozoans (Fig. 4a). Their co-expression in adult skeletogenic cells of asteroids31,33, ophiuroids31,70, and echinoids33,71, as well as the larval skeletogenic cells of ophiuroids, holothuroids, and echinoids, suggest they should be considered cell type-specific regulatory genes for these cell types within eleutherozoans. It has been shown that these two genes are activated by Ets1 in the skeletogenic GRN11, which also itself inputs into several skeletogenic differentiation genes (Fig. 1a). However, Ets1 does not activate skeletogenic genes in embryonic domains where it is also active. Another critical regulatory genes in this process is erg, which has been shown to be one of the first genes to be activated downstream of alx1 and ets111. Activation of erg establishes a subcircuit including erg, hex, and tgif, which serve to lockdown the regulatory state critical for skeletogenic cell specification. Thus, the combination of Alx1, Erg, Ets1, and Vegfr is a defining feature of the cell-type identity network in both adult and larval skeletogenic cells.
A key evolutionary event underlying the genetic and developmental individuation of adult and larval skeletogenic cells was the integration of tbrain into the cell-type identity network of larval skeletogenic cells in the stem lineage of eleutherozoans (Fig. 4a). Tbrain is not expressed in adult skeletogenic cells of asteroids33, ophiuroids70, and echinoids33. As our reconstructions suggest that tbrain was likely expressed in ancestral mesodermal cells of the eleutherozoan stem lineage (Fig. 2), we propose that tbrain was already present in ancestral eleutherozoan embryonic mesoderm and subsequently was integrated into the larval skeletogenic GRN. One putative mechanism that can accommodate this hypothesis is cell-type fusion, whereby regulatory genes already expressed in ancestral cell types or territories come to be co-expressed in a hybrid cell type1. In the case at hand, it would suggest that regulatory modules that respond to and were ancestrally under the control of tbrain could have become expressed in the larval skeletogenic cell type. This hypothesis could explain the shallow nature of tbrain wiring in the larval skeletogenic GRN (Fig. 1a). It remains to be seen whether other regulatory genes that were ancestrally expressed in embryonic mesoderm may have been integrated in a similar way into the larval skeletogenic GRN. Interestingly our analyses also suggest that tbrain subsequently became a cell-type specific component of the larval skeletogenic cells of camarodont euechinoids. Previous observations show that Ets1 protein directly activates the expression of tbrain in mesodermal cell types7,60. Our analyses provide support for this, as co-expression is considered the ancestral state (Node 3, Fig. 2). Other studies suggest that the restriction of Tbrain to larval skeletogenic cells in camarodont echinoids occurred via upregulation of the tbrain-repressor Erg by Ets1 in nonskeletogenic cells. This linkage must be an apomorphy of camarodont euechinoids as co-expression of erg and tbrain has been shown in cidaroid echinoids16, and Tbrain drives expression of erg in asteroids56. Furthermore, it is clear that tbrain has acquired larval skeletogenic cell-type specific functions, as the Tbrain protein sequence has itself evolved distinct DNA binding preferences since the divergence of asteroids and echinoids73 and exhibits little overlap of regulation of orthologous genes in early development of these clades74. Lastly, it should be noted that, when interpreted within the context of the convergent evolution hypothesis, tbrain would have had to be integrated independently into two larval skeletogenic GRNs, i.e., once in ophiuroids and once in echinozoans. Our hypothesis requires only a single tbrain integration event in the stem lineage of eleutherozoans.
The evolutionary relatedness of ophiuroid and echinoid larval skeletons, as well as the morphology of the pluteus larva itself, has long been debated in the literature, with various interpretations being given either in favor of convergent evolution25,26,30,31,35,75 or in favor of common ancestry22–24 (Fig. 4b). These arguments have been based, for instance, on the phylogenetic positions of ophiuroids and echinoids45, the developmental and structural differences in skeletal morphology27, and developmental gene-expression data13,29,30,35. On the other hand, and prior to contemporary resolution of the echinoderm phylogeny, some authors favored a common ancestry hypothesis, which at the time was supported by morphological analyses24, paleontological analyses76, and phylogenetic analyses23. Our results support the common ancestry hypothesis (Fig. 4b). Under this scenario it follows that the lineage leading to extant asteroids lost a larval skeletogenic cell (Fig. 4b). Interestingly, many regulatory genes shown to be important for specification of the euechinoid larval skeletogenic cell are also expressed in early asteroid development, including alx120, ets119,77, tbrain63,78, erg56, hex56, and tgif56. However, vegfr is not expressed31,71. As our analyses show, this suggests that loss of vegfr, which is a cell-type specific component of larval skeletogenic cells, during early development in the MRCA of extant asteroids, could have facilitated loss of a larval skeletogenic cell. Lastly, coincident with the evolution of eleutherozoan larval skeletogenic cells was the evolution of distinct isoforms of alx1 that are specifically expressed in larval skeletogenic cells49. These isoforms produce Alx1 proteins that are required for early skeletogenesis. Notably, these isoforms are not expressed early in the development of asteroids although they are present in the genome. This observation suggests a molecular mechanism by which a lineage of echinoderms could lose a larval skeleton, namely by the gain or loss of alternative splicing isoforms early in development49. Taken together these observations suggest molecular mechanisms that can rapidly bring about the loss of the larval skeletogenic cells, suggesting that a wholesale loss of larval skeleton in asteroids is a plausible evolutionary scenario.
In regards to the evolution of characters, it can be instructive to ask whether phylogenetic evidence suggests lineages are more likely to gain or lose a character during the course of evolution79. Analyses of extant eleutherozoans suggest that larval skeletons have been lost in early development of at least 20 species of echinoids and ophiuroids80,81. Surprisingly, vestigial larval skeletons occur frequently in the evolution of echinoderm larval forms25,82–84. In indirect developing holothurians, spicules are synthesized in the larva but are not elaborated, suggesting this character was lost or an elaborate skeleton failed to evolve in this lineage20. Morphologically speaking, all larval skeletogenic cells observed thus far synthesize triradiate spicules, in spite of the fact that echinoderm skeletogenic cells can readily produce a wide array of spicule morphologies85,86. This suggests that a shared developmental pathway may be utilized to synthesize early spicules in all lineages of echinoderms that possess larval skeletogenic cells. While our analyses do not strongly refute the convergent evolution of larval skeletogenic cells or the structures they synthesize, we only wish here to point out that future studies give due consideration to both evolutionary scenarios while undertaking comparative work analyzing the evolution of these structures and cell types, an argument that has also been suggested by others31,87.
It has been stated that heterochrony tinkers, but heterotopy creates88. Whereas heterochrony shifts developmental events in time relative to each other, heterotopy results in the spatial translocation of a developmental structure89. The origin of the larval skeletogenic cell is often cited as a classical example of heterochrony. It is also true then that adult skeletogenic cells became housed in a distinct developmental context and were subsequently elaborated into the iconic larval skeleton of echinoids and ophiuroids. In our view, heterotopy of the larval skeletogenic cell is often overlooked even though it has the potential to yield insight into how this cell-type evolved. Our cell-type duplication model predicts that transcription factors and signaling systems that were expressed in ancestral endomesodermal domains were likely integrated into the nascent larval skeletogenic cell in stem eleutherozoans. Such a model may help guide future interpretations of how these cell types evolved. Whether the elaborate larval skeletons of echinoids and ophiuroids are convergent or a product of common descent, it is clear that the distinct differences in morphology, development, and even families of differentiation genes underlying these two morphological structures had hundreds of millions of years to evolve. Therefore, the differences we see today may in fact be a product of the subsequent evolution that has occurred since the divergence long ago of these clades. To understand the evolutionary relatedness of these cells and of these morphological structures, we will need to bring to bear all tools at our disposal. By reconstructing character states driving changes in cell-type identity networks and ancestral cell types, cell-type phylogenetics is one such tool and has the capacity to inform very old questions.
Methods
Taxon selection, gene selection, and developmental timing
We selected taxa that had published gene-expression patterns during embryogenesis and larval development for five genes, alx1, erg, ets1, tbrain, and vegfr (vegfr-ig-10). In addition, these taxa have published 18S and 28S small subunit ribosomal RNA gene-sequence data which allowed us to time-calibrate our phylogeny using divergence time estimation. Sequences for both loci were available on Genbank for all genera in our analysis except for Apostichopus, for which only the 18s sequence was available. Analyses were run for each genus in order to allow us to maximize sampling with respect to these criteria. For instance there were no 28s sequences available on Genbank for Holothuria leucospilota, however, sequences for this gene were available for its congener Holothuria sanctori. Likewise, despite the fact gene expression patterns have been published for the congeners Patiria miniata and Patiria pectinifera we include only one asteroid, Patiria, in our analyses. In addition, though there is no 18S or 28S gene-sequence data available for the scutellid clypeasteroid Scaphechinus mirabilis, sequence data for these two genes were available for Echinodiscus, which, as a scutellid, belongs to the same family as Scaphechinus. For divergence time estimation, we thus used the sequence data from Echinodiscus to time calibrate the divergence between the neognathostomate and atelostomate irregular echinoids, which represents the divergence between Scaphechinus and Echinocardium in our analyses. For alx1 and ets1 sampling was good, and we were able to include twelve taxa in our analyses (Supplementary Data 1). For tbrain, we included all of the same taxa as for alx1 and ets1 except for the ophiuroid Amphipholis, for which there are no published expression patterns for tbrain. For vegfr, sampling is more limited, and gene expression data were available for seven genera, while for erg data were only available for six.
For gene selection, the number of genes included in the GRN for echinoderm skeletogenesis is large, and includes numerous transcription factors and even more differentiation genes. However, this GRN only pertains to euechinoid sea urchins, as this clade is the exclusive source of data for the GRN. We aimed to include genes where the spatial distribution of RNAs is known in both the adult and the early embryo in as many clades of echinoderms as possible. There are very few genes in the GRN where data both in the adult and larva exist in numerous taxa. We included alx1, erg, ets1, tbrain, and vegfr in our analyses mainly due to the spread of available data. For the other genes, data are piecemeal throughout developmental time and the phylogenetic tree. Thus we aimed to provide an analysis that included as many genes as possible for which there exists a phylogenetically broad sampling. Secondly, as far as specification of skeletogenic cells is concerned, it is clear from developmental studies that these four genes lie at key nodes of the GRN. On the other hand, we included tbrain due to its long known activity in different embryonic domains. This gene was included to provide an intriguing contrast to the other selected genes, which, relatively speaking, show domain-specific expression across all taxa analyzed.
Developmental timing varies across taxa, and careful selection of developmental timepoints is critical to obtain meaningful inter-taxon comparisons. In this study, we combined descriptions in the primary literature with analyses of spatial expression patterns to determine where a given gene was expressed. With the exception of sea stars, alx1 and vegfr are in all taxa expressed specifically in skeletogenic cells; and we used their activation and stabilization in skeletogenic precursors as a comparable developmental stage (e.g., mesenchyme blastula in euechinoids) for analysis in each taxon. We used this timepoint as a basis of developmental comparison, and thus collated spatial expression patterns for ets1, tbrain, and erg at these timepoints in each taxon. Broadly, these criteria resulted in the following developmental timepoints: mesenchyme blastula for euechinoids and ophiuroids, and late blastula/early gastrula for cidaroids, asteroids, and holothuroids.
Divergence time estimation
So that branch lengths of the phylogenetic trees onto which we reconstructed ancestral states reflected evolutionary time, we used fossil calibrated divergence time estimation to calibrate our phylogenetic tree. For our divergence time estimation analyses, we used two genes, 18S and 28S small subunit ribosomal RNA genes, which comprised the most complete dataset with respect to number of sites for the taxa included in our analyses. Sequences were concatenated and aligned using Clustal X90. The aligned matrix is 2919 base pairs long and is available in Supplementary File 1 at https://github.com/jthechino/Erkenbrack_-_Thompson. Each gene was treated as a separate partition, and for each of our two partitions, the best fitting model was determined using the Akaike Information Criterion in JmodelTest 2.0. For both partitions, the best model was identified as the GTR + I + Γ and the GTR + Γ identified as the second best model. Due to statistical issues associated with invariant sites91 we performed our analyses using the GTR + Γ model with four gamma categories.
Our divergence time analyses were run using the BEAST 2.347 software package using a constraint tree topology based off of recently published echinoderm phylogenies45,92 and calibrated using the seven constraints from the echinoderm fossil record (see “Calibration justification in divergence time analyses”). We used a relaxed clock lognormal model where the substitution rate at each branch is an independent draw from a lognormal distribution with parameters µ and σ247. To set priors on the parameters of our clock model, we first estimated the substitution rate per time unit with a strict molecular clock using the program BASEML in PAML93 and the root node calibrated at 500 MA. The substitution rate was found to be 0.016 + −0.001. This was used to calculate the parameters α = 1 and β = 62.5 of the gamma distribution used as the prior on the parameter µ following α = (0.016/0.016)2 and β = 0.016/(0.0162). The prior on σ2 was a gamma distribution with parameters α = 1 and β = 5. We used a birth-death prior on tree shape with a uniform prior of U(0,10000) on the birth rate. Model parameters were estimated using Markov Chain Monte Carlo (MCMC) for 20,000,000 iterations, sampling every 100th generation. Two separate analyses were run, and results were checked for convergence using the Tracer version 1.6 software94. The time tree showing 95% credible intervals on divergence times is shown in Supplementary Fig. 1 with 20% of the posterior sample discarded as burn in. A.xml file with details of analyses is in Supplementary File 1.
Ancestral state reconstruction
Ancestral state reconstruction models character evolution, in this case the evolution of gene expression patterns, as a Markov process, where the probability of an evolutionary change from one state to another is independent along each branch and depends only on the state at the beginning of each branch95–97. In reconstructing ancestral states, this method not only takes into account the topological relationships of taxa, but also branch lengths on the phylogeny. Because of uncertainty regarding the divergence times of the taxa included in our analyses, and thus uncertainty in branch lengths in our phylogenies, we used a Bayesian approach to explicitly integrate over this uncertainty42. We thus used a random sample of 10,000 time-calibrated trees from the posterior distribution of our divergence time estimation analyses as the input trees for our ancestral state reconstructions. For ancestral state reconstructions, we used the program BayesTraits V.342 run in the R software environment using the wrapper program btw (http://rgriff23.github.io/projects/btw.html)98.
For all analyses, branch lengths were scaled to have mean value of 0.1 as recommended by the BayesTraits manual. All analyses were initially run with a uniform prior between 0 and 1 on transition rates. Inspection of posterior distribution showed that this prior was truncating the posterior distribution, so analyses were re-run with a uniform prior between 0 and 2. For alx1, ets1, erg, and vegfr, characters were all scored as binary traits; tbrain, which displays a diversity of expression patterns throughout extant echinoderms, was scored as a multistate character with four character states. For all binary traits, the logarithm of the marginal likelihood was computed for each model using stepping stone sampling99 and the 2 times the Log Bayes Factor100 was computed to compare support for a single rate model, where q01 and q10 were constrained to be equal, and a two-rate model where each rate was allowed to take its own value101. For tbrain, Log Bayes Factors were computed to compare a single rate model to a multi-rate model. Results of Bayes Factor comparisons between one rate and multiple rate models are shown in Supplementary Table 1 and in all cases support one model over the other was negligible. Furthermore, analyses did not differ when analyses were run using either a one rate or multi-rate model (see “Sensitivity analyses”).
Bayesian ancestral state reconstructions use MCMC to estimate model parameters, and in particular the instantaneous transition rate for each character being analyzed. MCMC was run for 10,000,000 generations sampling every 1000 generations. Burn-in was 2,000,000 generations. Convergence was assessed by plotting and inspecting the sampled value per iteration, the probability density function for the posterior, and the autocorrelation for the Markov chain of the transition rate q01 using the traceplot(), densplot(), and autocorr.plot() functions in the CODA package102 in R. Results of ancestral state reconstructions using a single rate model are shown in Supplementary Tables 2–6 and Supplementary Figs. 2a, 3a, 4a, 5a, and 6a; while analyses using a two-rate, or multi-rate in the case of tbrain, are shown in Supplementary Tables 7–11 and Supplementary Figs. 2b, 3b, 4b, 5b and 6b. In most cases, inferred ancestral states did not differ dependent upon whether the model had multiple rates or a single rate. The exception to this was erg, where using a two-rate model resulted in slight support for mesodermal expression in the asterozoan MRCA, while the single-rate model returned equivocal results at this node (Supplementary Fig. 5). R code used to perform all analyses can be found at https://github.com/jthechino/Erkenbrack_-_Thompson.
Sensitivity analyses
In order to explore the sensitivity of our results to changes in model parameters, we ran a number of sensitivity analyses using different priors on the instantaneous transition rate q01. We initially used a uniform prior ranging from 0 to 2 on this transition rate (see “Ancestral state reconstruction”). To explore the sensitivity of a more diffuse prior, we ran our analyses with uniform priors of U(0, 20) and U(0, 200). To explore any changes in our results when analyses were run with less-diffuse priors, we ran our ancestral state reconstructions with a uniform prior of U(0, 0.2). Results of these sensitivity analyses are shown in Supplementary Tables 12–26 and Supplementary Figs. 7–11. Running analyses with wider priors reduced the confidence in inferred ancestral states (PPs approached unity at ancestral nodes), though this only slightly changed results for analyses of ets1 and alx1. In the case of these genes inferred ancestral states still supported the presence of an alx1 + and ets1 + skeletogenic cell and ets1 expression in the mesoderm at all ancestral nodes (Supplementary Figs. 7 and 8). Results were similar with respect to vegfr and erg, though the reduction in PP for all nodes was of greater magnitude (Supplementary Figs. 9 and 11). For tbrain, inferred PPs for ancestral states approached unity using the prior of U(0,20), though the inferred ancestral states did not differ from those with a prior of U(0,2), and still showed fairly strong support for the same inferred ancestral states (Supplementary Fig. 10b). The same trend was apparent to a greater extent using a prior of U(0,200), though results at the MRCAs of camarodonts, irregular echinoids, and cidaroids all still showed one state clearly favored over all others (Supplementary Fig. 10c).
Using a narrower prior (U(0, 0.2)) (Supplementary Figs. 7a, 8a, 9a, 10a, and 11a) resulted in slower modeled rates of character evolution, and more definitive inferences for ancestral states. This is not surprising, as lowering the modeled rate of evolution in ancestral state reconstruction analyses results in answers approaching a most-parsimonious reconstruction97. As a result, with a narrower prior, the most probable ancestral states inferred were the same as using a prior of U(0,2), but with stronger support for the inferred most likely ancestral states.
In addition to assessing sensitivity of our results to model parameters, we also explored the effects of taxon sampling on our results. This was particularly to assess reconstructions at the MRCA of eleutherozoans, and at the asterozoan MRCA. For ancestral state reconstructions of alx1, ets1, and tbrain we ran pruned analyses including only one taxon per class (four tips total). Results of these analyses are shown in Supplementary Tables 28–30 and Supplementary Fig. 12. In the case of alx1 and ets1, running analyses using only four tips did not alter interpretations at the MRCA of eleutherozoans, which still showed strong support for expression of alx1 in skeletogenic mesenchyme (Supplementary Fig. 12a), and ets1 in skeletogenic mesenchyme and non-skeletogenic mesoderm (Supplementary Fig. 12b); however, inferences at the MRCA of asterozoans were equivocal for both these genes (Supplementary Fig. 12a, b). Likewise, for tbrain, using only for tips resulted in ambiguous resolution at the MRCA of eleutherozoans, and ambiguous inferences at the MRCA of asterozoans and echinozoans (Supplementary Fig. 12c).
Bayes factor hypothesis testing
In order to compare support for different reconstructions of ancestral expression patterns for alx1, ets1, erg, and vegfr at the MRCA of asterozoans, we fixed the value at this node as either 0 or 1, corresponding to either expression in the mesoderm for alx1, erg, and ets1 or expression in the skeletogenic cell or NSM and skeletogenic cell for ets1 and erg or alx1, respectively. Bayes factors were then computed to estimate support for different models estimated using these fixed values. 2*ln Bayes Factors were calculated as 2*(ln[P|State = 0] − ln[P|State = 1] where ln[P|State] is the natural log of the marginal likelihood of the model calculated when the node representing the MRCA of asterozoans is fixed at either 0 or 1. Results were interpreted following the tables reported in Kass and Raftery100. Natural log marginal likelihoods were calculated in BayesTraits with the btw wrapper using stepping stone sampling99. MCMC for stepping stone sampling was run for 1000 stones with 100,000 iterations per stone. Bayes factors resulting from comparisons for each gene are shown in Supplementary Table 27. For both alx1 and vegfr, hypothesis testing using Bayes factors supported expression in the skeletogenic mesenchyme. For ets1 and erg Bayes factors supported the expression in the skeletogenic mesenchyme and nonskeletogenic mesoderm.
Calibration justification in divergence time analyses
Euechinoid-Cidaroid divergence—The divergence between cidaroid and euechinoid echinoids represents the oldest occurrence of a crown group echinoid in the fossil record, and thus the hard minimum on the divergence of the echinoid crown group. The oldest crown group echinoid is Eotiaris guadalupensis Thompson, 2017 from the Roadian of the Road Canyon Formation of West, Texas, USA103,104. Thompson et al.104 found this taxon to be a cidaroid using phylogenetic analyses, and the presence of two columns of interambulacral plates in an interambulacral area and a perignathic girdle of apophyses readily classify this species amongst the cidaroidea. The exact stratigraphic distribution of E. guadalupensis within the Roadian is unknown, so the top of the Roadian stage, 268.8 MYA, is used as the hard minimum for the divergence of the euechinoids and cidaroids. We round this to 269 for purposes of divergence time estimation. The maximum bound used for calibration is the bottom of the Viséan stage, or 346.7 MYA. The Viséan is home to abundant and diverse echinoid faunas including those of the Edwardsville Formation105, the Fort Payne Formation106, and the Molignée Formation107,108. The diversity and abundance of stem group echinoids in these faunas provide a taphonomic control; and despite the number of stem group echinoids known from these deposits, crown group echinoids are wanting, and thus the bottom of the Viséan is used as the maximum bound on the divergence.
Irregularia-Camarodonta divergence—The oldest irregular echinoid, Jesionekechinus hawkinsi, from the Sunrise Formation of New York Canyon, Nevada109 calibrates the hard minimum of the divergence between the Irregularia and the camarodont echinoids. J. hawkinsi was recorded from 15 to 30 ft below the Eoderoceras zone, which is equivalent to the unit G of Hallam110, which is the base of the Pleinsbachian Joker Peak member111. If J. hawkinsi is from below the Joker Peak member, at the youngest it must be from the New York Canyon Member, which underlies the Joker Peak Member. The age of the New York Canyon Member is Sinnemurian111, and given the imprecise stratigraphic position of J. hawkinsi, we use the top of the Sinemmurian, 190.8 MYA, as the hard minimum on the divergence between camarodonts and irregulars. The maximum bound on the divergence is set by the most diverse fauna of echinoids in the Triassic, from the Carnian St. Cassian Formation of Northern Italy112,113. This fauna is currently interpreted to consist entirely of cidaroids; though there are specimens with euechinoid-like morphologies known from disarticulated material, there are no putative irregulars. The age of the Cassian Formation is Carnian, and spans from the Julian 1 slightly into the Julian 2 ammonoid zones114. We use the top of the Julian 1 ammonoid zone as the lower constraint on the Irregularia-Camarodonta divergence, which is approximately 235 MYA.
Holothuroid-Echinoid divergence—There are a number of basal fossil holothuroids and echinoids in Ordovician strata115,116. The divergence between these two clades is calibrated by the oldest unequivocal fossil holothurian calcareous ring elements and body wall ossicles which are from the Red Orthoceras limestone of Sweden, which was found as a glacial erratic boulder in northern Germany116. These specimens were recovered from the Eoplacognathus suecicus conodont zone, which is itself within the Pseudoclamacograptus decorates graptolite zone. The top of the P. decorates zone is 463.97 MA, which we round to 464 MA for purposes of divergence time estimation. The maximum bound on the divergence is set by the Fezouata Lagerstätte, which yields a diverse and abundant echinoderm fauna117. Despite the wealth of echinoderms from the Fezouata, there have been no echinoid or holothurians fossils recovered. The Fezouata Shale is at the youngest Floian117, and thus we use the top of the Floian stage of the Ordovician as the maximum bound on this divergence, which is 470 MA.
Neognathostomata-Atelostomata divergence—Echinodiscus is a clypeasteroid echinoid, which is part of the larger clade the Neognathostomata. The clypeasteroids did not evolve and diversify until the Cenozoic92; however, they are paraphyletic with respect to the morphologically conservative cassiduloida92. Kroh and Smith92 found the Galeropygus, the Nucleolitidae, and the Clypeidae to be stem group neognathostomates but with low-bootstrap support. Barras118 additionally found the nucleolitids and clypeids to be amongst the neognathostomates (what he referred to as cassiduloids) in his 50% majority rule consensus tree. The stem group of the atelostomates comprises a number of species and genera previously known as the disasteroids. The phylogenetic position of these taxa seems to be sensitive to character choice and weighting scheme92,118. We thus use the first occurrence of stem neognathostomates in the fossil record to calibrate the minimum divergence between the atelostomates and neognathostomates. The oldest clypeid is Clypeus rostratus from the Toarcian D. levesquei ammonite zone of the Upper Lias of Walditch, Dorset. Barras118. The D. levesquei Zone is roughly equivalent to the D. pseudoradiosa and P. dispansum zones. The top of the D. pseudoradiosa zone is 174.71 MA. We round this to 174 MA, which we use as the minimum constraint on the Neognathostomata-Atelostomata divergence. The lower Jurassic has a rich echinoid fauna, however, irregular echinoids are rare. We thus use the base of the Jurassic, 201.3 MA, as the maximum bound on the neognathostomate and atelostomate divergence, which we round to 201 MA.
Asterozoan-Echinozoan divergence—Asterozoans are known from much earlier in the Ordovician fossil record than are crown-group echinozoans, and thus the divergence between asteroids and ophiuroids is calibrated by the oldest asterozoan known from the fossil record, Maydena roadsidensis119. M. roadsidensis co-occurs with the graptolite Psigraptus jacksoni in the Psigraptus zone, the top of which is dated as 481.67 MA. We round this to 481 MA, which is used as the hard minimum on the divergence of the asterozoans and echinozoans. The phylogenetic relationships amongst the early eleutherozoans are not well constrained, and thus the ancestry of echinozoans and asterozoans is unclear120. Echinozoans and asterozoans may have both evolved from an edrioasteroid ancestor, but though which edrioasteroids they evolved from are unknown. In order to account for this uncertainty, we use the oldest edrioasteroid, Stromatocystites walcotti, from the Cambrian Series 2121 as the maximum bound on the divergence. The maximum bound on the divergence is thus the base of Series 2 in the Cambrian Period, which is currently set at 521 MA.
Asteroid-Ophiuroid divergence—The asterozoans include ophiuroids, asteroids, and the extinct Early Paleozoic group the somasteroids122. As discussed above, the earliest asterozoan is Maydena roadsidensis119, which was noted by Jell119 as “likely to be part of the lineage leading to earliest ophiuroids such as Pradesura Spenger, 1951 and Eophiura Jaekel, 1903” (Jell119 p. 536). The ophiuroid Pradesura jacobi is the geologically oldest nonsomasteroid asterozoan other than M. roadsidensis119. P. jacobi is known from the St. Chinian Schist Formation, northeast of St. Chinian, la Croix-Rouge, southeast France122. While the phylogenetic position of M. roadsidensis has yet to be examined rigorously, P. jacobi was found to form a clade with other basal ophiuroids by Shackleton122. The age of P. jacobi is latest Tremadocian119. We thus use the top of the Tremadocian, 477.7 MA (rounded to 477 MA), as the hard minimum on the divergence between asteroids and ophiuroids. As previously mentioned, asterozoans can trace their ancestry to edrioasteroids. As for the Asterozoan-Echinozoan divergence, we thus use the occurrence of Stromatocystites walcotti and the base of the Cambrian Series 2, 521 MA, as the maximum bound on the prior for the Asteroid-Ophiuroid divergence.
Holothuriida-Neoholothuriida divergence—Holothuria belongs to the holothuroid family Holothuriidae, which was recently demonstrated based upon molecular data to form a clade with the Mesothuriidae, the Holothuriida123. The Holothuriida is the sister group to the Neoholothuriida, which contains the genus Apostichopus. A thorough discussion of the fossil calibrations for the holothuriidae can be found in Miller et al.123. The oldest putative holothuriid ossicles are known from the Wuchiapingian period of the Permian and may represent stem group holothuriids123. The oldest known holothuriid calcareous rings are of an undescribed species from the spinosus zone of the Early Ladinian Upper Muschelkalk of Baden-Württemberg, Germany124. The base of the Ladinian is 241.5 MA125 and we use the occurrence of these holothuriid calcareous rings as the hard minimum on the divergence between the holothuriids and neoholothuriids (rounded to 241); while the maximum age for the divergence is taken as Wuchiapingian in age, and is thus treated as 260 MA, approximately the age of the base of the Wuchiapingian126.
Statistics and reproducibility
All code and relevant data to reproduce our analyses are either included in this paper or are available at GitHub https://github.com/jthechino/Erkenbrack_-_Thompson.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Günter P. Wagner for insightful comments on an early version of this paper.
Author contributions
E.M.E. collated the data, interpreted the results, contributed the figures, and wrote the paper. J.R.T. ran the analyses, interpreted the results, contributed the figures, and wrote the paper.
Data availability
All data in this study are either available in the Supplement or have been previously published.
Code availability
Code used to conduct these analyses is available on GitHub at https://github.com/jthechino/Erkenbrack_-_Thompson.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eric M. Erkenbrack, Email: eric.erkenbrack@yale.edu
Jeffrey R. Thompson, Email: jeff.thompson@ucl.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0417-3.
References
- 1.Arendt D, et al. The origin and evolution of cell types. Nat. Rev. Genet. 2016;17:744–757. doi: 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- 2.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kin K, Nnamani MC, Lynch VJ, Michaelides E, Wagner GP. Cell-type phylogenetics and the origin of endometrial stromal cells. Cell Rep. 2015;10:1398–1409. doi: 10.1016/j.celrep.2015.01.062. [DOI] [PubMed] [Google Scholar]
- 4.Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat. Rev. Genet. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- 5.Erkenbrack EM, et al. The mammalian decidual cell evolved from a cellular stress response. PLoS Biol. 2018;16:e2005594. doi: 10.1371/journal.pbio.2005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner, G. P., Erkenbrack, E. M. & Love, A.C. Stress-induced evolutionary innovation: a mechanism for the origin of cell types. BioEssays41, 1–9 (2019). [DOI] [PMC free article] [PubMed]
- 7.Ochiai H, Sakamoto N, Momiyama A, Akasaka K, Yamamoto T. Analysis of cis-regulatory elements controlling spatio-temporal expression of T-brain gene in sea urchin, Hemicentrotus pulcherrimus. Mech. Dev. 2008;125:2–17. doi: 10.1016/j.mod.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Fuchikami T, et al. T-brain homologue (HpTb) is involved in the archenteron induction signals of micromere descendant cells in the sea urchin embryo. Development. 2002;129:5205–5216. doi: 10.1242/dev.129.22.5205. [DOI] [PubMed] [Google Scholar]
- 9.Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- 10.Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev. Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 11.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc. Natl Acad. Sci. USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 13.Dylus DV, et al. Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo. 2016;7:2. doi: 10.1186/s13227-015-0039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc. Natl Acad. Sci. USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erkenbrack EM, Davidson EH. Evolutionary rewiring of gene regulatory network linkages at divergence of the echinoid subclasses. Proc. Natl Acad. Sci. USA. 2015;112:E4075–E4084. doi: 10.1073/pnas.1509845112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erkenbrack EM, et al. Ancestral state reconstruction by comparative analysis of a GRN kernel operating in echinoderms. Dev. Genes Evol. 2016;226:37–45. doi: 10.1007/s00427-015-0527-y. [DOI] [PubMed] [Google Scholar]
- 17.Erkenbrack EM, Petsios E. A conserved role for VEGF signaling in specification of homologous mesenchymal cell types positioned at spatially distinct developmental addresses in early development of sea urchins. J. Exp. Zool. B Mol. Dev. Evol. 2017;328:423–432. doi: 10.1002/jez.b.22743. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki A, Kidachi Y, Yamaguchi M, Minokawa T. Larval mesenchyme cell specification in the primitive echinoid occurs independently of the double-negative gate. Development. 2014;141:2669–2679. doi: 10.1242/dev.104331. [DOI] [PubMed] [Google Scholar]
- 19.Koga H, et al. Functional evolution of Ets in echinoderms with focus on the evolution of echinoderm larval skeletons. Dev. Genes Evol. 2010;220:107–115. doi: 10.1007/s00427-010-0333-5. [DOI] [PubMed] [Google Scholar]
- 20.McCauley BS, Wright EP, Exner C, Kitazawa C, Hinman VF. Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. Evodevo. 2012;3:17. doi: 10.1186/2041-9139-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pennington JT, Strathmann RR. Consequences of the calcite skeletons of planktonic echinoderm larvae for orientation, swimming, and shape. Biol. Bull. 1990;179:121–133. doi: 10.2307/1541746. [DOI] [PubMed] [Google Scholar]
- 22.Smith, A. B. Echinoid Paleobiology. (Taylor & Francis, London, 1984).
- 23.Smith, A. B. Fossil evidence for the relationships of the extant echinodem classes and their times of divergence. In Echinoderm Phylogeny and Evolutionary Biology (eds C.R.C., Smith Paul, A. B.) pp. 85–97 (Clarendon Press, Oxford, 1988).
- 24.Hyman, L. H. The Invertebrates. Vol. 4 (McGraw-Hill, 1955).
- 25.Mortensen, T. H. Studies on the Developement and Larval Forms of Echinoderms. (GEC Gad, 1921).
- 26.De Beer, G. R. In The new systematics (ed. Huxley, J. S.) pp. 365–393 (Oxford University Press, Oxford, 1940).
- 27.Raff, R. A. F. K. G. et al. Molecular analysis of distant phylogenetic relationships in echinoderms. In Echinoderm Phylogeny and Evolutionary Biology (eds C. R. C., Smith Paul, A. B.) pp. 29–41 (Clarendon Press, Oxford, 1988).
- 28.Strathmann, R. R. Larvae, phylogeny, and von Baer's Law. In Echinoderm Phylogeny and Evolutionary Biology (eds C. R. C., Smith Paul, A. B.) pp. 53–68 (Oxford Scientific, Oxford, 1988).
- 29.Dylus DV, Czarkwiani A, Blowes LM, Elphick MR, Oliveri P. Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. Genome Biol. 2018;19:26. doi: 10.1186/s13059-018-1402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morino Y, Koga H, Wada H. The conserved genetic background for pluteus arm development in brittle stars and sea urchin. Evol. Dev. 2016;18:89–95. doi: 10.1111/ede.12174. [DOI] [PubMed] [Google Scholar]
- 31.Morino Y, et al. Heterochronic activation of VEGF signaling and the evolution of the skeleton in echinoderm pluteus larvae. Evol. Dev. 2012;14:428–436. doi: 10.1111/j.1525-142X.2012.00563.x. [DOI] [PubMed] [Google Scholar]
- 32.Bottjer DJ, Davidson EH, Peterson KJ, Cameron RA. Paleogenomics of echinoderms. Science. 2006;314:956–960. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- 33.Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc. Natl Acad. Sci. USA. 2008;105:6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaughn R, Garnhart N, Garey JR, Thomas WK, Livingston BT. Sequencing and analysis of the gastrula transcriptome of the brittle star Ophiocoma wendtii. Evodevo. 2012;3:19. doi: 10.1186/2041-9139-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shashikant Tanvi, Khor Jian Ming, Ettensohn Charles A. From genome to anatomy: The architecture and evolution of the skeletogenic gene regulatory network of sea urchins and other echinoderms. genesis. 2018;56(10):e23253. doi: 10.1002/dvg.23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minokawa T. Comparative studies on the skeletogenic mesenchyme of echinoids. Dev. Biol. 2017;427:212–218. doi: 10.1016/j.ydbio.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Hinman VF, Cheatle Jarvela AM. Developmental gene regulatory network evolution: insights from comparative studies in echinoderms. Genesis. 2014;52:193–207. doi: 10.1002/dvg.22757. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JR, et al. Paleogenomics of echinoids reveals an ancient origin for the double-negative specification of micromeres in sea urchins. Proc. Natl Acad. Sci. USA. 2017;114:5870–5877. doi: 10.1073/pnas.1610603114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wray GA, McClay DR. The origin of spicule-forming cells in a ‘primitive' sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development. 1988;103:305–315. doi: 10.1242/dev.103.2.305. [DOI] [PubMed] [Google Scholar]
- 40.Felsenstein J. Distance methods for inferring phylogenies: a justification. Evolution. 1984;38:16–24. doi: 10.1111/j.1558-5646.1984.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 41.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 42.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 43.Dunn CW, Zapata F, Munro C, Siebert S, Hejnol A. Pairwise comparisons across species are problematic when analyzing functional genomic data. Proc. Natl Acad. Sci. USA. 2018;115:E409–E417. doi: 10.1073/pnas.1707515115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Revell LJ, Mahler DL, Peres-Neto PR, Redelings BD. A new phylogenetic method for identifying exceptional phenotypic diversification. Evolution. 2012;66:135–146. doi: 10.1111/j.1558-5646.2011.01435.x. [DOI] [PubMed] [Google Scholar]
- 45.Telford M. J., Lowe C. J., Cameron C. B., Ortega-Martinez O., Aronowicz J., Oliveri P., Copley R. R. Phylogenomic analysis of echinoderm class relationships supports Asterozoa. Proceedings of the Royal Society B: Biological Sciences. 2014;281(1786):20140479–20140479. doi: 10.1098/rspb.2014.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith AB, et al. Testing the molecular clock: molecular and paleontological estimates of divergence times in the Echinoidea (Echinodermata) Mol. Biol. Evol. 2006;23:1832–1851. doi: 10.1093/molbev/msl039. [DOI] [PubMed] [Google Scholar]
- 47.Bouckaert R, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khor, J. M. & Ettensohn, C. A. Functional divergence of paralogous transcription factors supported the evolution of biomineralization in echinoderms. eLife610.7554/eLife.32728 (2017). [DOI] [PMC free article] [PubMed]
- 50.Erkenbrack EM, Davidson EH, Peter IS. Conserved regulatory state expression controlled by divergent developmental gene regulatory networks in echinoids. Development. 2018;145:1–11. doi: 10.1242/dev.167288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rottinger E, Besnardeau L, Lepage T. A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets. Development. 2004;131:1075–1087. doi: 10.1242/dev.01000. [DOI] [PubMed] [Google Scholar]
- 52.Yamazaki A, Minokawa T. Expession patterns of mesenchyme specification genes in two distantly related echinoids, Glyptocidaris crenularis and Echinocardium cordatum. Gene Expr. Patterns. 2015;17:87–97. doi: 10.1016/j.gep.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Yamazaki A, Furuzawa Y, Yamaguchi M. Conserved early expression patterns of micromere specification genes in two echinoid species belonging to the orders clypeasteroida and echinoida. Dev. Dyn. 2010;239:3391–3403. doi: 10.1002/dvdy.22476. [DOI] [PubMed] [Google Scholar]
- 54.Koga H, et al. Experimental approach reveals the role of alx1 in the evolution of the echinoderm larval skeleton. PLoS One. 2016;11:e0149067. doi: 10.1371/journal.pone.0149067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Materna SC, Ransick A, Li E, Davidson EH. Diversification of oral and aboral mesodermal regulatory states in pregastrular sea urchin embryos. Dev. Biol. 2013;375:92–104. doi: 10.1016/j.ydbio.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev. Biol. 2010;340:200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 57.Kurokawa D, et al. HpEts, an ets-related transcription factor implicated in primary mesenchyme cell differentiation in the sea urchin embryo. Mech. Dev. 1999;80:41–52. doi: 10.1016/S0925-4773(98)00192-0. [DOI] [PubMed] [Google Scholar]
- 58.Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus) Dev. Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 59.Sharma T, Ettensohn CA. Regulative deployment of the skeletogenic gene regulatory network during sea urchin development. Development. 2011;138:2581–2590. doi: 10.1242/dev.065193. [DOI] [PubMed] [Google Scholar]
- 60.Wahl ME, Hahn J, Gora K, Davidson EH, Oliveri P. The cis-regulatory system of the tbrain gene: Alternative use of multiple modules to promote skeletogenic expression in the sea urchin embryo. Dev. Biol. 2009;335:428–441. doi: 10.1016/j.ydbio.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev. Biol. 2007;312:584–595. doi: 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Croce J, Lhomond G, Lozano JC, Gache C. ske-T, a T-box gene expressed in the skeletogenic mesenchyme lineage of the sea urchin embryo. Mech. Dev. 2001;107:159–162. doi: 10.1016/S0925-4773(01)00470-1. [DOI] [PubMed] [Google Scholar]
- 63.Shoguchi E, Satoh N, Maruyama YK. A starfish homolog of mouse T-brain-1 is expressed in the archenteron of Asterina pectinifera embryos: possible involvement of two T-box genes in starfish gastrulation. Dev. Growth Differ. 2000;42:61–68. doi: 10.1046/j.1440-169x.2000.00481.x. [DOI] [PubMed] [Google Scholar]
- 64.Minemura K, Yamaguchi M, Minokawa T. Evolutionary modification of T-brain (tbr) expression patterns in sand dollar. Gene Expr. Patterns. 2009;9:468–474. doi: 10.1016/j.gep.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 65.Maruyama YK. A sea cucumber homolog of the mouse T-Brain-1 is expressed in the invaginated cells of the early gastrula in Holothuria leucospilota. Zool. Sci. 2000;17:383–387. doi: 10.2108/jsz.17.383. [DOI] [PubMed] [Google Scholar]
- 66.Duloquin L, Lhomond G, Gache C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development. 2007;134:2293–2302. doi: 10.1242/dev.005108. [DOI] [PubMed] [Google Scholar]
- 67.Rottinger E, et al. FGF signals guide migration of mesenchymal cells, control skeletal morphogenesis [corrected] and regulate gastrulation during sea urchin development. Development. 2008;135:353–365. doi: 10.1242/dev.014282. [DOI] [PubMed] [Google Scholar]
- 68.Adomako-Ankomah A, Ettensohn CA. Growth factors and early mesoderm morphogenesis: insights from the sea urchin embryo. Genesis. 2014;52:158–172. doi: 10.1002/dvg.22746. [DOI] [PubMed] [Google Scholar]
- 69.Saunders LR, McClay DR. Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development. 2014;141:1503–1513. doi: 10.1242/dev.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Czarkwiani A, Dylus DV, Oliveri P. Expression of skeletogenic genes during arm regeneration in the brittle star Amphiura filiformis. Gene Expr. Patterns. 2013;13:464–472. doi: 10.1016/j.gep.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao F, et al. Juvenile skeletogenesis in anciently diverged sea urchin clades. Dev. Biol. 2015;400:148–158. doi: 10.1016/j.ydbio.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 72.Tagawa K, Humphreys T, Satoh N. T-brain expression in the apical organ of hemichordate tornaria larvae suggests its evolutionary link to the vertebrate forebrain. J. Exp. Zool. 2000;288:23–31. doi: 10.1002/(SICI)1097-010X(20000415)288:1<23::AID-JEZ3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 73.Cheatle Jarvela AM, et al. Modular evolution of DNA-binding preference of a Tbrain transcription factor provides a mechanism for modifying gene regulatory networks. Mol. Biol. Evol. 2014;31:2672–2688. doi: 10.1093/molbev/msu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cary GA, Cheatle Jarvela AM, Francolini RD, Hinman VF. Genome-wide use of high- and low-affinity Tbrain transcription factor binding sites during echinoderm development. Proc. Natl Acad. Sci. USA. 2017;114:5854–5861. doi: 10.1073/pnas.1610611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hotchkiss FHC. Lovens law and adult ray homologies in echinoids, ophiuroids, edrioasteroids, and an ophiocistioid (echinodermata, eleutherozoa) Proc. Biol. Soc. Wash. 1995;108:401–435. [Google Scholar]
- 76.Paul CRCSAB. Radiation and phylogeny of echinoderms. Biol. Rev. 1984;59:443–481. doi: 10.1111/j.1469-185X.1984.tb00411.x. [DOI] [Google Scholar]
- 77.Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl Acad. Sci. USA. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl Acad. Sci. USA. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wagner, G. P. Homology, Genes, and Evolutionary Innovation. (Princeton University Press, Princeton, 2014).
- 80.Wray GA. The evolution of larval morphology during the post-Paleozoic radiation of echinoids. Paleobiology. 1992;18:258–287. doi: 10.1017/S0094837300010848. [DOI] [Google Scholar]
- 81.Emlet, R. B. Advances in Invertebrate Reproduction. Vo-l. 5 329–335 (Elsevier, Amsterdam, Netherlands, 1990).
- 82.Emlet RB. Larval spicules, cilia, and symmetry as remnants of indirect development in the direct developing sea urchin Heliocidaris erythrogramma. Dev. Biol. 1995;167:405–415. doi: 10.1006/dbio.1995.1037. [DOI] [PubMed] [Google Scholar]
- 83.Hendler G. An echinoderm vitellaria with a bilateral larval skeleton—evidence for the evolution of ophiuroid vitellariae from ophioplutei. Biol. Bull. 1982;163:431–437. doi: 10.2307/1541454. [DOI] [Google Scholar]
- 84.Selvakumaraswamy P, Byrne M. Vestigial ophiopluteal structures in the lecithotrophic larvae of Ophionereis schayeri (Ophiuroidea) Biol. Bull. 2000;198:379–386. doi: 10.2307/1542693. [DOI] [PubMed] [Google Scholar]
- 85.Knapp RT, Wu CH, Mobilia KC, Joester D. Recombinant sea urchin vascular endothelial growth factor directs single-crystal growth and branching in vitro. J. Am. Chem. Soc. 2012;134:17908–17911. doi: 10.1021/ja309024b. [DOI] [PubMed] [Google Scholar]
- 86.Comeau AD, Cameron CB. Ossicle development of the crinoid Florometra serratissima through larval stages. Can. J. Zool. 2016;95:183–192. doi: 10.1139/cjz-2016-0195. [DOI] [Google Scholar]
- 87.Wilt FH, Killian CE, Livingston BT. Development of calcareous skeletal elements in invertebrates. Differentiation. 2003;71:237–250. doi: 10.1046/j.1432-0436.2003.7104501.x. [DOI] [PubMed] [Google Scholar]
- 88.Hall, B. K. Evolutionary Developmental Biology. 2nd ed., (Kluwer Academic Publishers, Boston, 1999).
- 89.Zelditch MLFWL. Heterochrony and heterotopy: stabiity and innovation in the evolution of form. Paleobiology. 1996;22:241–254. doi: 10.1017/S0094837300016195. [DOI] [Google Scholar]
- 90.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 91.Yang, Z. Molecular Evolution: A Statistical Approach. (Oxford University Press, Oxford, 2014).
- 92.Kroh A, Smith AB. The phylogeny and classification of post-Palaeozoic echinoids. J. Syst. Palaeontol. 2010;8:147–212. doi: 10.1080/14772011003603556. [DOI] [Google Scholar]
- 93.Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst. Biol. 2007;56:453–466. doi: 10.1080/10635150701420643. [DOI] [PubMed] [Google Scholar]
- 94.Rambaut, A., Suchard, M. A. & Drummond, A. J. Tracer version 1.6 [computer program]. http://evolve.zoo.ox.ac.uk/ (2013).
- 95.Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. B Biol. Sci. 1994;255:37–45. doi: 10.1098/rspb.1994.0006. [DOI] [Google Scholar]
- 96.Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 1999;48:612–622. doi: 10.1080/106351599260184. [DOI] [Google Scholar]
- 97.Schluter, D., Price, T., Mooers, A. Ø. & Ludwig, D. Likelihood of ancestor states in adaptive radiation. Evolution 51, 1699–1711 (1997). [DOI] [PubMed]
- 98.BayesTraits Wrapper (btw) v. 2 (2018).
- 99.Xie W, Lewis PO, Fan Y, Kuo L, Chen M. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst. Biol. 2011;60:150–160. doi: 10.1093/sysbio/syq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kass RE, Raftery AE. Bayes factors. J. Am. Stat. Assoc. 1995;90:773–795. doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- 101.Mooers AOslash, Schluter D. Reconstructing ancestor states with maximum likelihood: support for one-and two-rate models. Syst. Biol. 1999;48:623–633. doi: 10.1080/106351599260193. [DOI] [Google Scholar]
- 102.Plummer M, Best N, Cowles K, Vines K. CODA: convergence diagnosis and output analysis for MCMC. R. News. 2006;6:7–11. [Google Scholar]
- 103.Thompson JR, Petsios E, Bottjer DJ. A diverse assemblage of Permian echinoids (Echinodermata, Echinoidea) and implications for character evolution in early crown group echinoids. J. Paleontol. 2017;91:767–780. doi: 10.1017/jpa.2016.158. [DOI] [Google Scholar]
- 104.Thompson, J. R. et al. Reorganization of sea urchin gene regulatory networks at least 268 million years ago as revealed by oldest fossil cidaroid echinoid. Sci. Rep.5, 15541 (2015). [DOI] [PMC free article] [PubMed]
- 105.Lane NG. Paleontology and paleoecology of the Crawfordsville fossil site (upper Osagian: Indiana) with sections by J. L. Matthews, E.G. Driscoll, and E. L. Yochelson. Univ. Calif. Publ. Geol. Sci. 1973;32:1–36. [Google Scholar]
- 106.Thompson JR, Ausich WI. Facies distribution and taphonomy of echinoids from the Fort Payne Formation (late Osagean, early Viséan, Mississippian) of Kentucky. J. Paleontol. 2016;90:239–249. doi: 10.1017/jpa.2016.46. [DOI] [Google Scholar]
- 107.Mottequin B. The “black marble” of Denée, a fossil conservation deposit from the Lower Carboniferous (Viséan) of southern Belgium. Geol. J. 2008;43:197–208. doi: 10.1002/gj.1102. [DOI] [Google Scholar]
- 108.Mottequin B, Poty E, Prestianni C. Catalogue of the types and illustrated specimens recovered from the “black marble” of Denée, a marine conservation-Lagerstätte from the Mississippian of southern Belgium. Geol. Belg. 2015;18:1–14. [Google Scholar]
- 109.Jesionek-Szymanska W. On a new pygasterid (Echinoidea) from the Jurassic (Middle Lias) of Nevada, U.S.A. Acta Palaeontol. Pol. 1970;15:411–419. [Google Scholar]
- 110.Hallam A. Observations on marine Lower Jurassic stratigraphy of North America, with special reference to the United States. Bull. Am. Assoc. Pet. Geol. 1965;49:1485–1501. [Google Scholar]
- 111.Taylor DG, Smith PL, Laws RA, Geux J. The stratigraphy and biofacies trends of the Lower Mesozoic Gabbs and Sunrise foramtiosn, west-central Nevada. Can. J. Earth Sci. 1983;20:1598–1608. doi: 10.1139/e83-149. [DOI] [Google Scholar]
- 112.Kier, P. M. Triassic Echinoids. 1–88 (Smithsonian Institution Press, Washington, D.C., 1977).
- 113.Kier, P. M. Echinoids fRom the Triassie (St. Cassian) of Italy, Their Lantern Supports, and A Revised Phylogeny of Triassic echinoids. (1984).
- 114.Dal Corso J, et al. Carbon isotope records reveal synchronicity between carbon cycle perturbation and the “Carnian Pluvial Event” in the Tethys realm (Late Triassic) Glob. Planet. Change. 2015;127:79–90. doi: 10.1016/j.gloplacha.2015.01.013. [DOI] [Google Scholar]
- 115.Smith AB, Savill JJ. Bromidechinus, a new Ordovician echinozoan (Echinodermata), and its bearing on the early history of echinoids. Earth Environ. Sci. Trans. R. Soc. Edinb. 2001;92:137–147. doi: 10.1017/S0263593300000109. [DOI] [Google Scholar]
- 116.Reich, M. The early evolution and diversification of holothurians (Echinozoa). In Echinoderms: Durham. Proceedings of the 12th International Echinoderm Conference, Durham, New Hampshire, USA, 7-11 August 2006 (eds Harris, L. G., Böttger, S. A., Walker, C. W. & Lesser, M. P.) 55–59 (Taylor & Francis Group, Durham, 2010).
- 117.Lefebvre B, et al. Palaeoecological aspects of the diversification of echinoderms in the Lower Ordovician of central Anti-Atlas, Morocco. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2016;460:97–121. doi: 10.1016/j.palaeo.2016.02.039. [DOI] [Google Scholar]
- 118.Barras, C. G. British Jurassic Irregular Echinoids. (The Palaeontographical Society, London, 2006).
- 119.Jell PA. A Tremadocian asterozoan from Tasmania and a late Llandovery edrioasteroid from Victoria. Alcheringa. 2014;38:528–540. doi: 10.1080/03115518.2014.911642. [DOI] [Google Scholar]
- 120.Smith AB, Jell PA. Cambrian edrioasteroids from Australia and the Origin of Starfishes. Memoirs of the Queensland. Museum. 1990;28:715–778. [Google Scholar]
- 121.Zamora S, et al. The Cambrian edrioasteroid Stromatocystites (Echinodermata): systematics, palaeogeography, and palaeoecology. Geobios. 2015;48:417–426. doi: 10.1016/j.geobios.2015.07.004. [DOI] [Google Scholar]
- 122.Shackleton JD. Skeletal homologies, phylogeny and classification of the earliest asterozoan echinoderms. J. Syst. Palaeontol. 2005;3:29–114. doi: 10.1017/S1477201905001525. [DOI] [Google Scholar]
- 123.Miller AK, et al. Molecular phylogeny of extant Holothuroidea (Echinodermata) Mol. Phylogenetics Evol. 2017;111:110–131. doi: 10.1016/j.ympev.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 124.Reich, M. The early evolution and diversification of holothurians (Echinozoa). In Echinoderms: Munchen: Proceedings of the 11th International Echinoderm Conference, 6-10 October 2003, Munich, Germany. (eds T. Henizeller & James H. Nebelsick) pp. 485–486 (Taylor and Francis Group, Munich, 2004).
- 125.Ogg, J. G. Triassic. In The Geologic Timescale 2012 (eds Gradstein, F. M., Ogg, J. G., Schmitz, M. & Ogg, G.) 681–730 (Elsevier, Amsterdam, Netherlands, 2012).
- 126.Henderson, C. M., Davydov, V. I. & Wardlaw, B. R. The Permian Period. In The Geologic Time Scale2012 (eds Gradstein, F. M., Ogg, J. G., Schmitz, M. & Ogg, G.) 653–679 (Elsevier, Amsterdam, Netherlands, 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data in this study are either available in the Supplement or have been previously published.
Code used to conduct these analyses is available on GitHub at https://github.com/jthechino/Erkenbrack_-_Thompson.