Abstract
Site-specific incorporation of multiple distinct noncanonical amino acids (ncAAs) into a protein is an emerging technology with tremendous potential, which relies on mutually orthogonal engineered aminoacyl-tRNA synthetase/tRNA pairs that suppress different nonsense/frameshift codons. So far, up to two distinct ncAAs have been incorporated into proteins expressed in E. coli, using archaea-derived tyrosyl and pyrrolysyl pairs. Here we report that the E. coli derived tryptophanyl pair can be combined with the archaeal tyrosyl or the pyrrolysyl pair in ATMW1 E. coli to incorporate two different ncAAs into one protein with high fidelity and efficiency. By combining all three orthogonal pairs, we further demonstrate simultaneous site-specific incorporation of three different ncAAs into one protein. To use this technology for chemoselectively labeling proteins with multiple distinct entities at predefined sites, we also sought to identify different bioconjugation handles that can be co-incorporated into proteins as ncAA-sidechains and subsequently functionalized through mutually compatible labeling chemistries. To this end, we show that the recently developed chemoselective rapid azo-coupling reaction (CRACR) directed to 5-hydroxytryptophan (5HTP) is compatible with strain-promoted azide-alkyne cycloaddition (SPAAC) targeted to p-azidophenylalanine (pAzF) and strain-promoted inverse electron-demand Diels-Alder cycloaddition (SPIEDAC) targeted to cyclopropene-lysine (CpK) for rapid, catalyst-free protein labeling at multiple sites. Combining these mutually orthogonal nonsense suppression systems and the mutually compatible bioconjugation handles they incorporate, we demonstrate site-specific labeling of recombinantly expressed proteins at up to three distinct sites.
Graphical Abstract
Introduction.
The ability to precisely label proteins with multiple distinct entities at predefined locations is highly desirable for numerous cross-disciplinary applications. A variety of different strategies have been developed for labeling proteins that target native residues, the termini, as well as peptide and protein tags appended to a recombinant protein.1 Co-translational site-specific incorporation of noncanonical amino acids (ncAA) enables precise placement of uniquely reactive chemical functionalities into proteins, which can be subsequently labeled using chemoselective bioconjugation reactions.2 Significant advantages of this strategy include the small size of the ncAAs relative to other genetically encoded tags, the flexibility of incorporating them into virtually any site of any target protein, and the large variety of unique chemical groups that can be genetically encoded using this technology. Incorporation of ncAAs into proteins in living cells is performed using engineered aminoacyl-tRNA synthetase (aaRS)/tRNA pairs that suppress nonsense or frameshift codons and do not cross-react with their counterparts from the host cell (i.e., orthogonal).2a-e Several different pairs have been developed for ncAA incorporation into proteins expressed in different domains of life.2a-e, 3 Using this technology, many chemoselective bioconjugation handles have been genetically encoded, including azides, alkynes, ketones, strained alkenes and alkynes, tetrazines, etc., which has enabled novel approaches to understand and manipulate protein function.2, 4
So far, the large majority of work using this technology has been restricted to the incorporation of a single ncAA into a polypeptide. However, the ability to incorporate multiple distinct bioconjugation handles into proteins, which can be independently functionalized with different entities, has the potential to facilitate many powerful applications, including the attachment of different probes for sophisticated biophysical studies, and the synthesis of advanced protein-based therapeutics and diagnostics. Such expression systems would require multiple different aaRS/tRNA pairs that do not cross-react with each other or with their host counterparts. It is important to remember that two pairs may cross-react at three different levels: ncAA-aaRS interaction, aaRS-tRNA interaction, and codon-anticodon interaction.5 These two pairs must be able to efficiently suppress two distinct nonsense or frameshift codons, and incorporate two different ncAAs that encode mutually compatible bioconjugation chemistries for their independent labeling. Using the M. jannaschii derived tyrosyl pair (MjTyr) and the methanosarcina-derived pyrrolysyl pair (Pyl), which are mutually orthogonal, it has indeed been possible to site-specifically incorporate two distinct ncAAs into proteins expressed in E. coli.6 MjTyr is typically used to suppress the UAG nonsense codon, while the more anticodon-permissive Pyl is assigned to UAA or AGGA codons.6a-e Taking advantage of bioconjugation handles that have been genetically encoded using these pairs, particularly those for strain-promoted azide-alkyne cycloaddition (SPAAC)7 and strain-promoted inverse electron-demand Diels-Alder cycloaddition (SPIEDAC),8 it has been possible to achieve site-specific dual labeling of proteins targeted to distinct ncAA residues.6a, 6c-e
Despite these exciting and empowering advances, further expansion of this technology is impeded by the unavailability of additional mutually orthogonal aaRS/tRNA pairs, as well as genetically encodable novel bioconjugation chemistries that are compatible with SPAAC and SPIEDAC. Recently, we developed a unique E. coli derived tryptophanyl-tRNA synthetase (EcTrpRS)/tRNA pair which can be used for ncAA incorporation in both E. coli and eukaryotes.9 We functionally substituted the endogenous EcTrp pair of E. coli with its yeast counterpart to generate the novel strain ATMW1. The “liberated” EcTrpRS/tRNA pair was then established as an orthogonal UGA suppressor in ATMW1, and was engineered to selectively charge a variety of tryptophan analogs, including 5-hydroxytryptophan (5HTP; Figure 1).9 Furthermore, we have developed chemoselective bioconjugation strategies that take advantage of the highly electron rich 5-hydroxyindole group of 5HTP.10 For example, based on the remarkably rapid coupling between 5-hydroxyindoles and various aromatic diazonium ions, we have developed the chemoselective rapid azo-coupling reaction (CRACR) for selective labeling of proteins at 5HTP residues. In this report, we show that 5HTP-directed CRACR is compatible with both SPAAC and SPIEDAC for chemoselective labeling of proteins at different sites. Next, we demonstrate that our UGA-suppressing EcTrpRS/tRNA pair can be orthogonally used with UAG-suppressing MjTyr or Pyl to site-specifically incorporate two distinct ncAAs into proteins expressed in ATMW1 E. coli. We used these new dual-ncAA mutagenesis platforms to express proteins co-incorporating 5HTP with p-azidophenylalanine (pAzF) or cyclopropene-lysine (CpK)(Figure 1), which were subsequently site-specifically double labeled using CRACR and SPAAC or SPIEDAC, respectively. Finally, we combined all three suppressors to site-specifically co-incorporate 5HTP,9 pAzF,11 and CpK12 into the same protein in response to three different stop codons and demonstrated their independent labeling with three distinct entities.
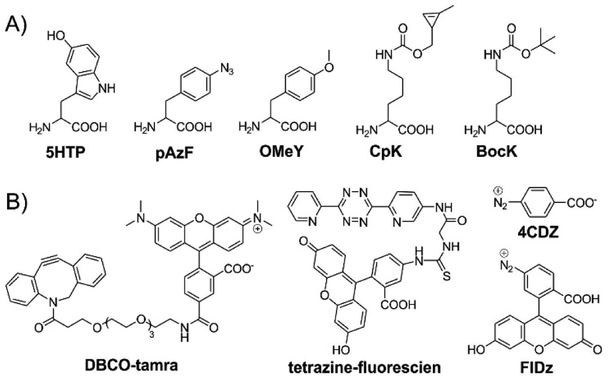
Figure 1:
Structures of the ncAAs (A) and labeling agents (B) used in this report.
Results
CRACR is compatible with SPAAC and SPIEDAC.
For site-specifically labeling proteins with different entities, distinct ncAAs with bioconjugation handles are needed which can be functionalized by chemistries that do not interfere with each other. So far, this has been achieved most successfully by combining ncAAs that can be labeled by SPAAC and SPIEDAC.5, 6c, 6e, 13 For example, proteins containing an azide and a cyclopropene sidechain have been sequentially labeled in one pot by a cyclooctyne and an electron-deficient tetrazine, respectively.5, 13 Distinctly reactive SPIEDAC handles have also been used to this end.6c, 6e Additionally, Cu(I)-catalyzed azide-alkyne cycloaddition and oxime conjugation have been shown to be compatible with SPIEDAC and SPAAC,6a, 6b, 14 respectively, but their scope is somewhat limited due to the requirement of toxic catalysts or low pH.
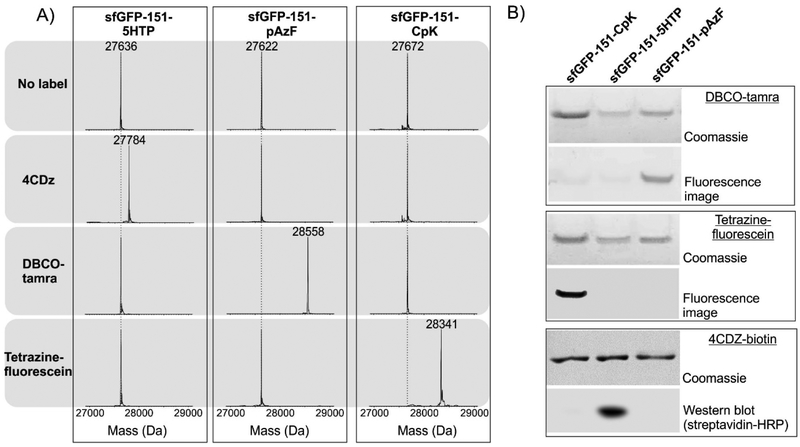
CRACR enables rapid labeling of proteins (>100 M−1s−1 for 4-carboxydiazonium; 4CDZ) at 5HTP residues under ambient conditions.10a To evaluate if CRACR is compatible with SPAAC and SPIEDAC mediated protein labeling, we expressed superfolder green fluorescent protein (sfGFP) incorporating 5HTP, CpK or pAzF at position 151. A previously reported polyspecific MjTyrRS/tRNA pair11b and the wild-type MbPylRS/tRNA pair12 were used to incorporate pAzF and CpK, respectively, in response to the UAG stop codons. 5HTP was incorporated using our recently reported EcTrpRS/tRNAUCA pair in the ATMW1 strain.9 To investigate whether the labeling conditions for each of these bioconjugation handles are compatible with each other, we subjected each of the three different sfGFP mutants to three different labeling conditions using DBCO-tamra, tetrazine-fluorescein, and 4CDZ (or 4CDZ-biotin)(Figure 1). Subsequent analysis by whole-protein ESI-MS analysis revealed that these reagents facilitate complete and selective labeling of sfGFP mutants harboring their corresponding bioconjugation partners (DBCO-pAzF, tetrazine-CpK, 4CDZ-5HTP), while leaving the other two mutants untouched (Figure 2A). We further resolved these proteins by SDS-PAGE and analyzed the labeling reaction by fluorescence imaging (for DBCO-TAMRA, and tetrazine-fluorescein) or Western blot (for 4CDZ-biotin, using streptavidin-HRP conjugate), further confirming selective labeling of each of the three mutants only with their intended reagents (Figure 2B). These observations establish that CRACR-mediated labeling of 5HTP, SPAAC-labeling of pAzF and SPIEDAC-labeling of CpK are mutually compatible. Therefore, it should be possible to distinctly functionalize these ncAA sidechains with multiple different entities if they could be simultaneously incorporated into one protein.
Figure 2.
CRACR mediated protein labeling is orthogonal to SPAAC and SPIEDAC. A) Treatment of three sfGFP mutants (incorporating 5HTP, pAzF or CpK at the 151 position) with 4CDZ, DBCO-tamra, and tetrazine-fluorescein followed by ESI-MS analysis reveals selective labeling of 5HTP, pAzF, and CpK by CRACR, SPAAC, and SPIEDAC, respectively. B) Analysis of protein samples from a similar experiment (4CDZ-biotin was used instead of 4CDZ) by SDS-PAGE followed by fluorescence imaging (anti-biotin Western blot for proteins treated with 4CDZ-biotin) further confirms selective labeling of each mutant with the intended reagent.
To facilitate the co-incorporation of 5HTP into proteins with pAzF and CpK, we next evaluated if the EcTrpRS/tRNAUCA pair can be combined with MjTyr or Pyl to suppress two distinct nonsense codons in a mutually orthogonal manner in our ATMW1 strain of E. coli. Unlike all other pairs (including MjTyr and Pyl) that typically use a UAG nonsense codon for ncAA incorporation, our EcTrpRS/tRNAUCA pair was developed as an efficient UGA-suppressor,9 making it intrinsically easier to combine it with other pairs for suppressing two distinct nonsense codons. Additionally, both MjTyr and Pyl have been used in regular E. coli strains as orthogonal UAG suppressors, suggesting that these do not cross-react with any of the endogenous pairs (including the EcTrp, which is used as an orthogonal pair in our engineered ATMW1 strain). However, it is important to note that in our expression system, the anticodon of tRNAEcTrp is altered to perform UGA suppression, and the pair is expressed at a significantly higher level relative to endogenous expression (to achieve high levels of nonsense suppression efficiency), which may compromise its mutual orthogonality with MjTyr and Pyl pairs.
A dual ncAA incorporation system using EcTrpRS/tRNAEcTrpUCA and PylRS/tRNAPylCUA.
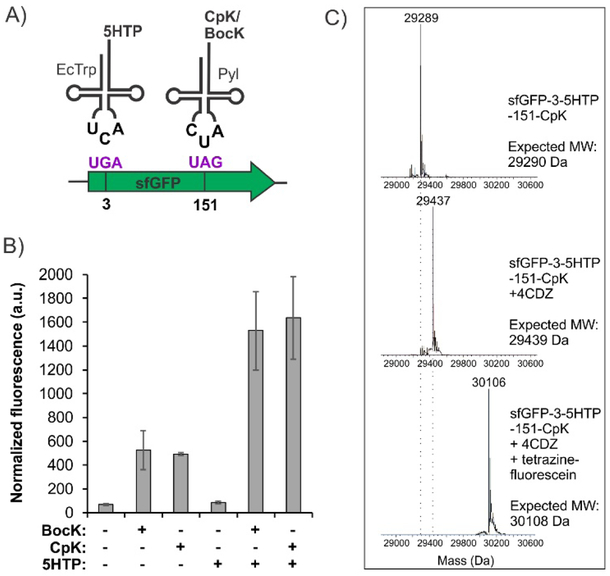
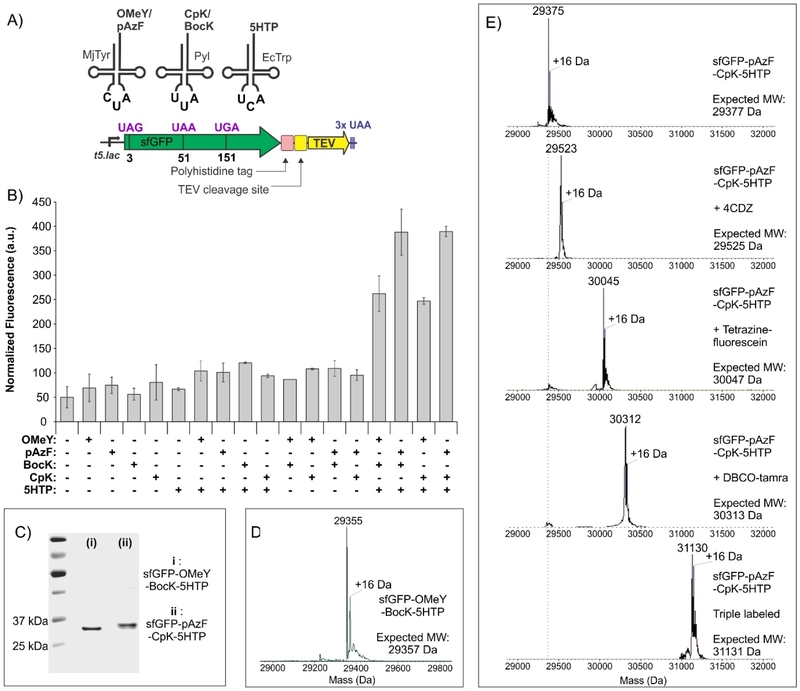
Due to its unique structural features, the pyrrolysyl (Pyl) pair is orthogonal in all domains of life. Its mutual orthogonality with other suppressor pairs has been the basis of all dual-ncAA incorporation systems developed so far. 5–6, 13–14 Consequently, we anticipated that it would also be orthogonal to our EcTrp suppressor pair in ATMW1 E. coli. To ensure that the EcTrp and the Pyl pairs can be simultaneously assigned to UGA and UAG codons with high fidelity, we constructed a dual suppressor plasmid expressing both of these pairs. Our previously reported polyspecific EcTrpRS (h14) and the wild-type MbPylRS were each expressed from tacI promoters, while the tRNAEcTrpUCA and tRNAPylCUA were expressed using separate proK promoters. This plasmid was co-transformed into ATMW1 E. coli strain with a sfGFP reporter that harbors a UGA codon at position 3 and a UAG codon at position 151 (Figure 3A). The expression of this sfGFP double mutant was monitored in rich media in the presence or absence of substrate ncAAs (BocK and CpK for Pyl, 5HTP for EcTrp) for both pairs. Robust reporter expression was observed when both substrates were present, while no sfGFP fluorescence was detected when both substrates were absent, or when 5HTP alone was present (Figure 3B). The latter observation is consistent with the fact that the Pyl pair is essentially silent in the absence of a substrate ncAA, causing no “leaky” UAG suppression. When only a Pyl-substrate (BocK or CpK) was present, weak reporter expression was observed due to the low levels of tryptophan-charging activity of the EcTrpRS mutant (h14) in the absence of 5HTP. We have previously shown that this mutant exclusively charges 5HTP when it is supplemented in the expression medium.9 We isolated full-length sfGFP-3–5HTP-151-BocK/CpK by immobilized metal ion chromatography (IMAC) using a C-terminal polyhistidine tag (17–20 mg/L) and analyzed the protein by SDS-PAGE and whole-protein ESI-MS (Figure 3C, S1), confirming successful incorporation of the intended ncAAs.
Figure 3.
A dual ncAA incorporation system using EcTrp(UCA) and Pyl(CUA) pairs in ATMW1 E. coli. A) Scheme of the dual ncAA incorporation experiment. B) Expression of sfGFP-3UGA-151UAG using this system in the presence of indicated ncAA substrates, measured by its characteristic in-cell fluorescence. C) Purified sfGFP-3-5HTP-151-CpK has the expected mass (top panel). Treatment with 4CDZ and tetrazine-fluorescein results in its one-pot quantitative double labeling, and confirms the feasibility of using CRACR and SPIEDAC for protein modification at two different sites.
The ability to site-specifically incorporate CpK and 5HTP into a protein with high fidelity and efficiency using the above dual ncAA incorporation system makes it possible to test whether these two bioconjugation handles can be used to attach two distinct labels using SPIEDAC and CRACR. Purified sfGFP-3–5HTP-151-CpK was subjected to sequential labeling first with 4CDZ, and then with tetrazine-fluorescein in the same pot under ambient conditions. ESI-MS analysis of the protein after each step revealed complete labeling with expected mass changes, confirming the suitability of these two chemoselective labeling strategies for facile site-specific dual labeling of recombinant proteins (Figure 3C).
A dual ncAA incorporation system using EcTrpRS/tRNAEcTrpUCA and MjTyrRS/tRNAMjTyrCUA.
Next, we investigated if the EcTrp pair can be combined with the MjTyr pair in a mutually orthogonal fashion. We built a dual suppressor plasmid, similar to the one described in the previous section, that expresses the EcTrpRS(h14)/tRNAEcTrpUCA and the MjTyrRS(polyspecific)/tRNAMjTyrCUA pairs (Figure S2A). However, a detailed characterization of its suppression behavior revealed a significant level of cross-reactivity between EcTrpRS and tRNAMjTyrCUA (Figure S2, S3). When this plasmid was used to express the sfGFP-3-UAG reporter, significant protein expression was observed in the absence of a substrate for MjTyrRS, and MS analysis of the resulting protein revealed the incorporation of tryptophan at the UAG codon (Figure S2B). However, OMeY was selectively incorporated at the UAG codon when it was supplemented in the growth medium, which shows that MjTyrRS can outcompete EcTrpRS for charging tRNAMjTyrCUA in the presence of its substrate ncAA. This cross-reactivity was surprising, given that the MjTyrRS/tRNAMjTyrCUA pair has been extensively used for ncAA mutagenesis in various E. coli strains with no evidence of cross-reactivity with the endogenous EcTrpRS.
We hypothesized that at its low endogenous expression levels in regular E. coli strains, EcTrpRS does not exhibit detectable levels of cross-reactivity toward tRNAMjTyrCUA. In contrast, the overexpression of EcTrpRS using a strong promoter (tacI) from a multi-copy plasmid in our system causes it to significantly cross-react with this non-cognate tRNA. Indeed, it has been previously reported that overexpression of an aaRS can result in its cross-reaction with non-cognate tRNAs.15 If correct, this hypothesis would predict a relief from cross-reactivity if we lower the expression level of EcTrpRS. To this end, we created an alternative double suppression plasmid, where the strong inducible tacI promoter was replaced with a much weaker constitutively active glnS promoter to drive EcTrpRS expression (Figure S4A). This plasmid facilitated the expression of the sfGFP-3-UAG reporter only in the presence of OMeY, the MjTyrRS substrate (Figure S4B). The lack of UAG suppression in the absence of a MjTyrRS-substrate confirms a relief from the aforementioned cross-reactivity, corroborating our hypothesis. We were also gratified to find that the reduced expression EcTrpRS did not significantly compromise its efficiency of 5HTP incorporation in response to UGA codons, but it drastically attenuated “leaky” incorporation of tryptophan in the absence of its substrate 5HTP: while the new double suppressor plasmid facilitated comparable levels of the sfGFP-3-UGA expression relative to its earlier counterpart in the presence of 5HTP, little reporter expression was observed in the absence of the ncAA (Figure S2C and S4C). These observations underscore the importance of expressing the aaRS/tRNA pairs at an optimal level for incorporating ncAAs with high fidelity and efficiency.
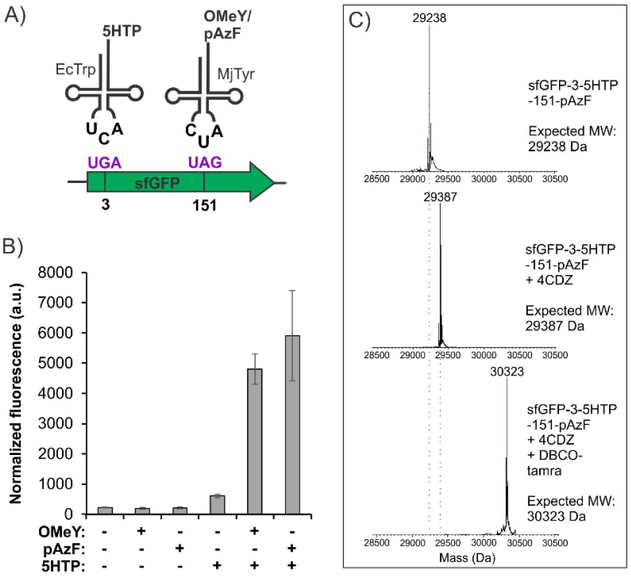
We then tested the incorporation of two distinct ncAAs into the sfGFP-3-UGA-151-UAG reporter using this optimized EcTrp(UCA)+MjTyr(CUA) double suppression system in ATMW1 E. coli. 5HTP was used as the substrate for EcTrpRS(h14), while OMeY or pAzF were tested for the polyspecific MjTyrRS mutant. We observed high levels of reporter expression only in the presence of both 5HTP and OMeY/pAzF (Figure 4B). Purification of the full-length double mutant using a C-terminal polyhistidine tag by IMAC (yields of 25–30 mg/L) followed by whole-protein ESI-MS analysis revealed the expected mass (Figure 4C and S1), validating incorporation of the two ncAAs at desired sites. The sfGFP-3–5HTP-151-pAzF also provides the opportunity to test if CRACR and SPAAC can be used together to attach two different labels on the same protein. Purified sfGFP-3–5HTP-151-pAzF was labeled first with 4CDZ, followed by DBCO-tamra under ambient conditions and analyzed by ESI-MS to demonstrate quantitative labeling of each ncAA residue (Figure 4C).
Figure 4.
A dual ncAA incorporation system using EcTrp(UCA) and MjTyr(CUA) pairs in ATMW1 E. coli. A) Scheme of the dual ncAA incorporation experiment. B) Expression of sfGFP-3UGA-151UAG using this system in the presence of indicated ncAA substrates, measured by its characteristic in-cell fluorescence. C) Purified sfGFP-3-5HTP-151-pAzF has the expected mass (top panel). Treatment with 4CDZ and DBCO-tamra results in its one-pot quantitative double labeling, and confirms the feasibility of using CRACR and SPAAC for protein modification at two different sites.
The ability to precisely functionalize antibodies and antibody fragments is highly important for numerous applications. Efficient site-specific dual modification of antibodies can facilitate the development of sophisticated diagnostics and therapeutics.1a, 4, 16 To explore the possibility of extending our dual labeling strategies described above to this important class of proteins, we selected the 5HTP+pAzF incorporation system using the EcTrpRS/tRNAEcTrpUCA + MjTyrRS/tRNAMjTyrCUA pairs, which was somewhat more efficient than the 5HTP+CpK incorporation system. The optimized double suppression plasmid harboring both pairs was co-transformed into ATMW1 E. coli with a plasmid expressing the Fab fragment of the anti-HER2 antibody Herceptin, where the heavy chain encoded two mutations: K169UGA and S202UAG (Figure 5A).17 We deliberately chose to incorporate both ncAAs into the same subunit of the antibody to highlight the robustness of our expression system. The wild-type Fab fragment was also expressed as a control. The double mutant Fab was expressed in the presence of 5HTP and pAzF and was purified from the periplasmic fraction by protein-A affinity chromatography. ESI-MS analysis of the whole antibody confirmed the incorporation of both ncAAs (Figure 5B and S5). When the purified double mutant Fab was treated with 4CDZ, or DBCO-tamra or both reagents, clean labeling of the corresponding sites was observed by whole-protein ESI-MS analysis (Figure 5B). Identical treatment of the wild-type Fab resulted in no modification (Figure S5), underscoring the selectivity of this double labeling strategy. We also showed that the Fab double mutant can be labeled with DBCO-tamra and fluorescein-diazonium (FlDZ) reagents to install two distinct fluorescent labels at two different sites (Figure 5C).
Figure 5.
Site-specific double labeling of anti-HER2 Fab using CRACR and SPAAC. A) The Fab was expressed incorporating 5HTP and pAzF into 169 and 202 sites of the heavy chain using the dual-ncAA incorporation system described in Figure 4. B) ESI-MS analysis of Fab-169-5HTP-202-pAzF (not reduced; HC and LC connected by a disulfide) shows the expected mass (top panel).Treatment of this Fab double mutant with 4CDZ or DBCO-tamra produces expected single modification, while quantitative double modification is observed when both are used. Identical treatment of the wild-type Fab leads to no modification (Supplementary Figure S4). C) Labeling Fab-169-5HTP-202-pAzF or wild-type Fab with DBCO-tamra or fluorescein-diazonium or both leads to expected labeling of the double mutant but not the wild-type, as revealed by reducing SDS-PAGE (HC and LC separates; each ~24 kDa) followed by fluorescence imaging.
Site-specific incorporation of three different ncAAs into one protein, and its chemoselective labeling with three distinct entities
Our work establishes EcTrp, MjTyr and Pyl as a set of three mutually orthogonal pairs in ATMW1 E. coli. We wondered if it would be possible to simultaneously use all three pairs to site-specifically incorporate 5HTP, pAzF and CpK into one protein. The compatible bioconjugation chemistries these encode should then enable the labeling of the resulting protein with three distinct entities. To our knowledge, site-specific incorporation of three different ncAAs into one protein in living cells has not yet been achieved, and the ability to do so would mark a major milestone for the ncAA mutagenesis technology. However, this would require the assignment of these three pairs to three distinct nonsense/frameshift codons. Since the suppression efficiency of nonsense codons are generally higher, we chose to simultaneously use the three nonsense codons to encode three distinct ncAAs. We envisioned assigning EcTrp, MjTyr and the Pyl pairs to suppress UGA, UAG, and UAA, respectively, to achieve site-specific incorporation of three different ncAAs, since Pyl has been previously used as a UAA-suppressor together with a UAG-suppressing MjTyr pair for dual ncAA incorporation.6a, 6d We confirmed that each of these three nonsense suppressors can facilitate efficient expression of sfGFP reporters encoding the appropriate nonsense codon at the 151 site in ATMW1 E. coli (Figure S6).
However, simultaneous reassignment of all three stop codons raises the important question of how to terminate translation. It is important to remember that at a reassigned nonsense codon, incorporation of the designated ncAA is not the only outcome; significant levels of termination is also observed. In particular, multiple consecutive stop codons have been shown to effectively terminate translation even in the presence of highly efficient suppressors.18 Although placing multiple consecutive stop codons at the end of a target gene offers an effective way to terminate its translation, the resulting protein would likely have a heterogeneous C-terminus from partial ncAA incorporation at these sites. To address this concern, we designed a novel expression system GTEV, where the desired recombinant protein is appended with a purification tag (polyhistidine in this case) at the C-terminus, followed by a tobacco-etch virus (TEV) protease cleavage sequence, the TEV protease,19 and three consecutive UAA stop codons (Figure S7A). We envisioned that upon expression, the C-terminal TEV protease will cleave itself out, leaving the desired recombinant protein with a clean C-terminus. We expressed wild-type sfGFP from the GTEV vector in ATMW1 E. coli and isolated it using the C-terminal polyhistidine tag by IMAC in high yield (140 mg/L; comparable with a non-GTEV expression system). Its SDS-PAGE and ESI-MS analysis showed only the desired protein, confirming efficient self-cleavage of the TEV protease (Figure S7B-C).
To achieve simultaneous incorporation of three different ncAAs, we needed a plasmid system for co-expressing all three suppressor pairs as well as a reporter gene in the same cell. To this end, we designed a two-plasmid expression system: 1) The aforementioned optimized dual suppressor plasmid encoding EcTrpRS(h14)/tRNAEcTrpUCA and the MjTyrRS(polyspecific)/tRNAMjTyrCUA pairs. 2) A GTEV vector for expressing a triple mutant sfGFP reporter (sfGFP-3UAG-51UAA-151UGA) that also encodes the MbPylRS/tRNAPylUUA pair. Co-transformation of these two plasmids into ATMW1 E. coli did not significantly alter its growth profile in the absence of induction (Figure S8). This strain was used to express the sfGFP reporter containing three different nonsense codons (Figure 6A) and full-length reporter expression was monitored in the presence or absence of ncAAs (5HTP for EcTrp, OMeY or pAzF for MjTyr, BocK or CpK for Pyl) by its characteristic fluorescence. Strong sfGFP expression was observed only when ncAA substrates for each of the three pairs were present in the medium, indicating successful incorporation of three ncAAs at desired sites (Figure 6B). The full-length sfGFP-3-OMeY-51-BocK-151–5HTP reporter was isolated by IMAC (yield 3 mg/L), and was analyzed by SDS-PAGE (Figure 6C) and whole-protein ESI-MS (Figure 6D) to reveal a mass consistent with the incorporation of the intended ncAAs at targeted sites. We also subjected this protein to tryptic digestion and analyzed the resulting peptides by HPLC-MS/MS analysis, to unambiguously confirm the incorporation of each ncAA at the desired sites (Figure S9-S11). The same expression system was also used to incorporate pAzF+CpK+5HTP into the sfGFP triple mutant reporter with similar yield (2 mg/L). When we treated this protein with either 4CDZ, DBCO-tamra, or tetrazine-fluorescein, separately, ESI-MS revealed complete single labeling in each case, confirming the presence of the three different bioconjugation handles which can be independently functionalized (Figure 6E). We then showed that all three ncAA residues can be labeled sequentially in one pot using CRACR, SPAAC and SPIEDAC to yield a triply functionalized protein (Figure 6E). The set of mutually compatible bioconjugation handles and the technology for their site-specific co-incorporation that we develop here should be valuable for numerous applications in chemical biology.
Figure 6.
Site-specific incorporation of three different ncAAs and their subsequent labeling. A) Scheme describing the triple ncAA incorporation experiment. B) Expression of sfGFP-3UAG-51-UAA-151UGA using this system in the presence of indicated ncAA substrates, measured by its characteristic in-cell fluorescence. C) SDS-PAGE analysis of purified sfGFP triple mutants incorporating either OMeY+BocK+5HTP or pAzF+CpK+5HTP. D) Whole-protein ESI-MS analysis of isolated sfGFP-OMeY-BocK-5HTP shows a mass consistent with the incorporation of the three different ncAAs at intended sites. E) Purified sfGFP-pAzF-CpK-5HTP also shows the expected mass (top panel). Its treatment with 4CDZ, tetrazine-fluorescein, and DBCO-tamra results in expected single labeling, while one-pot triple labeling is observed when all three labeling reagents are used.
Discussion.
Site-specific incorporation of ncAAs with bioconjugation handles provides a facile route to covalently attach a diverse variety of molecules onto proteins with remarkable precision. Extending this strategy to enable protein labeling at multiple chosen sites poses a multifaceted challenge that demand the development of both mutually compatible conjugation chemistries, and new routes for their site-directed incorporation into proteins. It has been possible to incorporate two different bioconjugation-ncAAs into one protein expressed in E. coli using MjTyr and Pyl pairs, and double label the resulting protein.6a-e Mutually compatible SPAAC and SPIEDAC chemistries have emerged as a facile strategy for such dual labeling under ambient catalyst-free conditions. Here we expand this toolbox by first demonstrating that our recently developed 5HTP-directed protein conjugation strategy CRACR is compatible with both SPAAC and SPIEDAC, owing to its fundamentally different labeling chemistry (Figure 2). We further show that the E. coli derived EcTrp pair, which we established as an orthogonal UGA-suppressor in the engineered E. coli strain ATMW1, can interface with MjTyr and Pyl pairs in a mutually orthogonal fashion to provide powerful new routes for incorporating multiple different bioconjugation-ncAAs into the same protein. We have previously demonstrated that the EcTrp pair can also be used in eukaryotic cells for site-specific incorporation of ncAAs in response to UAG or UGA codons.9 Consequently, it can be potentially combined with other orthogonal pairs in eukaryotic cells to site-specifically co-incorporate 5HTP and another bioconjugation-ncAA (such as CpK using Pyl, or pAzF using the E. coli derived tyrosyl pair),5, 13 enabling new routes for double labeling recombinant eukaryotic proteins – a direction that we are actively pursuing.
We also report the first example of site-specific incorporation of three distinct ncAAs into one protein expressed in a living cell. Development of EcTrp as a new orthogonal suppressor in the ATMW1 strain and its mutual orthogonality with MjTyr and Pyl pairs set the stage for their simultaneous use for triple ncAA incorporation. However, to achieve this, two key challenges needed addressing: 1) assigning these pairs to three distinct “blank” codons, and 2) developing a plasmid system that co-expresses all three orthogonal pairs. MjTyr and Pyl have been previously together in E. coli to suppress UAG and UAA, respectively.6a, 6d It appeared logical to combine the UGA-suppressing EcTrp pair with this dual-suppression system to build the first triple-suppression platform. To facilitate efficient termination of translation in an expression system where all three nonsense codons are reassigned, we took advantage of multiple consecutive stop codons. The UAA nonsense codon was used for this purpose, as the PylRS/tRNAPylUUA pair was the least efficient among the three different suppressors. To get around partial undesired ncAA incorporation at the C-terminal UAA codons, we also created a novel self-cleaving tag that leaves a clean C-terminus on the target protein. We also developed a plasmid system that co-expresses all three suppressor pairs to facilitate triple ncAA incorporation, without causing significant toxicity. Although the efficiency of incorporating three distinct ncAAs is not high (2–3 mg/L; ~2% of wild-type reporter), it can be improved by further optimization of this first-generation platform. The specific MjTyr, Pyl and the EcTrp pairs used here can be used to incorporate a variety of additional ncAAs with useful properties,2a, 2b, 2e further expanding the scope of the double and triple suppression systems described here.
Our work also highlights the need for carefully investigating multiple ncAA incorporation systems for any underlying cross-reactivity that can compromise fidelity. Nearly all dual-ncAA incorporation systems developed to date have taken advantage of the unique Pyl pair, which does not cross-react with other pairs owing to its distinctive structure. However, more caution must be taken when venturing beyond Pyl for creating new multi-ncAA incorporation systems. In addition to this work, we have also previously shown that in mammalian cells, an E. coli derived tyrosyl-tRNA synthetase can charge an E. coli derived leucyl-tRNACUA,5 which was unexpected given both pairs were derived from the same species. Here, we were able to overcome the mild cross-reactivity of EcTrpRS toward tRNAMjTyrCUA by simply reducing the expression level of EcTrpRS. This also underscores the importance of fine-tuning the expression levels of various components in complex multi-suppression systems, akin to the fine balance of endogenous aaRS/tRNA pairs that underlie the high fidelity of natural systems.
In this work, we have simultaneously reassigned all three nonsense codons and have thus reached the limit of the maximum number of distinct ncAAs that can be incorporated within the framework of the canonical triplet genetic code. However, exciting progress is being made to overcome this barrier and significantly expand the coding capacity for ncAAs in E. coli. For example, efforts are under way to reduce the number of triplet codons E. coli uses by global engineering of its genome.20 Such efforts can liberate additional triplet codons that can be simultaneously reassigned to encode more ncAAs. In an alternative approach, the Romesberg group have demonstrated the feasibility of using unnatural base pairs to encode ncAAs in E. coli.21 Use of an unnatural base pair can dramatically increase the number of available triplet codons that can be reassigned for ncAA incorporation. However, as these approaches overcome the limited selection of codons that are currently available for reassignment to ncAAs, an arsenal of mutually orthogonal ncAA incorporation systems would be needed to facilitate their simultaneous use to build a dramatically expanded genetic code. Our work offers a primer on how to approach this complex endeavor, reveals some of the underlying challenges, and provides novel strategies to overcome them.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by NIH grants 1R01GM118431 and 1R01GM117004 to E.W. and R01GM126220 and R01GM124319 to A.C. pRK793 was a gift from David Waugh (Addgene plasmid # 8827; http://n2t.net/addgene:8827; RRID:Addgene_8827)
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. Experimental methods, nucleotide sequences, supplementary figures and tables.
The authors declare no competing financial interest.
References.
- 1.(a).Agarwal P; Bertozzi CR, Site-specific antibody–drug conjugates: the nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjugate chemistry 2015, 26 (2), 176–192; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bloom S; Liu C; Kölmel DK; Qiao JX; Zhang Y; Poss MA; Ewing WR; MacMillan DW, Decarboxylative alkylation for site-selective bioconjugation of native proteins via oxidation potentials. Nature chemistry 2018, 10 (2), 205; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Boutureira O; Bernardes G. a. J., Advances in chemical protein modification. Chemical reviews 2015, 115 (5), 2174–2195; [DOI] [PubMed] [Google Scholar]; (d) deGruyter JN; Malins LR; Baran PS, Residue-Specific Peptide Modification: A Chemist’s Guide. Biochemistry 2017, 56 (30), 3863–3873; [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Koniev O; Wagner A, Developments and recent advancements in the field of endogenous amino acid selective bond forming reactions for bioconjugation. Chemical Society Reviews 2015, 44 (15), 5495–5551; [DOI] [PubMed] [Google Scholar]; (f) Lin S; Yang X; Jia S; Weeks AM; Hornsby M; Lee PS; Nichiporuk RV; Iavarone AT; Wells JA; Toste FD, Redox-based reagents for chemoselective methionine bioconjugation. Science 2017, 355 (6325), 597–602; [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Rosen CB; Francis MB, Targeting the N terminus for site-selective protein modification. Nature chemical biology 2017, 13 (7), 697–705; [DOI] [PubMed] [Google Scholar]; (h) Shih HW; Kamber DN; Prescher JA, Building better bioorthogonal reactions. Current opinion in chemical biology 2014, 21, 103–11; [DOI] [PubMed] [Google Scholar]; (i) Sletten EM; Bertozzi CR, Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angewandte Chemie (International ed. in English) 2009, 48 (38), 6974–98; [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Spicer CD; Davis BG, Selective chemical protein modification. Nature communications 2014, 5, ncomms5740; [DOI] [PubMed] [Google Scholar]; (k) Stephanopoulos N; Francis MB, Choosing an effective protein bioconjugation strategy. Nature chemical biology 2011, 7 (12), 876–84; [DOI] [PubMed] [Google Scholar]; (l) Los GV; Encell LP; McDougall MG; Hartzell DD; Karassina N; Zimprich C; Wood MG; Learish R; Ohana RF; Urh M, HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS chemical biology 2008, 3 (6), 373–382; [DOI] [PubMed] [Google Scholar]; (m) Keppler A; Gendreizig S; Gronemeyer T; Pick H; Vogel H; Johnsson K, A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nature biotechnology 2003, 21 (1), 86; [DOI] [PubMed] [Google Scholar]; (n) Gautier A; Juillerat A; Heinis C; Corrêa IR Jr; Kindermann M; Beaufils F; Johnsson K, An engineered protein tag for multiprotein labeling in living cells. Chemistry & biology 2008, 15 (2), 128–136; [DOI] [PubMed] [Google Scholar]; (o) Martin BR; Giepmans BN; Adams SR; Tsien RY, Mammalian cell–based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nature biotechnology 2005, 23 (10), 1308; [DOI] [PubMed] [Google Scholar]; (p) Fernández-Suárez M; Baruah H; Martínez-Hernández L; Xie KT; Baskin JM; Bertozzi CR; Ting AY, Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nature biotechnology 2007, 25 (12), 1483; [DOI] [PMC free article] [PubMed] [Google Scholar]; (q) Chen I; Howarth M; Lin W; Ting AY, Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nature methods 2005, 2 (2), 99; [DOI] [PubMed] [Google Scholar]; (r) Zhang C; Welborn M; Zhu T; Yang NJ; Santos MS; Van Voorhis T; Pentelute BL, π-Clamp-mediated cysteine conjugation. Nature chemistry 2016, 8 (2), 120; [DOI] [PMC free article] [PubMed] [Google Scholar]; (s) McKay CS; Finn MG, Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem Biol 2014, 21 (9), 1075–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a).Chin JW, Expanding and reprogramming the genetic code. Nature 2017, 550 (7674), 53; [DOI] [PubMed] [Google Scholar]; (b) Dumas A; Lercher L; Spicer CD; Davis BG, Designing logical codon reassignment–Expanding the chemistry in biology. Chemical Science 2015, 6 (1), 50–69; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Italia JS; Zheng Y; Kelemen RE; Erickson SB; Addy PS; Chatterjee A, Expanding the genetic code of mammalian cells. Biochemical Society transactions 2017, 45 (2), 555–562; [DOI] [PubMed] [Google Scholar]; (d) Mukai T; Lajoie MJ; Englert M; Söll D, Rewriting the genetic code. Annual review of microbiology 2017, 71, 557–577; [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Young DD; Schultz PG, Playing with the molecules of life. ACS chemical biology 2018, 13 (4), 854–870; [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lang K; Chin JW, Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chemical reviews 2014, 114 (9), 4764–4806; [DOI] [PubMed] [Google Scholar]; (g) Lang K; Chin JW, Bioorthogonal reactions for labeling proteins. ACS chemical biology 2014, 9 (1), 16–20. [DOI] [PubMed] [Google Scholar]
- 3.Vargas-Rodriguez O; Sevostyanova A; Söll D; Crnković A, Upgrading aminoacyl-tRNA synthetases for genetic code expansion. Current opinion in chemical biology 2018, 46, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a).Kim CH; Axup JY; Schultz PG, Protein conjugation with genetically encoded unnatural amino acids. Current opinion in chemical biology 2013, 17 (3), 412–9; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sun SB; Schultz PG; Kim CH, Therapeutic applications of an expanded genetic code. Chembiochem : a European journal of chemical biology 2014, 15 (12), 1721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y; Addy PS; Mukherjee R; Chatterjee A, Defining the current scope and limitations of dual noncanonical amino acid mutagenesis in mammalian cells. Chem Sci 2017, 8 (10), 7211–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a).Chatterjee A; Sun SB; Furman JL; Xiao H; Schultz PG, A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 2013, 52 (10), 1828–37; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Neumann H; Wang K; Davis L; Garcia-Alai M; Chin JW, Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 2010, 464 (7287), 441–4; [DOI] [PubMed] [Google Scholar]; (c) Sachdeva A; Wang K; Elliott T; Chin JW, Concerted, rapid, quantitative, and site-specific dual labeling of proteins. Journal of the American Chemical Society 2014, 136 (22), 7785–8; [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wan W; Huang Y; Wang Z; Russell WK; Pai PJ; Russell DH; Liu WR, A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angewandte Chemie (International ed. in English) 2010, 49 (18), 3211–4; [DOI] [PubMed] [Google Scholar]; (e) Wang K; Sachdeva A; Cox DJ; Wilf NM; Lang K; Wallace S; Mehl RA; Chin JW, Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem 2014, 6 (5), 393–403; [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Zheng Y; Gilgenast MJ; Hauc S; Chatterjee A, Capturing Post-Translational Modification-Triggered Protein–Protein Interactions Using Dual Noncanonical Amino Acid Mutagenesis. ACS chemical biology 2018, 13 (5), 1137–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a).Agard NJ; Prescher JA; Bertozzi CR, A strain-promoted [3+ 2] azide− alkyne cycloaddition for covalent modification of biomolecules in living systems. Journal of the American Chemical Society 2004, 126 (46), 15046–15047; [DOI] [PubMed] [Google Scholar]; (b) Jewett JC; Bertozzi CR, Cu-free click cycloaddition reactions in chemical biology. Chemical Society Reviews 2010, 39 (4), 1272–1279; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Jewett JC; Sletten EM; Bertozzi CR, Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. Journal of the American Chemical Society 2010, 132 (11), 3688–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a).Blackman ML; Royzen M; Fox JM, Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels− Alder reactivity. Journal of the American Chemical Society 2008, 130 (41), 13518–13519; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Devaraj NK; Weissleder R, Biomedical applications of tetrazine cycloadditions. Accounts of chemical research 2011, 44 (9), 816–827; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Kamber DN; Liang Y; Blizzard RJ; Liu F; Mehl RA; Houk K; Prescher JA, 1, 2, 4-triazines are versatile bioorthogonal reagents. Journal of the American Chemical Society 2015, 137 (26), 8388–8391; [DOI] [PubMed] [Google Scholar]; (d) Lang K; Davis L; Wallace S; Mahesh M; Cox DJ; Blackman ML; Fox JM; Chin JW, Genetic Encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions. Journal of the American Chemical Society 2012, 134 (25), 10317–20; [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Seitchik JL; Peeler JC; Taylor MT; Blackman ML; Rhoads TW; Cooley RB; Refakis C; Fox JM; Mehl RA, Genetically encoded tetrazine amino acid directs rapid site-specific in vivo bioorthogonal ligation with trans-cyclooctenes. Journal of the American Chemical Society 2012, 134 (6), 2898–901; [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Taylor MT; Blackman ML; Dmitrenko O; Fox JM, Design and synthesis of highly reactive dienophiles for the tetrazine-trans-cyclooctene ligation. Journal of the American Chemical Society 2011, 133 (25), 9646–9; [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Yang J; Šečkutė J; Cole CM; Devaraj NK, Live cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angewandte Chemie International Edition 2012, 51 (30), 7476–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Italia JS; Addy PS; Wrobel CJ; Crawford LA; Lajoie MJ; Zheng Y; Chatterjee A, An orthogonalized platform for genetic code expansion in both bacteria and eukaryotes. Nature chemical biology 2017, 13 (4), 446–450. [DOI] [PubMed] [Google Scholar]
- 10.(a).Addy PS; Erickson SB; Italia JS; Chatterjee A, A chemoselective rapid azo-coupling reaction (CRACR) for unclickable bioconjugation. Journal of the American Chemical Society 2017, 139 (34), 11670–11673; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Addy PS; Italia JS; Chatterjee A, An Oxidative Bioconjugation Strategy Targeted to a Genetically Encoded 5 hydroxytryptophan. Chembiochem : a European journal of chemical biology 2018, 19 (13), 1375–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a).Chin JW; Santoro SW; Martin AB; King DS; Wang L; Schultz PG, Addition of p-Azido-l-phenylalanine to the Genetic Code of Escherichia c oli. Journal of the American Chemical Society 2002, 124 (31), 9026–9027; [DOI] [PubMed] [Google Scholar]; (b) Young DD; Young TS; Jahnz M; Ahmad I; Spraggon G; Schultz PG, An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 2011, 50 (11), 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott TS; Townsley FM; Bianco A; Ernst RJ; Sachdeva A; Elsässer SJ; Davis L; Lang K; Pisa R; Greiss S, Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nature biotechnology 2014, 32 (5), 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y; Mukherjee R; Chin MA; Igo P; Gilgenast MJ; Chatterjee A, Expanding the scope of single and dual noncanonical amino acid mutagenesis in mammalian cells using orthogonal polyspecific leucyl-tRNA synthetases. Biochemistry 2018, 57 (4), 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao H; Chatterjee A; Choi SH; Bajjuri KM; Sinha SC; Schultz PG, Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angewandte Chemie (International ed. in English) 2013, 52 (52), 14080–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson R; Hoben P; Sumner-Smith M; Uemura H; Watson L; Soll D, Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science 1988, 242 (4885), 1548–1551. [DOI] [PubMed] [Google Scholar]
- 16.(a).Beck A; Goetsch L; Dumontet C; Corvaïa N, Strategies and challenges for the next generation of antibody-drug conjugates. Nature Reviews Drug Discovery 2017, 16 (5), 315–337; [DOI] [PubMed] [Google Scholar]; (b) Sievers EL; Senter PD, Antibody-drug conjugates in cancer therapy. Annual review of medicine 2013, 64, 15–29. [DOI] [PubMed] [Google Scholar]
- 17.(a).Hudis CA, Trastuzumab—mechanism of action and use in clinical practice. New England Journal of Medicine 2007, 357 (1), 39–51; [DOI] [PubMed] [Google Scholar]; (b) Hutchins BM; Kazane SA; Staflin K; Forsyth JS; Felding-Habermann B; Schultz PG; Smider VV, Site-specific coupling and sterically controlled formation of multimeric antibody fab fragments with unnatural amino acids. Journal of molecular biology 2011, 406 (4), 595–603; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hutchins BM; Kazane SA; Staflin K; Forsyth JS; Felding-Habermann B; Smider VV; Schultz PG, Selective formation of covalent protein heterodimers with an unnatural amino acid. Chem Biol 2011, 18 (3), 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y; Lajoie MJ; Italia JS; Chin MA; Church GM; Chatterjee A, Performance of optimized noncanonical amino acid mutagenesis systems in the absence of release factor 1. Molecular BioSystems 2016, 12 (6), 1746–1749. [DOI] [PubMed] [Google Scholar]
- 19.Kapust RB; Waugh DS, Controlled intracellular processing of fusion proteins by TEV protease. Protein expression and purification 2000, 19 (2), 312–318. [DOI] [PubMed] [Google Scholar]
- 20.(a).Ostrov N; Landon M; Guell M; Kuznetsov G; Teramoto J; Cervantes N; Zhou M; Singh K; Napolitano MG; Moosburner M, Design, synthesis, and testing toward a 57-codon genome. Science 2016, 353 (6301), 819–822; [DOI] [PubMed] [Google Scholar]; (b) Wang K; Fredens J; Brunner SF; Kim SH; Chia T; Chin JW, Defining synonymous codon compression schemes by genome recoding. Nature 2016, 539 (7627), 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a).Dien VT; Morris SE; Karadeema RJ; Romesberg FE, Expansion of the genetic code via expansion of the genetic alphabet. Current opinion in chemical biology 2018, 46, 196–202; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang Y; Ptacin JL; Fischer EC; Aerni HR; Caffaro CE; San Jose K; Feldman AW; Turner CR; Romesberg FE, A semi-synthetic organism that stores and retrieves increased genetic information. Nature 2017, 551 (7682), 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.