Abstract
Background
Accumulating evidence suggests that the intestinal microbiome may dramatically affect the outcomes of hematopoietic stem cell transplant (HSCT) recipients. Providing 16S ribosomal RNA based microbiome characterization in a clinically actionable time frame is currently problematic. Thus, determination of microbial metabolites as surrogates for microbiome composition could offer practical biomarkers.
Methods
Longitudinal fecal specimens (n = 451) were collected from 44 patients before HSCT through 100 days after transplantation, as well as 1-time samples from healthy volunteers (n = 18) as controls. Microbiota composition was determined using 16S ribosomal RNA V4 sequencing. Fecal indole and butyrate levels were determined using liquid chromatography tandem mass spectrometry.
Results
Among HSCT recipients, both fecal indole and butyrate levels correlated with the Shannon diversity index at baseline (P = .02 and P = .002, respectively) and directly after transplantation (P = .006 and P < .001, respectively). Samples with high butyrate levels were enriched for Clostridiales, whereas samples containing high indole were also enriched for Bacteroidales. A lower Shannon diversity index at the time of engraftment was associated with increased incidence of acute intestinal graft-vs-host disease (iGVHD) (P = .02) and transplant-related deaths (P = .03). Although fecal metabolites were not associated with acute iGVHD or overall survival, patients contracting bloodstream infections within 30 days after transplantation had significantly lower levels of fecal butyrate (P = .03).
Conclusions
Longitudinal analysis of fecal microbiome and metabolites after HSCT identified butyrate and indole as potential surrogate markers for microbial diversity and specific taxa. Further studies are needed to ascertain whether fecal metabolites can be used as biomarkers of acute iGVHD or bacteremia after HSCT.
Keywords: butyrate, graft-vs-host disease (GVHD), hematopoietic stem cell transplant (HSCT), indole, microbiome
Accumulating evidence suggests that intestinal microbiome composition may dramatically impact the outcomes of hematopoietic stem cell transplant (HSCT) patients, however real-time monitoring of the microbiome using current sequencing technologies is clinically impractical. Researchers therefore evaluated the fecal excretion patterns of intestinal microbial metabolites indole and butyrate, the bacterial species associated with these changes, and how this relates to clinical outcomes in a cohort of HSCT patients in order to understand the potential for using fecal metabolites as surrogate markers for microbiome characterization.
Previous investigations indicate that microbial diversity and community structure have important health implications, including resistance against infection [1–5]. Moreover, metabolites derived from intestinal microbiota are often markedly altered in human disease [6–10]. Thus, the understanding of microbial metabolite patterns, the species associated with these changes, and their interactions with the host and clinical consequences pose substantial opportunities to develop assays that use metabolites as surrogate markers for microbiome characterization. Examples of metabolites with important roles in human health and disease are short-chain fatty acids, such as butyrate, and tryptophan metabolites, such as indole [9, 10].
Butyrate, the main energy substrate for colonocytes [11, 12], is of therapeutic interest because it promotes intestinal health [9]. The capacity to produce butyrate from complex carbohydrates is generally attributed to gram-positive anaerobic bacteria that inhabit the colon. Two of the major butyrate producing groups are Faecalibacterium (clostridial cluster IV) and Eubacterium rectale/Roseburia spp. (clostridial cluster XIVa) [13–15]. Previous studies in animal models have shown that a decrease in intestinal butyrate after allogeneic hematopoietic stem cell transplantation (HSCT) is correlated with adverse outcomes [7]. Specifically, reduced butyrate in intestinal epithelial cells resulted in decreased histone acetylation, whereas exogenous administration of butyrate restored intestinal epithelial cell junction integrity and diminished graft-vs-host disease (GVHD) [7]. Furthermore, addition of high-butyrate producing bacteria also mitigated GVHD [7]. In 2018, it was also reported that higher levels of butyrate producing fecal microbiota were associated with increased resistance against lower respiratory viral infections in allogeneic HSCT recipients [14].
Indole is an important bacterial quorum signaling molecule exclusively catabolized from tryptophan by commensal bacteria that express tryptophanase [16]. Indole plays a crucial role in maintaining epithelial barrier function, promoting immune tolerance, and protecting against pathogens [17]. A variety of bacteria such as Fusobacterium spp., Escherichia coli, Bacteroides spp., and Enterococcus faecalis have the ability to convert tryptophan into indole and its derivatives [16]. Low levels of 3-indoxyl sulfate (3-IS) (a major conjugate of indole) in the urine after allogeneic stem-cell transplantation (allo-SCT) were associated with significantly higher transplant-related mortality (TRM), intestinal GVHD (iGVHD) being the primary cause [18]. That study not only revealed a prognostic tool for poor outcomes after allo-SCT, but it also showed that urinary 3-IS levels could be predictive of gut microbiome composition [18].
Real-time monitoring of microbiome composition by sequencing based techniques in the clinical setting is challenging [5, 19–24] because samples must be batched to allow sequencing to be cost-effective, and time and expertise are needed for data analyses. Because low microbial diversity has been associated with increased GVHD and mortality rates after HSCT, we hypothesized that loss of microbial diversity results in decreased production of intestinal bacterial derived metabolites that are relevant to mucosal integrity and immune tolerance. Thus, we sought to determine whether measuring stool metabolites using liquid chromatography tandem mass spectrometry (commonly used in clinical laboratories of larger hospitals or tertiary care centers) could be used to monitor microbiome diversity in the clinical setting, and whether these metabolite measurements could be correlated with complications associated with loss of microbial diversity.
METHODS
Patients, Specimens, and Clinical Data Collection
For this cohort study, 50 adult patients undergoing HSCT were enrolled between May 2014 and November 2015 at the University of Texas MD Anderson Cancer Center (MDACC). The study protocol was approved by the MDACC Institutional Review Board, and written consent was obtained from all patients. As controls, we studied stool samples from 18 healthy adult volunteers (aged 18–45 years) without significant medical illness, history of antibiotic exposure, use of immunosuppressive medications, or recent travel; samples were obtained under an institutional review board–approved protocol at the University of Texas Health Science Center. Characteristics of the HSCT cohort are reported in Table 1. Eligibility criteria, clinical metadata, and definitions are reported in the Supplementary Material. (Data are publicly available in the National Center for Biotechnology Information Sequence Read Archive SRA accession number PRJNA528754.)
Table 1.
Clinical Characteristics of Hematopoietic Stem Cell Transplant Recipients
| Characteristics | Patients, %. (No.)a |
|---|---|
| Age, median (range), y | 58.5 (20–72) |
| Male-female ratio, no. | 29:15 |
| Donor age, median (range), y | 58 (13–69) |
| Comorbidity index,b median (range) | 3 (0–10) |
| Underlying disease | |
| Myeloid | 79.5 (35) |
| AML/MDS | 65.9 (29) |
| CML/MPD | 11.4 (5) |
| Aplastic anemia | 2.3 (1) |
| Lymphoid | 20.4 (9) |
| ALL | 13.6 (6) |
| Lymphoma | 6.8 (3) |
| Disease status | |
| Remission | 40.9 (18) |
| Active/advanced | 59.1 (26) |
| Type of SCT | |
| Donor relation | |
| MUD | 68.2 (30) |
| MRD | 27.3 (12) |
| Otherc | 4.5 (2) |
| Cell type | |
| Cord blood (HPC-C) | 4.5 (2) |
| Bone marrow (HPC-M) | 36.4 (16) |
| Peripheral blood (HPC-A) | 59.1 (26) |
| Conditioning intensity | |
| Myeloablative | 97.7 (43) |
| Nonmyeloablative | 2.3 (1) |
| Prophylaxis | |
| Tacrolimus-MMF | 11.4 (5) |
| Tacrolimus-methotrexate | 88.6 (39) |
| CMV status (donor-recipient) | |
| Negative-negative | 9.1 (4) |
| Negative-positive | 45.5 (20) |
| Positive-negative | 6.8 (3) |
| Positive-positive | 34.1 (15) |
| Unknown | 4.5 (2) |
| Sex match/mismatch (donor-recipient) | |
| Female-male | 13.6 (6) |
| Female-female | 9.1 (4) |
| Male-female | 13.6 (6) |
| Male-male | 36.4 (16) |
| Unknown | 27.3 (12) |
| Bacteremia within 30 d | 34 (15) |
| Acute iGVHD (6 mo) | 36.4 (16) |
| Stage 1 | 25.6 (12) |
| Stage 3 | 7.0 (3) |
| Stage 4 | 2.3 (1) |
| Acute liver GVHD | 4.5 (2) |
| Stage 1 | 2.3 (1) |
| Stage 3 | 2.3 (1) |
| Acute skin GVHD | 54.5 (24) |
| Stage 1 | 20.4 (9) |
| Stage 2 | 15.9 (7) |
| Stage 3 | 13.6 (6) |
| Stage 4 | 4.5 (2) |
Abbreviations: ALL, acute lymphoid leukemia; AML acute myeloid leukemia; CML chronic myelogenous leukemia; CMV, cytomegalovirus; GVHD, graft-vs-host disease; HPC-A, hematopoietic progenitor cells-apheresis; HPC-C, hematopoietic progenitor cell-cord; HPC-M, hematopoietic progenitor cell-marrow; iGVHD, intestinal GVHD; MDS myelodysplastic syndrome; MMF mycophenolate mofetil; MPD, myeloproliferative disorder; MRD, matched related donor; MUD, matched unrelated donor; SCT stem cell transplantation.
aData represent percentage of cohort (no. of patients) unless otherwise specified.
bSorror comorbidity index [31].
c“Other” denotes 1- or 2-allele HLA-mismatched unrelated donors.
Microbiome Community Analyses
DNA was extracted from stool samples, and the 16S ribosomal RNA V4 region was amplified and sequenced using an Illumina MiSeq platform and a 2 × 250–base pair paired-end protocol adapted from methods developed for the National Institutes of Health Human Microbiome Project, as described elsewhere [2, 25]. In this process, 16S ribosomal RNA gene sequences were assigned into operational taxonomic units (OTUs) using the UPARSE pipeline and alignment to the SILVA SSURef_NR99_119 database at 97% sequence identity [26]. Microbiome analyses were conducted with R software (R Core Team 2015; version 3.2.2), using the phyloseq package to calculate α- and β-diversity metrics and characterize microbiome community profiles [27].
Analysis of Fecal Indole and Butyrate
Inpatient samples (n = 252; weeks 1–4) were analyzed for fecal indole and butyrate levels as determined using liquid chromatography tandem mass spectrometry at the Pharmaceutical Science Facility, Institute for Applied Cancer Science at MDACC. A Waters Xevo TQ-S triple quadrupole mass spectrometer coupled to a Waters Acquity UPLC Classic System was operated in selected reaction monitoring mode for data collection. A Phenomenex Kinetex PFP 2.6-µm, 100 × 2-mm analytical column was used. Mobile phase A was 0.1% formic acid in water, and mobile phase B (MPB) was 0.1% formic acid in 80:20 acetonitrile-methanol. The chromatographic method included a mobile phase flow rate of 0.5 mL/min, a column temperature of 60°C, a sample temperature of 5°C, an injection volume of 10 µL and a gradient elution program as follows: 0–0.5 minutes, 10% MPB; 0.5–2 minutes; 10%–95% MPB; 2–3 minutes, 95% MPB; 3–3.5 minutes, 95%–10% MPB; and 3.5–5 minutes, 10% MPB. Negative mode electrospray ionization was used for the measurement of 7-hydroxy-coumarin, while positive mode was used for butyrate and indole. The selected reaction monitoring transitions monitored and collision energies used were 89.15 > 43.15, 118.10 > 91.04, and 161.0 > 133.1 mass-to-charge ratio (m/z) and 15, 22, 22 V, respectively, for butyrate, indole, and 7-hydroxycoumarin. The dwell time was 0.158 second and the cone voltage was 25 V for all compounds measured.
Statistical Analyses
Pairwise test and linear correlation analyses were performed in GraphPad Prism 6 (GraphPad Software) for significance testing and plotting. Box plots, β-diversity biplots, principal coordinate analysis, hierarchical clustering analyses, and respective statistical analyses were executed in the user interface ATIMA (Agile Toolkit for Incisive Microbial Analyses; http://atima.jplab.net)/. LefSe analyses were completed using the Galaxy web application and workflow framework (https://huttenhower.sph.harvard.edu/galaxy/). Linear mixed models were used to assess changes in metabolite abundance over time and their association with acute iGVHD status. Clinical covariates, microbiome variables, and metabolite abundances were tested for association with the cause-specific hazard of acute iGVHD, using the Fine-Gray method [28], and tested for association with overall survival and TRM rate, using univariate and multivariable Cox proportional hazards models. Additional statistical methods are described in the Supplementary Material.
RESULTS
Patients and Clinical Data
Patient characteristics and outcomes can be found in Table 1. The study included 1-time samples from healthy volunteers (n = 18) as controls as well as longitudinally tracked HSCT recipients (n = 50). Of the 50 patients, 6 withdrew from the study at some point during the 14-week sampling period and thus were excluded from analyses. Stool samples were collected from each patient before HSCT, at the time of conditioning (baseline sample, visit 0), then twice weekly in weeks 1–4, once a week in weeks 5–8, and once a week in weeks 10, 12, and 14 after transplantation, for a total of 451 samples.
The most common underlying disease among the HSCT recipients was acute myeloid leukemia or myelodysplastic syndrome (n = 29; 65.9%). Sixty-eight percent of HSCT recipients received grafts from matched unrelated donors, and 27% received grafts from matched related donors. The majority of patients received myeloablative conditioning (97.7%) and GVHD prophylaxis with tacrolimus and methotrexate (88.6%). Patients with neutropenia received antimicrobial prophylaxis with a fluoroquinolone (98%). If neutropenic fever developed, patients received cefepime (88%), piperacillin tazobactam (18%), or meropenem (50%) coupled with vancomycin as empiric regimens. Twenty participants (45%) had bloodstream infection (BSI) in the 14 weeks after transplantation, 15 within the first 30 days . Of those 20 patients, 15 (34%) had gram-positive and 8 (18%) had gram-negative bacteremia. Three patients had multiple BSIs.
Significant Differences in Microbial Diversity and Fecal Metabolites Between Healthy Volunteers and Baseline HSCT Samples
We compared the gut microbiome composition between healthy individuals and the HSCT cohort at baseline. Compared with healthy individuals, HSCT recipients had significantly lower microbial diversity at baseline, according to both the total number of observed OTUs (P < .001) and the Shannon diversity index (P < .001) (Figure 1A). Differences in microbial communities (genera) among healthy volunteers and HSCT recipients at baseline could also be detected via β-diversity biplots (Figure 1B). The variation between the 2 groups was driven by the abundance of Pseudobutyrivibrio in healthy volunteers, and Bacteroides, Enterobacter, and Akkermansia in HSCT recipients at baseline. This was further examined by comparing the relative abundance of specific genera at baseline; levels were significantly higher for Pseudobutyrivibrio and Subdoligranulum in healthy individuals and for Enterococcus in HSCT recipients (Figure 1C). Our findings are in contrast to a previous report in which HSCT recipients who did not contract severe GVHD maintained stool microbiota broadly similar to healthy donors [29].
Figure 1.
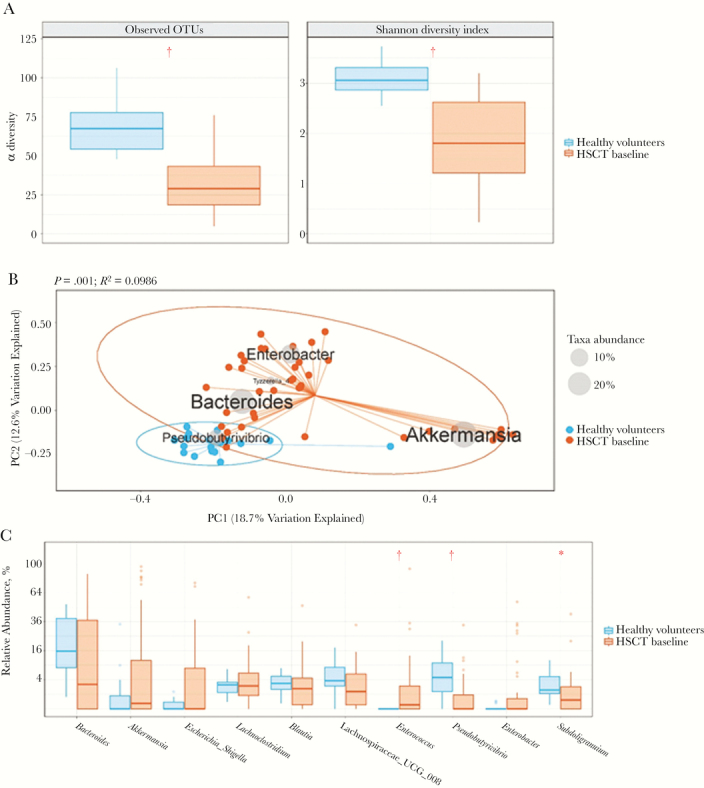
Differences in microbial diversity between healthy volunteers and hematopoietic stem cell transplant (HSCT) recipients at baseline. A, Box plots of the number of observed operational taxonomic units (OTUs) and the Shannon diversity index for each group are shown, where blue represents 1-time samples from healthy volunteers and orange, samples from HSCT recipients at baseline. †P < .001 (nonparametric Mann-Whitney U test). B, Principal coordinate analyses using Bray-Curtis distances. Biplot displays the dominant taxa (listed on the diagram where average genera abundance across samples is denoted by size of the gray sphere in inlaid legend), which explain the variation seen in the principal component analysis plot. Again, blue represents 1-time samples from healthy volunteers, and orange, samples from HSCT recipients at baseline. Permutational multivariate analysis of variance test was used to determine P and R2 values for the differences between the 2 groups. C, Box plots of the differences in relative abundance of the top 10 most abundant genera, with a minimum average abundance of >0.01. P values were determined using the nonparametric Mann-Whitney U test and were adjusted to account for multiple testing using the Benjamini-Hochberg correction. *P < .01; †P < .001.
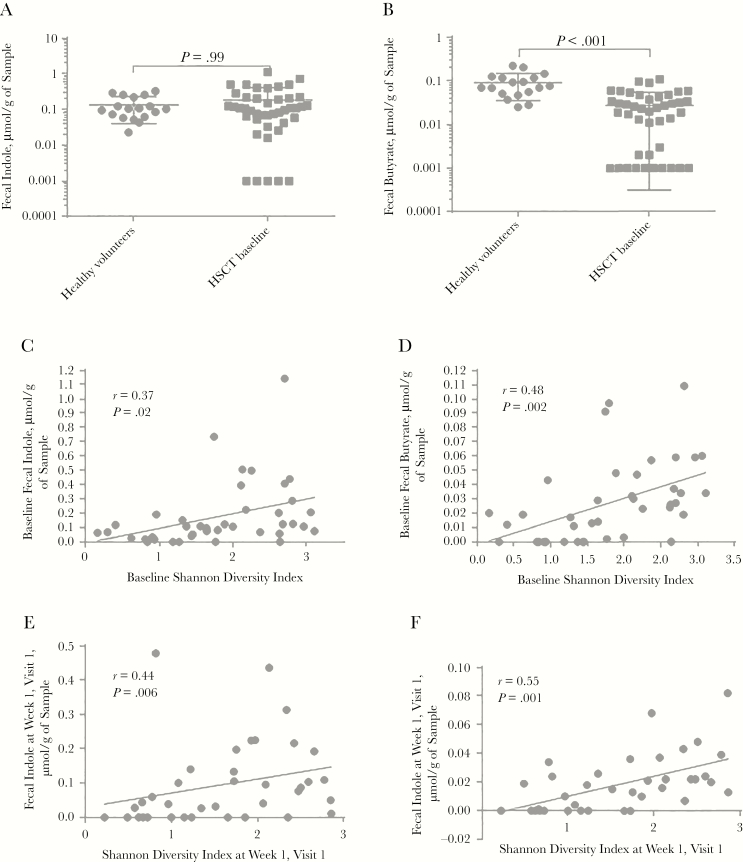
To determine whether there were differences in microbial metabolites among the 2 study groups, we next determined fecal indole and butyrate levels. Although there was no difference in fecal indole levels between healthy volunteers and baseline samples of HSCT recipients (Figure 2A), the latter had significantly lower butyrate levels at baseline (Figure 2B).
Figure 2.
Fecal metabolite differences and correlations in healthy volunteers and hematopoietic stem cell transplant (HSCT) recipients at baseline. A, B, Fecal indole (A) and butyrate (B) levels in healthy volunteers and HSCT recipients at baseline. P values refer to pairwise comparisons using Mann-Whitney U test. For the purpose of visualization, metabolite abundance is expressed in log scale, and samples with an abundance of zero were set to 0.001. C–F, Longitudinal HSCT recipient cohort. Fecal indole (C, E) and butyrate (D, F) levels at baseline and first sample after transplantation, respectively, are correlated with the Shannon diversity index. Pearson r and P values for the linear regression coefficient are shown.
Fecal Butyrate and Indole Levels in HSCT Recipients Correlated With Microbial Diversity
We next sought to determine whether microbial diversity was correlated with fecal indole and butyrate levels in our HSCT recipient cohort. Indeed, using linear regression analyses, we found that both fecal indole and butyrate levels were correlated with the Shannon diversity index (r = 0.37 and P = .02 for indole; r = 0.48 and P = .002 for butyrate) and the total number of observed OTUs (r = 0.41 and P = .009 for indole; r = 0.49 and P = .001 for butyrate) at baseline (Figure 2C and 2D and Supplementary Figure 1). The correlations between microbial diversity and fecal metabolites were strengthened when we analyzed the sample directly after transplantation rather than at baseline; higher Shannon diversity indices (r = 0.44 and P = .006 for indole; r = 0.55 and P < .001 for butyrate) and number of observed OTUs (r = 0.45 and P = .005 for indole; r = 0.67 and P < .001 for butyrate) were associated with higher fecal indole and butyrate measurements (Figure 2E and 2F and Supplementary Figure 1).
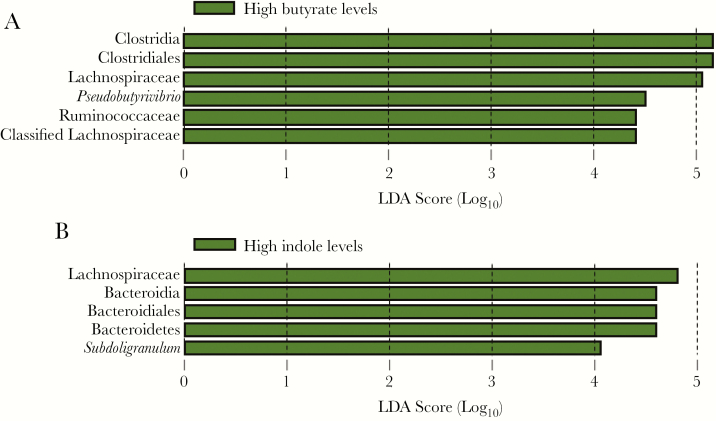
We next used the LefSe algorithm to look for taxa differences associated with fecal butyrate levels at baseline and found that samples with high levels of butyrate (median for all samples, >0.02 μmol/g) were enriched for the order Clostridiales, including the families Lachnospiraceae and Ruminococcaceae and the genus Pseudobutyrivibrio (Figure 3A). Furthermore, although the samples with higher levels of butyrate collected the week after transplantation (week 1, visit 1) remained enriched for the same taxa, these samples were now enriched for several other taxa, to include Eggerthella and Erysipelotrichaceae (Supplementary Figure 2A). In addition, patient samples with low levels of butyrate were now enriched for Enterococcus and Lactobacillus.
Figure 3.
Linear discriminant analysis (LDA) scoring taxa associations with levels of fecal butyrate and indole. Histograms represent LDA scores for differentially abundant features among groups. The threshold on the logarithmic LDA score for discriminative features was set to 4.0. A, Taxa differences associated with high levels of butyrate (>0.02 μmol/g) at baseline [1]. B, Taxa enriched in samples with high indole levels (>0.1 μmol/g) at baseline.
When the same approach was used to assess taxa differences associated with fecal indole levels, the order Bacteroidales, the Lachnospiraceae family, and the genus Subdoligranulum were enriched in samples with higher indole levels at baseline (median for all samples, >0.1 μmol/g) at baseline (Figure 3B). Again marked changes occurred between baseline and the first week after transplantations, when samples with higher indole levels at week 1, visit 2, after transplantation were now also enriched for the Ruminococcaceae and Erysiopelotrichaceae families. Similar to patient samples with low levels of butyrate, those with low levels of indole were also enriched for Enterococcaceae and Lactobacillales (Supplementary Figure 2B). It is of interest that not all of the taxa positively associated with indole levels are known to produce indole, such as Erysiopelotrichia [16].
Because the LefSe algorithm performs only binary, or categorical, analyses, we performed regression analyses, taking into consideration all samples from baseline through week 4, visit 1 (approximately 8 samples per patient) to determine which genera were positively or negatively correlated with fecal metabolites across time points. Blautia was positively correlated with fecal butyrate levels, whereas Enterococcus and Lactobacillus were negatively correlated with these levels (Supplementary Figure 3). Akkermansia was positively correlated with fecal indole levels, and Enterococcus, Lactobacillus, and Streptococcus were negatively correlated.
Fecal Microbiome and Metabolite Correlations With Acute iGVHD in HSCT Recipients
We next wanted to determine whether our study showed the same microbiota associations with GVHD as had been found at other institutions [13, 17, 23, 24, 26, 30]. Of the 44 patients, 16 (36%) experienced acute iGVHD, 5 (11%) had progression of their underlying hematologic disease, and 3 (7%) died during the study (Supplementary Figure 4). The remaining patients were censored at the end of the study, with a time to last follow-up of 8 months (182 days) for acute iGVHD analyses (Supplementary Figure 4). The median time to diagnosis of acute iGVHD was 53 days. We found no significant associations between clinical covariates and iGVHD-specific hazard to include patient age, donor age, comorbidity index [31], disease status, donor relation, cell-type, sex mismatch, disease category, and cytomegalovirus status (Table 2). However, we did find the Shannon diversity index of the fecal samples at the time of engraftment to be significantly associated with the incidence of acute iGVHD (Table 2), as has been reported elsewhere [5, 23]. Moreover, we found only 1 taxa at the time of engraftment, Coriobacteriia, to be negatively correlated with the incidence of acute iGVHD (regression coefficient = −0.572; P = .046) (Supplementary Table 1).
Table 2.
Clinical and Microbiome Predictors of Acute Gastrointestinal Graft-vs-Host Diseasea
| Predictor | Coefficient | Exponentiated Coefficient | P Value |
|---|---|---|---|
| Clinical covariates | |||
| Patient age | −0.014 | 0.99 | .35 |
| Donor age | 0.009 | 1.01 | .62 |
| Sorror comorbidity index | 0.092 | 1.10 | .45 |
| Disease status | 0.258 | 1.29 | .60 |
| Remission (reference standard) | … | … | … |
| Active/advanced | 0.258 | 1.29 | .60 |
| Donor relation | |||
| MUD (reference standard) | … | … | … |
| MRD | 0.254 | 1.29 | .63 |
| Other | 0.374 | 1.45 | .67 |
| Cell type | |||
| HPC-A (reference standard) | … | … | … |
| HPC-C | 0.227 | 1.26 | .80 |
| HPC-M | −0.197 | 0.82 | .72 |
| Sex mismatch | |||
| No mismatch (reference standard) | … | … | … |
| Mismatch | −1.14 | 0.32 | .25 |
| Underlying disease | |||
| Myeloid (reference standard) | … | … | … |
| Lymphoid | 0.693 | 2.00 | .17 |
| CMV status (donor and recipient) | |||
| No positive results (reference standard) | … | … | … |
| Any positive results | 0.745 | 2.11 | .47 |
| Microbiome and metabolite measures | |||
| Shannon diversity index | |||
| Baseline | −0.318 | 0.727 | .24 |
| Engraftment | −0.612 | 0.542 | .02b |
| Indole | |||
| Continuous | |||
| Baseline | −0.599 | 0.55 | .59 |
| 2.1 wk | 0.167 | 1.18 | .91 |
| Binary | |||
| Baseline | −0.293 | 0.75 | .56 |
| 2.1 wk | −0.299 | 0.74 | .57 |
| Butyrate | |||
| Continuous | |||
| Baseline | −6.45 | 0.002 | .45 |
| week 2, visit 1 | 11.0 | 58230 | .42 |
| Binary | |||
| Baseline | −0.058 | 0.94 | .91 |
| week 2, visit 1 | 0.351 | 1.42 | .53 |
Abbreviations: CMV, cytomegalovirus; GVHD, graft-vs-host disease; HPC-A, hematopoietic progenitor cells-apheresis; HPC-C, hematopoietic progenitor cell-cord; HPC-M, hematopoietic progenitor cell-marrow; MRD, matched related donor; MUD, matched unrelated donor.
aTable 2 summarizes the competing risk regression results obtained using the Fine-Gray method with the clinical and microbiome-based covariates tested for association to the GVHD-specific hazard. For the binary analyses, indole and butyrate levels were categorized as either less than or at least the median value.
bSignificant at P < .05.
In light of the relationship between low microbial diversity and GVHD incidence [13, 17, 24] and our finding that fecal butyrate and indole levels could be correlated with microbial diversity, we sought to determine whether butyrate and indole levels were associated with acute iGVHD occurrence within our cohort. We did not identify a correlation between fecal butyrate or indole levels at baseline or the second week after transplantation (samples closest to engraftment for most patients) and the incidence of acute iGVHD, whether we used the continuous measurements of the fecal metabolites or stratified the patients by the median into high- and low-level groups (Table 2). Although we found a statistically significant decline in both metabolite levels, iGVHD status was not significantly associated with butyrate or indole levels over time (Supplementary Figure 5).
Fecal Microbiome and Metabolite Correlations With Other Clinical Outcomes in HSCT Recipients
Of the 44 patients, 22 (50%) died within 2 years. Based on the Kaplan-Meier estimate, the median survival time was 692 days (1.90 years), and the median follow-up time 713 days (1.95 years) (Supplementary Figure 6). Of the clinical and microbiome-related covariates tested at baseline and engraftment, only disease status and higher baseline indole levels were associated at univariate analysis with increased risk of death (P = .02 and P = .047, respectively). However, in a multivariable Cox model adjusting for diseases status, there was no statistical association between the baseline indole measurement and overall survival (Supplementary Table 2). No taxa were found to be significantly associated with overall survival (Supplementary Table 3).
Although we had only 5 deaths occurring within 6 months without progression of the underlying disease, we also analyzed microbial diversity and fecal indole level associations with the TRM rate (Supplementary Table 4). The Shannon diversity index of stool samples at the time of engraftment (analyzed as a continuous variable) was significantly associated with TRM (coefficient = −1.44; P = .02). Despite the significance of this finding being based on such few events, this is in line with prior studies [19]. We also found baseline fecal indole levels to be positively associated with TRM (coefficient = 1.74; P = .040), but the significance is marginal and the directionality of the data is hard to reconcile with the findings of Weber et al [18], who reported that low urinary 3-IS levels were associated with significantly increased risk of TRM [18].
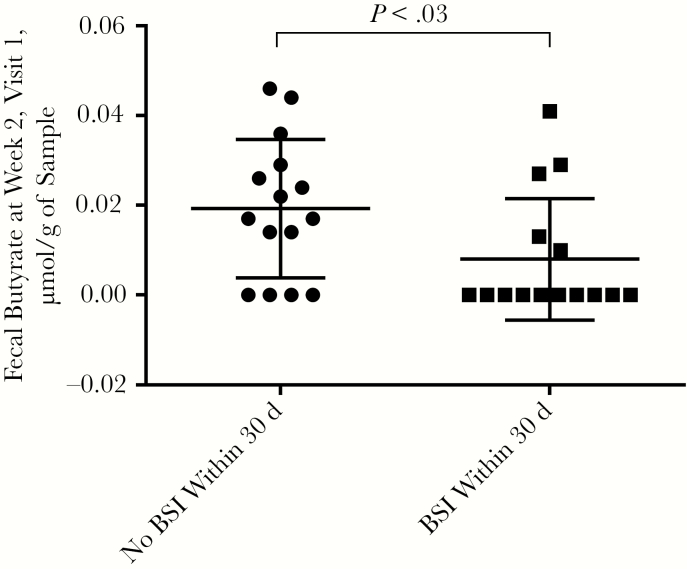
In light of emerging data regarding close links between the gastrointestinal microbiome and infection [2, 5, 19, 32], we analyzed baseline, week 1, and week 2 microbiome and metabolite measurements and their association with infection. Although patients with BSI within 30 days after transplantation had significantly lower levels of butyrate at week 2, visit 1 (P = .03) (Figure 4), there were no differences in Shannon diversity indices, specific taxa, or indole levels at week 2, visit 1, among patients who did or did not have a BSI within 30 days after transplantation.
Figure 4.
Fecal butyrate levels 2 weeks after transplantation are significantly decreased among patients in whom bloodstream infection (BSI) develops within 30 days after transplantation. All data points represent fecal butyrate levels at week 2, visit 1, and are segmented according to BSI status. (P value determined with Mann-Whitney U test.)
DISCUSSION
There is an increasing appreciation of the role of commensal microbiota role in influencing the health of the patients with cancer, which is generating interest in bringing microbiome analyses to the clinical arena [33–35]. In the current study, fecal butyrate and indole levels in HSCT recipients was correlated with microbial diversity, as well as the abundances of specific taxa at baseline and directly after transplantation (Figure 2 and 3). Thus, fecal indole and butyrate could be used as surrogate markers for microbial diversity or specific taxa. Given the associations found in previous studies between microbial diversity or particular taxa and key clinical outcomes [36, 37], metabolite measurements could potentially be applicable in the clinical setting, given that these assays are amenable to a shorter turnaround time than metagenomic sequencing.
Although we could not directly associate the levels with acute iGVHD outcomes or survival when adjusting for significant clinical factors (Table 2 and Supplementary Material), we believe that the relatively small number of patients studied and/or inherent difficulties associated with volatile metabolite measurements from the feces is likely to have affected our ability to identify significant correlations between the fecal microbiome and metabolites with HSCT outcomes. Nevertheless, it remains a possibility that butyrate levels in the actual colonic mucosa, rather than measurements from the feces, are the important factor to consider for GVHD outcomes. For example, Mathewson et al [7] observed a lack of changes in the amounts of luminal butyrate, despite microbiome shifts after allogeneic bone marrow transplantation, but they found that butyrate decreases occurred in the intestinal tissue, leading to decreased histone acetylation. However, unlike Mathewson et al [7], we found that fecal indole and butyrate were correlated with microbial α diversity and specific taxa of the stool samples reported herein (Figure 2 and 3).
We did, however, find the Shannon diversity index and Coriobacteriaceae at engraftment to be negatively associated with acute iGVHD outcomes (Supplementary Table 1). A 2017 study found Coriobacteriaceae in donor and recipient microbiota to be positively associated with acute iGVHD [30], and another study suggested that suggested that Coriobacteriaceae contributed to the benefits of exercise in preventing disease, improving health outcomes, and regulating the intestinal environment [38]. Thus, the definitive role of Coriobacteriaceae in intestinal health has yet to be determined and will need to be explored further.
A previous report found that members of the Lachnospiraceae and Ruminococcaceae families are associated with high urinary 3-IS levels in patients during days 0–10 after allo-SCT [18]. We saw similar taxa associated with high indole levels in stool samples (Figure 3 and Supplementary Figure 2). Bacilli were enriched in samples with low fecal indole levels, as was previously reported for low urinary 3-IS levels [39]. An analogous trend occurred in butyrate fecal levels; high butyrate levels were associated with groups of bacteria within the Clostridia class, and low butyrate levels were enriched for groups of bacteria within the Bacilli class. Of note, Blautia, Bacteroidaceae, Lachnospiraceae, and Ruminococcus, whose losses were previously reported to be associated with acute GVHD or GVHD-related death, were taxa associated with high indole and butyrate measurements in this study. Similarly, increases in Enterococcus and Streptococcus, which were negatively associated with fecal butyrate and indole levels in the current study, have also been reported to be associated with acute GVHD or GVHD-related death (Supplementary Figure 3) [20, 21, 23, 24, 29, 30].
Considering infection outcomes, lower levels of stool butyrate shortly after transplantation were associated with the development of a BSI within 30 days after transplantation (Figure 4). This is consistent with findings of studies showing that depletion of butyrate-producing Clostridia after antibiotic treatment elevates mucosal colonic lumen oxygen and promotes aerobic pathogen expansion [40]. Such expansion could lead to intestinal domination by pathogenic bacteria with subsequent translocation across damaged epithelium [5]. Inasmuch as dietary interventions can facilitate butyrate production, these data suggest that a “prebiotic” approach could help ameliorate serious infections and other adverse outcomes in HSCT recipients, and this is the subject of an ongoing clinical trial (https://clinicaltrials.gov/ct2/show/NCT02763033).
Metabolite measurement by mass spectrometry is susceptible to factors that can affect accuracy and precision. To minimize these effects, we included an internal standard to all sample runs and conducted absolute quantitation by including a calibration curve constructed by spiking known concentrations of external reference standards. A limitation was that we did not determine the external standard curve in a null sample matrix devoid of the compounds of interest. When matrix effects were evaluated using the instrument standard (7-hydroxy-coumarin), the stool sample matrix suppressed the output signal, potentially resulting in an underestimate of the true concentrations. In our study fecal butyrate measurements were approximately 10-fold less than previously reported [41] and is likely to reflect the use of antibiotics and chemotherapy, which can reduce the overall biomass of metabolite-producing microbes within the gut, or be a consequence of the method. Nevertheless, because we diluted samples to ensure measurement in the linear range of the calibration curve, the sample-to-sample comparisons of the measured metabolites are most likely accurate.
In conclusion, we report the first longitudinal analyses correlating fecal butyrate and indole concentrations with microbiome diversity after HSCT in patients with hematologic cancer and demonstrating associations with clinical outcomes, including bacteremia. Our findings further solidify the association of α diversity at the time of engraftment with acute iGVHD. Our results suggest using fecal microbial metabolites as surrogate markers for low microbial diversity or particular taxa. Further studies in larger cohorts in multiple institutions will need to be performed to confirm the predictive capabilities of fecal metabolites as surrogate biomarkers for microbial diversity and specific taxa before and after transplantation. In addition, further investigations on how these metabolites affect host physiology and clinical outcomes in HSCT recipients may offer appropriate targets for therapeutic interventions by modulating defined bacterial species, the enzymes required to produce the metabolites, or exogenous administration of the metabolites themselves.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Author contributions. J. R. G. P., D. P. S., R. F. C., A. M. A., R. R. J., D. P. K., E. J. S., S. A. S. and P. C. O. were involved in the conception and design of the study; P. V. S., D. P. S., L. G. C., and R. F. C. were responsible for enrollment of patients and collection of clinical samples; J. R. G. P., D. P. S., C. E. B., J. S. I., and G. R. captured clinical data; L. G. C., E. F., L. V., and P. L. L. were involved in fecal extracts for liquid chromatography tandem mass spectrometry and fecal metabolite measurements; J. R. G. P., C. B. P., F. M., and P .C. O. performed microbiota analyses; J. R. G. P. and C. B. P. contributed to statistical analyses; C. B. P. performed linear mixed models; J. R. G. P., A. M. A., R. R. J., R. R. J., S. A. S., and P. C. O. wrote the manuscript; and all authors read and corrected the final draft.
Financial support. This work was supported by the MD Anderson Cancer Center (MDACC) (S. A. S. and P. C. O.; Multidisciplinary Research Program funding to D. P. K. and E. J. S.; and Odyssey Fellowship Program support to J. R. G. P.); the CFP Foundation (J. R. G. P.); and the American Cancer Society (Mentored Research Scholar Grants in Applied and Clinical Research, MRSG-16-152-01-CCE to D.P.S.) The Grant–Biostatistics Shared Resource (C.B.P.) and the Proteomics and Metabolomics Core Facility (L. V. and P. L. L.) are supported by the National Cancer Institute, National Institutes of Health Cancer Center Support Grant (grant 4P30CA016672-41. The Proteomics and Metabolomics Core Facility (L. V. and P. L. L.) is also supported by the Cancer Prevention and Research Institute of Texas (grant RP130397), and the National Institutes of Health (grant 1S10OD012304-01).
Potential conflict of interest. R. R. J. reports personal fees from Merck, Seres Therapeutics, MicrobiomeDx, and Ziopharm Oncology outside the submitted work. D. P. K. reports being a consultant for Astellas Pharma, Amplyx Pharmaceuticals, Mayne Pharma, Merck, and Cidara Therapeutics and receiving honoraria from Merck, Gilead Sciences, and United Medical. P. C. O. reports receiving research funding and consultant fees from Merck and being a consultant to Jaguar Pharmaceuticals, Pfizer, and Singulex. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galloway-Peña JR, Smith DP, Sahasrabhojane P, et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer 2016; 122:2186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Montassier E, Al-Ghalith GA, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med 2016; 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh P, Teal TK, Marsh TL, et al. Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 2015; 3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bachmann M, Meissner C, Pfeilschifter J, Mühl H. Cooperation between the bacterial-derived short-chain fatty acid butyrate and interleukin-22 detected in human Caco2 colon epithelial/carcinoma cells. Biofactors 2017; 43:283–92. [DOI] [PubMed] [Google Scholar]
- 7. Mathewson ND, Jenq R, Mathew AV, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 2016; 17:505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sugimoto S, Naganuma M, Kanai T. Indole compounds may be promising medicines for ulcerative colitis. J Gastroenterol 2016; 51:853–61. [DOI] [PubMed] [Google Scholar]
- 9. Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med 2016; 8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan J, McKenzie C, Potamitis M, et al. The role of short-chain fatty acids in health and disease. Adv Immunol 2014; 121:91–119. [DOI] [PubMed] [Google Scholar]
- 11. Duncan SH, Barcenilla A, Stewart CS, et al. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 2002; 68:5186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther 1998; 12:499–507. [DOI] [PubMed] [Google Scholar]
- 13. Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009; 294:1–8. [DOI] [PubMed] [Google Scholar]
- 14. Haak BW, Littmann ER, Chaubard JL, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 2018; 131:2978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 2014; 5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 2010; 34:426–44. [DOI] [PubMed] [Google Scholar]
- 17. Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A 2010; 107:228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weber D, Oefner PJ, Hiergeist A, et al. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 2015; 126:1723–8. [DOI] [PubMed] [Google Scholar]
- 19. Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014; 124:1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biagi E, Zama D, Nastasi C, et al. Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone Marrow Transplant 2015; 50:992–8. [DOI] [PubMed] [Google Scholar]
- 21. Jenq RR, Taur Y, Devlin SM, et al. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant 2015; 21:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Y, Chen Y, Liu H, Depei W. Alterations of intestinal microbiota correlate with the emergence of acute graft-versus-host disease in patients undergoing allogeneic hematopoietic stem cell transplantation. Blood 2015;126:3145. [Google Scholar]
- 23. Holler E, Butzhammer P, Schmid K, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant 2014; 20:640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jenq RR, Ubeda C, Taur Y, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med 2012; 209:903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013; 41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. doi:10.1080/01621459.1999.10474144 [Google Scholar]
- 29. Golob JL, Pergam SA, Srinivasan S, et al. Stool microbiota at neutrophil recovery is predictive for severe acute graft vs host disease after hematopoietic cell transplantation. Clin Infect Dis 2017; 65:1984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu C, Frank DN, Horch M, et al. Associations between acute gastrointestinal GvHD and the baseline gut microbiota of allogeneic hematopoietic stem cell transplant recipients and donors. Bone Marrow Transplant 2017; 52:1643–50. [DOI] [PubMed] [Google Scholar]
- 31. Sorror Maris MB, Storb R,_, et al. Hematopoietic cell transplantation (HCT)-specific comorbidity index: a new tool for risk assessment before allogeneic HCT. Blood 2005; 106:2912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mancini N, Greco R, Pasciuta R, et al. Enteric microbiome markers as early predictors of clinical outcome in allogeneic hematopoietic stem cell transplant: results of a prospective study in adult patients. Open Forum Infect Dis 2017; 4:ofx215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galloway-Peña JR, Jenq RR, Shelburne SA. Can consideration of the microbiome improve antimicrobial utilization and treatment outcomes in the oncology patient? Clin Cancer Res 2017; 23:3263–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shono Y, Docampo MD, Peled JU, et al. Intestinal microbiota-related effects on graft-versus-host disease. Int J Hematol 2015; 101:428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taur Y, Jenq RR, Ubeda C, et al. Role of intestinal microbiota in transplantation outcomes. Best Pract Res Clin Haematol 2015; 28:155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shono Y, van den Brink MRM. Gut microbiota injury in allogeneic haematopoietic stem cell transplantation. Nat Rev Cancer 2018; 18:283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Galloway-Peña J, Brumlow C, Shelburne S. Impact of the microbiota on bacterial infections during cancer treatment. Trends Microbiol 2017; 25:992–1004. [DOI] [PubMed] [Google Scholar]
- 38. Zhao X, Zhang Z, Hu B, et al. Response of gut microbiota to metabolite changes induced by endurance exercise. Front Microbiol 2018; 9:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jenq RR. How’s your microbiota? let’s check your urine. Blood 2015; 126:1641–2. [DOI] [PubMed] [Google Scholar]
- 40. Rivera-Chávez F, Zhang LF, Faber F, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 2016; 19:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Torii T, Kanemitsu K, Wada T, et al. Measurement of short-chain fatty acids in human faeces using high-performance liquid chromatography: specimen stability. Ann Clin Biochem 2010; 47:447–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.