Significance
Spontaneous accumulation of somatic DNA mutations has been hypothesized as a cause of both cancer and aging. However, detecting mutations in normal, noncancer cells is challenging, because mutations accumulate independently in each cell. Using an advanced single-cell whole-genome sequencing method, we characterized the landscape of mutations in human B lymphocytes as a function of age, from newborns to centenarians. Mutations were found to increase significantly with age, with mutational signatures correlating with those previously observed in B cell cancers. Mutation distribution across the genome indicates potentially adverse functional effects in old age.
Keywords: aging, somatic DNA mutation, B lymphocyte, single-cell whole-genome sequencing, functional genome
Abstract
Accumulation of mutations in somatic cells has been implicated as a cause of aging since the 1950s. However, attempts to establish a causal relationship between somatic mutations and aging have been constrained by the lack of methods to directly identify mutational events in primary human tissues. Here we provide genome-wide mutation frequencies and spectra of human B lymphocytes from healthy individuals across the entire human lifespan using a highly accurate single-cell whole-genome sequencing method. We found that the number of somatic mutations increases from <500 per cell in newborns to >3,000 per cell in centenarians. We discovered mutational hotspot regions, some of which, as expected, were located at Ig genes associated with somatic hypermutation (SHM). B cell–specific mutation signatures associated with development, aging, or SHM were found. The SHM signature strongly correlated with the signature found in human B cell tumors, indicating that potential cancer-causing events are already present even in B cells of healthy individuals. We also identified multiple mutations in sequence features relevant to cellular function (i.e., transcribed genes and gene regulatory regions). Such mutations increased significantly during aging, but only at approximately one-half the rate of the genome average, indicating selection against mutations that impact B cell function. This full characterization of the landscape of somatic mutations in human B lymphocytes indicates that spontaneous somatic mutations accumulating with age can be deleterious and may contribute to both the increased risk for leukemia and the functional decline of B lymphocytes in the elderly.
To accurately detect a full complement of mutations in somatic cells is a major technical challenge because of the random nature and very low abundance of most somatic mutations. This means that sequencing DNA from cell populations will show the germ line genotype rather than de novo somatic mutations (1). The solution to this problem, single-cell sequencing, is hampered by the high error rate of the genome amplification procedures required for single-cell genomics (2–4). We recently developed a highly accurate single-cell multiple displacement amplification (SCMDA) procedure to comprehensively determine the full spectrum of base substitutions in a single somatic cell, and then validated the procedure by comparing mutations identified from SCMDA-amplified single cells with those in unamplified DNA from clones of the same cell population (5).
Here we used SCMDA to assess mutation accumulation as a function of age in human B lymphocytes from healthy individuals varying in age from 0 (newborns) to >100 y (centenarians). Mutation accumulation in these cells could be a causal factor in both the age-related increase in the incidence of B cell leukemia (6) and the observed age-related functional decline, such as loss of B cell activation (7). We sequenced the genome of 56 single B lymphocytes and that of their corresponding 14 bulk DNAs using the remaining peripheral blood mononuclear cells (PBMCs) (SI Appendix, Figs. S1 and S2). The average sequencing depth across the genome was 27.9 ± 2.9 for the single cells and 25.3 ± 2.3 for the bulk DNAs. For single cells, 51.4 ± 4.7% of the genome was covered at a depth of ≥20×; for bulk DNAs, this coverage was 78.6 ± 5.1% (SI Appendix, Table S1). Somatic single nucleotide variants (SNVs) in single cells were identified at a depth of ≥20× using SCcaller, a software tool developed specifically for single-cell SNV detection by filtering out potential artifacts due to whole-genome amplification (5). Germ line variants were filtered out using the bulk DNA of the same donors. Using Sanger sequencing, we confirmed 62 of the 65 mutations selected at random (95.4%), indicating the high specificity of the variant calls (SI Appendix, Fig. S3 and Table S2). Because all somatic SNVs should be heterozygous due to the extremely low likelihood that mutations occur at the exact same site on both alleles, heterozygous germ line SNP calling in the same cell is a good control for measuring sensitivity, which appeared to be on average 60% (SI Appendix, Table S3). That is, 60% of the heterozygous SNPs identified in bulk DNA were also identified in a single cell. The less than 100% sensitivity is a consequence of the stringent filtering, which avoids false-positives at the cost of occasionally missing a true variant.
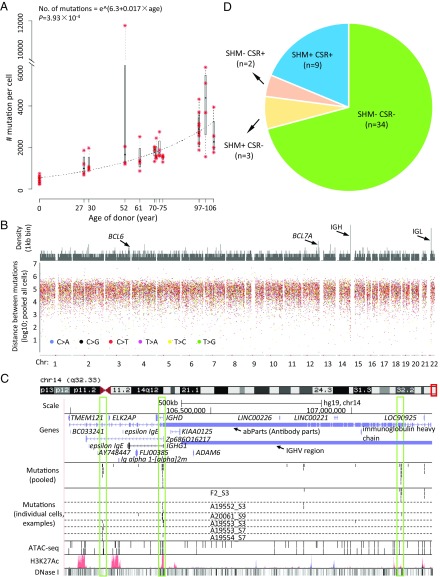
After adjusting for genomic coverage and sensitivity, the estimated numbers of SNVs per cell ranged from 237 to 5,862, with one extreme outlier of 11,765 (Fig. 1A and SI Appendix, Table S3). The median number of mutations per cell was found to increase significantly with the age of the donor (P = 3.93 × 10−4; Fig. 1A), with 463.4 in the newborns (n = 8 cells from two donors), 1,181.9 in the 27- to 30-y-olds (n = 8 cells from two donors), 2,101.7 in the 52- to 75-y-olds (n = 24 from six donors), and 3,127.0 in the 97- to 106-y-olds (n = 16 from four donors).
Fig. 1.
Somatic mutations accumulate with age in human B lymphocytes. (A) The number of somatic mutations per genome as a function of age. Red asterisks indicate individual cells. Each box corresponds to summary statistics of four cells from one donor. Regression was performed on the median numbers of mutations of the four cells from each donor, using exponential model nonlinear least squares regression. (B) Rainfall plot illustrating distances of neighboring mutations. (Top) Density of mutations (56 cells pooled) in kilobase bins. (Bottom) Distances of each mutation to its closest other mutation. (C) Mutation hotspots in Ig H chain regions visualized using the UCSC Genome Browser. In the mutation panels, each bar represents one SNV. In the ATAC sequencing (ATAC-seq) panel, obtained from pooled B cells from one young individual and one old individual, each bar represents one open chromatin peak. Other panels were obtained from annotations of the UCSC Genome Browser. The three vertical boxes highlight three hotspots identified in this region. (D) The SHM and CSR status of the 48 adult B cells; all eight B cells from cord blood are SHM−CSR−. As expected, the majority of adult B cells are classified into two categories: SHM−CSR− (primarily naïve B cells) and SHM+CSR+ (memory B cells). A few cases of SHM+CSR− and SHM−CSR+ cells are known as nonswitched memory B cells and GC-independent memory B cells, respectively.
While virtually all mutations were unique for each cell, a total of 24 recurrent mutations (i.e., shared between cells) were identified. These recurrent mutations were present only in cells from the same donor, as validated by Sanger sequencing of the recurrent mutations in all samples (SI Appendix, Fig. S4 and Table S2). The cells that share mutations most likely derive from the same ancestor hematopoietic stem cell in which the mutation originally occurred. Of note, the recurrent mutations were mostly found in elderly donors (P = 0.0115, permutation test, one-tailed, 20,000 permutations), in keeping with the observed decrease in the number of active lineages of hematopoietic stem cells during aging (8). However, the numbers of recurrent mutations were small, and none occurred in the cancer driver genes used to identify clonal hematopoiesis of indeterminate potential (CHIP) (9); we also applied the same method as described in ref. 9 and found that none of our 14 donors displayed CHIP.
While the observed increase in mutation frequency with age is in keeping with previously reported data on other cell types in both humans and mice (10–14), accurate numbers obtained by single-cell sequencing are sparse and have not been thoroughly validated (2). In terms of validation, B lymphocytes offer the advantage of endogenous control loci in the form of mutational hotspots at Ig genes (15). On pooling all mutations of the 56 cells, the large majority of mutations were distributed randomly across the genome (SI Appendix, Fig. S5 A–C). However, we noted already at the megabase level a substantial enrichment of mutations at one end of chromosome 14 (SI Appendix, Fig. S5 B and C), which harbors Ig gene sequences (15). At the kilobase level, we identified 24 mutational hotspots, defined as regions with multiple mutations separated by <5 kb from their nearest neighbor (Fig. 1B and SI Appendix, Table S4) (16–18). The 24 mutational hotspots are composed of 4–12 mutations in regions of 469–8,970 bp corresponding to a mutation frequency of 2.79 × 10−3 per bp per cell, in agreement with previous findings as determined from cloned, PCR-amplified target regions (19, 20); the average genome-wide mutation frequency is only approximately 10−7 per bp.
Of the 24 hotspots (SI Appendix, Fig. S6), only 5 were located in or close to Ig gene regions, including 3 in the Ig H chain region on chromosome 14 (Fig. 1C), 1 in the Ig L chain region, and 1 downstream of this region on chromosome 22 (SI Appendix, Fig. S7). The other 19 hotspots are likely off-target effects of activation-induced cytidine deaminase (AID) (15), the enzyme that initiates somatic hypermutation (SHM). Some of these hotspots have been described previously, including BCL6 (B cell CLL/lymphoma 6) (21) and BCL7A (BCL tumor suppressor 7A) (22). To our knowledge, to date nobody has demonstrated B lymphocyte mutational hotspots collectively across a single genome. However, in comparison with a mouse AID (mAID) ChIP sequencing dataset (23), we found substantial overlap of the hotspot regions that we identified with genes in the mAID dataset (odds ratio = 1.93) (SI Appendix, Fig. S8). While not significant (P = 0.134, one-tailed Fisher’s exact test), this finding contributes to the notion that the hotspots that we found are AID off-targets. Indeed, this conclusion is strengthened by the fact that approximately one-half of the 24 hotspots have been reported in other studies as associated with human lymphoma, leukemia, or AID off-target loci in mouse B cells (21, 22, 24–28) (SI Appendix, Table S4). Moreover, the hotspots significantly overlap with or are close to open chromatin regions, transcription start sites, and transcription factor (TF) binding sites associated with AID binding (SI Appendix, Table S4).
Based on the presence or absence of mutations in the five Ig hotspots, we distinguished SHM+ from SHM− cells. Due to the very low spontaneous mutation frequency (∼10−7 per bp) and the small size of the five hotspot areas (kilobase level per area), the chance of false-positive classification of a real SHM− cell as SHM+ is very low (i.e., 1.6%). In addition to SHM, we also identified class switch recombination (CSR) in some B cells, allowing us to distinguish memory B cells (SHM+ and CSR+) from the SHM−/CSR− B cells, primarily naïve B cells (Fig. 1D and SI Appendix, Fig. S9). Of note, the SHM+ cells had a significantly higher mutation frequency than the SHM− cells (P = 5.19 × 10−5) (SI Appendix, Fig. S10A). A significant age-related increase in mutations in both cell types was observed when analyzing all cells together with both aging and SHM as variables (P = 3.07 × 10−4 for aging). An age-related increase was also statistically significant when testing SHM− cells alone, with aging as the sole variable (P = 1.08 × 10−4) (SI Appendix, Fig. S10B).
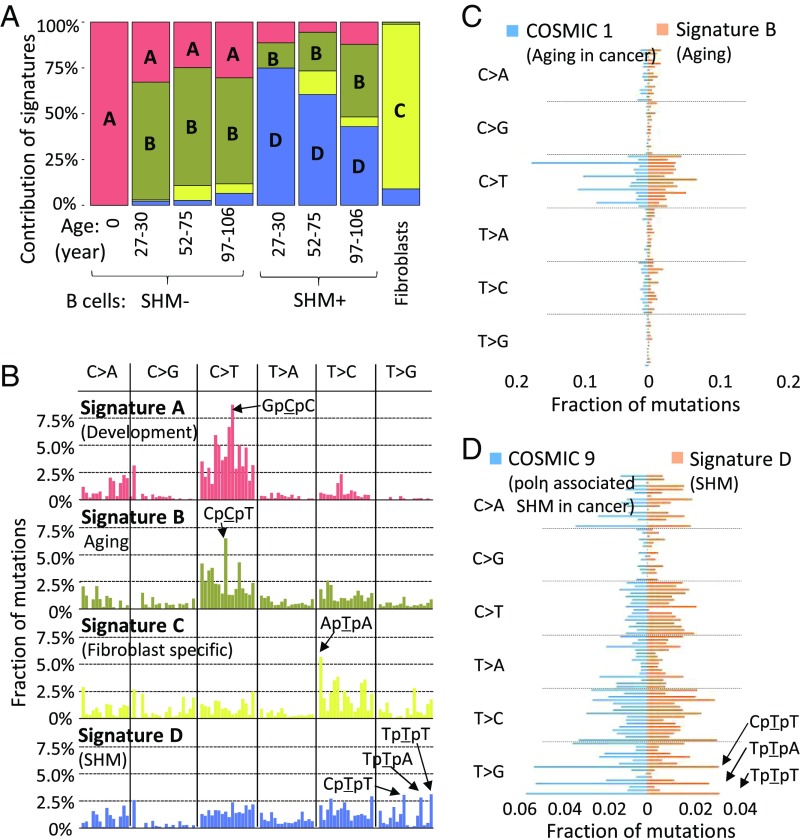
We next analyzed the mutation spectra. In addition to the affected base (SI Appendix, Fig. S11 A and B), we also included the two flanking bases in the analysis. This allowed us to comparatively analyze mutation spectra as a function of age and SHM status. The results were compared with the spectrum that we previously obtained for human fibroblasts (5) (SI Appendix, Fig. S11C). Using nonnegative matrix factorization (17, 29), we decomposed the mutation spectra into mutation signatures, designated signatures A–D (Fig. 2 A and B). Based on these signatures, we were able to (i) show that somatic mutations in B lymphocytes (signatures A, B, and D) are substantially different from those in fibroblasts (signature C) and (ii) identify mutations specifically associated with development (signature A), aging (signature B; P = 9.25 × 10−4 with age) (SI Appendix, Fig. S12), and SHM (signature D).
Fig. 2.
Mutation signatures. (A) Contributions of signatures A, B, and D to all mutations in B lymphocytes in the different age groups. Signature C is from spontaneous mutations previously obtained for human primary fibroblasts. (B) Mutation signatures in the context of their flanking base pairs (in alphabetical order, e.g., in the first column C > A category from left to right, ApCpA > ApApA to TpCpT > TpApT). (C) Comparison between signature B and COSMIC1. The fractions of mutations are presented in alphabetical order (e.g., in the first column C > A category from top down, ApCpA > ApApA to TpCpT > TpApT). (D) Comparison between signature D and COSMIC 9.
Importantly, these signatures observed in normal B lymphocytes from healthy human donors were very similar to mutation signatures obtained from human cancers in the Catalogue of Somatic Mutations in Cancer (COSMIC) (30–32) (SI Appendix, Fig. S11D). Our aging signature B was found to correlate highly with the cancer aging signatures COSMIC1 and COSMIC5 in many cancer types (Spearman’s correlation coefficients ρ = 0.80 and 0.74 separately) (Fig. 2C and SI Appendix, Fig. S11D). Our developmental signature A also correlates with COSMIC1 (ρ = 0.78), as does signature B. These results suggest that the mechanistic origin of these signatures even in tumors firmly resides in the normal process of age-related mutation accumulation. The “aging signature” appeared mostly as a result of spontaneous cytosine deamination (33). The latter was also observed in the mutation signature of a childhood cancer, pilocytic astrocytoma (COSMIC19), resulting in a high correlation (ρ = 0.81) with our signature B (SI Appendix, Fig. S11D). We also observed a high correlation between the SHM signature D and the chronic lymphocytic leukemia and malignant B cell lymphoma signature COSMIC9 (ρ = 0.81) (Fig. 2D). In signature D as well as in the COSMIC9 signature, we noted that mutations occur at both C/G and A/T base pairs, suggesting that more than one mechanism is involved in the generation of these mutations. Indeed, AID preferentially leads to C > T, while A/T bases appear to be especially susceptible to polymerase η errors during hypermutation (34, 35). These results were confirmed by refitting the known COSMIC signatures to the observed somatic mutations in normal B lymphocytes, using the “deconstructSigs” package in R (36); that is, COSMIC9 was found only in SHM+ cells, and COSMIC5 and COSMIC19 were found in all cells (SI Appendix, Fig. S13). Together with the observation of off-target SHM hotspots, these results strongly support the hypothesis that somatic hypermutation contributes significantly to the development of B cell cancers (15).
An important question that we were now able to directly address is whether somatic mutation accumulation during aging can have a functional impact other than increasing cancer risk. For this purpose, we subdivided all mutations into two groups: those found and those not found in the functional part of the genome. Mutations in the functional genome were defined as those occurring in the transcribed exome or its regulatory regions. To identify the transcribed exome, we performed RNA sequencing of purified bulk B lymphocytes (SI Appendix, Fig. S14 and Table S5). As gene regulatory regions, we considered both promoters of active genes and open chromatin regions (e.g., TF binding regions) identified by ATAC sequencing in the same bulk B cells (SI Appendix, Fig. S15 and Table S5).
Of the median 9.2 ± 6.8 nonsynonymous mutations per cell in the transcribed B cell exome of the ≥97-y-olds (Table 1 and SI Appendix, Table S6), 6.5 ± 5.5 (71%) were functionally deleterious according to SIFT and PROVEAN (37, 38) (SI Appendix, Table S7). Many more potentially functional mutations occurred in noncoding regions of the genome (Table 1 and SI Appendix, Table S6). When analyzing the TF binding regions reported by ENCODE (39) in our B lymphocytes, we found that mutations in these regions increased with age, from a median of 79.6 per cell to 435.9 per cell (P = 8.79 × 10−6). More specifically, when analyzing our ATAC data specific for B lymphocytes, we found an almost fivefold age-related increase in the number of mutations in active open chromatin regions (likely to be TF binding regions specific in B cells), from 5.4 to 24.5 per cell (P = 0.0199). Even more mutations were found collectively in proximate promoter (from −1,500 to +500 bp of transcription start sites), 5′ UTR, and 3′ UTR regions, with 56.9 ± 26.9, 4.9 ± 5.7, and 13.6 ± 6.5 per cell, respectively, in the oldest subjects. Thus, in the functional part of the B cell, the median number of genome mutations increased from 27.0 ± 18.3 per cell in newborns to 85.1 ± 36.9 per cell in >97-y-olds (P = 0.000878, exponential model, nonlinear least squares regression).
Table 1.
Average ± SD number of functional SNVs per cell
| Class* | Age | |||
| 0 y | 27–30 y | 52–75 y | 97–106 y | |
| Total in the genome | 463.4 ± 179.0 | 1,181.9 ± 484.7 | 2,101.7 ± 2,106.1 | 3,127.0 ± 1,237.1 |
| Nonsynonymous | 2.2 ± 4.1 | 1.7 ± 2.5 | 5.4 ± 5.3 | 9.2 ± 6.8 |
| Loss-of-function SNVs† | 0.4 ± 1.1 | 0.0 ± 0.0 | 0.6 ± 1.7 | 0.5 ± 1.4 |
| Damaging‡ | 1.3 ± 2.8 | 1.7 ± 2.5 | 4.0 ± 5.0 | 6.5 ± 5.5 |
| Open chromatin region§ | 5.4 ± 5.8 | 11.5 ± 4.4 | 17.6 ± 24.9 | 24.5 ± 15.8 |
| Promoter | 17.7 ± 10.1 | 19.6 ± 7.4 | 33.1 ± 34.9 | 56.9 ± 26.9 |
| 5′ UTR | 2.1 ± 3.5 | 2.5 ± 4.0 | 3.9 ± 5.7 | 4.9 ± 5.7 |
| 3′ UTR | 4.6 ± 5.9 | 11.0 ± 11.9 | 10.9 ± 11.2 | 13.6 ± 6.5 |
Except for the genome total, only SNVs transcribed or in open chromatin regions are reported. All transcribed and nontranscribed SNVs are reported in SI Appendix, Table S6.
Loss-of-function SNVs, including stop gain, stop loss, and splicing alterations.
SNVs annotated as damaging by SIFT or deleterious by PROVEAN.
Identified using ATAC sequencing of B lymphocytes.
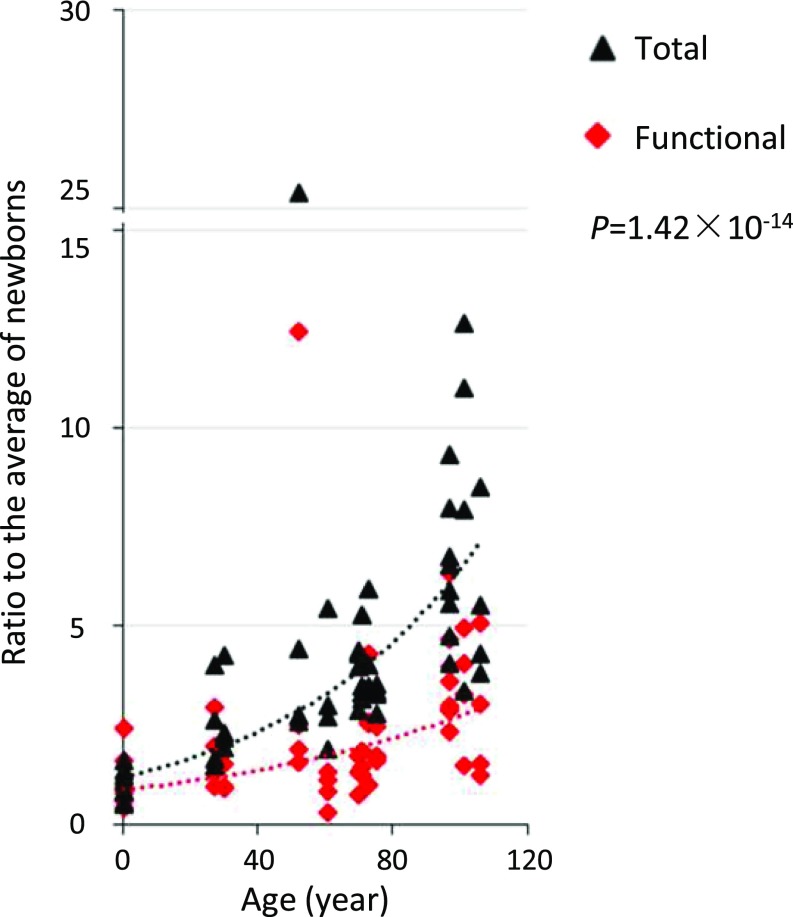
We next tested whether mutations in the functional genome were also functionally impactful. To do this, we compared the pace of age-related accumulation of mutations in the functional genome with that of mutations in the genome overall. The results indicated a highly significant slower pace of potentially functional mutations (P = 1.42 × 10−14) (Fig. 3). In this case, we used all protein-coding genes, since we could not ascertain the origin of the mutations, which could have been hematopoietic stem cells with other genes active than those in the mature B lymphocytes. These results indicate protection against deleterious mutations in the functional genome during human aging, suggesting that many random somatic mutations are damaging to cellular function.
Fig. 3.
Accumulation of mutations in the functional genome and genome overall during aging. Each data point represents the ratio of the number of mutations per cell to the average of the mutation number per cell in newborn B lymphocytes (functional genome and whole genome calculated separately). The ratios of the functional genome are in red, and those in the genome overall are in black. P values are two-tailed and were estimated using the Wilcoxon signed-rank test.
The significantly lower rate of mutation accumulation in functional genomic regions than the genome-wide average suggests that the B cells and their ancestral stem cells that harbored such mutations may have died or been at a growth disadvantage. This is in keeping with the discovery of a decreased number of lymphoid hematopoietic lineages during aging (8) and may underlie the CHIP phenomenon found in a fraction of elderly persons (40). However, our data show that many mutations in the functional genome remain and continue to accumulate. While not lethal, such mutations may still contribute to the well-documented age-related decline in function that has been observed for many cell types in humans and experimental animals. As we and others have reported previously, this effect may be mediated by increased cell-to-cell transcriptional variability (41–43).
In summary, this comprehensive characterization of the landscape of somatic mutations in B lymphocytes across the entire human age range shows a highly significant age-related increase of mutations. Mutation signatures in these normal B cells, which are significantly different from those in human primary fibroblasts (5), were found to correlate with those previously observed in human cancers, most notably B cell leukemia. This finding underscores the increasing realization that age is the most important risk factor for cancer. A substantial fraction of all mutations was found to occur in sequence features that are functionally active in B lymphocytes. These mutations were subject to negative selection, as evidenced by their much slower rate of accumulation with age. Thus, somatic mutations may contribute to functional decline in aging through depletion of cells or accumulation of cells with slightly deleterious mutations. Of note, in this study, we only analyzed base substitution mutations. Other types of mutations, such as small INDELs, copy number variations, and structural variations, are much less frequent but may still contribute to a gradual age-related deterioration of the functional somatic genome. In principle, we could test for such mutations in our single-cell whole-genome sequences. Reliable computational tools for that purpose, equally sensitive and accurate as our current tool for base substitutions, are currently under development. Our present results provide the foundation for finally testing the somatic mutation theory of aging by studying multiple human organs and tissues for the accumulation of potentially functional mutations and their causal relationship to age-related functional decline and disease.
Materials and Methods
More detailed information is provided in SI Appendix.
Human Specimens.
The collection of blood for B lymphocyte isolation and whole-genome sequencing was approved by the Einstein-Montefiore Institutional Review Board. Two frozen human umbilical cord blood mononuclear cell samples were obtained from AllCells and STEMCELL Technologies. Informed consent was obtained from all the donors. The donors in this study were healthy with no history of cancer. Age, sex, and other donor characteristics are reported in SI Appendix, Table S1.
B Lymphocyte Isolation.
For each donor, PBMCs were isolated using Ficoll-Paque PLUS solution (GE Healthcare) according to the manufacturer’s instructions by the Molecular Cytogenetics Core at the Albert Einstein College of Medicine. In brief, fresh peripheral blood was collected in ACD solution A and solution B anticoagulant tubes and then diluted with PBS and 2 mM EDTA (ratio 1:1; Gibco). Subsequently, the blood mixture was layered onto Ficoll-Paque PLUS. After centrifugation, lymphocyte cells remained at the plasma–Ficoll-Paque PLUS interface and were carefully transferred to a new tube and washed twice with 1× PBS. These PBMCs were resuspended with MACS buffer (Miltenyi Biotec).
B lymphocytes were isolated using MACS separation (Kit 130–050301; Miltenyi Biotec). In brief, a total of 107 total PBMCs were resuspended in 80 µL of buffer and mixed with 20 µL of CD19 MicroBeads (Miltenyi Biotec). The mixture was incubated at 2–8 °C for 15 min, washed, and centrifuged. Cell pellets were resuspended and applied onto the column, which was placed in the magnetic field of a suitable MACS separator. Unlabeled cells were washed away with washing buffer and collected for T cell isolation or discarded. Labeled B lymphocytes on the column were eluted into a collection tube.
To validate the purity of B lymphocytes, cells were fluorescently stained with CD19-FITC (130–113-730; Miltenyi Biotec). Cell debris and dead cells were excluded from the analysis based on DAPI fluorescence; an example is shown in SI Appendix, Fig. S2A.
Single B Lymphocyte Isolation.
Single B lymphocytes were collected using the CellRaft array (Cell Microsystems) following a protocol that was modified from our previous procedure (5). In brief, a CellRaft array was rinsed with medium and then coated with gelatin 2% in water (G1393-100ML; Sigma-Aldrich) at 37 °C for 1 h to help the B lymphocytes attach (44, 45). After the superfluous gelatin solution was rinsed off the array, approximately 5,000 B lymphocytes in 3 mL of medium were added to the CellRaft array, followed by incubation for 3 h at 37 °C in 3% O2 and 10% CO2. The array was then washed twice with 1 mL of PBS to remove floating cells, and 3 mL of fresh complete medium was added. Single rafts containing one cell (as observed by microscopy using a 10× objective; SI Appendix, Fig. S2B) were transferred using the magnetic wand supplied with the CellRaft system into 0.2-mL PCR tubes containing 2.5 µL of PBS. We verified that there was only one raft in a PCR tube using a magnifying glass. Tubes containing single rafts were immediately frozen on crushed dry ice and kept at −80 °C until use.
Single-Cell Multiple Displacement Amplification.
We amplified multiple single B lymphocytes from each of 14 humans according to our SCMDA protocol (5). In brief, 1 µL of Exo-Resistant random primer (Thermo Fisher Scientific) was added to the cell, followed immediately by 3 µL of lysis buffer (400 mM KOH, 100 mM DTT, and 10 mM EDTA). Cell lysis and DNA denaturation were performed on ice for 10 min. Then 3 µL of stop buffer (400 mM HCl and 600 mM Tris⋅HCl pH 7.5) was added to neutralize the lysis buffer. Finally, 32 µL of Master Mix containing 30 µL of MDA reaction buffer and 2 µL of Phi29 polymerase (REPLI-g UltraFast Mini Kit; Qiagen) were added. SCMDA was carried out for 1.5 h at 30 °C, 3 min at 65 °C, and holding at 4 °C until purification. SCMDA product was purified using AMPureXP-beads (Beckman Coulter), and the concentration was measured with the Qubit High-Sensitivity dsDNA Kit (Thermo Fisher Scientific). Meanwhile, 1 ng of human genomic DNA in 2.5 µL of PBS was amplified as a positive control, and 2.5 µL of PBS without any template was amplified as a negative control. All single-cell amplicons of sufficient yield were subjected to a locus dropout test (5), after which a total of four amplicons per donor that passed the test were prepared for whole-genome sequencing.
Bulk DNA Extraction.
Total DNA was collected from the cell pellets remaining after Ficoll density centrifugation for B lymphocyte isolation using the DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s specifications. The concentrations of DNA were quantified using the Qubit High-Sensitivity dsDNA Kit, and the qualities of DNA were evaluated with 1% agarose gel electrophoresis.
DNA Library Preparation and Whole-Genome Sequencing.
The libraries of 14 bulk DNAs and 56 single B lymphocyte MDA products were prepared using the TruSeq Nano DNA HT Sample Prep Kit (Illumina) by Novogene. In brief, 1.0 μg of DNA per sample as input was fragmented by sonication to a size of 350 bp, and DNA fragments were end-polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing with further PCR amplification. PCR products were purified (AMPure XP System; Beckman Coulter), and libraries were analyzed for size distribution using an Agilent 2100 Bioanalyzer and quantified by real-time PCR. All libraries were sequenced on the Illumina HiSeq X Ten sequencing platform, and paired-end reads, 2 × 150 bp, were generated by Novogene.
Data Analyses and Availability.
All data analyses are described in SI Appendix. The sequencing data have been deposited in the dbGaP database (accession no. phs001808.v1.p1).
Supplementary Material
Acknowledgments
We thank the Molecular Cytogenetics Core and the Epigenomics Core at the Albert Einstein College of Medicine for B cell isolation and RNA/ATAC sequencing, respectively. We thank Drs. Bernice Morrow, Sofiya Milman, and Nir Barzilai for sharing human samples and assisting us with the human subjects part of the research. This study was supported by the National Institutes of Health (Grants P01 AG017242 to J.V., P01 AG047200 to J.V., and K99 AG056656 to X.D.), Einstein’s Nathan Shock Center of Excellence in Basic Biology of Aging (Grant P30 AG038072), and the Paul F. Glenn Center for the Biology of Human Aging.
Footnotes
Conflict of interest statement: L.Z., X.D., M.L., A.Y.M., and J.V. are cofounders of SingulOmics Corp.
This article is a PNAS Direct Submission.
Data deposition: The raw sequencing data reported in this paper have been deposited in the dbGaP database, www.ncbi.nlm.nih.gov/gap (accession no. phs001808.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902510116/-/DCSupplemental.
References
- 1.Gundry M, Vijg J. Direct mutation analysis by high-throughput sequencing: From germline to low-abundant, somatic variants. Mutat Res. 2012;729:1–15. doi: 10.1016/j.mrfmmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 3.Gregory MT, et al. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res. 2016;44:e22. doi: 10.1093/nar/gkv915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C, et al. Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI) Science. 2017;356:189–194. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong X, et al. Accurate identification of single-nucleotide variants in whole-genome–amplified single cells. Nat Methods. 2017;14:491–493. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gribben JG. Chronic lymphocytic leukemia: Planning for an aging population. Expert Rev Anticancer Ther. 2010;10:1389–1394. doi: 10.1586/era.10.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frasca D, Blomberg BB. B cell function and influenza vaccine responses in healthy aging and disease. Curr Opin Immunol. 2014;29:112–118. doi: 10.1016/j.coi.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat Rev Immunol. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- 9.Jaiswal S, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vijg J, Suh Y. Genome instability and aging. Annu Rev Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- 11.Dollé ME, et al. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- 12.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodato MA, et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science. 2018;359:555–559. doi: 10.1126/science.aao4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae T, et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science. 2018;359:550–555. doi: 10.1126/science.aan8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. Evidence for mutation showers. Proc Natl Acad Sci USA. 2007;104:8403–8408. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lora D. 2016 ClusteredMutations: Location and visualization of clustered somatic mutations. Available at https://rdrr.io/cran/ClusteredMutations/. Accessed March 31, 2019.
- 19.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–2070. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- 20.Yeap LS, et al. Sequence-intrinsic mechanisms that target AID mutational outcomes on antibody genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 22.Kato L, et al. Nonimmunoglobulin target loci of activation-induced cytidine deaminase (AID) share unique features with immunoglobulin genes. Proc Natl Acad Sci USA. 2012;109:2479–2484. doi: 10.1073/pnas.1120791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamane A, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns A, et al. Whole-genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHVmut and IgHVunmut subgroups. Leukemia. 2018;32:332–342. doi: 10.1038/leu.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwindt H, et al. Chromosomal translocations fusing the BCL6 gene to different partner loci are recurrent in primary central nervous system lymphoma and may be associated with aberrant somatic hypermutation or defective class switch recombination. J Neuropathol Exp Neurol. 2006;65:776–782. doi: 10.1097/01.jnen.0000229988.48042.ae. [DOI] [PubMed] [Google Scholar]
- 27.Warzocha K, et al. Identification of two lymphotoxin beta isoforms expressed in human lymphoid cell lines and non-Hodgkin’s lymphomas. Biochem Biophys Res Commun. 1997;238:273–276. doi: 10.1006/bbrc.1997.7277. [DOI] [PubMed] [Google Scholar]
- 28.Ramsay AJ, et al. Frequent somatic mutations in components of the RNA processing machinery in chronic lymphocytic leukemia. Leukemia. 2013;27:1600–1603. doi: 10.1038/leu.2012.344. [DOI] [PubMed] [Google Scholar]
- 29.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: Inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–3675. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forbes SA, et al. COSMIC: Somatic cancer genetics at high resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421, and erratum (2013) 502:258. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faili A, et al. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J Exp Med. 2004;199:265–270. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuberger MS, Rada C. Somatic hypermutation: Activation-induced deaminase for C/G followed by polymerase eta for A/T. J Exp Med. 2007;204:7–10. doi: 10.1084/jem.20062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 38.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Jimenez CP, et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355:1433–1436. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enge M, et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell. 2017;171:321–330 e14. doi: 10.1016/j.cell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arencibia I, Sundqvist KG. Collagen receptor on T lymphocytes and the control of lymphocyte motility. Eur J Immunol. 1989;19:929–934. doi: 10.1002/eji.1830190521. [DOI] [PubMed] [Google Scholar]
- 45.Cornelissen CG, et al. Fibronectin coating of oxygenator membranes enhances endothelial cell attachment. Biomed Eng Online. 2013;12:7. doi: 10.1186/1475-925X-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.