Abstract
Background
One of the most controversial problems for liver transplantation in patients affected by hepatocellular carcinoma (HCC) remains the lack of an oncologic staging system to predict cancer recurrence after liver transplantation (LT). We analyzed allelic imbalance (AI) in 19 microsatellites, and assessed the post-LT HCC recurrence risk.
Material/Methods
Seventy-one patients were included; 18 had tumor recurrence within 5 years post-transplant. Molecular analysis was done in the primary HCC and peripheral blood samples: a total of 19 microsatellites was used to assess AI. Specific AI was evaluated when outside of range value between 0.66 and 1.5. Based on data in the literature, we grouped the 19 microsatellites into 4 panels. We calculated the fractional allelic imbalance (FAI) to make comparisons between different panels including different subsets of microsatellites.
Results
We report that AI was associated with HCC recurrence in 3 main loci (D3S2303, D9S251, and D9S254). Tumor recurrence was associated only with 2 specific panels with 9 microsatellites previously reported to be associated with high risk for HCC recurrence. Our data show that fractional allelic imbalance (FAI) index has good negative ability to predict HCC recurrence (Panel 2: negative predictive value of 95%).
Conclusions
AI analysis could have prognostic value in risk management of HCC recurrence after LT, especially for early recurrence.
MeSH Keywords: Allelic Imbalance; Carcinoma, Hepatocellular; Liver Transplantation; Treatment Outcome
Background
Over the last 20 years hepatocellular carcinoma (HCC) has become one of the most common indications for liver transplantation (LT). Data from the European Liver Transplant Registry from 1988 to 2015 show that cancer represents 16.5% of overall causes of LT, and 87.3% are HCC-related. Survival analysis demonstrates that long-term survival (5 or more years after LT) of this specific subset of patients is lower than that of recipients with other diseases [1], as also reported by the Organ Procurement and Transplantation Network in the USA [2].
In a seminal paper published in 1997, Mazzaferro et al. established what came to be called the Milan criteria, an attempt to standardize criteria for transplantation of patients with HCC. They reported that patients with either 1 tumor with a diameter ≤5 cm, or patients with 2/3 tumors each with a diameter ≤3 cm, had a recurrence-free survival (RFS) rate at 5 years of 75% and 83%, respectively [3]. The Milan criteria have been validated by several groups, and were adopted by the United Network for Organ Sharing UNOS staging system for allocating organs, although the criteria have also been criticized for being too restrictive and for limiting the prognostic information to only radiologic appearance, without considering other clinical, molecular, or pathologic data [4]. Other research groups have tried to extend the Milan criteria, some based on dimensional features [5–8], while others have considered microvascular invasion [9,10] or tumor grade [11], or evaluating biochemical parameters [12–14]. Mazzaferro et al. tried to simplify data, first with the Metroticket Paradigm [15], and then by proposing the Up-to-7 Criteria [16].
Nonetheless, the debate over the criteria for transplantation of HCC patients is still far from over. Some recent studies have defined loss of heterozygosity (LOH) or allelic imbalance (AI) according to the level of gene damage and genomic instability, and explored whether a panel of tumor-suppressor gene markers could be useful in predicting recurrence with an index of cumulative mutational damage (fractional allelic imbalance, FAI) [17–19].
Other studies have reported that panels including specific microsatellites were significantly correlated with the post-transplant recurrence-free survival (RFS), but none achieved perfect discrimination alone [20,21], and proposed a rational approach using FAI in conjunction with models based on clinical data to expand the conventional LT selection policies [22].
Recently, Schmidt, and Marsh, at UPMC, proposed integrating data from FAI and macrovascular invasion, developing a novel staging system predicting HCC recurrence after LT (Pittsburgh staging system) [23].
Our study aimed to analyze the microsatellite LOHs to assess the risk of tumor recurrence after LT in recipients with HCC, in order to help move the field away from morphometric criteria to ones that are based on molecular information on tumor biology.
Material and Methods
Patients
The study population included 71 orthotopic LT recipients affected by HCC at IRCCS – ISMETT, in Palermo, Italy between 2006 and 2012. Severity of chronic liver disease was based on the Model for End-stage Liver Disease score (MELD). Histopathological findings were used to determine the tumor stage, classified by the tumor-node-metastasis (TNM) staging system. To monitor hepatic recurrence or distant metastasis, all patients were routinely followed up for at least 3 years with blood tests, magnetic resonance imaging (MRI), and/or computerized tomography (CT) scans. ISMETT’s Institutional Research Review Board and local Ethics Committee approved the research protocol (approval number IRRB/26/11, signed by Mariolina Crisci, 5th December 2016).
Microsatellite analysis
Genomic DNA was extracted from whole blood, formalin-fixed paraffin-embedded (FFPE), amplified using Type-it Microsatellite PCR kit (QIAgen), and separated by capillary electrophoresis on the 3500 Genetic Analyzer (ThermoFisher). A total of 19 microsatellites were used to assess AI, which included 9 loci previously reported as significantly associated with a high tumor recurrence risk after LT [18, 24], 6 loci that may be associated with recurrence but were excluded from the UPMC group [23], and an additional 4 loci suggested as indicative of recurrence [23].
Identification of microsatellites panels
Based on data in the literature, we grouped the analyzed microsatellites into 4 different panels (Table 1) to assess the association between FAI and the presence of tumor recurrence. Panel 1 (9 loci) included the 9 microsatellites previously reported as associated with a high risk for HCC recurrence [24]. Panel 2 (15 loci) included an additional 6 microsatellites that may be associated with HCC recurrence [20]. Panel 3 (13 loci) included, in addition to the previous 9 loci, another 4 microsatellites suggested to be indicative of HCC recurrence (personal communication by Dr. Marsh, from UPMC). Panel 4 (19 loci) included all selected microsatellites (Table 1).
Table 1.
Microsatellites and panels.
| Gene | Locus | Microsatellite | Panel 1 | Panel 2 | Panel 3 | Panel 4 |
|---|---|---|---|---|---|---|
| L-myc | 1p33 | D1S162 | x | x | ||
| L-myc | 1p34 | MYCL.5NT | x | x | x | x |
| L-myc | 1p35 | D1S1161 | x | x | ||
| CMM | 1p36 | D1S407 | x | x | x | x |
| OGG1 | 3p24 | D3S2303 | x | x | ||
| VHL | 3p26 | D3S1539 | x | x | x | x |
| APC | 5q21 | D5S615 | x | x | x | x |
| MCC | 5p21 | D5S592 | x | x | ||
| PTCH | 9q22 | D9S252 | x | x | ||
| CDKN2A/p16 | 9p21 | D9S251 | x | x | x | x |
| CDKN2A/p16 | 9p21 | D9S254 | x | x | ||
| PTEN | 10q23 | D10S520 | x | x | ||
| PTEN | 10q23 | D101173 | x | x | ||
| TP53 | 17p13 | D17S1289 | x | x | x | x |
| TP53 | 17p13 | D17S974 | x | x | x | x |
| TP53 | 17p13 | TP53 L1 | x | x | x | x |
| TP53 | 17p23 | D17S786 | x | x | ||
| TP53 | 17p23 | D17S516 | x | x | ||
| DCC/SMAD4 | 18q21 | D18S814 | x | x | x | x |
Allelic imbalance analysis
An AI analysis investigated and compared healthy tissue (peripheral blood samples) and diseased tissue in the primary HCC. Microsatellite markers were designated as informative/heterozygous (2 different alleles/peaks) or non-informative/homozygous (1 allele/peak excluded for frequency calculation of AI). For any informative microsatellite, signal intensity in tumor DNA was compared with those of the corresponding normal DNA; the allele ratio (AR) between 2 allele peaks for each marker for each sample (peak height of allele 1/peak height of allele 2) was calculated, and AI was subsequently obtained as the ratio between the AR of the healthy sample and the AR of the diseased sample. Alleles were considered to be in balance when AI values were between 0.66 and 1.50 [21]. Two different grades of LOH were considered: 1) presence of LOH when AI values were either 0.50–0.65 or 1.51–2.00; 2) high-level LOH when AI values were either below 0.50 or greater than 2.00, respectively [18–29].
To make comparisons between different panels including different subsets of microsatellites, we calculated the FAI as the proportion of informative (heterozygous) microsatellites that exhibit LOH over the total number of informative loci.
Statistical analysis
Continuous variables are expressed as mean and inter-quartile range (IQR), while categorical variables are expressed as frequency and percentage. Differences in the rates of HCC recurrence were tested by Fisher’s exact test or Pearson’s chi-square test without Yates correction, as appropriate; odds ratio (OR) was also used to assess the relationships. Predictive performances of different microsatellite panels were evaluated in terms of negative predictive value (NPV), positive predictive value (PPV), sensitivity, and specificity; the best discriminating cut-off value (Youden’s index) was established for the FAI. GraphPad Prism 6, MedCal, and EpiTools were used for all statistical analyses.
Results
Clinical data and HCC recurrence
Of the 71 patients, 59 (83%) were male, and HCV infection was the main cause of liver disease. TNM classification of HCC was available in 52 patients. Forty-eight patients had multiple tumors involving 1 or both liver lobes, and 29 had microvascular invasion. Forty-four patients were classified as beyond the Milan criteria after histological examination of the explanted liver. Clinicopathological data are summarized in Table 2.
Table 2.
Clinical and pathological data: Association between clinical data and HCC recurrence. Fisher’s exact test and Pearson’s chi-square (chi-square without Yates correction) were used, as appropriate.
| Clinical and pathological data | Overall | No HCC Recurrence | HCC Recurrence | p-Value |
|---|---|---|---|---|
| n | 71 | 53 | 18 | |
| Male Gender, no. (%) | 59 (83.1) | 44 (83.0) | 15 (83.3) | 1.000 |
| Age, median [IQR] | 59.00 [53.5, 63.0] | 59.0 [52.0, 63.0] | 59.0 [56.0, 61.8] | 0.706 |
| HCC etiology, no (%) | 0.216 | |||
| HBV | 14 (19.7) | 13 (24.5) | 1 (5.6) | |
| HBV+HCV | 1 (1.4) | 1 (1.9) | 0 (0.0) | |
| HCV | 49 (69.0) | 35 (66.0) | 14 (77.8) | |
| Other | 7 (9.9) | 4 (7.5) | 3 (16.7) | |
| MELD, median [IQR] | 12.00 [9.0, 15.0] | 12.00 [9.0, 15.0] | 11.5 [9.0, 14.8] | 0.801 |
| HCC nodules, no. (%) | 0.128 | |||
| 1 | 23 (32.4) | 19 (35.8) | 4 (22.2) | |
| 2 | 19 (26.8) | 16 (30.2) | 3 (16.7) | |
| 3+ | 29 (40.8) | 18 (34.0) | 11 (61.1) | |
| Largest nodule size (cm), median [IQR] | 2.50 [2.0, 3.5] | 2.5 [1.7, 3.4] | 2.7 [2.2, 3.8] | 0.331 |
| Histologic grade G2–G3, no. (%) | 31 (43.7) | 23 (43.4) | 8 (44.4) | 0.938 |
| Vascular invasion, no. (%) | 29 (40.8) | 20 (37.7) | 9 (50.0) | 0.360 |
| TNM (%) | 0.047 | |||
| T1 | 10 (14.1) | 10 (18.9) | 0 (0.0) | |
| T2–T3 | 42 (59.2) | 32 (60.4) | 10 (55.6) | |
| NA | 19 (26.8) | 11 (20.8) | 8 (44.4) | |
| Milan = Out (%) | 44 (62.0) | 36 (67.9) | 8 (44.4) | 0.076 |
During the median follow-up period of 5.26 years, 18 patients (25%) developed intrahepatic and/or extrahepatic HCC recurrence, with most of the lesions already detectable within the first 2 years. Sites of tumor recurrence were liver (2), lung (5), abdominal cavity (4), bone (2), and multiple sites, in addition to the liver (5) [25–27]. No significant difference was found in clinical parameters, except for TNM Stage (Table 2).
Association between AI and HCC recurrence
Based on data in the literature [18,20,24], we decided to analyze 19 loci situated within or adjacent to specific genes of interest. We found the presence of AI in at least 1 locus in 90% of patients with HCC recurrence compared with 74.5% of patients without recurrence. Significant association was found for D3S2303 (p=0.048) considering the presence of LOH (Table 3A), and D1S407 (p=0.006) D9S251 (p=0.02), D1S162 (p=0.005), D5S592 (p=0.005), D9S254 (p=0.002) and D10S520 (p=0.04) considering high-level LOH (Table 3B).
Table 3.
Univariate Cox models for microsatellites association with hepatocellular carcinoma recurrence. (A) Patients with loss of heterozygosis (LOH). (B) Patients with high-level loss of heterozygosis.
| (A) | exp(coef) [confint] | p | Code |
|---|---|---|---|
| D1S407.LOH1 | 0.74 [0.10, 5.68] | 0.7740 | |
| MYCL1.LOH1 | 1.92 [0.55, 6.70] | 0.3036 | |
| D3S1539.LOH1 | 2.06 [0.81, 5.23] | 0.1273 | |
| D5S615.LOH1 | 2.07 [0.77, 5.57] | 0.1493 | |
| D9S251.LOH1 | 2.55 [0.91, 7.19] | 0.0763 | . |
| D17S1289.LOH1 | 0.88 [0.25, 3.17] | 0.8492 | |
| D17S974.LOH1 | 0.62 [0.08, 4.72] | 0.6471 | |
| TP53.LOH1 | 3.11 [0.34, 28.23] | 0.3136 | |
| D18S814.LOH1 | 1.46 [0.50, 4.27] | 0.4916 | |
| D1S162.LOH1 | 0.86 [0.25, 2.94] | 0.8066 | |
| D1S1161.LOH1 | 1.00 [0.33, 3.07] | 0.9991 | |
| D17S516.LOH1 | 0.00 [0.00, Inf] | 0.9988 | |
| D17S786.LOH1 | 3.40 [0.44, 26.20] | 0.2407 | |
| D3S2303.LOH1 | 6.45 [1.77, 23.55] | 0.0048 | ** |
| D5S592.LOH1 | 3.33 [0.43, 25.79] | 0.2485 | |
| D9S254.LOH1 | 1.77 [0.55, 5.77] | 0.3398 | |
| D9S252.LOH1 | 1.28 [0.29, 5.73] | 0.7456 | |
| D10S1173.LOH1 | 2.52 [0.80, 7.98] | 0.1157 | |
| D10S520.LOH1 | 2.14 [0.61, 7.52] | 0.2355 | |
| D1S407.HighLOH1 | 49.50 [3.10, 791.37] | 0.0058 | ** |
| D3S1539.HighLOH1 | 1.42 [0.47, 4.33] | 0.5329 | |
| D5S615.HighLOH1 | 2.06 [0.75, 5.67] | 0.1630 | |
| D9S251.HighLOH1 | 4.58 [1.27, 16.45] | 0.0198 | * |
| D17S1289.HighLOH1 | 4.26 [0.93, 19.43] | 0.0612 | . |
| D17S974.HighLOH1 | 1.31 [0.17, 9.96] | 0.7916 | |
| TP53.HighLOH1 | 10.49 [0.95, 115.73] | 0.0550 | . |
| D18S814.HighLOH1 | 2.67 [0.75, 9.51] | 0.1286 | |
| D1S162.HighLOH1 | 31.50 [2.86, 347.37] | 0.0049 | ** |
| D1S1161.HighLOH1 | 2.36 [0.68, 8.23] | 0.1781 | |
| D17S786.HighLOH1 | 8159967418361.53 [0.00, Inf] | 0.9999 | |
| D5S592.HighLOH1 | 53.50 [3.35, 855.32] | 0.0049 | ** |
| D9S254.HighLOH1 | 8.61 [2.24, 33.07] | 0.0017 | ** |
| D10S1173.HighLOH1 | 3.83 [0.50, 29.31] | 0.1956 | |
| D10S520.HighLOH1 | 3.78 [1.07, 13.37] | 0.0387 | * |
Evaluation of specific panels and association with HCC recurrence
Descriptive analysis of the different panels showed a relevant grade of informativeness in Panel 2 (Table 4). We made a diagnostic test evaluation of each panel to assess the association between AI and the risk of HCC recurrence. No association was found between LOH and HCC recurrence, whereas a significant association was found in all panels considering high-level LOH, and the Panel 2 performance was better than the others (Table 5). The high significance of Panel 2 is further strengthened and supported by FAI analysis, which proves that a high proportion of informative microsatellites exhibit LOH over the total number of informative loci in this panel (Table 5).
Table 4.
Descriptive analysis of microsatellites panels fractional allelic imbalance (FAI) and loss of heterozygosity (LOH).
| Panel 1 | Panel 2 | Panel 3 | Panel 4 | |
|---|---|---|---|---|
| n | 71 | 71 | 71 | 71 |
|
| ||||
| Informativeness | ||||
| Median | 77.8 | 93.3 | 76.9 | 73.7 |
| IQR | 66.7–77.8 | 86.7–100.0 | 61.5–76.9 | 68.4–78.9 |
|
| ||||
| Presence of LOH, n (%) | 48 (67.6) | 51 (71.8) | 53 (74.6) | 56 (78.9) |
|
| ||||
| FAI | ||||
| Mean | 0.19 | 0.13 | 0.18 | 0.16 |
| Std. dev. | 0.18 | 0.12 | 0.15 | 0.14 |
| Median | 0.17 | 0.12 | 0.17 | 0.13 |
| IQR | 0.00–0.31 | 0–0.18 | 0.04–0.25 | 0.07–0.21 |
| Range | 0.00–0.62 | 0–0.53 | 0–0.58 | 0–0.67 |
|
| ||||
| Presence of high-level LOH, no. (%) | 31 (43.7) | 34 (47.9) | 31 (43.7) | 34 (47.9) |
|
| ||||
| High-level of FAI | ||||
| Mean | 0.09 | 0.05 | 0.07 | 0.06 |
| Std. dev. | 0.12 | 0.07 | 0.10 | 0.08 |
| Median | 0.00 | 0.00 | 0.00 | 0.00 |
| IQR | 0–0.14 | 0–0.07 | 0–0.11 | 0–0.08 |
| Range | 0–0.5 | 0–0.30 | 0–0.42 | 0–0.40 |
Table 5.
Univariate Cox models to assess the association between AI and the risk of HCC recurrence.
| At least one microsatellite | ||||
|---|---|---|---|---|
| exp(coef) [confint] | p | Code | Concordance | |
| Panel 1 | 2.95 [0.86, 10.09] | 0.0841 | . | 0.5961 |
| Panel 2 | 4.06 [0.94, 17.52] | 0.0602 | . | 0.6092 |
| Panel 3 | 2.01 [0.59, 6.86] | 0.2663 | 0.5552 | |
| Panel 4 | 2.69 [0.62, 11.59] | 0.1848 | 0.5683 | |
| At least one microsatellite (high-level) | ||||
| exp(coef) [confint] | p | Code | Concordance | |
| Panel 1 | 2.79 [1.11, 7.01] | 0.0286 | * | 0.6282 |
| Panel 2 | 4.12 [1.49, 11.35] | 0.0062 | ** | 0.672 |
| Panel 3 | 2.79 [1.11, 7.01] | 0.0286 | * | 0.6282 |
| Panel 4 | 4.12 [1.49, 11.35] | 0.0062 | ** | 0.672 |
| Fractional allelic imbalance | ||||
| exp(coef) [confint] | p | Code | Concordance | |
| Panel 1 FAI | 11.64 [1.11, 122.15] | 0.0407 | * | 0.6379 |
| Panel 2 FAI | 127.58 [4.60, 3535.44] | 0.0042 | ** | 0.6737 |
| Panel 3 FAI | 10.04 [0.70, 143.33] | 0.0891 | . | 0.6172 |
| Panel 4 FAI | 31.82 [2.08, 487.06] | 0.0129 | * | 0.6627 |
| Fractional allelic imbalance (high-level) | ||||
| exp(coef) [confint] | p | Code | Concordance | |
| Panel 1 FAI | 148.58 [9.39, 2351.75] | 0.0004 | *** | 0.6825 |
| Panel 2 FAI | 12736.03 [192.99, 840485.78] | <0.0001 | *** | 0.7255 |
| Panel 3 FAI | 1784.22 [40.74, 78135.33] | 0.0001 | *** | 0.6914 |
| Panel 4 FAI | 16309.40 [298.76, 890346.50] | <0.0001 | *** | 0.7395 |
Association of AI and time to HCC recurrence
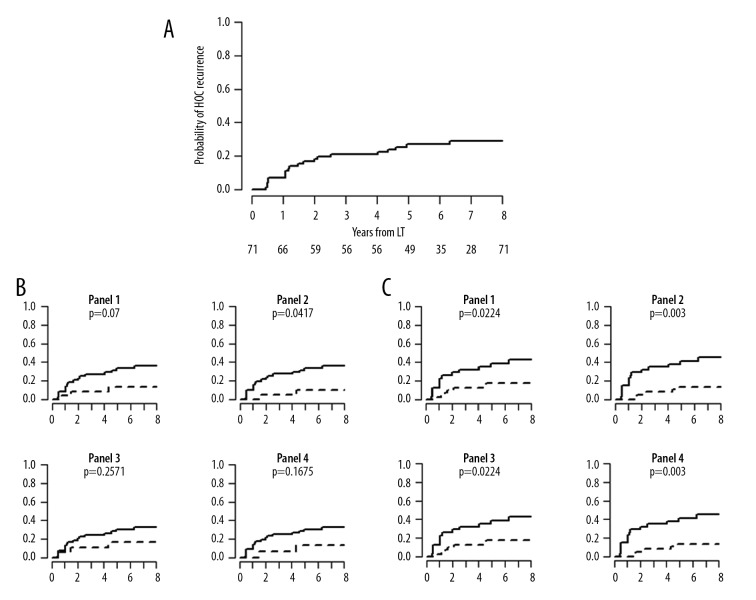
After proving the possible association between AI and HCC recurrence, we focused our attention on time to recurrence. Kaplan-Meier curves were performed, proving that LOH, as expected, has no significant correlation with time to recurrence, whereas high-level LOH is strongly correlated with particularly high performance (Figure 1).
Figure 1.
Kaplan-Meier time to recurrence curves. (A) Overall study population. (B) Patients with loss of heterozygosis. (C) Patients with high-level loss of heterozygosis.
Association of frequency of AI with early HCC recurrence
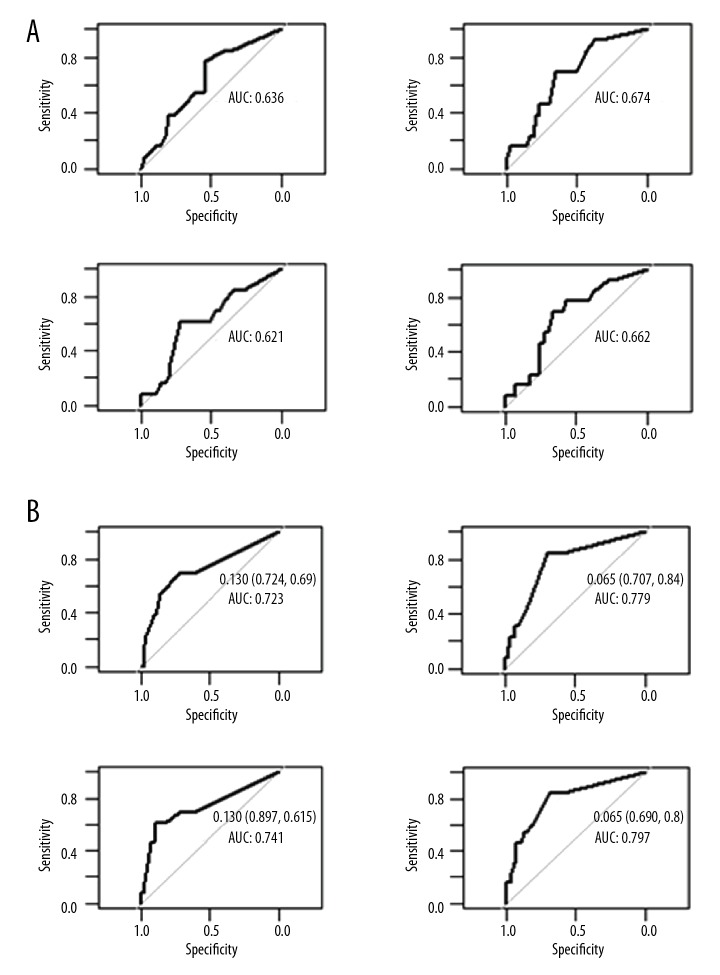
To further extend our analysis, we assessed whether AI could have a prognostic value in early HCC recurrence detection. Thus, we focused our attention on the 14 patients (11 of whom were classified beyond the Milan criteria after pathology examination) with recurrence within 2 years of LT. We found that the FAI index for high-level LOH has good predictive negative performance. In particular, Panel 2 reaches a negative predictive value (NPV) of 95% due to its high sensitivity, with a loss of specificity and positive predictive value (PPV) lower than 35% (Table 6). This was confirmed by receiver operating characteristic (ROC) curve analysis (Figure 2).
Table 6.
Predictive performances of early recurrence panels.
| Loss of heterozygosis | |||||
|---|---|---|---|---|---|
| Se | Sp | PPV | NPV | Acc | |
| Panel 1 FAI | 0.8462 | 0.3621 | 0.2292 | 0.913 | 0.4507 |
| Panel 2 FAI | 0.9231 | 0.3276 | 0.2353 | 0.95 | 0.4366 |
| Panel 3 FAI | 0.8462 | 0.2759 | 0.2075 | 0.8889 | 0.3803 |
| Panel 4 FAI | 0.9231 | 0.2414 | 0.2143 | 0.9333 | 0.3662 |
| High-level loss of heterozygosis | |||||
| Se | Sp | PPV | NPV | Acc | |
| Panel 1 FAI | 0.6923 | 0.6207 | 0.2903 | 0.9 | 0.6338 |
| Panel 2 FAI | 0.8462 | 0.6034 | 0.3235 | 0.9459 | 0.6479 |
| Panel 3 FAI | 0.6923 | 0.6207 | 0.2903 | 0.9 | 0.6338 |
| Panel 4 FAI | 0.8462 | 0.6034 | 0.3235 | 0.9459 | 0.6479 |
Figure 2.
ROC curve for prediction of early recurrence. (A) Patients with loss of heterozygosis. (B) Patients with high-level loss of heterozygosis.
Association of AI with other predictors of HCC recurrence
Finally, we focused our attention on any possible correlation between AI and other clinical or pathological parameters available in our dataset that could be associated with HCC recurrence [5–16]. We evaluated the association between AI and 3 different poor-prognosis HCC indicators: G2/G3 grading, vascular invasion, and Milan criteria. No single microsatellite showed any association with these 3 variables, neither considering presence of LOH nor considering high-level LOH.
As previously reported, 31 patients were graded as having G2/G3 HCC; of these, 18 were graded G2, only 1 was graded G3, and the remaining 12 were labelled G2–G3 by the pathologist. Importantly, only 5 of the 13 highest graded (G3+G2–G3) were inside the Milan criteria, and only 1 of these 5 had an early recurrence. Only Panel 2 and Panel 4 succeeded in detecting this recurrence, considering both presence of LOH (Sp=0.5, Se=1, PPV=0.3, NPV=1) and high-level LOH (Sp=0.75, Se=1, PPV=0.5, NPV=1), thus confirming in this small subset the performance observed in the overall study population.
Discussion
HCC recurrence after LT is one of the most important causes of mortality and morbidity [28]. At present, there are conflicting recommendations about what tumor characteristics are responsible for HCC recurrence, and the clinical and radiological criteria (e.g., Milan criteria, Up-to-7 criteria) seem to be inadequate because they provide characteristics of the tumor that do not always correspond to the actual pathological characteristics [16]. Several studies have identified microvascular invasion as the strongest independent predictor factor of recurrence [29,30], associating it with other poor prognostic factors that, except for AFP, can only be known on histological analysis of the explanted liver, or need procedures that are invasive, not easily available, and lack adequate accuracy (e.g., liver biopsy) [31]. Scores based exclusively on radiological or biochemical parameters are not adequate to establish the risk of HCC recurrence, and a number of authors have attempted to combine the 2 methods. Mazzaferro et al. redesigned the Metroticket Paradigm [15], in which patients are assessed on the basis of the combination of AFP levels, and number and size of nodules [32]. A Belgian research group developed a model to predict the likelihood of recurrence based on 4 parameters: Time, Radiological-response, AFP, and INflammation (TRAIN) [33]. Similarly, the MoRAL (Model of Recurrence After Liver transplantation) score combines the biochemical features of AFP and neutrophil-to-lymphocyte ratio with the radiological feature of tumor dimension (tumor size ≥3 cm) [34]. In the present retrospective study, we evaluated the relationship between prognostic clinicopathological factors and HCC recurrence, but no statistically significant association was found except for TNM stage (Table 2; p=0.047).
To date, however, it has not been possible to identify sufficiently reliable markers of the biological behavior of HCC. Therefore, the attention of researchers is increasingly shifting from the analysis of clinical and radiological characteristics to the analysis of genetic mutations typical of this neoplasm. In this context, several studies in the last few years have mentioned AI (LOH) as a possible predictor of HCC recurrence [18,20,21,24]. Marsh et al. developed a multivariate model (artificial neural network – ANN) based on 5 risk factors (sex, tumor number, tumor size, lobar tumor distribution, and grade of vascular invasion) that could predict the risk of tumor recurrence [35]. This model, in association with the study of AI based on tissue microdissection genotyping in 9 microsatellites at 6 genomic loci [18,20], had an accuracy of 100%, with a discriminatory power of 85% in the predicted 3-year recurrence outcome [36]. In addition, a recent meta-analysis of 41 eligible studies noted the importance of identifying serological (pre-LT AFP and pre-LT DCP) and molecular biomarkers able to predict HCC recurrence [37]. In our study, we found a significant association between AI in specific microsatellite loci and the risk of HCC recurrence, validating for the first time in a European population the results of several other studies [18,20,22,24,35]. A statistically significant association with tumor recurrence was found for D3S2303 (gene OGG1; p=0.048) considering the presence of LOH (Table 3A), and D1S407 (gene CMM; p=0.006) D9S251 (gene CDKN2A/p16; p=0.02), D1S162 (gene L-myc; p=0.005), D5S592 (gene MCC; p=0.005), D9S254 (gene CDKN2A/p16; p=0.002), and D10S520 (gene PTEN; p=0.04) considering high-level LOH. Thus, we confirmed that the LOH frequency on 9p21 locus (D9S251 and D9S254, CDKN2A gene) is associated with recurrence [21].
Many retrospective studies have used different and arbitrary cumulative mutational damage indexes (e.g., FAI) to investigate the relationship between AI and HCC recurrence, and identified specific discriminatory values between true positives and false positives using ROC curve analysis constructed using results obtained in each study [18,20–23]. Our data show that the FAI index for high-level LOH in a specific panel of microsatellites (Panel 2) has a good negative predictive performance, with a 95% NPV, to identify early-HCC recurrence. Unfortunately, the PPV for each analyzed panel cannot be considered satisfactory. Thus, our analysis confirms the existence of an association between HCC recurrence and LOH in specific loci (specifically high-level LOH), proving at the same time that AI should not be considered as a positive predictor of tumor recurrence, but its absence can be considered as a negative one.
Of note, in contrast to previously research, our study did not analyze the AI between neoplasia and healthy fragments of liver parenchyma, but obtained similar results by comparing very limited quantities of neoplastic tissue with whole blood, thus reducing the potential invasiveness of the analysis. In fact, previous studies have indicated that the use of minimally invasive laparoscopic techniques allows the successful resection of HCC nodules without increasing the risk of hepatic failure or peri-operative complications [38,39]. Thus, such surgical approaches could be proposed not only as a bridge-therapy for LT, but also as a useful diagnostic tool to evaluate AI between neoplastic tissue and whole blood [40]. The scenarios that would lead to a similar combined use of minimally invasive surgery and AI to predict patients at greater risk of recurrence could change the priorities in organ allocation procedures and would ensure tailored post-transplant therapies (e.g., changes of immunosuppression regimens).
Our study has several limitations. This was an analysis with a limited number of patients, and we could not evaluate the effect of pre-transplantation treatments on survival and recurrence (e.g., radiofrequency thermal ablation and trans-arterial chemoembolization). Unfortunately, there was no donor and recipient allelotyping, and we could not distinguish whether the tumor recurrence represents metastasis of the first cancer or a donor-derived de novo cancer, as proposed by recent studies [25–27]. Moreover, HCC genotyping can be complicated by interpretative difficulties due to heterogeneity of tumor tissues from a pathological specimen containing various subpopulations of cells. Unfortunately, we could not perform microdissection, and DNA from nonmalignant cells could have contaminated the DNA extracted from these samples. Finally, working with FFPE tissues of the native liver, and DNA extraction and amplification might be affected by tissue fixation time and progressive DNA degradation in archival tissue blocks.
In the post-LT setting, FAI could provide useful information to adapt and personalize therapies (e.g., immunosuppression protocols), thus reducing the risk of recurrence. However, the application of molecular markers needs further evaluation before being used as selection criteria for LT. We intend to validate the HCC recurrence prediction model based on the use of specific microsatellites (especially D9S251) in a future study in order to verify its concrete predictive ability and to identify patients at lower risk of HCC recurrence, combining clinicopathological, radiological, and genomic data, easily available in the pre-LT setting. Moreover, using a larger number of patients with more complete clinical information, we intend to study the association of AI in each locus with clinicopathological characteristics and to analyze its biological role in HCC development, progression, and aggressiveness.
Conclusions
One of the important ethical implications of LT for HCC, which is the subject of lively debate in the transplantation community and beyond, is that ability to predict which recipients will successfully complete the post-transplant course. Transparency and clinical clarity are 2 of the cornerstones in sharing experiences with the medical community at large. We observed that the FAI for a high-level LOH has a good NPV for tumor recurrence within 2 years after transplant (95%). These data confirm a relevant role of the CDKN2A gene for HCC recurrence. Our data suggest that the information obtained by AI analysis can be useful, and has a prognostic application in risk management of HCC recurrence in patients who have undergone LT, especially in early tumor recurrence. One of the important ethical implications of LT for HCC, which is the subject of lively debate in the transplantation community and beyond, is the ability to predict which recipients will successfully complete the post-transplant course. Developing new protocols for immunosuppressive regimen and surgical decision-making is paramount for continued success in this delicate field of medicine.
Acknowledgments
The authors would like to thank Warren Blumberg, of ISMETT’s Language Services Department, for help in editing this paper.
Abbreviations
- AI
allelic imbalance
- AR
allelic ratio
- AUC
area under the ROC curve
- CT
computerized tomography
- FAI
fractional allelic imbalance
- FFPE
formalin-fixed paraffin-embedded
- HCC
hepatocellular carcinoma
- LOH
loss of heterozygosis
- LT
liver transplantation
- MELD
model for end-stage liver disease
- MRI
magnetic resonance imaging
- NPV
negative predictive value
- OR
odds ratio
- PPV
positive predictive value
- RFS
recurrence-free survival
- ROC
receiver operative characteristic
- TNM
tumor-node-metastasis
Footnotes
Source of support: Departmental sources
References
- 1.European Liver Transplant Registry. Available: http://www.eltr.org/Overall-indication-and-results.html.
- 2.Organ Procurement and Transplantation Network. Available: https://optn.transplant.hrsa.gov/data/view-data-reports/national-data/
- 3.Mazzaferro V, Regalia E, Doci R, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693–99. doi: 10.1056/NEJM199603143341104. [DOI] [PubMed] [Google Scholar]
- 4.Marsh JW, Schmidt C. The Milan criteria: no room on the metro for the king? Liver Transpl. 2010;16:252–55. doi: 10.1002/lt.22037. [DOI] [PubMed] [Google Scholar]
- 5.Yao FY, Ferrell L, Bass NM, et al. Liver transplantation for hepatocellular carcinoma: Expansion of the tumor size limits does not adversely impact survival. Hepatology. 2001;33:1394–403. doi: 10.1053/jhep.2001.24563. [DOI] [PubMed] [Google Scholar]
- 6.Herrero JI, Sangro B, Quiroga J, et al. Influence of tumor characteristics on the outcome of liver transplantation among patients with liver cirrhosis and hepatocellular carcinoma. Liver Transpl. 2001;7:631–36. doi: 10.1053/jlts.2001.25458. [DOI] [PubMed] [Google Scholar]
- 7.Merli M, Nicolini G, Gentili F, et al. Predictive factors of outcome after liver transplantation in patients with cirrhosis and hepatocellular carcinoma. Transplant Proc. 2005;37:2535–40. doi: 10.1016/j.transproceed.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 8.Ito T, Takada Y, Ueda M, et al. Expansion of selection criteria for patients with hepatocellular carcinoma in living donor liver transplantation. Liver Transpl. 2007;13:1637–44. doi: 10.1002/lt.21281. [DOI] [PubMed] [Google Scholar]
- 9.Lai Q, Merli M, Ginanni Corradini S, et al. Predictive factors of recurrence of hepatocellular carcinoma after liver transplantation: A multivariate analysis. Transplant Proc. 2009;41:1306–9. doi: 10.1016/j.transproceed.2009.03.094. [DOI] [PubMed] [Google Scholar]
- 10.Marsh JW, Dvorchik I, Bonham CA, Iwatsuki S. Is the pathologic TNM staging system for patients with hepatoma predictive of outcome? Cancer. 2000;88:538–43. doi: 10.1002/(sici)1097-0142(20000201)88:3<538::aid-cncr7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 11.Cillo U, Vitale A, Bassanello M, et al. Liver transplantation for the treatment of moderately or well-differentiated hepatocellular carcinoma. Ann Surg. 2004;239:150–59. doi: 10.1097/01.sla.0000109146.72827.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang SH, Suh KS, Lee HW, et al. A revised scoring system utilizing serum alphafetoprotein levels to expand candidates for living donor transplantation in hepatocellular carcinoma. Surgery. 2007;141:598–609. doi: 10.1016/j.surg.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Cillo U, Navaglia F, Vitale A, et al. Clinical significance of alpha-fetoprotein mRNA in blood of patients with hepatocellular carcinoma. Clin Chim Acta. 2004;347:129–38. doi: 10.1016/j.cccn.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 14.Cillo U, Vitale A, Navaglia F, et al. Role of blood AFP mRNA and tumor grade in the preoperative prognostic evaluation of patients with hepatocellular carcinoma. World J Gastroenterol. 2005;11:6920–25. doi: 10.3748/wjg.v11.i44.6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazzaferro V. Results of liver transplantation: With or without Milan criteria? Liver Transpl. 2007;13:S44–47. doi: 10.1002/lt.21330. [DOI] [PubMed] [Google Scholar]
- 16.Mazzaferro V, Llovet JM, Miceli R, et al. Metroticket Investigator Study Group. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: A retrospective, exploratory analysis. Lancet Oncol. 2009;10:35–43. doi: 10.1016/S1470-2045(08)70284-5. [DOI] [PubMed] [Google Scholar]
- 17.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–46. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 18.Marsh JW, Finkelstein SD, Demetris AJ, et al. Genotyping of hepatocellular carcinoma in liver transplant recipients adds predictive power for determining recurrence-free survival. Liver Transpl. 2003;9:664–71. doi: 10.1053/jlts.2003.50144. [DOI] [PubMed] [Google Scholar]
- 19.Woo HG, Park ES, Thorgeirsson SS, Kim YJ. Exploring genomic profiles of hepatocellular carcinoma. Mol Carcinog. 2011;50:235–43. doi: 10.1002/mc.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkelstein SD, Marsh W, Demetris AJ, et al. Microdissection-based allelotyping discriminates de novo tumor from intrahepatic spread in hepatocellular carcinoma. Hepatology. 2003;37:871–79. doi: 10.1053/jhep.2003.50134. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz M, Dvorchik I, Roayaie S, et al. Liver transplantation for hepatocellular carcinoma: extension of indications based on molecular markers. J Hepatol. 2008;49:581–88. doi: 10.1016/j.jhep.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dvorchik I, Schwartz M, Fiel MI, et al. Fractional allelic imbalance could allow for the development of an equitable transplant selection policy for patients with hepatocellular carcinoma. Liver Transpl. 2008;14:443–50. doi: 10.1002/lt.21393. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt C, Marsh JW. Molecular signature for HCC: Role in predicting outcomes after liver transplant and selection for potential adjuvant treatment. Curr Opin Organ Transplant. 2010;15:277–82. doi: 10.1097/MOT.0b013e328339837b. [DOI] [PubMed] [Google Scholar]
- 24.Marsh JW, Dvorchik I. Liver organ allocation for hepatocellular carcinoma: Are we sure? Liver Transpl. 2003;9:693–93. doi: 10.1053/jlts.2003.50086. [DOI] [PubMed] [Google Scholar]
- 25.Altimari A, Gruppioni E, Fiorentino M, et al. Genomic allelotyping for distinction of recurrent and de novo hepatocellular carcinoma after orthotopic liver transplantation. Diagn Mol Pathol. 2005;14:34–38. doi: 10.1097/01.pas.0000143609.85487.36. [DOI] [PubMed] [Google Scholar]
- 26.Morita K, Taketomi A, Soejima Y, et al. De novo hepatocellular carcinoma in a liver graft with sustained hepatitis C virus clearance after living donor liver transplantation. Liver Transpl. 2009;15:1412–16. doi: 10.1002/lt.21894. [DOI] [PubMed] [Google Scholar]
- 27.Vernadakis S, Poetsch M, Weber F, et al. Donor origin de novo HCC in a noncirrhotic liver allograft 3 years after liver transplantation. Transpl Int. 2010;23:341–43. doi: 10.1111/j.1432-2277.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- 28.Zavaglia C, De Carlis L, Alberti AB, et al. Predictors of long-term survival after liver transplantation for hepatocellular carcinoma. Am J Gastroenterol. 2005;100:2708–16. doi: 10.1111/j.1572-0241.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- 29.Yao FY. Selection criteria for liver transplantation in patients with hepatocellular carcinoma: Beyond tumor size and number? Liver Transpl. 2006;12:1189–91. doi: 10.1002/lt.20853. [DOI] [PubMed] [Google Scholar]
- 30.Dudek K, Kornasiewicz O, Remiszewski P, et al. Impact of tumor characteristic on the outcome of liver transplantation in patients with hepatocellular carcinoma. Transplant Proc. 2009;41:3135–37. doi: 10.1016/j.transproceed.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Takamori R, Wong LL, Dang C, Wong L. Needle-tract implantation from hepatocellular cancer: Is needle biopsy of the liver always necessary? Liver Transpl. 2000;6:67–72. doi: 10.1002/lt.500060103. [DOI] [PubMed] [Google Scholar]
- 32.Mazzaferro V, Sposito C, Zhou J, et al. Metroticket 2.0 model for analysis of competing risks of death after liver transplantation for hepatocellular carcinoma. Gastroenterology. 2018;154:128–39. doi: 10.1053/j.gastro.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 33.Lai Q, Nicolini D, Inostroza Nunez M, et al. A novel prognostic index in patients with hepatocellular cancer waiting for liver transplantation: Time-Radiological-response-Alpha-fetoprotein-INflammation (TRAIN) score. Ann Surg. 2016;264:787–96. doi: 10.1097/SLA.0000000000001881. [DOI] [PubMed] [Google Scholar]
- 34.Halazun KJ, Najjar M, Abdelmessih RM, et al. Recurrence after liver transplantation for hepatocellular carcinoma: A new MORAL to the story. Ann Surg. 2017;265:557–64. doi: 10.1097/SLA.0000000000001966. [DOI] [PubMed] [Google Scholar]
- 35.Marsh JW, Dvorchik I, Subotin M, et al. The prediction of risk of recurrence and time to recurrence of hepatocellular carcinoma after orthotopic liver transplantation: A pilot study. Hepatology. 1997;26:444–50. doi: 10.1002/hep.510260227. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Luna H, Vargas HE, Byrne T, Rakela J. Artificial neural network and tissue genotyping of hepatocellular carcinoma in liver-transplant recipients: Prediction of recurrence. Transplantation. 2005;79:1737–40. doi: 10.1097/01.tp.0000161794.32007.d1. [DOI] [PubMed] [Google Scholar]
- 37.Pommergaard HC, Burcharth J, Rosenberg J, Rasmussen A. Serologic and molecular biomarkers for recurrence of hepatocellular carcinoma after liver transplantation: A systematic review and meta-analysis. Transplant Rev (Orlando) 2016;30:171–77. doi: 10.1016/j.trre.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Gruttadauria S, Tropea A, Pagano D, et al. Mini-invasive approach contributes to expand the indication for liver resection for hepatocellular carcinoma without increasing the incidence of posthepatectomy liver failure and other perioperative complications: A single-center analysis. J Laparoendosc Adv Surg Tech A. 2016;26:439–46. doi: 10.1089/lap.2016.0134. [DOI] [PubMed] [Google Scholar]
- 39.Gruttadauria S, Pagano D, Tropea A, et al. Laparoscopic approach for thermoablation microwave in the treatment of hepatocellular carcinoma: A single-center experience. J Laparoendosc Adv Surg Tech A. 2016;26:808–11. doi: 10.1089/lap.2016.0373. [DOI] [PubMed] [Google Scholar]
- 40.Gruttadauria S, di Francesco F, Spada M. Fifty-six-month survival after liver transplantation in a patient with more than one-hundred hepatocellular carcinoma nodules. Transpl Int. 2012;9:e101–3. doi: 10.1111/j.1432-2277.2012.01507.x. [DOI] [PubMed] [Google Scholar]