Abstract
Osteoarthritis (OA) classification in the knee is most commonly done with radiographs using the 0–4 Kellgren Lawrence (KL) grading system where 0 is normal, 1 shows doubtful signs of OA, 2 is mild OA, 3 is moderate OA, and 4 is severe OA. KL grading is widely used for clinical assessment and diagnosis of OA, usually on a high volume of radiographs, making its automation highly relevant. We propose a fully automated algorithm for the detection of OA using KL gradings with a state-of-the-art neural network. Four thousand four hundred ninety bilateral PA fixed-flexion knee radiographs were collected from the Osteoarthritis Initiative dataset (age = 61.2 ± 9.2 years, BMI = 32.8 ± 15.9 kg/m2, 42/58 male/female split) for six different time points. The left and right knee joints were localized using a U-net model. These localized images were used to train an ensemble of DenseNet neural network architectures for the prediction of OA severity. This ensemble of DenseNets’ testing sensitivity rates of no OA, mild, moderate, and severe OA were 83.7, 70.2, 68.9, and 86.0% respectively. The corresponding specificity rates were 86.1, 83.8, 97.1, and 99.1%. Using saliency maps, we confirmed that the neural networks producing these results were in fact selecting the correct osteoarthritic features used in detection. These results suggest the use of our automatic classifier to assist radiologists in making more accurate and precise diagnosis with the increasing volume of radiographic image being taken in clinic.
Keywords: Osteoarthritis, Radiographs, Neural networks, Machine learning
Introduction
Osteoarthritic degenerative joint disease is a leading cause of chronic disabilities in the USA. OA symptoms include stiffness, limited joint function, and pain which lead to a decrease in quality of life. OA primarily affects weight-bearing joints with the knee and hip joints being the most common sites. Pain clearly is one of the most important outcome measures in OA and is measured using questionnaires and patient-reported outcomes (PROMs), such as the Knee Outcomes in Osteoarthritis Scores (KOOS) [1] or Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) [2]. The ultimate clinical outcome of osteoarthritis is often total joint replacement in the hip or knee, which is effective in managing symptoms and reversing loss of function in most patients, but is costly, not without risk of complications, and only effective for a limited length of time [3].
In an effort to develop quantitative biomarkers for OA and fill the void that exists for diagnosing, monitoring, and assessing the extent of whole joint degeneration in OA, the past decade has been marked by a greatly increased role of imaging for OA. OA assessment and diagnosis are most commonly done with radiographs (x-rays) using the 0–4 Kellgren Lawrence (KL) grading system where 0 is normal, 1 shows doubtful signs of OA with potential abnormality, 2 demonstrates definite osteophytes (mild OA), 3 shows definite joint space narrowing (moderate OA), and 4 is severe joint space narrowing with subchondral sclerosis and bony deformity (severe OA) [4, 5]. Although this is a class-based method, the grading system does represent a continuous progression of OA, beginning with osteophytes and ending with deformation of the bone. KL grading is widely used for clinical assessment and diagnosis of OA, usually on a high volume of radiographs; however, it is still subject to inter- and intra-user variability, making its automation highly relevant [6].
Automation of several computer vision tasks has been accelerated in the last few years by the usage of artificial intelligence (AI) and machine learning techniques [7, 8] with the availability of large amounts of annotated data and processing power. Using the concepts of transforming data to knowledge by the observation of examples, supervised learning can today accomplish challenges never demonstrated before [9–12].
OA research was recently marked by the curation of public datasets in which imaging plays a central role. Large-scale epidemiologic trials such as the Osteoarthritis Initiative (OAI) provide a rich array of structural and functional features of musculoskeletal tissues, which have shed light on disease etiology, and long-range outcomes in OA. This extraordinary data availability opens a variety of possibility in applying machine learning to the study of OA.
In addition to the data availability, the now available processing power is a completely new concept that has changed the machine learning field. Conventional machine learning techniques were limited in their ability to process natural data in their raw form. For decades, constructing a pattern recognition or machine learning system required careful engineering and considerable domain expertise to design feature extractors able to transform raw data, such as the pixel values of an image, into a suitable internal representation or feature vector from which the classifier could detect patterns in the input. In contrast, representation learning is a set of novel methods that allows a machine to be fed with raw data and to automatically discover the best representations of the information hidden in the data needed to accomplish a specified task [13].
Deep learning neural networks are representation learning methods characterized by the usage of multiple, simple, but non-linear units to build several interconnected layers. Each layer aggregates the information at increasing levels of abstraction starting with simple image elements, such as edges or contrast, to more complex and semantic aggregations, uncovering latent patterns able to accomplish pattern recognition tasks [14]. The key aspect of deep learning is that these models are not designed to solve a specific task, but are adaptive to the specific problems, learning directly from the data and using a general-purpose learning procedure.
In this study, we aim to capitalize these recent advancements in deep learning field to develop a method for the automatic knee x-ray inspection and staging of OA severity based on KL grading.
Materials and Methods
Subjects
Four thousand five hundred four bilateral PA fixed-flexion knee radiographs were collected from the OAI dataset (https://oai.epi-ucsf.org/) (age = 61.2 ± 9.2, BMI = 28.6 ± 4.8, male:female = 1886:2618). Subjects were collected from six different time points (baseline, 12, 36, 48, 72, 96 months) for both left and right knees resulting in a total of 39,593 images. Each of these cases was graded by skilled radiologists involved in the development of the OAI dataset. A handful of these cases were graded multiple times, in which case the modal grade was selected.
In order to preprocess the radiographs to be fed into the machine learning models, they needed to be split into left and right knees and then localized around the knee joint in order to provide for a smaller, more precise field of view for the neural network to learn from. The x-rays were split into left and right knees by dividing the image directly in the middle and then flipping the left side of the image so that it appears to be facing the same way as the right. This alleviates the algorithm of having to learn an additional feature of side when learning. From these split images, cropped localization bounding boxes around the knee were made using a 2-D cross-correlation template matching method (where the template was made using an average of two manually cropped knee joint images) across 500 images, implemented in MATLAB (Mathworks, Natick, MA). These 500 bounding boxes were then quality checked to ensure the template had correctly extracted the knee joint. This resulted in 450 usable localized knee joints, which were then used to train a U-Net network, described in a previous musculoskeletal tissue segmentation study [15], to extract the bounding box around the knee joint created by the template matching method. The U-Net was trained for only 10 epochs using a cross entropy loss function and learning rate of 1e−4 with Adam optimizer. The results from this model were then manually quality checked on a new set of 500 knee images. Four hundred ninety-eight of the bounding boxes produced by the U-Net correctly localized the knee, showing improvement over the base template matching method.
The trained U-Net was applied to the available OAI dataset, resulting in 39,593 joint localized radiographs, which were then resized to 500 × 500 images for model building.
Model Architecture
Data was divided with a 65/20/15 split resulting in 25,873 training images, 7779 validation image, and 5941 testing images. In order ensure generalizability of model performances, the training/validation/testing split was made such that subjects in testing were completely different from those used in training and validation, resulting in 3883 unique subjects in training and validation and 621 unique subjects in testing (Table 1 has the demographic breakdown per group).
Table 1.
Demographic information for training, validation, and testing datasets as well as KL score distributions
| Age | BMI | Gender | KL: | 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|---|---|---|---|
| Training | 61.2 ± 9.2 | 28.6 ± 4.8 | 1886 M 2618 F | 10,391 | 4714 | 6263 | 3467 | 1038 | |
| Validation | 61.3 ± 9.2 | 28.7 ± 4.8 | 1639 M 2242 F | 3112 | 1507 | 1877 | 997 | 286 | |
| Testing | 60.2 ± 9.1 | 28.2 ± 4.8 | 246 M 375 F | 2541 | 1293 | 1281 | 660 | 166 |
Per recommendation of our internal clinical radiologist (TL) and to assist the neural networks learning, KL grades of 0–1 were grouped to represent x-rays with “no OA” since the clinical response for these two grades are usually the same, where KL 2–4 still represent mild, moderate, and severe OA, respectively. Given this grouping and the large imbalance between overall KL gradings (Table 1 shows the initial KL distribution between training, validation, and testing), KL scores of 3 or 4 were augmented with random rotations and translations (3 times for grade 3, 6 times for grade 4) to allow for a more balanced training dataset between scores, resulting in an additional 16,629 augmented images for training.
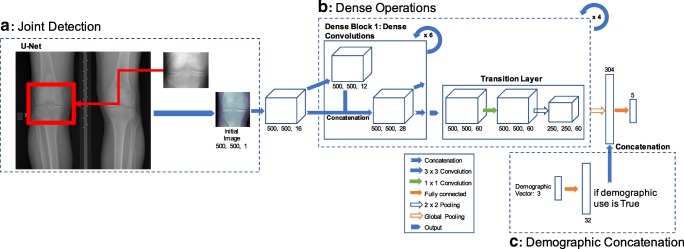
We used an ensemble of different model checkpoints from different variations of the DenseNet neural network architectures to make OA assessments [16, 17] based on their performances with the validation dataset. The DenseNet neural network architecture consists of a series of concatenating previous layers’ feature maps together, but keeping the same number of feature maps generated by each convolution functions the same. This sequence of operations is called a dense block (Fig. 1b). DenseNets consist of multiple dense blocks stringed together. This novel approach allows for learning from feature maps in previous layers while keeping the number of learnable parameters low (which is desirable to avoid over-fitting the model).
Fig. 1.
Pipeline for machine learning algorithm. a Template pattern matching using an average of two manually cropped knee joints. b DenseNet architecture with potential for addition of demographic vector. c Fully connected layer for the transformation of the demographic vector in 32 feature vector
All DenseNets were trained on the training dataset with a learning rate of 1e−4, 0–1 input image normalization, cross entropy loss function, growth rate of 12, block depth of 6, for 20 epochs on a NVIDIA Titan X GPU, implemented in native TensorFlow (Google, Mountain View, CA). The choice of these learning parameters (cross entropy loss function, growth rate, block depth, learning rate) was decided via grid hyper-parameter search based on performance on the validation dataset ranging our search with values that had previously been experimented within the Huang et al. paper and other DenseNet implementations. The difference between the architectures was the decision on whether or not to include demographic information to the network (Fig. 1b shows the traditional DenseNet architecture with the demographic variation). For the demographic inclusion DenseNet, age, sex, and race were fed in as a 3-dimensional vector and then multiplied element-wise by a 32-dimensional weight vector (simply a trainable fully connected layer). BMI was not included due to a number of subjects missing this piece of information. This 32-dimensional layer was then concatenated onto the flattened image output of the DenseNet (Fig. 1c). The ensemble of these DenseNets was made by averaging the softmax logit outputs of different model checkpoints from the two DenseNet architectures. Softmax is a linear mapping of outputs to the 0–1 range, displayed in Eq. 1, where x is a given input node, ak is the raw model output for the kth class channel of input node x, and K is the total number of channels in the output layer, which also represents the number of classes in the classification problem. Predictions are made by taking the position of the maximal softmax value.
| 1 |
The model’s resulting performances were compared to other efforts towards automatic KL grading using a chi-squared two-sampled proportion test at the α < 0.05 level.
Clinical Radiologist Review and Saliency Maps
To fully assess the performance of our ensemble model, we conducted a single-blind study by selecting 54 random cases that the neural network classified differently than the original radiologists’ grading and showed them to our internal clinical radiologist. A musculoskeletal radiologist with more than 20 years of experience (TL) was presented the image and two grades (the original radiologist grading and the algorithmic grading) without knowing which grade corresponded to the source and was asked to select the grade he agreed most with or, if he disagreed with both, what grade he would give it. This allowed for a more in-depth assessment of the accuracy rate of our algorithm.
We also did a case-by-case examination with our clinical radiologist on 12 subjects which the algorithm misclassified to better understand which features the algorithm may have been missing. Additionally, to asses which features the algorithm was using for its decision-making, saliency/heat maps of each input image were generated by taking the derivative of the outputted models’ logits with respect to the input image and then visualizing the derivative mapping [18]. Higher values of this derivative for a pixel suggest that the given pixel was influential in the calculations made by the neural network.
Results
The U-net for knee joint localization worked on 98.3% of the 1000 randomly sampled cases. In order to keep the fully automatic nature of this classification pipeline, the incorrectly localized images were kept in the modeling dataset. Left and right knee localization and resizing for an inputted DICOM image takes 1.49 s to generate.
The optimal DenseNet models used for the ensemble learning were a standard DenseNet after 15 epochs (DN15) and DenseNet with demographic input vector after 8 epochs (DN+DEM8) (the validation sensitivity and specificity accuracy results can be viewed in Table 2). The resulting softmax logit outputs of these two models were averaged to make the final predictions. This ensemble of DenseNets’ testing sensitivity rates of no OA, mild, moderate, and severe OA was 83.7, 70.2, 68.9, and 86.0% respectively. The corresponding specificity rates were 86.1, 83.8, 97.1, and 99.1%. The testing confusion matrix can be viewed in Fig. 2 and a full breakdown of training, validation, and testing Specificity and sensitivity ensemble results can be viewed in Table 3. OA severity prediction from the inputted 500 × 500 knee localized image takes 7.38 s, making the entire prediction pipeline just under 9 s for a given image.
Table 2.
Validation set specificity and sensitivity accuracy for the two DenseNet models used for ensemble building (DN DenseNet, DN+DEM DenseNet with demographic information)
| No OA (%) | Mild OA (%) | Moderate OA (%) | Severe OA (%) | ||
|---|---|---|---|---|---|
| DN15 | Sensitivity | 84.4 | 72.1 | 69.4.4 | 83.1 |
| Specificity | 86.8 | 84.01 | 97.2 | 99.2 | |
| DN-DEM8 | Sensitivity | 85.8 | 71.11 | 69.7 | 80.19 |
| Specificity | 85.18 | 84.4 | 97.8 | 99.6 | |
Fig. 2.
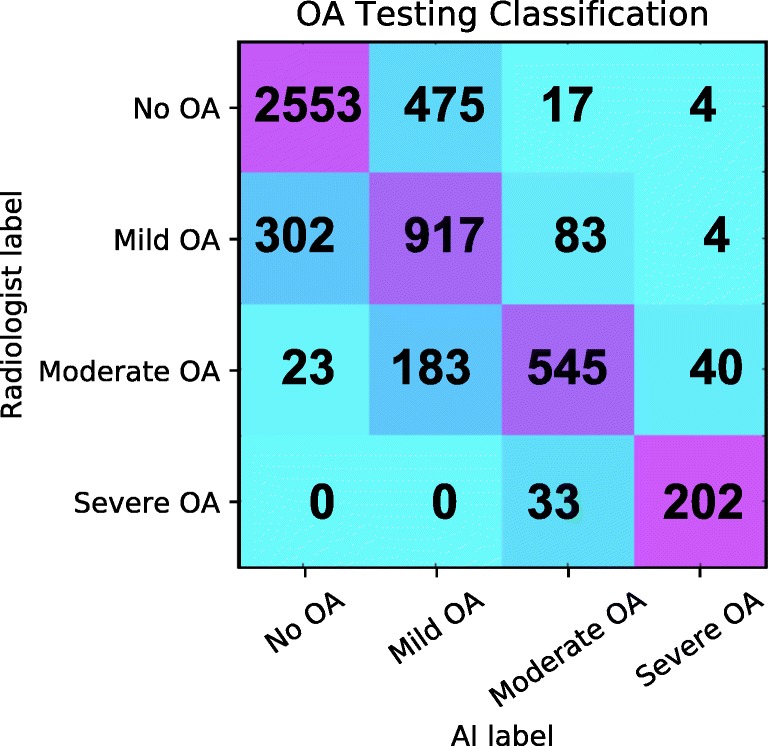
Confusion matrix for OA: true labels are the rows while the predicted labels are the columns. The diagonal represents the number of correct predictions for that class. The total number of subjects in each group can be obtained by summing that respective row. Darker squares represent a higher percentage of that group classified for a predicted label
Table 3.
Specificity and sensitivity of the ensemble DenseNet model for the training, validation, and testing datasets
| No OA (%) | Mild OA (%) | Moderate OA (%) | Severe OA (%) | ||
|---|---|---|---|---|---|
| Training | Sensitivity | 96.4 | 99.5 | 97.1 | 100 |
| Specificity | 99.9 | 96.8 | 99.9 | 99.9 | |
| Validation | Sensitivity | 83.8 | 74.2 | 75.9 | 87.6 |
| Specificity | 88.5 | 84.3 | 97.2 | 99.4 | |
| Testing | Sensitivity | 83.7 | 70.2 | 68.9 | 86.0 |
| Specificity | 86.1 | 83.8 | 97.1 | 99.1 | |
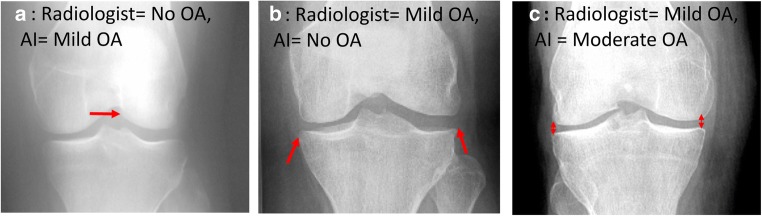
Of the 5941 testing cases, 1639 predications disagreed, 54 of which were sampled and given to our clinical radiologists to evaluate which grading they agreed most with. The internal radiologist agreed with original radiologist grading for 33 cases (61.1%), the neural network grading for 16 cases (29.6%), and neither score for 5 cases (9.3%). Specific cases where the internal radiologist agreed with the original radiologist grading can be viewed in Fig. 3a, b and a case where the radiologist agreed with the algorithm can be viewed in Fig. 3c. Of the 49 cases that the internal radiologist agreed with either the initial grading or the AI, there were 26 cases of no OA, 4 cases of mild OA, 16 cases of moderate OA, and 3 cases of severe OA. For no OA, the internal radiologist agreed with the AI grading 9.1% of the time, 75% for mild OA, 56.3% for moderate OA, and 66.7% for severe OA. However, due to the small sample sizes between the KL groups, it is difficult to say if these inter-agreement rates are statistically significant. It should also be noted that the internal radiologist did mention that a number of the provided 54 cases were particularly tough to select a correct grading for.
Fig. 3.
Examples for misclassification: (red arrows represent features of interest) a Correct grading by the radiologist, misinterpretation by the AI. This subject was graded as normal by the radiologist; however, related to a mildly oblique projection, there appears to be a prominent bony structure in the intercondylar notch that could be interpreted as an osteophyte which would result in a mild OA grade. b Correct grading by the radiologist, misinterpretation by the AI. This was graded as mild OA given small osteophytes on either side of the tibial plateau. These osteophytes appear insignificant and could have therefore potentially also been graded as no OA. c Incorrect grading by the radiologist, correct grading by the AI. This was graded as mild OA by the radiologist; however, there is lateral femoro-tibial joint space narrowing, marked by the fact that the lateral joint space should be more narrow than the medial joint space, consistent with moderate OA, which is what the algorithm graded it as moderate OA
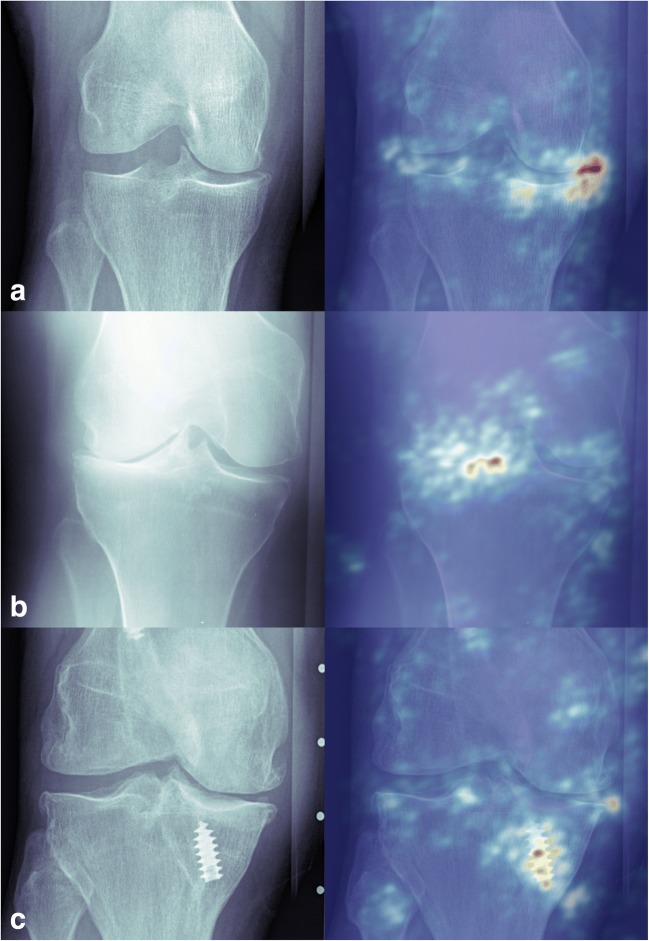
Saliency maps were then used to examine which features of the input image the algorithm was identifying as important for decision-making. In cases were the algorithm and radiologists agreed, relevant features such as osteophytes (Fig. 4a) and joint space narrowing (Fig. 4b) were identified. For cases that the algorithm misclassified the OA grading, many times it was due to the presence of hardware in the knee (Fig. 4c).
Fig. 4.
Saliency map examples. a Example of a knee with mild OA that the model also predicted as mild OA. The saliency map has highlighted the relevant osteophytes. b Knee with severe OA that the model also predicted as severe OA. The saliency map has highlighted the relevant joint space narrowing. c Knee with moderate OA that the model classified as mild OA. This misclassification was likely made due to that fact the model was assigning high importance to the screw instead of features within the joint
Discussion
Our study’s results show insight into the application of deep neural networks within the field of musculoskeletal research. It also shows consistency as well as improvement over a previous study. Antony et al. conducted a similar project for automatic KL prediction with a different neural network architecture (more closely resembling an ImageNet) using data from the OAI and Multicenter Osteoarthritis Study [19, 20]. From their reported confusion matrix, sensitivity of 82.3, 29.7, 71.7, and 78.1% and specificity of 68.1, 83.9, 97.4, and 98.7% were reported for no OA, mild, moderate, and severe OA, respectively. Using a chi-squared contingency test across OA groups, it was found with statistical significance that our model had higher sensitivity (p < 0.0001) for mild OA and higher specificity for no OA classification (p < 0.0001) than Antony et al.’s work, but lower specificity for mild OA (p = 0.004). All other comparison metrics had no statistically significant differences between results at the α < 0.05 level.
Additionally, the single-blind experiment with our internal radiologists highlights the inter-observer reliability of KL classification, which has been reported to range from 0.51 to 0.89 [5, 21, 22]. This part of the study also suggests that our reported classification accuracies are potentially 30% higher, which would add around 7.5% to each group classification getting us closer to meeting the upper ends of inter-observer agreement reported for KL classification. It is again difficult to definitively make these claims given the relatively small number of cases reviewed by our internal radiologist. It is however reassuring to see that most of the misclassifications (shown in the Fig. 2 confusion matrix) are being made with adjacent groups (i.e., the majority of moderate OA misclassifications are for mild OA).
While our neural network model does show very promising performance advantages compared to manual and previous automatic KL grading schemes, there is room for improvement in accuracy outside of the “ground truth” KL values being somewhat subjective to user preference. A large number of misclassifications came from subjects with hardware in the knee. While an algorithm is ideally generalizable enough to deal with these cases along with normal cases together, a potential solution would be to have another machine learning algorithm to detect the presence of hardware and then build two separate classifiers to assess OA progression in “hardware present” and normal cases.
Conclusion
Using state-of-the-art convolutional neural networks with novel implementations of their ensemble learning and inclusion of demographic variables directly into the network, we were able to produce a precise automatic classifier for the assessment of OA in knee radiographs. Additionally, our ensembled models have shown the ability to correctly identify relevant features within a knee radiograph that are used to make OA assessments through the analysis of saliency maps. Both of these achievements are essential for the clinical translation of this type of algorithm to assist radiologists in making more accurate and precise diagnosis with the increasing volume of radiographic images being taken in clinic.
Funding Information
This project was supported by Grant Numbers P50 AR060752 (SM), R01AR046905 (SM), K99AR070902 (VP), and R61AR073552 (SM/VP) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, United States of America (NIH-NIAMS) and Arthritis Foundation, Trial 6157. The Titan X Pascal used for this research was donated by the NVIDIA Corporation.
Compliance with Ethical Standards
Disclaimer
This content is solely the responsibility of the authors and does not necessarily reflect the views of the NIH-NIAMS or Arthritis Foundation.
References
- 1.Roos EM, Roos HP, Lohmander LS, Ekdahl C, Beynnon BD. Knee Injury and osteoarthritis outcome score (KOOS)—development of a self-administered outcome measure. J Orthop Sports Phys Ther. 1998;28(2):88–96. doi: 10.2519/jospt.1998.28.2.88. [DOI] [PubMed] [Google Scholar]
- 2.Ackerman I. Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) Aust J Physiother. 2009;55(3):213. doi: 10.1016/S0004-9514(09)70088-1. [DOI] [PubMed] [Google Scholar]
- 3.Pelletier JP, Cooper C, Peterfy C, Reginster JY, Brandi ML, Bruyère O, Chapurlat R, Cicuttini F, Conaghan PG, Doherty M, Genant H, Giacovelli G, Hochberg MC, Hunter DJ, Kanis JA, Kloppenburg M, Laredo JD, McAlindon T, Nevitt M, Raynauld JP, Rizzoli R, Zilkens C, Roemer FW, Martel-Pelletier J, Guermazi A. What is the predictive value of MRI for the occurrence of knee replacement surgery in knee osteoarthritis? Ann Rheum Dis. 2013;72(10):1594–1604. doi: 10.1136/annrheumdis-2013-203631. [DOI] [PubMed] [Google Scholar]
- 4.Kohn MD, Sassoon AA, Fernando ND. Classifications in brief: Kellgren-Lawrence classification of osteoarthritis. Clin Orthop Relat Res. 2016;474(8):1886–1893. doi: 10.1007/s11999-016-4732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16(4):494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunther KP, Sun Y. Reliability of radiographic assessment in hip and knee osteoarthritis. Osteoarthr Cartil. 1999;7(2):239–246. doi: 10.1053/joca.1998.0152. [DOI] [PubMed] [Google Scholar]
- 7.Kallenberg M, Petersen K, Nielsen M, Ng AY, Diao P, Igel C, Vachon CM, Holland K, Winkel RR, Karssemeijer N, Lillholm M. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging. 2016;35(5):1322–1331. doi: 10.1109/TMI.2016.2532122. [DOI] [PubMed] [Google Scholar]
- 8.Lee H, Grosse R, Ranganath R, Ng AY. Unsupervised learning of hierarchical representations with convolutional deep belief networks. Commun ACM. 2011;54(10):95–103. doi: 10.1145/2001269.2001295. [DOI] [Google Scholar]
- 9.Li XG, Hong CF, Yang YN, Wu XH: Deep neural networks for syllable based acoustic modeling in Chinese speech recognition. Asia-Pacific Signal and Information Processing Association Annual Summit and Conference (Apsipa), 2013
- 10.Hinton G, Deng L, Yu D, Dahl G, Mohamed AR, Jaitly N, Senior A, Vanhoucke V, Nguyen P, Sainath T, Kingsbury B. Deep neural networks for acoustic modeling in speech recognition. IEEE Signal Process Mag. 2012;29(6):82–97. doi: 10.1109/MSP.2012.2205597. [DOI] [Google Scholar]
- 11.Pan J, Liu C, Wang ZG, Hu Y, Jiang H: Investigation of Deep Neural Networks (Dnn) for Large Vocabulary Continuous Speech Recognition: Why Dnn Surpasses Gmms in Acoustic Modeling. 2012 8th International Symposium on Chinese Spoken Language Processing, 2012, pp 301–305
- 12.Leung MKK, Delong A, Alipanahi B, Frey BJ. Machine learning in genomic medicine: a review of computational problems and data sets. Proc IEEE. 2016;104(1):176–197. doi: 10.1109/JPROC.2015.2494198. [DOI] [Google Scholar]
- 13.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 14.Bengio Y, Lee H. Editorial introduction to the neural networks special issue on deep learning of representations. Neural Netw. 2015;64:1–3. doi: 10.1016/j.neunet.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Norman B, Pedoia V, Majumdar S: Use of 2D U-net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology:172322, 2018 [DOI] [PMC free article] [PubMed]
- 16.Gao Huang ZL, Kilian Q: Weinberger, Laurens van der Maaten. Densely connected convolutional networks. ARXIV;eprint arXiv:1608.06993:12, 2016
- 17.Cheng JAB, Mark JL: The relative performance of ensemble methods with deep convolutional neural networks for image classification. arXiv:170401664, 2017 [DOI] [PMC free article] [PubMed]
- 18.Karen Simonyan AV, Zisserman A: Deep inside convolutional networks: visualising image classification models and saliency maps. ARXIV;1312.6034v2, 2014
- 19.Joseph Antony KM, Connor NEO, Moran K: Quantifying radiographic knee osteoarthritis severity using deep convolutional neural networks. Pattern Recogn:1195–1200, 2016
- 20.UCSF. Multicenter Osteoarthritis Study (MOST)
- 21.Wright RW, Group M Osteoarthritis classification scales: interobserver reliability and arthroscopic correlation. J Bone Joint Surg Am. 2014;96(14):1145–1151. doi: 10.2106/JBJS.M.00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gossec L, Jordan JM, Mazzuca SA, Lam MA, Suarez-Almazor ME, Renner JB, Lopez-Olivo MA, Hawker G, Dougados M, Maillefert JF, OARSI-OMERACT task force "total articular replacement as outcome measure in OA" Comparative evaluation of three semi-quantitative radiographic grading techniques for knee osteoarthritis in terms of validity and reproducibility in 1759 X-rays: report of the OARSI-OMERACT task force. Osteoarthr Cartil. 2008;16(7):742–748. doi: 10.1016/j.joca.2008.02.021. [DOI] [PubMed] [Google Scholar]