Abstract
Metal tolerance proteins (MTPs) play an important role in the transport of metals at the cellular, tissue and whole plant levels. In the present study, 11 MTP genes were identified and these clustered in three major sub-families Fe/Zn-MTP, Zn-MTP, and Mn-MTP, and seven groups, which are similar to the grouping of MTP genes in both Arabidopsis and rice. Vitis vinifera metal tolerance proteins (VvMTP) ranged from 366 to 1092 amino acids, were predicted to be located in the cell vacuole, and had four to six putative TMDs, except for VvtMTP12 and VvMTP1. The VvMTPs had putative cation diffusion facilitator (CDF) domains and the putative Mn-MTPs also had zinc transporter dimerization domains (ZD-domains). V. vinifera Mn-MTPs had gene structures and motif distributions similar to those of the Fe/Zn-MTP and Zn-MTP sub-families. The upstream regions of VvMTP genes had variable frequencies of cis-regulatory elements that could indicate regulation at different developmental stages and/or differential regulation in response to stress. Comparison of the VvMTP coding sequences with known miRNAs found in various plant species indicated the presence of 13 putative miRNAs, with 7 of these associated with VvMTPs. Temporal and spatial expression profiling indicates a potential role for VvMTP genes during growth and development in grape plants, as well as the involvement of these genes in plant responses to environmental stress, especially osmotic stress. The data generated from this study provides a basis for further investigation of the roles of MTP genes in grapes.
Keywords: Bioinformatics, Cation diffusion facilitator, Gene expression, Heavy metals, Protein structure
Introduction
Certain metal cations, e.g., Mn+2, Zn+2, Fe+2, Cu+2, Co+2, and Ni+2, are essential for many cellular and physiological functions in plants, including photosynthesis, DNA replication, protein processing, electron transport in the chloroplasts and mitochondria, and numerous other metabolic processes. They are important cofactors for many regulatory proteins and enzymes (Ricachenevsky et al. 2013; Yuan et al. 2012) and a deficiency in essential metal ions can have negative impacts on plant growth and development (Marschner 2011). The accumulation of excessive amounts of metal ions within plant cells can result in toxicity and growth inhibition (Cambrollé et al. 2015; Thomine and Vert 2013). In contrast, non-essential metals, such as Cd+2, Cr+2, Pb+2, Al+2, and Hg, have no known functions in plants and are toxic at very low levels, having negative impacts on plant growth and development (Gill et al. 2013; Hayat et al. 2012; Wang et al. 2013). In addition to their negative effects in plants, some non-essential metals can also threaten human health when they accumulate in crop plants used for food.
The presence of heavy metals in soils such as groundwater supplies is recognized as a significant global environmental issue. Non-essential toxic metals, e.g., Pb+2, can have a high residence time in soils (e.g., 150–5000 years). When taken up by crop plants grown on contaminated soils, they can pose a threat to human health for many years (Yang et al. 2005). Consumption of foods from plants grown on soils contaminated with toxic metal ions has been linked to diseases in humans, including several types of cancer (Singh et al. 2016).
Plants have complex mechanisms to regulate cellular concentrations of metal ions, including controlling the uptake and movement of metal ions at both cell and tissue levels, and the chelation and sequestration/detoxification of metal ions within cells (Clemens et al. 2002; Hall 2002). The vacuole is the main site of detoxification/sequestration and storage of excess metal ions in plant cells (Singh et al. 2011), and the tonoplast contains many metal ion (Me) transporter proteins from different transporter families. Cation diffusion facilitators (CDFs) have been identified in both prokaryotes and eukaryotes (Singh et al. 2016), and are mainly Me2+/H+ counter ion transport proteins involved in the transport of Cd2+, Fe2+, Zn2+, Mn2+, Co2+, or Ni2+ from the cytosol into organelles or out of cells (Gustin et al. 2011; Migocka et al. 2015; Montanini et al. 2007; Ricachenevsky et al. 2013). The CDF family of metal ion transport proteins contains three main sub-families: (1) Mn-CDFs, (2) Fe/Zn-CDFs and (3) zinc CDFs transporting Zn and other metal ions, but not Fe or Mn (Montanini et al. 2007). Most CDF proteins have four to six transmembrane domains (TMDs) and cytoplasmic C-terminal domains (CTDs) (Kolaj-Robin et al. 2015; Lu et al. 2009; Lu and Fu 2007). They normally have six predicted multiple transmembrane domains interconnected by extra- and intracellular interconnecting loops, with one cytosolic loop usually containing a histidine-rich domain (Haney et al. 2005; Montanini et al. 2007).
The CDFs found in plant cells are generally named metal tolerance proteins (MTPs) (Fu et al. 2017), with the MTPs being divided into seven phylogenetic groups. The Zn-CDFs are placed in groups 1 (MTP1–MTP4), 5 (MTP5) and 12 (MTP12), the Fe/Zn-CDFs in groups 6 (MTP6) and 7 (MTP7), and groups 8 (MTP8) and 9 (MTP9–MTP11) contain the Mn-CDFs (Gustin et al. 2011).
The group 1 CDFs have the ability to transport not only Zn but also other metallic ions, namely: Co (AtMTP3 and HvMTP1 from Hordeum vulgare), Cd (CsMTP1 and CsMTP4 from Cucumis sativus, OsMTP1 from Oryza sativa, CitMTP1 from Citrus sinensis), Cu (CitMTP1), Ni (OsMTP1) and Fe (OsMTP1), into the vacuole (Arrivault et al. 2006; Blaudez et al. 2003; Fu et al. 2017; Kobae et al. 2004; Menguer et al. 2013; Migocka et al. 2015; Shahzad et al. 2010; Yuan et al. 2012). The MTP8 proteins from Stylosanthes hamata (ShMTP8), Oryza sativa (OsMTP8.1), Cucumis sativus (CsMTP8), Hordeum vulgare (HvMTP8.1 and HvMTP8.2), Citrus sinensis (CitMTP8 and CitMTP8.1), Triticum aestivum (TaMTP8) and Arabidopsis thaliana (AtMTP8) transport Mn into the vacuole or Golgi apparatus (Chen et al. 2013; Delhaize et al. 2003; Eroglu et al. 2016; Fu et al. 2017; Migocka et al. 2015; Vatansever et al. 2017). While MTPs in Arabidopsis thaliana have received much attention, MTPs in other plant species are less well understood (Ueno et al. 2015). AtMTP11 is involved in Mn tolerance and is localized in endosome vesicles (Delhaize et al. 2003; Peiter et al. 2007). AtMTP12 has 14 TMDs, forms a heterodimer with AtMTP5, and is involved in the transport of Zn into Golgi bodies (Fujiwara et al. 2015). MTPs in plants are named according to their similarities to members of the A. thaliana MTP families (Gustin et al. 2011).
Grape (Vitis vinifera L.) is an important fruit crop and sequencing of its entire genome is completed, facilitating classification and comparative genomics (Jaillon et al. 2007). The present whole genome association study was carried out to identify the MTP gene family in grape and interpret their sequences. To help understand the possible functions of grape MTPs, the expression of MTP genes was investigated with respect to developmental stage and exposure to environmental stress, using a microarray data approach. This study aims to provide information important for understanding the relevance of MTPs for the growth and development of this important crop plant.
Materials and methods
Identification of MTP gene family in grape
To determine grape MTP gene family members, the protein sequences of A. thaliana (AT2G46800.1, AT3G58810.1, AT2G29410.1, AT2G47830.1, AT2G04620.1, AT2G39450.1, AT1G79520.2, AT1G16310.1, AT1G51610.1, AT3G58060.1, AT3G12100.1, AT3G61940.1) (Fu et al. 2017) and Oryza sativa (Os05g38670, Os04g23180, Os02g58580, Os03g12530, Os01g62070, Os05g03780, Os02g53490, Os08g32650, Os01g03914 and Os03g22550) (Vatansever et al. 2017) were obtained and these sequences are compared to the grape genome, using the tBLASTn method (Goodstein et al. 2011) to search the phytozome (https://phytozome.jgi.doe.gov/) database. The hidden Markov model (HMM) profile of the cation efflux domain (PF01545) was acquired from the Pfam database (http://pfam.xfam.org) and was used to validate the presence of cation efflux domains in putative MTPs using the hmmscan tool (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan).
Phylogenetic analysis and nomenclature
Multiple sequences were aligned using ClustalX 2.0.8 (Thompson et al. 1994) and studies of phylogenetic relationships were carried out using the MEGA 5.2 with neighbor-joining (NJ) method and 1000 bootstrap replicates (Felsenstein 1985). The recognized MTP genes from grape were named VvtMTP genes based on their phylogenetic distribution and sequence similarities with MTPs in Arabidopsis and rice, and the similarities of sequences to AtMTPs were confirmed using MatGAT software.
Sequence analysis of MTP proteins
The molecular weights (kDa) and isoelectric points (pI) of MTP proteins were determined using ProtParam (http://web.expasy.org/protparam) (Gasteiger et al. 2005) and the prediction of protein transmembrane helices is determined using protter [http://wlab.ethz.ch/protter/start; (Omasits et al. 2013]. The subcellular localization of proteins was predicted using the Plant-mPLoc server [http://csbio.sjtu.edu.cn/bioinf/plantmulti/; (Hall 2002)].
Conserved motifs were predicted using the MEME (http://meme-suite.org/tools/meme) tool with the following parameters; 60 ≥ widths ≥ 5 and the maximum number of motifs 6 (Bailey et al. 2009), and the functionalities of these motifs were determined using the hmmscan tool (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). Identified MTP sequences were aligned using ClustalW, and identity remnants were found. Predicted 3D protein structure models for VvMTPs were constructed using Phyre2, by homology modeling under the intensive model (http://sbg.bio.ic.ac.uk/phyre2/) (Kelley et al. 2015), using profile–profile matching and predicted secondary structure. The quality and reliability of the models were evaluated by Ramachandran plot analysis using the VADAR server (http://vadar.wishartlab.com/).
Chromosomal location and duplication
The grape genome database was used to identify the chromosomal positions of the VvMTP genes and MapChart (https://www.wur.nl/en/show/MapChart-2.32.htm) was used to generate the location images. Identification of segmental duplication was performed by searching for genome duplications using PGDD (http://chibba.agtec.uga.edu/duplication/ database of plants). The presence of genes on the same chromosome, with the highest spacing of 20 genes, were considered as tandem duplication with coverage of > 75% and similarity > 75% in aligned sequences (Liu et al. 2014; Ozyigit et al. 2016).
Gene structure, promoter analysis and predicting microRNA target sites
The gene structure display server (GSDS, http://gsds.cbi.pku.edu.cn/) was used to analyze the exon–intron distributions and splicing phases of VvMTP genes. Splicing is divided into three phases, based upon the location of splicing. Phase 1 is where splicing occurs in the first nucleotide after the codon. The second phase occurs in the second nucleotide after the codon and zero phases are also generated in the third nucleotide after the codon (Sharp 1981). The promoter sequence for each VvMTP gene was defined as 1500 bp upstream from the start codon. Promoter analysis, using PlantCARE database (http://www.bioinformatics.psb.ugent.be/webtools/plantcare/html/), identified all the cis-regulatory elements (CREs). The CDS sequences of genes were examined for VvMTP-targeted miRNAs present in the psRNATarget database (http://plantgrn.noble.org/psRNATarget/) schema V2, using the following parameters: max expectation 3 and target accessibility (UPE) 25.
Gene expression analysis using microarray data
To investigate the developmental expression profile of the VvMTP genes, the Ensembl plant database (https://plants.ensembl.org/index.html) was used to find corresponding gene IDs for given VvMTPs. Using the Grape eFP Browser (http://bar.utoronto.ca/efp_grape/cgi-bin/efpWeb.cgi), data from 54 plant specimens, of green and woody tissues and organs at various developmental stages, were extracted based on the array expression atlases for VvMTPs.
The MTP gene family in Arabidopsis, which had homology to VvMTPs, was used for expression profiling of MTPs in response to abiotic stresses, using the Affymetrix Arabidopsis ATH1 Genome Array microarray data in the NCBI Gene Expression Omnibus (GEO) database. The accession numbers identified in the GEO database included salinity (GSE5623), drought (GSE5624), osmotic (GSE5622), cold (GSE5621), genotoxic (GSE5625), UV (GSE5626), wounding (GSE5627), heat (GSE5628) stress and control (GSE5620) genes. The RMA (Robust Multi-array Average) algorithm was used to normalize the consequent unprocessed data (CEL files) using the RMAexpress 1.0.5 software and log2 transformation. The Probe Match tool in the NetAffx Analysis Center was used to identify probe sets corresponding to AtMTP genes and changes in the transcriptional rates of various MTP genes in plants under stress were measured.
Hierarchical clustering of developmental and stress-associated gene expression data was performed using Pearson correlation and a complete linkage algorithm, and heat maps were generated using Mev4.0 software (Saeed et al. 2006).
Results
Identification of MTP gene families in grape and phylogenetic tree construction and analysis
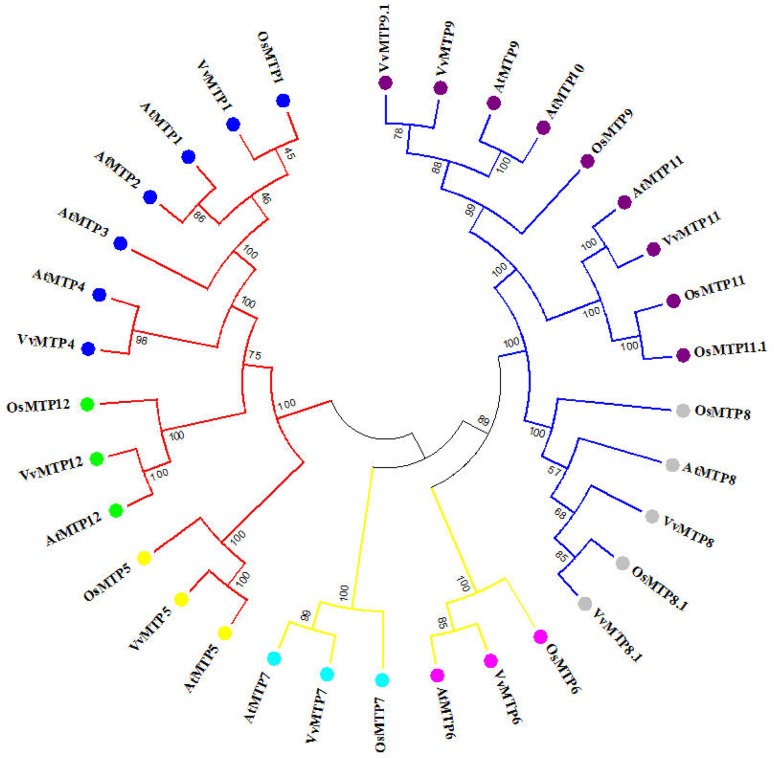
To identify the MTP genes present in the grape genome, a homology search for 12 MTP proteins found in A. thaliana (Fu et al. 2017) and 10 MTPs found in O. sativa (Vatansever et al. 2017) was performed using tBLASTn. Subsequent HMM verification resulted in 14 MTP sequences that contained the cation efflux domain in the grape genome. Three protein sequences (GSVIVG01029613001, GSVIVG01029603001, and GSVIVG01029622001) were removed due to short protein lengths and a low E values, because defective sequences and domains make phylogenetic studies difficult (Jiang et al. 2010), leaving 11 MTP proteins identified in the grape genome. To examine the evolutionary relationships of this protein family, a phylogenetic tree of grape, Arabidopsis, and rice MTPs was constructed (Fig. 1). According to sequence similarities (MatGAT software) with the Arabidopsis and rice MTP families, the MTPs of grape were, respectively, named VvMTP1 to VvMTP12. The GSVIVG01026116001 and GSVIVG01036746001 sequences had higher similarities to AtMTP9 than AtMTP10 and were named VvMTP9 and VvMTP9.1, and two orthologs of VvMTP8 were found that corresponded to AtMTP8. According to the classification described by Montanini et al. (2007), the VvMTPs were divided into three sub-families Mn-MTP, Fe/Zn-MTP, and Zn-MTP that were similar to the AtMTPs and OsMTPs. VvMTP1, VvMTP4, VvMTP5, and VvMTP12 all belonged to Zn-MTP family; VvMTP6 and VvMTP7 to Fe/Zn-MTP and VvMTP8, VvMTP8.1, VvMTP9, VvMTP9.1 and VvMTP11 to Mn-MTP.
Fig. 1.
A phylogenetic tree based on protein sequences of MTP family members of grape, rice, and Arabidopsis, and constructed using the neighbor-joining (NJ) method and MEGA 5.2 software. The identified proteins were classified into three sub-families (Fe/Zn-MTPs, Mn-MTPs, and Zn-MTPs) and seven groups based upon previous reports and their phylogenetic relationships. The Zn-MTP sub-family (red line) contains 1: MTP1 to MTP4 (blue), 5: MTP5 (yellow), and 12: MTP12 (green) groups; the Mn-CDF sub-family (blue line) contains 8: MTP8 (gray), and 9: MTP9 to MTP11 (violet) groups; the Zn/Fe-CDF sub-family (yellow line) contains 6: MTP6 (pink), and 7: MTP7 (light blue) groups. Bootstrap values are indicated (1000 replicates)
Sequence analysis of grape MTP proteins
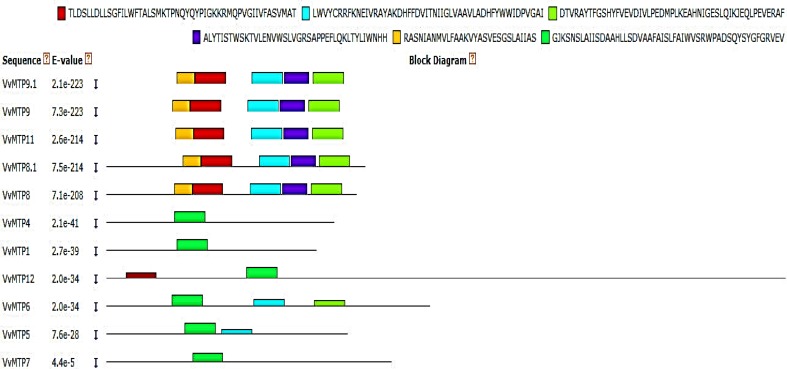
The putative VvMTP genes were predicted to encode proteins ranging from 366 to 1092 amino acids. The molecular weights and pIs of these grape proteins ranged from 37.09 to 123.68 kDa and 4.90–8.59, respectively (Table 1). All proteins were predicted to accumulate in the vacuole and to have four to six TMDs with cytosolic N and C termini, except for VvtMTP12 and VvMTP1 that had 12 and 7 predicted TMDs, respectively. The six conserved motifs predicted in VvMTP proteins, using the MEME tool, (Fig. 2) varied in size, with 1–3 and 6 being 50 amino acids, whereas motifs 4 and 5 were 40 and 29 residues, respectively. The predicted motif sequences are: motif 1 TLDSLLDLLSGFILWFTALSMKTPNQYQYPIGKKRMQPVGIIVFASVMAT, motif 2 LWVYCRRFKNEIVRAYAKDHFFDVITNIIGLVAAVLADHFYWWIDPVGAI, motif 3 DTVRAYTFGSHYFVEVDIVLPEDMPLKEAHNIGESLQIKJEQLPEVERAF, motif 4 ALYTISTWSKTVLENVWSLVGRSAPPEFLQKLTYLIWNHH, motif 5 RASNIANMVLFAAKVYASVESGSLAIIAS, and motif 6 GJKSNSLAIISDAAHLLSDVAAFAISLFAIWVSRWPADSQYSYGFGRVEV. Motifs 1, 2 and 6 were associated with the cation efflux domain (cation_efflux; PF01545) and motif 3 with the zinc transporter dimerization domain (ZT_dimer; PF16916), while motifs 4 and 5 were not assigned by the hmmscan tool. Highly similar motifs are expected to have similar functions. VvMTPs belong to the Mn-MTP group contained five of the six motif sequences, namely two cation_efflux (1 and 2) motifs, and a ZT_dimer motif. VvMTP6 contained two cation_efflux motifs (2 and 6) and a ZT_dimer motif. VvMTP5 contained two cation_efflux motifs (2 and 6) and VvMTP12 contained two cation_efflux motifs (1 and 6). VvMTP1, VvMTP4, and VvMTP7 had only one cation_efflux motif (6) that no one of Mn-MTP group members has this motif. The VvMTP sequences were aligned using ClustalW and the localization of the six motifs is showed with different color rectangles in Fig. 3. The motifs 1 and 5 had a slight overlap and the motif 6 completely spans the area containing motifs 1 and 5. It is noteworthy that the Gly (G) in motifs 1 and 6 was preserved in all VvMTP sequences.
Table 1.
MTP proteins information for grape
| Gene | Accession number | Peptide length | PI | MW(kDa) | No. of TMDs N to C | Subcellular localization |
|---|---|---|---|---|---|---|
| VvMTP6 | GSVIVG01010968001 | 520 | 6.90 | 56.91 | 4/out to out | Vacuole |
| VvMTP11 | GSVIVG01016640001 | 399 | 4.90 | 45.12 | 6/into in | Vacuole |
| VvMTP1 | GSVIVG01019690001 | 388 | 6.45 | 37.09 | 7/out to in | Vacuole |
| VvMTP4 | GSVIVG01024865001 | 366 | 6.18 | 40.90 | 6/into in | Vacuole |
| VvMTP8.1 | GSVIVG01025383001 | 416 | 5.44 | 47.07 | 4/into in | Vacuole |
| VvMTP9 | GSVIVG01026116001 | 423 | 8.24 | 48.56 | 5/into out | Vacuole |
| VvMTP7 | GSVIVG01029626001 | 459 | 8.59 | 50.59 | 4/into in | Vacuole |
| VvMTP12 | GSVIVG01032551001 | 1092 | 7.31 | 123.68 | 12/out to out | Vacuole |
| VvMTP8 | GSVIVG01033471001 | 403 | 5.93 | 45.62 | 5/into out | Vacuole |
| VvMTP5 | GSVIVG01033491001 | 388 | 6.50 | 43.45 | 6/into in | Vacuole |
| VvMTP9.1 | GSVIVG01036746001 | 400 | 6.12 | 45.73 | 6/into in |
Cell membrane Vacuole |
Gene (gene name based on phylogenetic distribution with sequence similarity to Arabidopsis and rice MTPs; MW (predicted molecular weight); pI (predicted isoelectric point); predicted TMDs (no. of the transmembrane domains); in (cytoplasmic) or out (extracellular) predicted from N to C terminus
Fig. 2.
Block diagram outlining the six most highly conserved motif sequences in the 11 putative VvMTP proteins. The motifs 1, 2 and 6 were associated with the cation efflux domain (Cation_efflux; PF01545) and motif 3 is related to the zinc transporter dimerization domain (ZT_dimer; PF16916), while motifs 4 and 5 did not associate with any motifs
Fig. 3.
Multiple alignments of putative VvMTP protein sequences and localization of the six motifs are shown with rectangles in various colors; motif 1 with violet, motif 2 with yellow, motif 3 with green, motif 4 with gray, motif 5 with red and motif 6 with blue. The relevant percentage identity is shaded in gray
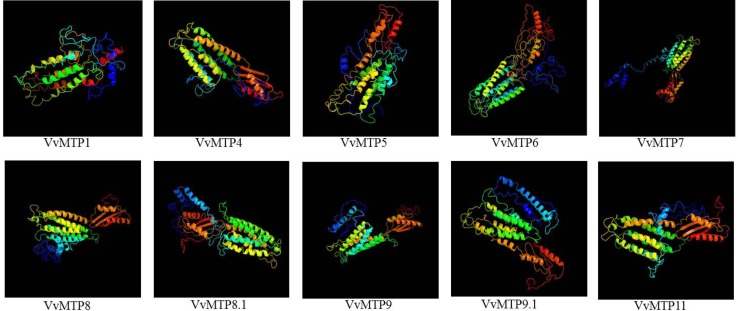
Homology modeling of putative VvMTP proteins
Ten VvMTPs were modeled using Phyre2 (Fig. 4). Due to the presence of numerous undefined residues in the amino acid sequence, VvMTP12 was not modeled for the reasons detailed above. Models were predicted using the maximum coverage heuristics (MCH) of alignment between templates and query, percent positive substitutions and a confidence level for the queried sequences. All of VvMTP (except VvMTP12) were modeled at > 90% confidence level. Two known examples, c2qfiB (from the structure of the zinc transporter yiip) and c3j1zP (from the cation efflux protein transmembrane domain-like family), were used for the modeling of VvMTP4, VvMTP5, VvMTP7, VvMTP8, and VvMTP9. For the other VvMTPs, in addition to the above-known examples, other known examples were also used: d2qfia2 (from the inward-facing conformation of the zinc transporter yiip by 2 cryo-electron microscopy) to model VvMTP1; d1ng0a (from the cation efflux protein cytoplasmic domain like), c5ensA (from the rhodamine bound structure of bacterial efflux pump), d1c8na (from the Tombusviridae-like VP family), and c1ng0A (from the three-dimensional structure of cocksfoot mottle virus at 2.7a2 resolution) to model VvMTP6; c2enkA (from the structure of a putative DNA-binding domain of the 2 human solute carrier family 30 zinc transporter protein) to model VvMTP8.1; c5gasN (from the Thermus thermophilus v/a-ATPase, conformation 2) to model VvMTP9.1; and d1d4ua1 (from DNA repair factor XPA DNA- and RPA-binding domain, C-terminal subdomain) to model VvMTP11. The analysis of modeled structure of VvMTP proteins using Ramachandran plot showed that 61–85% of residues were in the allowed region, 10–24% of residues were in the generous region, and 2–7% of residues were in the outside region indicating the reliability of the VvMTP structural models (Table 2).
Fig. 4.
predicted, using the Phyre2 server intensive model, 3D models of grape MTP proteins. The structure of VvMTP12 could not reliably predict because of nonspecific sequential residues. Models were colored using rainbow from the N → C terminus
Table 2.
VvMTPs were modeled using Phyre2
| VvMTPs (query) | Template proteins using for modeled |
|---|---|
| VvMTP4 | c2qfiB, c3j1zP |
| VvMTP5 | c2qfiB, c3j1zP |
| VvMTP7 | c2qfiB, c3j1zP |
| VvMTP8 | c2qfiB, c3j1zP |
| VvMTP9 | c2qfiB, c3j1zP |
| VvMTP1 | c2qfiB, c3j1zP, d2qfia2 |
| VvMTP6 | c2qfiB, c3j1zP, d1ng0a, c5ensA, d1c8na, c1ng0A |
| VvMTP8.1 | c2qfiB, c3j1zP, c2enkA, |
| VvMTP9.1 | c2qfiB, c3j1zP, c5gasN |
| VvMTP11 | c2qfiB, c3j1zP, d1d4ua1 |
Models were predicted using the maximum coverage heuristics (MCH) of alignment between templates and query, c2qfiB (from the structure of the zinc transporter yiip) and c3j1zP (from the cation efflux protein transmembrane domain-like family), d2qfia2 (from the inward-facing conformation of the zinc transporter yiip by 2 cryo-electron microscopy), d1ng0a (from the cation efflux protein cytoplasmic domain-like), c5ensA (from the rhodamine bound structure of bacterial efflux pump), d1c8na (from the Tombusviridae-like VP family), c1ng0A (from the three-dimensional structure of cocksfoot mottle virus at 2.7a2 resolution), c2enkA (from the structure of a putative DNA-binding domain of the 2 human solute carrier family 30 zinc transporter protein), c5gasN (from the Thermus thermophilus v/a-ATPase, conformation 2), d1d4ua1 (from DNA repair factor XPA DNA- and RPA-binding domain, C-terminal subdomain)
Sequence analysis of putative MTP genes
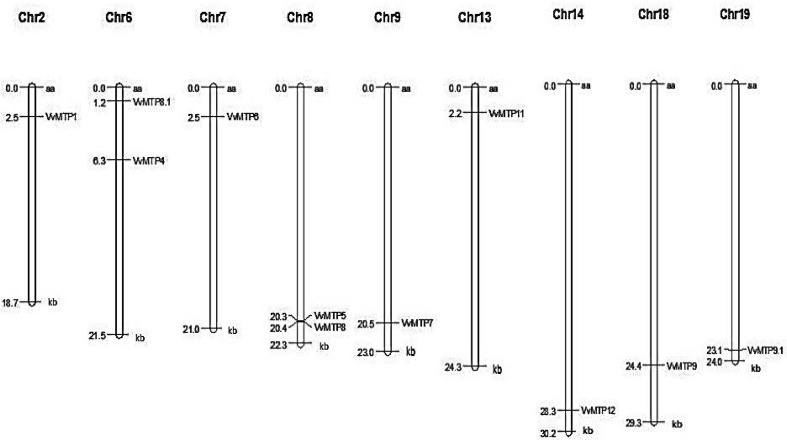
Mapping of the VvMTP genes onto grape chromosomes utilized MapInspect software (Fig. 5), with the sizes of the grape chromosomes being obtained from Wang et al. (2014). Analysis showed that the putative VvMTP genes are distributed on 9 of the 19 grape chromosomes. The locations of VvMTP1, VvMTP6, VvMTP7, VvMTP11, VvMTP12, VvMTP9 and VvMTP9.1 were mapped to chromosomes 2, 7, 9, 13, 14, 18 and 19, respectively. VvMTP4 and VvMTP8.1 were mapped to chromosome 6, and VvMTP5 and VvMTP8 to chromosome 8.
Fig. 5.
Distribution of grape MTP genes in 9 out of the 19 chromosomes
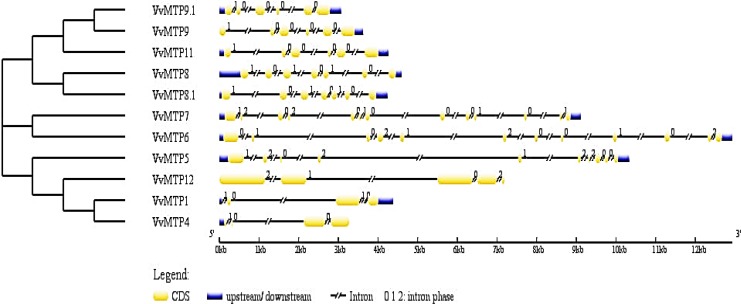
There was no tandem or segmental duplication between any VvMTP genes. Investigation of the exon–intron structure of the VvMTP gene family showed that these genes contained 4–13 exons. The VvMTPs in the same group and then in the same sub-family showed a similar gene structure, particularly the Mn-MTP sub-family (Fig. 6). Exon–intron structure in Mn-MTP group had six or seven exons, with the highest number of exons and introns found in the Zn/Fe-MTP.
Fig. 6.
Diagram of VvMTP gene structures according to the phylogenetic relationship. Coding DNA sequences of VvMTP are shown with yellow boxes. Thick blue lines at either terminal of the genes show UTRs (untranslated regions). Thin lines show introns. The numbers are for the splicing phase
Intron splicing in the VvMTPs of grape would, in general, involve all three phases, except for Mn-MTP sub-family that had zero and first splicing phases only. At the DNA level, the VvMTP genes, VvMTP6, VvMTP5, and VvMTP7, were the largest being approximately 13, 10 and 9 kb, respectively, and having the greatest number of introns.
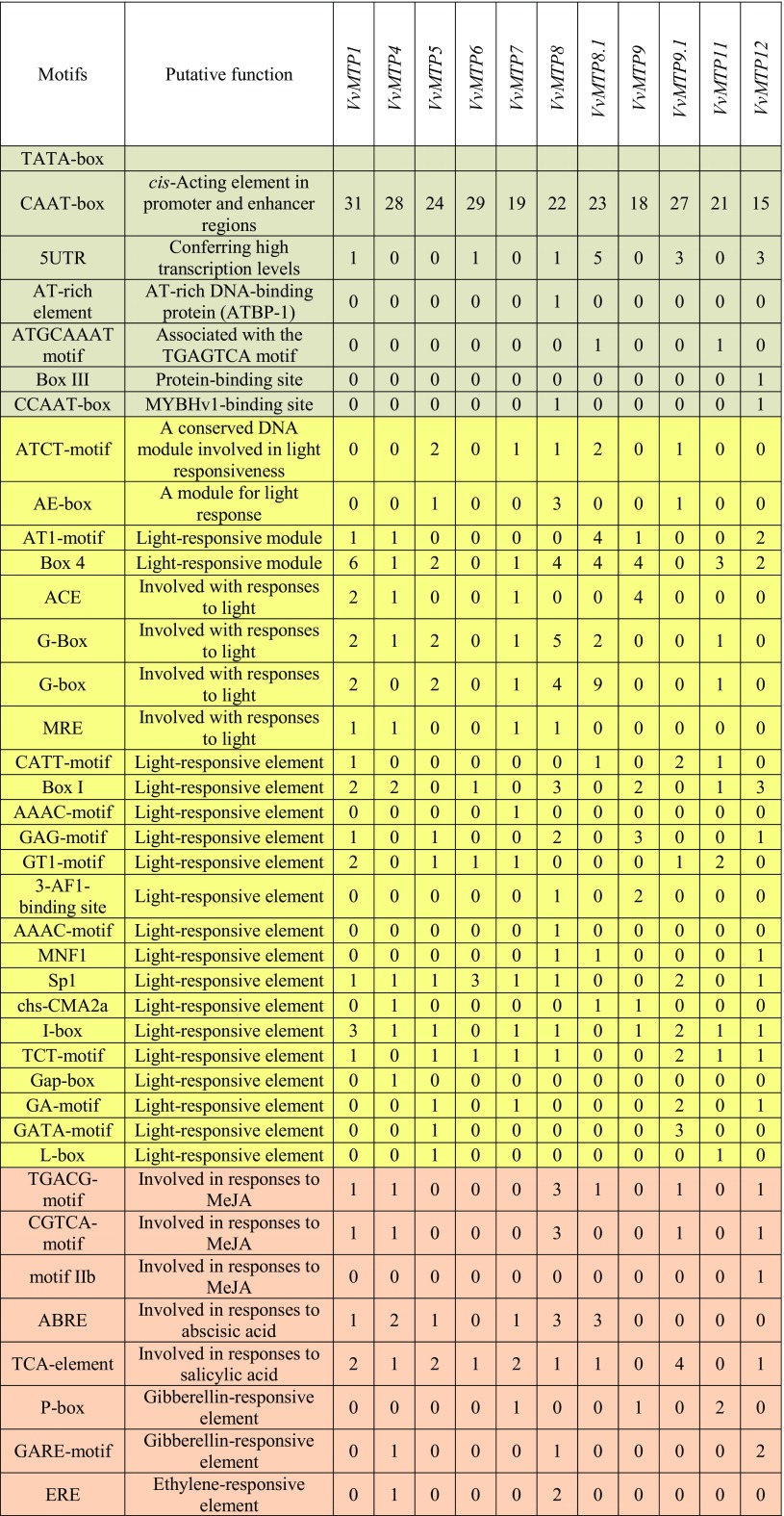
To understand the mechanism of VvMTP gene transcription, a search for cis-regulatory elements (CREs) in the 1.5 kb upstream of these genes was conducted using the PlantCare database. The result of promoter analyses, which excluded the unknown motifs, identified 54 different CREs in the upstream regions of VvMTP genes that had the potential to regulate gene expression in response a total of five groups of factors, both internal and external, i.e., general regulatory elements, light regulated, developmentally regulated, phytohormone responsive, and environmental stress responsive (Table 3). The frequency of these elements in the regulatory region of each corresponding gene, as well as their overall frequency in family members, is very diverse. Elements such as CAAT- and TATA-boxes (the numbers are not shown) were frequently found in all of the VvMTP genes. Most CREs in the VvMTP genes were related to light-responsive elements and are displayed in yellow in Table 3. Phytohormone-responsive CREs were also identified in the upstream region of VvMTP that could be involved in MeJA, abscisic acid, salicylic acid, gibberellin, ethylene, and auxin-mediated responses. A TCA element, which participates in salicylic acid responses, was shared in most VvMTP promoters, expect for VvMTP9 and VvMTP11, with this element more abundant than other phytohormone-responsive CREs. CREs involved in the regulation of developmental process, i.e., the violet and Skn-1_motif, which is necessary for the expression of genes involved in endosperm development was frequently found in all VvMTP genes expect for VvMTP6. Stress-related CREs, i.e., the pink and circadian CREs (involved in diurnal rhythm regulation), HSE (involved in high-temperature stress responses) and ARE (a regulatory element required for the initiation of anaerobic metabolism), were found in most VvMTP, expect for VvMTP9, VvMTP12, and VvMTP9, respectively. 5UTR (conferring high transcription levels), AT-rich elements (AT-rich DNA-binding protein ATBP-1), the ATGCAAAT motif (associated with the TGAGTCA motif), Box III (protein-binding site) and CCAAT-box (MYBHv1-binding site) were other CREs also found in VvMTPs.
Table 3.
Frequency and function of cis-regulatory elements (CREs) in the promoter regions of VvMTP genes
CREs of VvMTPs are divided into five groups depending on their putative major cellular functions, i.e., general regulatory elements (Gray), light responsive (yellow), regulation of plant development (violet), phytohormone responsive (orange), environmental stress responsive (pink)
The results of VvMTP coding sequence analysis comparison to miRNAs of various plant species identified 13 miRNAs from seven VvMTPs (Table 4) and seven of VvMTPs were potential targets for cleavage inhibition. VvMTP1 and VvMTP12 were targets for three miRNAs (bdi-miR5180a, bdi-miR5180b and ptc-miR473b) and (aly-miR158b-5p, ath-miR398b-5p, ath-miR398c-5p), respectively. Two VvMTPs were targeted by two individual miRNAs; aly-miR156 h-3p and smo-miR1099 for VvMTP8, and ptc-miR6456 and osa-miR1882e-3p for VvMTP9.1. VvMTP11, VvMTP7 and VvMTP8.1 were targeted by one miRNAs, ppt-miR1221-5p, osa-miR2875 and ppt-miR1076-5p, respectively.
Table 4.
Prediction of miRNAs for VvMTP transcripts
| miRNA Acc. | Target gene | Alignment | Inhibition |
|---|---|---|---|
| bdi-miR5180a | VvMTP1 |
miRNA 21UCAAGUUUUGACUCUGUGAAU 1 Target473 UCUUCAGGAUUGAGAUACUUG 93 |
Cleavage |
| bdi-miR5180b | VvMTP1 |
miRNA 21 UCAAGUUUUGACUCUGUGAAU 1 Target 473 UCUUCAGGAUUGAGAUACUUG 493 |
Cleavage |
| ptc-miR473b | VvMTP1 |
miRNA 20 ACCUUCGGGACUCCCUCUCG 1 Target 947 UGGAGGUUCUGAUGGAGAGC 966 |
Cleavage |
| ppt-miR1221-5p | VvMTP11 |
miRNA 21 AAACUGGGACGUGUGGUAGGU 1 Target 852 UCUGGCCUUGUACACCAUCCG 872 |
Cleavage |
| aly-miR158b-5p | VvMTP12 |
miRNA 20 AAAGGUUUUAACAUCUGUUU 1 Target 35 CUUCUAUGAUUGUGGAUAAA 54 |
Cleavage |
| ath-miR398b-5p | VvMTP12 |
miRNA 21 CACACAAGAGUAUAGUUGGGA 1 Target 1425 UGGGGUUGUUAUAUCAACCCU 1445 |
Cleavage |
| ath-miR398c-5p | VvMTP12 |
miRNA 21 CACACAAGAGUAUAGUUGGGA 1 Target 1425 UGGGGUUGUUAUAUCAACCCU 1445 |
Cleavage |
| aly-miR156 h-3p | VvMTP8 |
miRNA 21 CCACCGUCUUCCUUUCUCUCG 1 Target 186 CUUGGUUGAAGGAGAGAGAGA 206 |
Cleavage |
| smo-miR1099 | VvMTP8 |
miRNA 21 CUGUUUUUGUGGUAACGAUAU 1 Target 9 UGUAAAAACACCAUUGUUGUC 29 |
Cleavage |
| osa-miR2875 | VvMTP7 |
miRNA 24 AUAUUUGACAUAUACUGACAUUUA 1 Target 1058 UUGAUGCUCUAUAUGAUUGUAAAA 081 |
Cleavage |
| ppt-miR1076-5p | VvMTP8.1 |
miRNA 21 GUUAAUAGCGUGGAACACUAU 1 Target 982 AAAUUGACGUAUCUUGUGAUA 1002 |
Cleavage |
| osa-miR1882e-3p | VvMTP9.1 |
miRNA 24 GAUCUAAUGCAGGUUCUAGUAAAG 1 Target 742 AGGGCAUAUGCUCAAGAUCAUUUC 765 |
Cleavage |
| ptc-miR6456 | VvMTP9.1 |
miRNA 21 CCUAGAUUACCUUCCUGAGUU 1 Target 225 GAAGCUACUGGAAGGGUUCAA 245 |
Cleavage |
Expression profiles of VvMTP genes
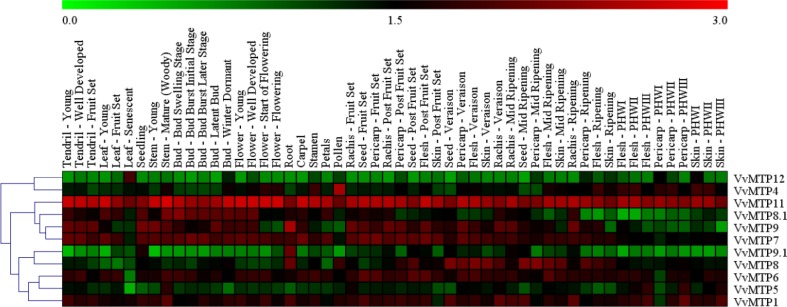
Expression patterns of VvMTP genes in a total of 54 grape tissues/organs including bud, carpel, petal, stamen, rachis, pericarp, flesh, skin, pollen, flower, leaf, root, seed, seedling, stem, and tendril were examined under different growth phases. A detailed analysis of VvMTP expression profiles (Fig. 7) showed that VvMTP11 had the highest expression levels in all tissues/organs, suggesting a key role(s) throughout plant development. The expression level of VvMTP12 appeared to be mostly involved in leaf senescence, and expression was generally low in other tissues/organs, potentially indicating a specific role in leaf senescence. VvMTP4 had uniformly moderate expression level in all of the plant tissues/organs except for pollen, which highlighted it has a significant role in male reproductive organ development. VvMTP6 and VvMTP7, from the Zn/Fe-CDF group, showed generally both high and uniform transcript levels at all stages of plant developmental, except in senescent leaves where transcript levels were low. Similarly, transcription of VvMTP5 was lowest in senescent leaves, which is the opposite trend to that observed for VvMTP12. VvMTP7 also showed low expression levels in pollen, which was the opposite of VvMTP4 that had high expression in pollen. With the exception of roots, very low levels of expression of VvMTP9.1 were observed in other tissue types at all stages of development. VvMTP1 had a moderate expression level in most tissues/organs, but expression was generally higher in mature organs, e.g., the pericarp at post-harvest withering III, the rachis at ripening and the stem at maturity (Woody). VvMTP8, VvMTP8.1 and VvMTP9 expressions were quite variable in terms of tissue/organs and developmental stages. The fundamental knowledge in regard to the role of VvMTP genes during the developmental stages of grape was obtained through these data.
Fig. 7.
MTP genes in grape and their expression in 54 specimens of green and woody tissues, and organs at various growth phases. The genes that have the same profiles for all arrays are grouped on the left using a hierarchical clustering method. The intensity of expression is defined in the colored bar at top of the chart. The scale bar represents log10 values from 0 to 3. The color bar outlines proportional expression values, the lowest (green), medium (black) and the highest (red)
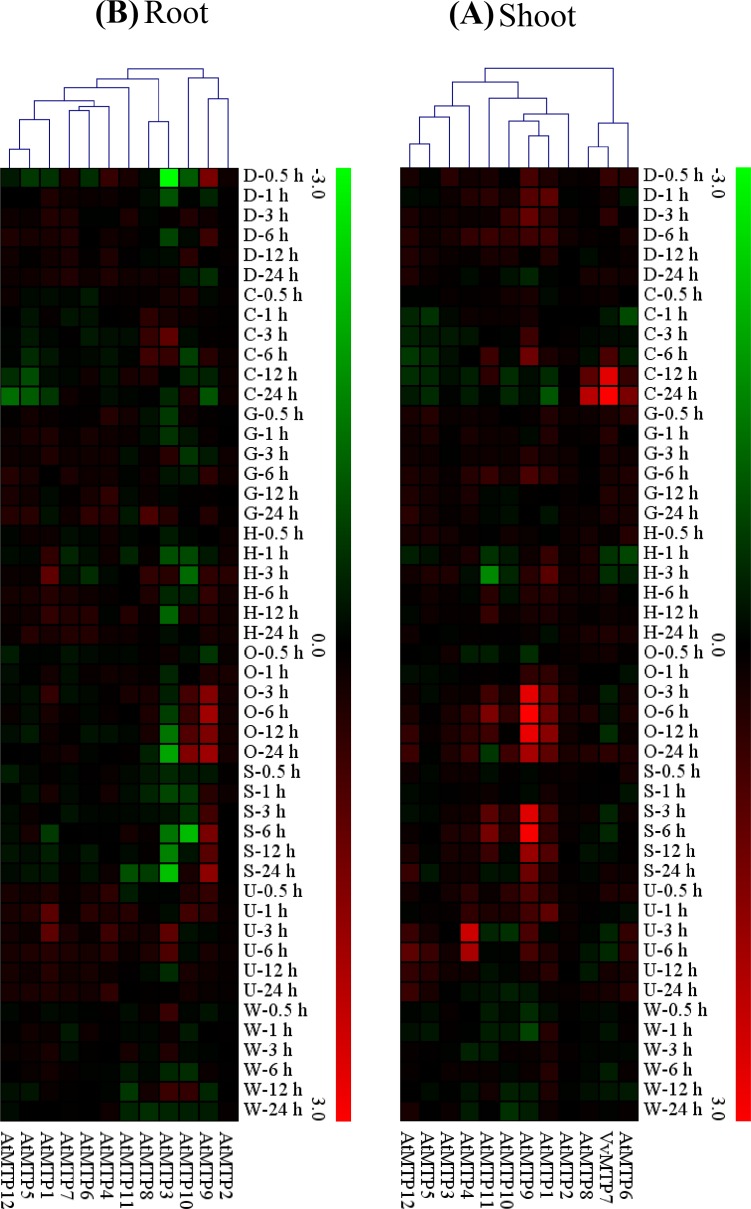
Despite the availability of grape microarray data for abiotic stress responses such as salinity and drought, a search for the proprietary ID probe for the VvMTP genes was not successful on the Affymetrix site. Therefore, the expression of VvMTP homologous genes in Arabidopsis was used to explore putative VvMTP function. This approach has been used for expression analysis of the NAC gene family in pigeon pea plants in response to (Satheesh et al. 2014) and for expression of DERB gene family in apple (Zhao et al. 2012). The expression of Arabidopsis MTP genes was determined under various abiotic stresses in both root and shoot tissues. Identification of differences in expression using Arabidopsis as a model relied on two key factors and included the type and duration of stress exposure (Fig. 8a, b). The highest level of expression was for MTP9, in response to osmotic stress, in both shoots and roots, at 3, 12 and 24 h post-stress. Upregulation in shoot tissues was higher compared to roots. Upregulation of MTP9 was high in response salinity and UV stress in shoots at 3–24 h and 0.5–6 h, respectively. MTP9 was highly expressed in response to salinity at 6 h and 24 h in roots. AtMTP9 has two orthologs in grape, VvMTP9, and VvMTP9.1, and as expected these two genes were upregulated in response to osmotic shock, salinity, and UV. Upregulation of MTP3, MTP7, and MTP11 in shoots and MTP1 in roots was observed under drought stress at all time points. Other MTPs varied with respect to expression, up- and downregulation being very much stress dependent, and this illustrates the many different roles genes in MTP family could play in plants the face of various abiotic stressors.
Fig. 8.
Expression patterns of putative MTP homologous genes in the shoots (a) and (b) roots of Arabidopsis plants under salinity, drought, osmotic, high and low temperatures, genotoxic, and UV and physical treatments, after hours of stress (0.5, 1, 3, 6, 12 and 24 h). Fold differences are shown as log2 value. Genes that have the same profiles throughout arrays are grouped on the top using a hierarchical clustering method. The color bar right indicates the levels of relative expression, the lowest (green), medium (black) and the highest (red)
Discussion
In this investigation, we detected 11 MTP genes in grape that have the cation efflux domain in their corresponding protein sequences. According to their phylogenetic relationships, the VvMTP proteins were divided into three sub-families following the classification system previously reported (Montanini et al. 2007). Phylogenetic relationships can be used to deduce the configuration and practical roles of genes among species (Takahashi et al. 2012; Vatansever et al. 2017). While orthologous genes found in different species, in each phylogenetic cluster, can be same, paralogous genes may have new biological functions relative to their ancestor genes (Guo et al. 2008). In the present study, we concluded that VvMTPs might be functionally similar to their associated homologs in Arabidopsis and rice. VvMTPs were more similar to AtMTP orthologous genes than OsMTPs, but no AtMTP2, AtMTP3, and AtMTP10 were detected in the grape genome. However, orthologs of VvMTP8 and VvMTP9 corresponding to AtMTP8 and AtMTP9 were identified and named VvMTP8, VvMTP8.1, and VvMTP9, VvMTP9.1, respectively. The orthologs of VvMTP8 have been reported in citrus, rice, maize, cucumber and barley (Fu et al. 2017; Gustin et al. 2011; Migocka et al. 2015; Pedas et al. 2014; Ricachenevsky et al. 2013). Most of the VvMTP proteins contained four to six putative transmembrane domains (TMDs), which is in accordance with previous reports on MTPs families in plant species (Kolaj-Robin et al. 2015; Lu et al. 2009; Lu and Fu 2007), with the exception of VvMT12 that is similar to CitMTP12 and contains 12 TMDs (Fu et al. 2017). All of the VvMTPs were predicted to localize to the vacuolar, as do the MTPs of wheat (Vatansever et al. 2017). AtMTP1 is involved in the transport of excess Zn into vacuoles and controls cellular Zn homeostasis (Kobae et al. 2004), while AtMTP3 also has a role of the vacuolar detoxification of Zn and/or Co (Arrivault et al. 2006). Migocka et al. (2015) suggested that many Zn transporters can in fact function as proteinaceous metal transporters that facilitate vacuolar detoxification of metals.
MTP8 and MTP9 are involved in detoxification of Mn by aiding in the intracellular scavenging of surplus Mn and facilitating storage in the vacuole (Migocka et al. 2015). Members of the Mn-MTP sub-family contain the same five conserved motif sequences, with one of these motifs similar to motifs found in Zn carrier proteins. This motif is involved in the formation of a dimerization zone, which is important as these proteins form homo-dimers during Zn transport (Kolaj-Robin et al. 2015; Lu et al. 2009). The Zn transporter dimerization domain has also been identified in some MTPs found in wheat (Vatansever et al. 2017).
Alignment of sequences showed that glycine was conserved in motifs 1 and 6, and this could indicate a specific function for this amino acid in the cation efflux domain. Motif 5 overlapped with both motifs 1 and 6. Although no specific function was defined for motif 5, it could have a similar function to the cation efflux domains. For the homology modeling of the VvMTP proteins, the structures of the zinc transporters (2QFI and 3J1Z) were utilized templates, as these were also used for homology modeling of MTPs in wheat (Vatansever et al. 2017). The quality of the structural models generated in this study was validated indicating that this information could also be used for a proteomic survey of grape MTPs.
Members of the VvMTP gene family are scattered over nine chromosomes, with two pairs of genes located on each of two chromosomes, but no duplication was detected between these genes, which may be related to the asexual reproduction of grape (Jiang et al. 2015).
Insights into the exon–intron structure can supply additional information to back phylogenetic groupings (Shiu and Bleecker 2003) because exon–intron structure divergence can play a pivotal role in the evolutionary development of gene families (Zhang et al. 2012). Results of the present study showed a significant correlation between phylogeny and exon/intron structure among VvMTPs. A degree of similarity can also be observed inside groups and sub-families, and the VvMTPs, showing both similar gene structures and intron phases within the same group and the same sub-family; this was very obvious for the Mn-MTP sub-family.
Each VvMTP gene promoter had a unique combination of CREs that may control gene expression in terms of time, location, and response to external stimuli. A comprehensive study of these regulatory elements and gene expression control would be useful for determining the role of a gene. The CAAT- and TATA-boxes were two usual CREs found in the upstream zones of VvMTP genes at a high frequency. The CAAT-box creates a binding site for RNA TFs and also is a regulatory motif that controls the expression of associated genes (Laloum et al. 2013), and the TATA-box is an element involved in transcription (Bae et al. 2015). Most CREs in VvMTP genes were related to light-responsive elements and a high frequency of these elements was also found in the CREs of MTP gene family members of wheat (Vatansever et al. 2017). This finding means that the expression of MTP genes could often be regulated to some extent by light. Light-dependent reactions in plants are complex and light can influence many developmental and physiological processes, such as seedling photomorphogenesis, phototropism, diurnal rhythms, and flower initiation (López-Ochoa et al. 2007). The above findings suggest that combinations of different CREs, instead of a single element, are important regulators of potentially light-regulated promoters (Chattopadhyay et al. 1998; Puente et al. 1996).
In the present study, VvMTP transcripts that are potential targets of miRNAs were investigated with respect to possible the post-transcriptional control mechanisms, miRNAs identified corresponding to seven VvMTPs. The miRNAs perform a regulatory role through not only by targeting specific mRNAs for degradation but also by suppressing the expression of the target gene (Bartel 2004; Carrington and Ambros 2003). Many metabolic pathways and processes associated with plant growth and development, signal transduction, and responses to biotic and abiotic stresses, including heavy metal stress, can be regulated by miRNAs (Gielen et al. 2012; Lv et al. 2012). Bioinformatic methods represent a useful and cost-effective way to investigate potential miRNA interactions associated with specific gene families (Jones-Rhoades et al. 2006). In previous studies, the involvement of miR158 in Cd stress in Brassica napus was demonstrated (Zhou et al. 2012) and miR398 has been shown to play a role in the responses of plants including A. thaliana, M. truncatula, O. sativa, N. tabacum and P. vulgaris to Cu, Fe, Mn, Al and Al2O3 nanoparticle exposure (Burklew et al. 2012; Lima et al. 2011; Sunkar et al. 2006; Valdés-López et al. 2010; Zhou et al. 2008a, b). In addition, miR156 has been shown to influence plant development and metal (Cd, Al, Mn, and As) detoxification processes, in B. napus, O. sativa, Glycine soja, P. vulgaris, and Brassica juncea (Ding et al. 2011; Huang et al. 2010; Lima et al. 2011; Srivastava et al. 2012; Valdés-López et al. 2010; Xie et al. 2007; Yu et al. 2012; Zeng et al. 2012; Zhou et al. 2012). The miR473 was also found to be important for drought responsiveness in Populus plants (Shuai et al. 2013) and miR1221-3p was shown to upregulate the WCOR413 cold acclimation gene in Physcomitrella patens plants exposed to drought stress (Wan et al. 2011). The above findings support a possible role for the miRNAs identified in the grape genome in the present study to play roles of metal tolerance in grape plants.
The main role of MTP genes is resistance to metals, and the expression analysis showed that these genes have different expressions at different stages of growth and development. Expression profiles of BrrMTP genes in roots and leaves tissues of Turnip demonstrated different expression patterns that are similar to this experiment (Li et al. 2018).
By analyzing promoters for members of VvMTP gene family, we identified several tissue-related and light-responsive elements that may play roles in developmental processes in grape plants. Previous studies of rice OsMTP11, a Mn-specific transporter, showed that this gene was widely expressed in various tissues throughout rice during plant development (Zhang and Liu 2017), and the OsMTP1 gene was expressed at particularly high levels in mature leaves and stems (Yuan et al. 2012). These findings and similar predicted expression patterns detailed in the present study suggest that VvMTP11 may be involved in several aspects of the development of grape plants, and that VvMTP1 may also be important in mature grape tissues.
Information of Arabidopsis MTP genes was used to study the VvMTPs. The expression patterns of the MTP gene family under different stresses may reflect differences in the type and number (composition) of CRE regulatory elements in the promoter region of the genes, which leads to different genes responding at different times and different stressors (Vatansever et al. 2017).
In this study, different MTPs showed a different response to various stress conditions and the highest level expression was observed for MTP9 in response to osmotic stress. The potential role of BrrMTP family of turnips was analyzed in the presence of different metal ions and the specific gene differentially expressed under different metal treatments. Some genes upregulated or downregulated with each metal ion and gene in the same cluster did not show similar expression changes to the same metal treatments (Li et al. 2018). Overexpression of cucumber MTP9 in A. thaliana caused a significant tolerance to cadmium and surplus manganese, and transfer of the CsMTP9 gene to yeast resulted in detoxification of high concentrations of Cd2+ and Mn2+ (Migocka et al. 2015). In the present study, the most relevant gene expression for MTP9 was reported in osmotic stress. According to the results of this study, it is suggested that the product of the MTP9 gene may increase the accumulation of Mn and Cd in plant shoots and is useful for phytoremediation of soils contaminated with Mn or Cd metals (Migocka et al. 2015).
In general, MTP3, MTP7, MTP11, MTP1, and MTP9 are all upregulated in plants exposed to stresses, e.g., high salinity, osmotic stress, and drought. Salinity and drought are two of the most important abiotic stresses that limit the production of food crops worldwide, and plant responses to salinity and drought are often similar (Ahmad 2016). In addition, metal toxicity can also induce similar responses in plants to salinity, as salinity causes both hyper-osmotic and ionic stresses in plants and reduces agricultural production (Chinnusamy et al. 2005; Li et al. 2010). Increasing the expression levels of MTP-related genes in plants exposed to salinity or drought might reduce to some extent the damaging effects of these stressors.
Conclusions
In this survey, 11 MTP genes were found in the grape genome and the putative protein properties, evolutionary relationships, gene structure, chromosome location, gene expression patterns and promoter functions were investigated using the tools of bioinformatics. The phylogenetic study showed VvMTP genes are clustered into three sub-families and seven groups, similarly to the MTP genes found in Arabidopsis and rice. Within each sub-family, gene structures and motifs distributions were also similar, particularly in the Mn-CDFs. The orthogonal expression pattern of VvMTP genes in Arabidopsis indicates that the main role of the MTP genes is in responses to environmental stress, especially osmotic stress, which apparently requires the expression of MTP genes that are generally thought to be involved in reducing the impacts on plants of toxic metal exposure. The expression of VvMTP genes in different tissues/organs and different stages of plant growth and development also demonstrated wide-ranging roles for VvMTPs in grape plants. This study provides important fundamental information on the putative functions of grape MTP genes, and will help efforts towards their functional characterization.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Mojtaba Kordrostami, Email: kordrostami009@gmail.com.
Mohammad Anwar Hossain, Email: anwargpb@bau.edu.bd.
References
- Ahmad P. Water stress and crop plants: a sustainable approach. New York: Wiley-Blackwell; 2016. [Google Scholar]
- Arrivault S, Senger T, Krämer U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006;46:861–879. doi: 10.1111/j.1365-313X.2006.02746.x. [DOI] [PubMed] [Google Scholar]
- Bae S-H, Han HW, Moon J. Functional analysis of the molecular interactions of TATA box-containing genes and essential genes. PLoS One. 2015;10:e0120848. doi: 10.1371/journal.pone.0120848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blaudez D, Kohler A, Martin F, Sanders D, Chalot M. Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell. 2003;15:2911–2928. doi: 10.1105/tpc.017541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burklew CE, Ashlock J, Winfrey WB, Zhang B. Effects of aluminum oxide nanoparticles on the growth, development, and microRNA expression of tobacco (Nicotiana tabacum) PLoS One. 2012;7:e34783. doi: 10.1371/journal.pone.0034783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambrollé J, García J, Figueroa M, Cantos M. Evaluating wild grapevine tolerance to copper toxicity. Chemosphere. 2015;120:171–178. doi: 10.1016/j.chemosphere.2014.06.044. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998;15:69–77. doi: 10.1046/j.1365-313x.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Chen Z, et al. Mn tolerance in rice is mediated by MTP8. 1, a member of the cation diffusion facilitator family. J Exp Bot. 2013;64:4375–4387. doi: 10.1093/jxb/ert243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Jagendorf A, Zhu J-K. Understanding and improving salt tolerance in plants. Crop Sci. 2005;45:437–448. [Google Scholar]
- Clemens S, Palmgren MG, Krämer U. A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci. 2002;7:309–315. doi: 10.1016/s1360-1385(02)02295-1. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR. Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell. 2003;15:1131–1142. doi: 10.1105/tpc.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chen Z, Zhu C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa) J Exp Bot. 2011;62:3563–3573. doi: 10.1093/jxb/err046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu S, Meier B, von Wirén N, Peiter E. The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol. 2016;170:1030–1045. doi: 10.1104/pp.15.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fu X-Z, et al. Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene. 2017;629:1–8. doi: 10.1016/j.gene.2017.07.072. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Kawachi M, Sato Y, Mori H, Kutsuna N, Hasezawa S, Maeshima M. A high molecular mass zinc transporter MTP12 forms a functional heteromeric complex with MTP5 in the Golgi in Arabidopsis thaliana. FEBS J. 2015;282:1965–1979. doi: 10.1111/febs.13252. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. The proteomics protocols handbook. Berlin: Springer; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [Google Scholar]
- Gielen H, Remans T, Vangronsveld J, Cuypers A. MicroRNAs in metal stress: specific roles or secondary responses? Int J Mol Sci. 2012;13:15826–15847. doi: 10.3390/ijms131215826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Hasanuzzaman M, Nahar K, Macovei A, Tuteja N. Importance of nitric oxide in cadmium stress tolerance in crop plants. Plant Physiol Biochem. 2013;63:254–261. doi: 10.1016/j.plaphy.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2011;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, et al. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J Genet Genom. 2008;35:105–118. doi: 10.1016/S1673-8527(08)60016-8. [DOI] [PubMed] [Google Scholar]
- Gustin JL, Zanis MJ, Salt DE. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol Biol. 2011;11:76. doi: 10.1186/1471-2148-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. Cellular mechanisms for heavy metal detoxification and tolerance. J Exp Bot. 2002;53:1–11. [PubMed] [Google Scholar]
- Haney CJ, Grass G, Franke S, Rensing C. New developments in the understanding of the cation diffusion facilitator family. J Ind Microbiol Biotechnol. 2005;32:215–226. doi: 10.1007/s10295-005-0224-3. [DOI] [PubMed] [Google Scholar]
- Hayat S, Khalique G, Irfan M, Wani AS, Tripathi BN, Ahmad A. Physiological changes induced by chromium stress in plants: an overview. Protoplasma. 2012;249:599–611. doi: 10.1007/s00709-011-0331-0. [DOI] [PubMed] [Google Scholar]
- Huang SQ, Xiang AL, Che LL, Chen S, Li H, Song JB, Yang ZM. A set of miRNAs from Brassica napus in response to sulfate deficiency and cadmium stress. Plant Biotechnol J. 2010;8:887–899. doi: 10.1111/j.1467-7652.2010.00517.x. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiang S-Y, Ma Z, Ramachandran S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol Biol. 2010;10:79. doi: 10.1186/1471-2148-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, et al. Genome-wide analysis of HD-Zip genes in grape (Vitis vinifera) Tree Genet Genomes. 2015;11:827. [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 2004;45:1749–1758. doi: 10.1093/pcp/pci015. [DOI] [PubMed] [Google Scholar]
- Kolaj-Robin O, Russell D, Hayes KA, Pembroke JT, Soulimane T. Cation diffusion facilitator family: structure and function. FEBS Lett. 2015;589:1283–1295. doi: 10.1016/j.febslet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013;18:157–166. doi: 10.1016/j.tplants.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Li Q, Cai S, Mo C, Chu B, Peng L, Yang F. Toxic effects of heavy metals and their accumulation in vegetables grown in a saline soil. Ecotoxicol Environ Saf. 2010;73:84–88. doi: 10.1016/j.ecoenv.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Li X, Wu Y, Li B, He W, Yang Y, Yang Y. Genome-wide identification and expression analysis of the cation diffusion facilitator gene family in turnip under diverse metal ion stresses. Front Genet. 2018;9:103. doi: 10.3389/fgene.2018.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima J, Arenhart R, Margis-Pinheiro M, Margis R. Aluminum triggers broad changes in microRNA expression in rice roots. Genet Mol Res. 2011;10:2817–2832. doi: 10.4238/2011.November.10.4. [DOI] [PubMed] [Google Scholar]
- Liu Z, et al. Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis) DNA Res. 2014;21:379–396. doi: 10.1093/dnares/dsu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ochoa L, Acevedo-Hernández G, Martínez-Hernández A, Argüello-Astorga G, Herrera-Estrella L. Structural relationships between diverse cis-acting elements are critical for the functional properties of a rbcS minimal light regulatory unit. J Exp Bot. 2007;58:4397–4406. doi: 10.1093/jxb/erm307. [DOI] [PubMed] [Google Scholar]
- Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317:1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- Lu M, Chai J, Fu D. Structural basis for autoregulation of the zinc transporter YiiP. Nat Struct Mol Biol. 2009;16:1063. doi: 10.1038/nsmb.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Nie X, Wang L, Du X, Biradar SS, Jia X, Weining S. Identification and characterization of microRNAs from barley (Hordeum vulgare L.) by high-throughput sequencing. Int J Mol Sci. 2012;13:2973–2984. doi: 10.3390/ijms13032973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Marschner’s mineral nutrition of higher plants. 3. Cambridge: Academic; 2011. [Google Scholar]
- Menguer PK, Farthing E, Peaston KA, Ricachenevsky FK, Fett JP, Williams LE. Functional analysis of the rice vacuolar zinc transporter OsMTP1. J Exp Bot. 2013;64:2871–2883. doi: 10.1093/jxb/ert136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migocka M, et al. Cucumber metal tolerance protein CsMTP9 is a plasma membrane H+-coupled antiporter involved in the Mn2+ and Cd2+ efflux from root cells. Plant J. 2015;84:1045–1058. doi: 10.1111/tpj.13056. [DOI] [PubMed] [Google Scholar]
- Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. Phylogenetic and functional analysis of the cation diffusion facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genom. 2007;8:107. doi: 10.1186/1471-2164-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2013;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- Ozyigit II, Filiz E, Vatansever R, Kurtoglu KY, Koc I, Öztürk MX, Anjum NA. Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front Plant Sci. 2016;7:301. doi: 10.3389/fpls.2016.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedas P, Stokholm MS, Hegelund JN, Ladegård AH, Schjoerring JK, Husted S. Golgi localized barley MTP8 proteins facilitate Mn transport. PLoS One. 2014;9:e113759. doi: 10.1371/journal.pone.0113759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, et al. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc Natl Acad Sci. 2007;104:8532–8537. doi: 10.1073/pnas.0609507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Ricachenevsky FK, Menguer PK, Sperotto RA, Williams LE, Fett JP. Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front Plant Sci. 2013;4:144. doi: 10.3389/fpls.2013.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Satheesh V, Jagannadham PTK, Chidambaranathan P, Jain P, Srinivasan R. NAC transcription factor genes: genome-wide identification, phylogenetic, motif and cis-regulatory element analysis in pigeonpea (Cajanus cajan (L.) Millsp.) Mol Biol Rep. 2014;41:7763–7773. doi: 10.1007/s11033-014-3669-5. [DOI] [PubMed] [Google Scholar]
- Shahzad Z, Gosti F, Frérot H, Lacombe E, Roosens N, Saumitou-Laprade P, Berthomieu P. The five AhMTP1 zinc transporters undergo different evolutionary fates towards adaptive evolution to zinc tolerance in Arabidopsis halleri. PLoS Genet. 2010;6:e1000911. doi: 10.1371/journal.pgen.1000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. Speculations on RNA splicing. Cell. 1981;23:643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai P, Liang D, Zhang Z, Yin W, Xia X. Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genom. 2013;14:233. doi: 10.1186/1471-2164-14-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Anandhan S, Singh S, Patade VY, Ahmed Z, Pande V. Metallothionein-like gene from Cicer microphyllum is regulated by multiple abiotic stresses. Protoplasma. 2011;248:839–847. doi: 10.1007/s00709-010-0249-y. [DOI] [PubMed] [Google Scholar]
- Singh S, Parihar P, Singh R, Singh VP, Prasad SM. Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and ionomics. Front Plant Sci. 2016;6:1143. doi: 10.3389/fpls.2015.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Srivastava AK, Suprasanna P, D’souza S. Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. J Exp Bot. 2012;64:303–315. doi: 10.1093/jxb/ers333. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu J-K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Buchner P, Yoshimoto N, Hawkesford MJ, Shiu S-H. Evolutionary relationships and functional diversity of plant sulfate transporters. Front Plant Sci. 2012;2:119. doi: 10.3389/fpls.2011.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Vert G. Iron transport in plants: better be safe than sorry. Curr Opin Plant Biol. 2013;16:322–327. doi: 10.1016/j.pbi.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno D, et al. A polarly localized transporter for efficient manganese uptake in rice. Nat Plants. 2015;1:15170. doi: 10.1038/nplants.2015.170. [DOI] [PubMed] [Google Scholar]
- Valdés-López O, Yang SS, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernández G. MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 2010;187:805–818. doi: 10.1111/j.1469-8137.2010.03320.x. [DOI] [PubMed] [Google Scholar]
- Vatansever R, Filiz E, Eroglu S. Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification. Biometals. 2017;30:217–235. doi: 10.1007/s10534-017-9997-x. [DOI] [PubMed] [Google Scholar]
- Wan P, et al. Computational analysis of drought stress-associated miRNAs and miRNA co-regulation network in Physcomitrella patens. Genom Proteomics Bioinform. 2011;9:37–44. doi: 10.1016/S1672-0229(11)60006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Q, Tao W, Ping M, Z-c Li, Ling Y. Quantitative trait loci for mercury tolerance in rice seedlings. Rice Sci. 2013;20:238–242. [Google Scholar]
- Wang M, et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic Res. 2014;1:14016. doi: 10.1038/hortres.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett. 2007;581:1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- Yang X, Feng Y, He Z, Stoffella PJ. Molecular mechanisms of heavy metal hyperaccumulation and phytoremediation. J Trace Elem Med Biol. 2005;18:339–353. doi: 10.1016/j.jtemb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Yu LJ, et al. Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa) New Phytol. 2012;195:97–112. doi: 10.1111/j.1469-8137.2012.04154.x. [DOI] [PubMed] [Google Scholar]
- Yuan L, Yang S, Liu B, Zhang M, Wu K. Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 2012;31:67–79. doi: 10.1007/s00299-011-1140-9. [DOI] [PubMed] [Google Scholar]
- Zeng Q-Y, Yang C-Y, Ma Q-B, Li X-P, Dong W-W, Nian H. Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 2012;12:182. doi: 10.1186/1471-2229-12-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liu B. Identification of a rice metal tolerance protein OsMTP11 as a manganese transporter. PLoS One. 2017;12:e0174987. doi: 10.1371/journal.pone.0174987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao M, Singer SD, Fei Z, Wang H, Wang X. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS One. 2012;7:e44465. doi: 10.1371/journal.pone.0044465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Liang D, Wang P, Liu J, Ma F. Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol Genet Genom. 2012;287:423–436. doi: 10.1007/s00438-012-0687-7. [DOI] [PubMed] [Google Scholar]
- Zhou ZS, Huang SQ, Yang ZM. Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem Biophys Res Commun. 2008;374:538–542. doi: 10.1016/j.bbrc.2008.07.083. [DOI] [PubMed] [Google Scholar]
- Zhou ZS, Wang SJ, Yang ZM. Biological detection and analysis of mercury toxicity to alfalfa (Medicago sativa) plants. Chemosphere. 2008;70:1500–1509. doi: 10.1016/j.chemosphere.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Zhou ZS, Zeng HQ, Liu ZP, Yang ZM. Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 2012;35:86–99. doi: 10.1111/j.1365-3040.2011.02418.x. [DOI] [PubMed] [Google Scholar]