Significance
The adaptive immune system is able to protect us from a large variety of pathogens, even ones it has not seen yet. Can predicting the future pathogen distribution help in protection? We find that a combination of probabilistic forecasting and occasional sampling of the current environment reduces infection costs—a scheme easily implemented by the memory repertoire. The proposed theoretical framework offers a modular recipe for updating the memory repertoire, which quantitatively predicts the strength of the immune response in flu-vaccination experiments, unlike other update schemes. It also links the observed early life dynamics of the memory pool to the sparseness properties of the pathogen distribution and competitive receptor dynamics for pathogens.
Keywords: immune repertoire, Bayesian prediction, biophysics, immune memory, stochastic dynamics
Abstract
An adaptive agent predicting the future state of an environment must weigh trust in new observations against prior experiences. In this light, we propose a view of the adaptive immune system as a dynamic Bayesian machinery that updates its memory repertoire by balancing evidence from new pathogen encounters against past experience of infection to predict and prepare for future threats. This framework links the observed initial rapid increase of the memory pool early in life followed by a midlife plateau to the ease of learning salient features of sparse environments. We also derive a modulated memory pool update rule in agreement with current vaccine-response experiments. Our results suggest that pathogenic environments are sparse and that memory repertoires significantly decrease infection costs, even with moderate sampling. The predicted optimal update scheme maps onto commonly considered competitive dynamics for antigen receptors.
All living systems sense the environment, learn from the past, and adapt predictively to prepare for the future. Their task is challenging because environments change constantly, and it is impossible to sample them completely. Thus, a key question is how much weight should be given to new observations vs. accumulated past experience. Because evidence from the world is generally uncertain, it is convenient to cast this problem in the language of probabilistic inference where past experience is encapsulated in a prior probability distribution which is updated according to sampled evidence. This framework has been successfully used to understand aspects of cellular (1–4) and neural (5–8) sensing. Here, we propose that the dynamics of the adaptive immune repertoires of vertebrates can be similarly understood as a system for probabilistic inference of pathogen statistics.
The adaptive immune system relies on a diverse repertoire of B- and T-cell receptors to protect the host organism from a wide range of pathogens. These receptors are expressed on clones of receptor-carrying cells present in varying copy numbers. A defining feature of the adaptive immune system is its ability to change its clone composition throughout the lifetime of an individual, in particular via the formation of memory repertoires of B and T cells following pathogen encounters (9–14). In detail, after a proliferation event that follows successful recognition of a foreign antigen, some cells of the newly expanded clone acquire a memory phenotype. These cells make up the memory repertoire compartment that is governed by its own homeostasis, separate from the inexperienced naive cells from which they came. Upon reinfection by a similar antigen, memory guarantees a fast immune response. With time, our immune repertoire thus becomes specific to the history of infections and adapted to the environments we live in. However, the commitment of part of the repertoire to maintaining memory must be balanced against the need to also provide broad protection from as-yet-unseen threats. What is more, memory will lose its usefulness over time as pathogens evolve to evade recognition.
How much benefit can immunological memory provide to an organism? How much memory should be kept to minimize harm from infections? How much should each pathogen encounter affect the distribution of receptor clones? To answer these questions, we extend a framework for predicting optimal repertoires given pathogen statistics (15) by explicitly considering the inference of pathogen frequencies as a Bayesian forecasting problem (16). We derive the optimal repertoire dynamics in a temporally varying environment. This approach can complement more mechanistic studies of the dynamics and regulation of immune responses (12, 17–22) by revealing adaptive rationales underlying particular features of the dynamics. In particular, we link the amount of memory production to the variability of the environment and show that there exists an optimal time scale for memory attrition. Additionally, we demonstrate how biologically realistic population dynamics can approximate the optimal inference process and analyze conditions under which memory provides a benefit. Comparing predictions of our theory to experiment, we argue for a view in which the adaptive immune system can be interpreted as a machinery for learning a highly sparse distribution of antigens.
Theory of Optimal Immune Prediction
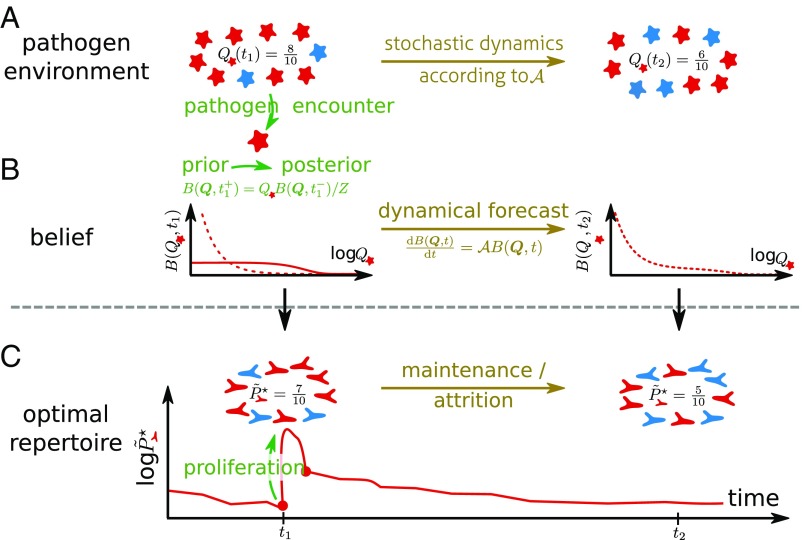
The pathogenic environment is enormous, and the immune system can only sample it sparsely, as pathogens enter into contact with it at some rate . We consider an antigenic space of different pathogens with time-varying frequencies . These frequencies are unknown to the organism and evolve stochastically. Their dynamics is formally described by a Fokker–Planck operator encoding how pathogenic frequencies change (Fig. 1A and Materials and Methods). We reason that the immune system should efficiently use the information available through these encounters, along with prior knowledge of how pathogens evolve encoded in the system dynamics, to build an internal representation of the environment (Fig. 1B). Biologically, we can think about this representation as being encoded in the composition of the adaptive immune repertoire (the size and specificity of naive and memory lymphocyte clones), but generally further cellular memory mechanisms might also contribute. Based on this representation of the world, the immune system should organize its defenses to minimize harm from future infections (Fig. 1C).
Fig. 1.
Sketch of a model of immune repertoire dynamics as a sequential inference process about a time-varying pathogen distribution. (A) The organism lives in a pathogenic environment with frequencies of different pathogen strains that change over time. (B) Past pathogen encounters provide an avenue for the immune system to learn the pathogen distribution. Using sequential Bayesian inference provides an optimal way to update the beliefs about the frequencies of different pathogens over time. (C) Based on its beliefs about the prevalence of pathogens, the optimal immune dynamics allocates lymphocytes across different pathogens to minimize the expected harm from infections. Broadly, the more frequent a pathogen is, the more the organism should be covered. This resource allocation maps the changes in beliefs to the changes in the repertoire composition.
How could the immune system leverage a representation of beliefs about pathogen frequencies to provide effective immunity? Each lymphocyte (B or T cell) of the adaptive immune system expresses on its surface a single receptor out of possible receptors. This receptor endows the lymphocyte with the ability to specifically recognize pathogens (labeled ) with probability . The immune repertoire is defined by frequencies of these receptors across the lymphocyte population, denoted by . These frequencies sum up to one, which implies a resource allocation trade-off between the different receptor types—having more of one in the repertoire implies having less of others. How much harm an infection inflicts depends on how much resources the immune system has devoted to fighting the infection, i.e., the fraction of the repertoire specific to antigen , which we will refer to as the coverage of the antigen. Given the pathogen frequencies and repertoire distribution , the mean harm cause by the next infection is given by , where is decreasing function of the fraction of the repertoire specific to the infection (15). The host organism does not know Q with certainty, but has an internal belief about the frequencies learned through sampling during previous infections. An optimal immune system can then distribute its resources to minimize the expected harm of the next infection:
| [1] |
where are the expected frequencies of pathogens. Note that, because in our framework the state of the immune system does not affect the stochastic dynamics of the pathogenic environment, this strategy is the best possible one, even for minimizing harm against all future infections, provided that the repertoire is allowed to revise its composition instantaneously. Although the function may be complicated, it generally implies that receptors that are specific to frequent infections (high ) should be well represented in the optimal repertoire (high ) (ref. 15 and Materials and Methods). In this framework, we have assumed that infections, their clearing by the immune system, and the subsequent update of the repertoire are all fast compared with changes in the environment, which occur over a time , and to the mean time between pathogenic encounters (). In our optimization, we keep the specificity of the receptors () fixed, assuming that this specificity is set independently by biophysical constraints and by the need to allow discrimination between antigens, especially between self and foreign ones (12, 23).
The internal representation of the environment can be regarded as a system of beliefs, or guesses, about pathogen frequencies. Formally, these beliefs can be represented in the form of a probability distribution function over pathogen frequencies, which the host implicitly computes using all of the information it has garnered over time. Optimally, these beliefs are computed by the rules of Bayesian sequential forecasting, by combining the memory of past encounters with knowledge of the stochastic rules under which the pathogenic environment evolves (Materials and Methods). Optimally, the belief distribution should be initialized at birth to reflect the steady-state distribution of the dynamics,
| [2] |
where . Upon encountering a pathogen at time , the prior belief distribution is combined with the likelihood of the observed pathogen to compute the postencounter belief according to Bayes rule (16):
| [3] |
Between encounters, the immune system should continue to update its beliefs by forecasting how pathogen frequencies change with time. The optimal way to do so is to project the old belief distribution forward in time using (16)
| [4] |
This prediction step, which is performed in the absence of any new information, relies on the immune system “knowing,” i.e., having learned over long evolutionary time scales, the probability laws governing the stochastic evolution of the environment—but not, of course, the actual path that it takes. In Results, we show how Eqs. 2–4 can be turned from abstract belief updates into dynamical equations for a well-adapting immune repertoire.
The Bayesian forecasting framework provides a broad account of the possible adaptive value of many features of the adaptive immune system without the need for additional assumptions. Immune memory formed after a pathogenic challenge is explained as an increase in optimal protection level resulting from an increase in estimated pathogen frequency, following Eq. 3. Attrition of immune memory is also adaptive, because it allows the immune repertoire to forget about previously seen pathogens which it should do in a dynamically changing environment (Eq. 4). Lastly, some of the biases in the recombination machinery and initial selection mechanisms (24) represent an evolutionary prior (Eq. 2) which tilts the naive repertoire toward important regions of antigenic space.
We are proposing an interpretive framework for understanding adaptive immunity as a scheme of sequential inference. This view provides two key insights. First, it confirms the intuition that new experience should be balanced against previous memory and against unknown threats in order for adaptive immunity to work well. Second, it suggests a particular dynamics of implicit belief updates that can globally reorganize the immune repertoire to minimize harm from the pathogenic environment. Going beyond these broad ideas, in Results, we analyze in detail a model for optimal immune prediction in which all these statements can be made mathematically precise. We also show a plausible implementation that the immune system could follow to approximate optimal Bayesian inference, and we compare the resulting dynamics with specific features of the adaptive immune system.
Results
A Lymphocyte Dynamics for Approximating Optimal Sequential Inference.
For concreteness, we consider a drift-diffusion model of environmental change (Eq. 11). The drift-diffusion model, while clearly a much-simplified model of real evolution, captures two key features of changing pathogenic environments: the coexistence of diverse pathogens and the temporal turnover of dominant pathogen strains. The aim of this model of pathogen evolution is not to provide a realistic description of short-term pathogen dynamics within a population such as during an epidemic, but rather to capture overall features of the long-term dynamics of many pathogens over a host’s lifetime. The drift-diffusion model is mathematically equivalent to a classical neutral stochastic evolution of pathogens (25) driven by genetic drift happening on a characteristic time scale and immigration from an external pool with immigration parameters (Eq. 11). Generally, pathogens are under selective pressure to evade host immunity, and strains are replaced faster than under the sole action of genetic drift. Matching the time scale of pathogen change to those observed experimentally, the model then provides a simple, effective description of the pathogen dynamics.
Can a plausible dynamics of lymphocyte receptor clones approximate the optimal repertoire dynamics? In particular, is there an approximate autonomous dynamics for the repertoire composition, which does not require access to the full latent high-dimensional belief distribution ? Consider the simple case of a logarithmic cost function, , and uniquely specific receptors, , in which the optimal match between the receptor and pathogen distributions (Eq. 1) is the identity, (ref. 15 and Materials and Methods).
For the pathogen dynamics of Eq. 10, we show, using a decomposition of into a mixture of Dirichlet distributions (SI Appendix, section 1B), that there exists an approximate but autonomous dynamics for the number of lymphocyte receptors . It takes the following simple form (parallel to Eqs. 2–4):
| [5] |
| [6] |
| [7] |
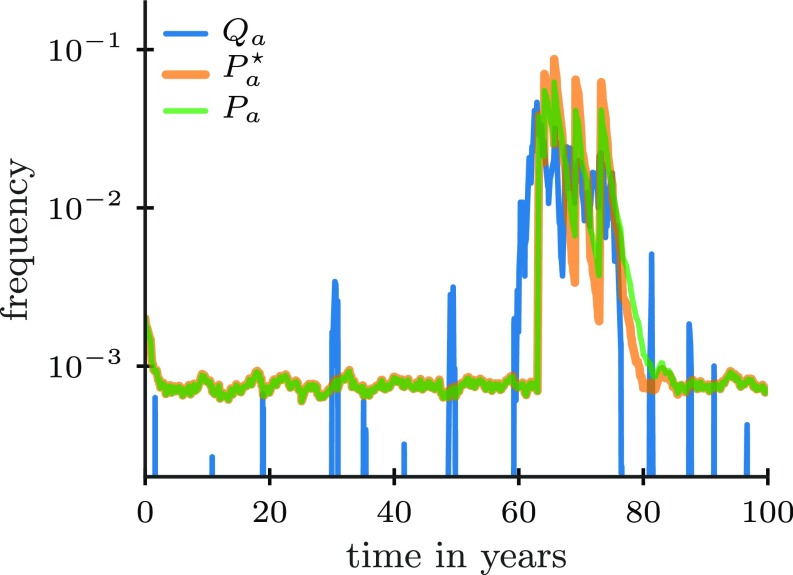
where is a scale factor controlling the total population size and where . Here and in the following, we use to denote the -norm of a vector . We compared these approximate dynamics to an exact solution computed by spectrally expanding the generator of the stochastic dynamics (SI Appendix, section 1C) and found that they approximate the optimal dynamics closely (Fig. 2).
Fig. 2.
A plausible repertoire dynamics can implement approximate Bayesian inference. Frequency of a pathogen over time (blue line) in a dynamically changing environment along with the exact optimal repertoire dynamics (orange line) and the approximate dynamics based on Eqs. 5–7 (, green line). The approximate dynamics closely follows the exact Bayesian dynamics implemented as described in SI Appendix, section 1C (, orange line). Parameters are as follows: , , year, /year, , and .
These dynamics have a plausible biological implementation. Each pathogen encounter leads to a fixed increment of the number of specific lymphocytes (Eq. 6), implying a regulation mechanism that controls the number of cell divisions upon clonal expansion as a function of precursor frequency. Through thymic output, the repertoire starts at and is renewed by the naive repertoire—encoded in (Eqs. 5 and 7). By definition, this naive repertoire is optimally tuned to the baseline frequencies of pathogens . The total number of lymphocytes regulates the time scale of cell decay and renewal (Eq. 7), which can be done through some global homeostatic mechanisms.
To gain intuition about what the dynamics of Eqs. 5–7 mean, let us define the rescaled variables . Eq. 6 increases by 1 whenever pathogen is sampled. can thus be interpreted as counting the number of times the different pathogens were sampled and their initial value as pseudocounts corresponding to the prior belief in the pathogen frequencies. In a changing environment, experience gained from previous encounters eventually loses its predictive power, and thus Eq. 7 discounts these old encounters, giving more relative weight to recent ones. If there are no new pathogen encounters, eventually relaxes back to the prior .
The assumption that can be relaxed, provided that the nonlinear relation between and (Eq. 1) remains approximately invertible, leading to autonomous dynamics of the lymphocyte clone sizes similar to Eqs. 5–7 (SI Appendix, section 2C).
Learnability of Pathogen Distribution Implies a Sparse Pathogenic Landscape.
The immune system must be prepared to protect us not just from one pathogen but a whole distribution of them. Even restricting recognition to short peptides and accounting for cross-reactivity (26), estimates based on precursor frequencies for common viruses give an effective antigen environment of size (27). How can the immune system learn anything useful about such a high-dimensional distribution from a limited number of pathogenic encounters? Naively, one might expect that the number of samples needed to learn the distribution of pathogens must be larger than of the number of pathogens, i.e., , where is the time over which learning takes place. Although little is known about the receptor–antigen encounter rate , this estimate suggests that the pathogenic environment is not easily learnable, and therefore memory has limited utility.
This apparent paradox can be resolved by the fact that the pathogenic environment may be sparse, meaning that only a small fraction of the possible pathogens are present at any given time. In our model of the pathogen dynamics, this sparsity is controlled by the parameter . In the scenario that we are considering, typical pathogen landscapes are drawn from the steady-state distribution of the immigration-drift dynamics, which is a Dirichlet distribution parametrized by (SI Appendix, section 1, Eq. S1). When is small, the distribution is peaked at , meaning that pathogen is absent a majority of the time. For instance, for uniform , the effective number of pathogens present at any given time is (SI Appendix, section 3C). Since the system only needs to learn about the pathogens that are present, the condition for efficient learning should naively be , which is much easier to achieve realistically for small .
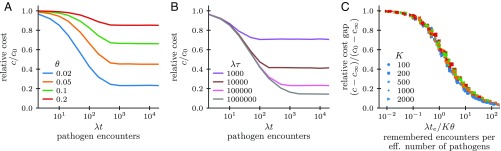
Our theory can be used to quantify the benefit of memory as a function of the different immunological parameters. We compute the optimized cost function and study how it decreases as a function of age, , relative to the cost at birth , as the organism learns from pathogen encounters. This relative cost also depends on the encounter rate , the size of the pathogenic space , the sparsity of pathogenic space , and time scale of change in the environment . Fig. 3A shows that the benefit of memory increases with pathogen sparsity—when is small, even a few encounters suffice to seed enough memory to reduce the cost of future infections. The cost saturates with age to a value , either because memory approaches optimality or because memory eventually gets discarded and renewed as the environment changes. Fast-changing environments lead to an earlier and higher saturation of the cost with age (Fig. 3B) since learning and prediction are limited by decorrelation of the environment. The pathogen dynamics is sped up when there are strong selection pressures to evade immunity. Faster dynamics decrease learning efficiency and in turn reduce selective pressures. The effective time scale should in practice be set by a coevolutionary balance between both effects.
Fig. 3.
Advantage of immunological memory depends on sufficient sampling. The mean expected cost of an infection in a changing environment is a function of the age of the organism , the time scale on which the environment changes, and the sparsity of the environment. (A) Relative cost as a function of age for environments with different sparsity (for fixed , K = 2,000). (B) Relative cost as a function of age for environments changing more or less rapidly (for fixed , K = 2,000). (C) Collapse of the data by plotting the relative cost gap against the number of samples per effective (eff.) dimension. Simulations with logarithmically spaced parameters in the ranges from 1 to 5,000, from 1 to 1,000,000, from 100 to 2,000, and from 0.01 to 0.2 (color-coded as in A). Cost is , and receptors are assumed to be uniquely specific, .
Analytical arguments show that in the limit of few samples the relative cost achievable in a static environment scales as (SI Appendix, section 3). In general, we find that the cost is a function of , where the effective time is defined via with being the vector of the encounter counts discussed above (SI Appendix, section 1D for derivation from Eqs. 6 and 7). Plotted in terms of this variable, the relative cost gap as function of time, , collapses onto a single curve for all parameter choices (Fig. 3C). Fig. 3C shows that the cost drops by a factor of , when . Thus, there is a substantial benefit to memory already when the effective number of encounters is comparable to the effective number of pathogens. At young ages (small ) or with slowly changing environments (large ), , and so this condition is simply , i.e., the total number of encounters should be comparable to effective number of pathogens that are present.
Optimal Attrition Time Scale.
Our theory suggests that there is an optimal time scale for forgetting about old infections which is related to the time scale over which the environment varies. Eq. 7 shows that memory should optimally be discounted on an effective time scale . Comparing this to the slowest time scale of environmental variation, (SI Appendix, Eq. 37), where , we have
| [8] |
The time scale on which old memories should be forgotten scales with the environmental correlation time scale. The two time scales are equivalent when the immune system has little information about the pathogenic environment (). Given the long time scales over which many relevant pathogens change, immune memory should generally be long-lived (with the time scale of decay being of the order of years or decades). Indeed, despite the relatively short life span of memory cells (28), constant balanced turnover keeps elevated levels of protection for decades after an infection, even in the absence of persistent antigens (29–31).
Interestingly, our theory predicts that memory should be discounted more quickly when the immune system has gathered more information (larger ). Using the mean-field equations for from the previous section, we can derive how the memory time scales at steady state at high sampling rate. Using that for large times holds in the high sampling rate limit, one can simplify the mean-field result to in steady state (). Combined with Eq. 8, follows, which shows that a larger sampling rate leads to a faster discounting of past evidence. This is reminiscent of results in optimal cellular signaling where there are similar trade-offs between noise averaging and responsiveness to changes in the input signal (32).
Memory Production in Sparse Environments Should Be Large and Decrease with Prior Exposure.
The theory can be used to make quantitative and testable predictions about the change in the level of protection that should follow a pathogen encounter. Consider an infection cost function that depends as a power law on the coverage, , with a cost exponent that sets how much attention the immune system should pay to recognizing rare threats. (Below, we will use the shorthand to indicate logarithmic cost.) Cost functions of this form can be motivated by considering the time to recognition of an exponentially growing antigen population by the immune system (15) or, alternatively, by considering the time delay of the expansion of the precursor cells to some fixed number of effector cells (SI Appendix, section 4).
In the simplest model for repertoire updates, recognition of pathogens leads to proliferation proportionally to the number of specific precursor cells, followed by a homeostatic decrease of the memory pool (18, 33). Thus, the fold change where , are times just before and after the encounter. By contrast, our Bayesian theory predicts that the fold change upon encountering pathogen should be
| [9] |
where depends on prior expectations about the antigenic environment and previous pathogen encounters (Materials and Methods). Setting gives the result for a logarithmic cost function.
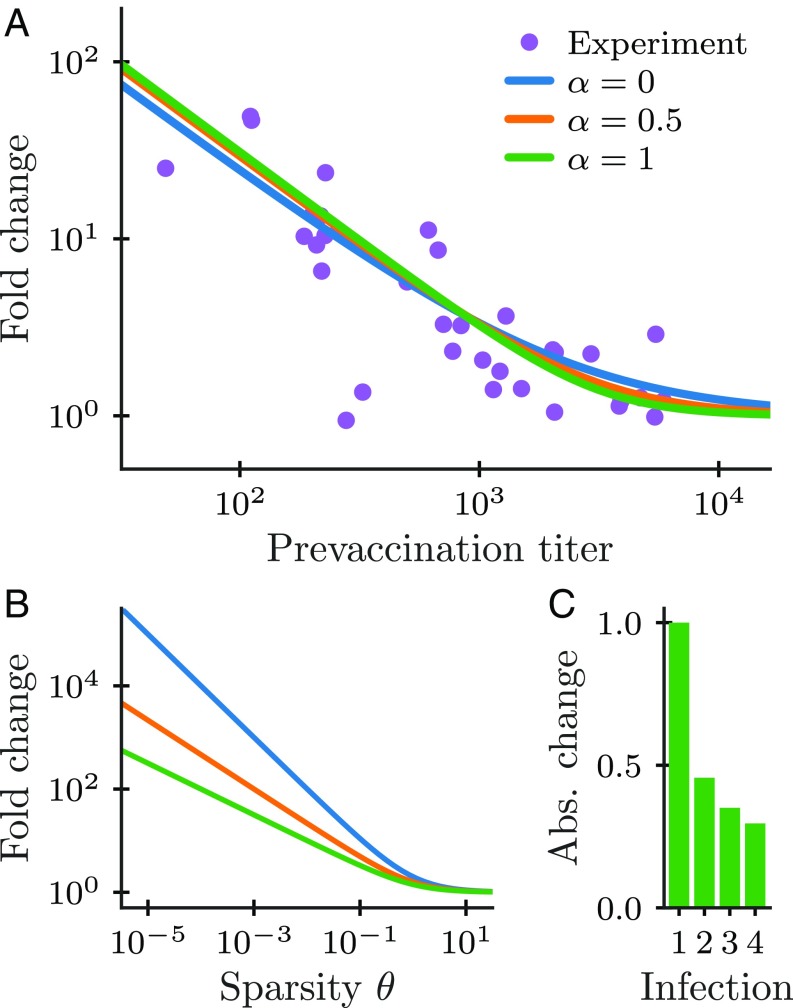
To understand this prediction, first consider the effect of a primary infection on a naive repertoire, , , and , where the receptors are uniquely specific (). In this case, (SI Appendix, section 2A) and Eq. 9 predicts a fold change of . We have argued previously that their learnability implies that pathogenic environments are sparse, i.e., . Therefore, we predict that primary antigenic encounters should lead to a large memory production. Experimentally, memory production typically leads to the proliferation of antigen-specific cells by a factor of 100- to 1,000-fold (14), in qualitative agreement with this prediction. Turning the argument around, such a large increase in protection upon an encounter is only adaptive in highly sparse environments. Quantitatively, it implies a sparsity parameter (here, taking for definiteness) (Fig. 4B). Combined with the estimate (27), this suggests that the effective number of pathogens at any given time ranges from to .
Fig. 4.
Changes in protection levels upon infections for cost functions ( indicates logarithmic cost). (A) The prediction of the Bayesian model (Eq. 9 for different ) closely fits experimental data on antibody titers, while a constant fold-change model (which would correspond to a straight horizontal line) does not. Data on prevaccination and postvaccination antibody titers against stem and head hemagglutinin epitopes following a booster vaccination with inactivated H5N1 in humans from ref. 34. (B) Optimal fold change of coverage for for all . The fold change increases with the sparsity of the environment controlled by parameter . (C) Absolute (Abs.) change in coverage for primary infection and reinfections (for ), normalized such that the change for the primary infection is 1 and neglecting attrition.
To test Eq. 9 on immunological data, we fit the Bayesian update model to experiments reporting fold changes in antigen titers upon booster vaccinations against influenza from ref. 34 (Fig. 4A) using least squares. Titers correspond to the concentration of antibodies that are specific to the antigen and can thus be viewed as an experimental estimate of . The optimal Bayesian strategy explains the data, accounting for the larger boosting at small prevaccination titers and showing no increase for large titers, while the proportional model predicts constant boosting for all titers. Similar experimental results have been reported for antibody titers before and after a shingles vaccination (35). Mechanistic models have been proposed to explain how the population dynamics of expanding lymphocytes might give rise to nonproportional boosting for both B and T cells (22, 33, 36, 37).
Interestingly, for T cells, Quiel et al. (38) have shown that fold expansion to peak cell numbers in an adoptive transfer experiment depends on the initial number of T cells as a power law with exponent . That scaling, which is for the peak expansion, predicts more expansion at high precursor number than Eq. 9, which is for memory production. This implies a nonlinear relationship between peak T-cell level and memory production, which further suppresses memory production at high precursor numbers. This prediction might be checked in experiments measuring memory production after infection clearance, as well as the expansion peak.
Long-Term Dynamics of a Well-Adapting Repertoire.
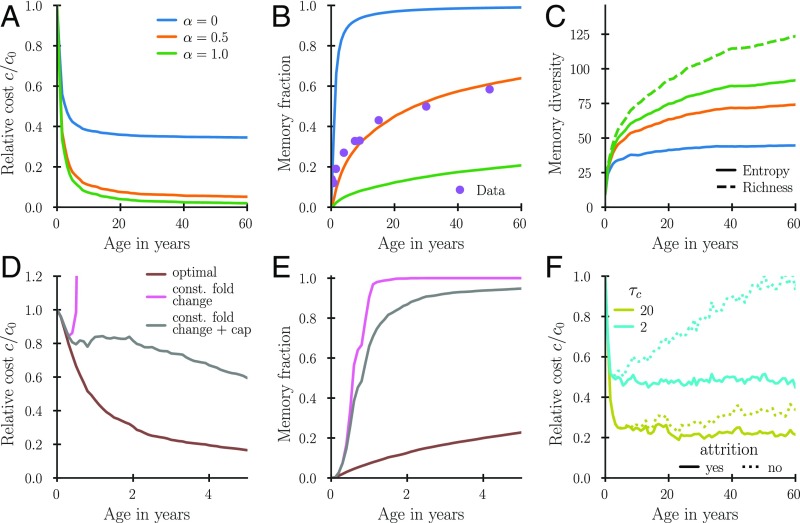
Our model makes predictions for the dynamics of growth and attrition of memory over time, with consequences for immunity and for the diversity of the immune repertoire. We quantify the dynamics in terms of a memory fraction defined as a sum of the coverage fractions over all previously encountered pathogens . The memory fraction measures the size of memory relative to the size of the whole immune repertoire. Early in life, every infection is new, and even modest increases in the memory fraction lead to large drops in infection susceptibility (measured by the expected cost of new infections in Fig. 5A). At the same time, the memory fraction increases rapidly (Fig. 5B), but the growth of memory slows as subsequent infections lead to less memory production following the optimal fold-change rule in Eq. 9 and as attrition begins to play a role. The fraction of the repertoire devoted to memory in midlife is largely determined by how the cost of infections scales with coverage. The observed memory fraction of at midlife suggests a cost exponent of (Fig. 5B). The diversity of the memory repertoire increases with time at a rate that slows with age (quantified in Fig. 5C by richness, which measures the number of unique specificities and the Shannon entropy of the repertoire frequency distribution).
Fig. 5.
Relative cost (A and D), memory-cell fraction (B and E), and memory diversity (C) as a function of age. A–C show long-term dynamics for a repertoire following optimal Bayesian update dynamics for three different . Memory diversity is plotted as richness, i.e., the number of unique memory specificities, as well as the exponential of Shannon entropy of the memory compartment defined for a probability distribution with as . D and E compare the optimal Bayesian dynamics with a constant fold change update by a factor of 30 and the same proportional update but with a cap to of the total fold expansion for any clone. (F) Comparison of the cost as a function of age for the optimal dynamics (solid lines) and a dynamics without attrition (dotted lines) for two environmental correlation time scales (SI Appendix, section 1C). Parameters are as follows: encounter rate , antigen space dimensionality , and antigen sparsity . In A–E, we used , and in D–F, . To reduce fluctuations, the statistics were averaged over multiple runs of the dynamics. Data in B are from refs. 39 and 40.
To gain insight into these dynamics of our model, we average the stochastic equations over the statistics of pathogen encounters. We show in SI Appendix, section 2B that this mean-field approximation yields a differential equation for the population fraction of different clones with two opposing contributions which balance alignment of the immune repertoire with the current pathogenic environment (i.e., memory production) against alignment with the long-term mean environment (i.e., attrition). Interestingly, the mean-field equation broadly coincides with dynamics that were proposed in ref. 15 to self-organize an optimal immune repertoire. The essential difference here is that the time scale of learning slows down with increasing experience following the rules of optimal sequential update in Eq. 9.
We then asked which features of the proposed repertoire dynamics are most relevant to ensure its effectiveness. How important is the negative correlation between fold expansion and prior immune levels, and how important is attrition? Furthermore, if the immune system follows Bayesian dynamics, it must have integrated on an evolutionary time scale a prior about composition and evolution of the pathogen environment through the parameters and —however, the prior may be inaccurate. How robust is the benefit of memory to imperfections of the host’s prior assumptions about pathogen evolution? To answer these questions, we compare the long-term immune-repertoire dynamics using the optimal Bayesian scheme to other simplified schemes. We find that a constant fold expansion dynamics quickly leads to very suboptimal repertoire compositions (Fig. 5D, pink line), since the exponential amplification of cells specific to recurrent threats quickly leads to a very large fraction of the repertoire consisting of memory of those pathogens (Fig. 5E, pink line). This suboptimality persists, even if we assume that some global regulation caps the constant fold expansion such that no individual receptor clone can take over all of the repertoire (Fig. 5 D and E, gray line). Thus, negative feedback in T-cell expansion to individual antigens is very important to maintain a properly balanced diverse repertoire. In contrast, within dynamics with a negative correlation, the precise levels of updating do not need to be finely tuned to the environmental statistics: Varying the assumed sparsity of the pathogen distribution, which controls fold expansion upon primary infection in the optimal dynamics, leads to a relatively modest deterioration of the convergence speed of the learning process (SI Appendix, Fig. S2A) and does not matter asymptotically (SI Appendix, Fig. S2B). Attrition does not matter at young age, but can play an important role for long-term adaptation to relatively rapidly changing pathogen distributions (Fig. 5F). However, the attrition time scale need not be finely tuned to get close to optimal dynamics (SI Appendix, Fig. S3).
Adapting a Cross-Reactive Repertoire.
Above, we described adaptation of immune repertoires in terms of changes in the effective coverage , where the cross-reactivity matrix reflects the ability of each receptor to recognize many antigens and also the propensity of each antigen to bind to many receptors (26). Because of cross-reactivity, each pathogen encounter should result in the expansion of not just one but potentially many receptor clones. Here, we ask how the optimal immune response is distributed among clones with different affinities.
Following Perelson and Oster (41), we will represent the interaction of receptors and antigens by embedding both in a multidimensional metric recognition “shape space,” where receptors are points surrounded by recognition balls. Antigens that fall within a ball’s radius will be recognized by the corresponding receptor. In this presentation, and are the coordinates of antigens and receptors, respectively, and their recognition propensity depends on their distance, .
Earlier sections have already discussed the optimal dynamics of the coverage , which is a convolution of the cross-reactivity matrix with the receptor clone distribution . Thus, the optimal dynamics of the clone distribution can be derived by deconvolving the cross-reactivity subject to the constraint that cannot be negative. Carrying out this analysis in SI Appendix, section 2C reveals a general qualitative phenomenon—competitive exclusion between clones expressed in the repertoire and their close neighbors within the cross-reactivity radius (SI Appendix, Fig. S4, blue line). This exclusion is not an assumption of the model, but rather stems from the optimal Bayesian theory. Given a receptor clone that covers one region of antigenic shape space, the global likelihood of detecting infections increases by placing other clones to cover other regions. This can be shown analytically when cross-reactivity is limited, memory updates are small in magnitude, and the pathogen distribution is assumed to be uncorrelated (SI Appendix, section 2C).
In general, the frequencies of pathogens might be correlated in antigenic space, for example, because mutations from a dominant strain give rise to new neighboring strains. An optimally adapting immune system should incorporate such correlations as a prior probability favoring smoothness of the pathogen distribution. Such priors work their way through the optimal belief update scheme that we have described and weaken the competitive exclusion between clones with overlapping cross-reactivity (SI Appendix, Fig. S4, orange line).
In general, when cross-reactivity is wide or the required clone fraction update is large, numerical analysis shows that achieving optimally predictive immunity after a pathogen encounter requires a global reorganization of the entire repertoire (SI Appendix, Fig. S5, blue line). There is no plausible mechanism for such a large-scale reorganization since it would involve up- and down-regulation, even of unspecific clones. However, in SI Appendix, section 2C we show that the optimal update can be well-approximated by changes just to the populations of specific clones with pathogen-binding propensities that exceed a threshold. The optimizing dynamics with this constraint exhibit strong competitive exclusion, where only the highest-affinity clones proliferate, while nearby clones with lower affinity are depleted from the repertoire (SI Appendix, Fig. S5, orange line). The local update rule provides protection that comes within 1% of the cost achievable by the best global update. Thus, reorganization of pathogen-specific receptor clone populations following an infection, as seen in vertebrates, can suffice to achieve near-optimal predictive adaptation of the immune repertoire.
Discussion
The adaptive immune system has long been viewed as a system for learning the pathogenic environment (10). We developed a mathematical framework in which this notion can be made precise. In particular, we derived a procedure for inferring the frequencies of pathogens undergoing an immigration-drift dynamics and showed how such inference might approximately be performed by a plausible population dynamics of lymphocyte clones. Additionally, we analyzed how quickly the immune system can learn about its environment and find that the antigenic environment must be effectively sparse to be learnable with a realistic rate of pathogen encounters.
The optimal repertoire dynamics recapitulate a number of properties of real adaptive immune systems. Two repertoire compartments emerge from the theory: memory, which encodes lived experience, and naive, which encodes prior expectations. Memory is effective in reducing harm from infection despite the high dimensionality of pathogenic space; having encountered circulating pathogens only once on average can reduce the overall cost of infections by half. The first encounter of a naive individual with a pathogen leads to a large response, which increases protection levels by several orders of magnitude. Memory production depends on prior protection levels and eventually is predicted to saturate, as seen in vaccination experiments (34). This saturation is predicted to lead to a sublinear increase with age of the fraction of the total repertoire taken up by memory, consistent with observations in human cohorts (39, 40).
Our work makes a number of concrete predictions amenable to further testing. We make quantitative predictions about how much memory should be formed following an infection, as a function of number of pathogens effectively present in the environment and the number of previous infections. These dependencies might be tested by using a comparative approach, which relates the amount of formed memory in different species to how many pathogens to which they are susceptible. Our model also makes predictions about changes in the size and diversity of the memory compartment with aging, which might be tested in future repertoire-sequencing studies similar to those done by Britanova et al. (42), by sorting cells into naive and memory types before sequencing. Ultimately, we hope that our work might help motivate studies with longitudinal tracking of the long-term repertoire dynamics in model organisms living in controlled pathogenic environments.
Our framework can be extended to incorporate additional constraints on immune-system function or further aspects of pathogen evolution. An example of a constraint would be to introduce a maximal rate of change in the optimization to model maximal cellular division and death rates. A more complex pathogen dynamics might model explicitly their mutational dynamics in antigenic space. Such dynamics will lead to correlations in the pathogen distribution, which we showed will influence the structure of the optimal conjugate repertoire. In particular, the optimal response should spread around the currently dominant antigens to also provide protection against potential future mutations. Hypermutations in B cells may play a role in this diversification, in addition to their known function of generating receptors with increased affinity for antigens of current interest. It would also be interesting to extend our framework to other immune-defense mechanisms, including innate immunity, where the role of memory has received recent attention (43).
Although our study was motivated by the adaptive immune system, some of our main results extend to other statistical inference problems. We have extended earlier results on exactly computable solutions to the stochastic filtering problem for Wright–Fisher diffusion processes (44) to derive an efficient approximate inference procedure. This procedure might be of use in other contexts where changing distributions must be inferred from samples at different time points, e.g., in population genetics. Additionally, we have derived the convergence rate for Bayesian inference of categorical distributions in high dimensions in the undersampled regime, showing that effectively sparse distributions can be inferred much more quickly. These results add to the growing literature on high-dimensional inference from few samples (45, 46), which has arisen in the context of the big-data revolution.
We propose that the adaptive immune system balances integration of new evidence against prior knowledge, while discounting previous observations to account for environmental change. Similar frameworks have been developed for other biological systems. In neuroscience, leaky integration of cues has been proposed as an adaptive mechanism to discount old observations in change-point detection tasks (47, 48), and close-to-optimal accumulation and discounting of evidence has been reported in a behavioral study of rat decision-making in dynamic environments (49). Inference from temporally sparse sampling has been considered in the framework of infotaxis, which is relevant for olfactory navigation (50). In the context of immunity, related ideas about inference and prediction of pathogen dynamics have been used to predict flu-strain and cancer-neoantigen evolution in silico (51, 52). Finally, ideas similar to those developed here could be used in ecology or microbiome studies to reconstruct evolutionary or ecological trajectories of population dynamics from incomplete sampling of data at a finite number of time points, e.g., from animal sightings or metagenomics.
Materials and Methods
Modeling Pathogen Dynamics by an Immigration-Drift Process.
In our model, we describe the stochastic dynamics of the pathogenic environment (Fig. 1A) by a Fokker–Planck equation for the conditional probability distribution
| [10] |
where is a differential operator acting on that controls the dynamics. For concreteness, we consider a population that changes due to genetic drift and immigration from an external reservoir, which we describe by a Wright–Fisher diffusion equation (53, 54)
| [11] |
where sets the time scale of dynamics, is a -dimensional vector of immigration rates, and is the Kronecker delta, which is 1 if and 0 otherwise. To efficiently simulate trajectories according to this dynamics, we sample the new distribution of frequencies directly from the transition density of the stochastic process as described in SI Appendix, section 1E. This dynamics retains key features of real pathogen environments. First, at a given point in time, the environment contains many different pathogens with different frequencies determined by genetic drift and immigration. Second, the dominant pathogens change over time, such as is the case for many viruses, e.g., the flu or HIV.
Minimizing the Cost of Infection.
To solve the optimization problem Eq. 1 analytically, a set of necessary conditions for optimality, the so-called Karush–Kuhn–Tucker conditions, can be derived. When all receptors are present at a nonzero frequency in the optimal repertoire , these conditions imply (15)
| [12] |
where is set by the condition . If we further simplify the problem by assuming that there is no cross-reactivity between different pathogens and by considering power-law cost functions, then this simplifies to the explicit solution
| [13] |
where is a normalization constant. Other cases are discussed in detail in ref. 15, including how to solve the optimization problem numerically by using a projected gradient algorithm in the general case.
Change in Protection upon a Pathogen Encounter.
The inference dynamics induces via the mapping from to (Eq. 1) a dynamics of an optimally adapting immune repertoire. To get intuition we derive how the coverage changes in a simple setting in which Eq. 13 holds (further cases are considered in SI Appendix, section 2).
Rewriting Eq. 6 in terms of the expected pathogen frequencies , we obtain an update equation for the expected frequencies upon encounter of antigen as
| [14] |
where to simplify notations, we use , and where . By using Eq. 13, it follows that coverages are updated as
| [15] |
Defining and neglecting the change in normalization which is of order relative to the update size, we obtain Eq. 9. To fit the dataset, we note that a proportional rescaling of by a factor can be subsumed within the model by redefining . Therefore, the scaling of to an antibody titer can be subsumed within .
Supplementary Material
Acknowledgments
The work was supported by European Research Council Starting Grant 306312, a Lewis–Sigler fellowship (to A.M.), Simons Foundation Mathematical Modeling of Living Systems Grant 400425, and NSF Grant PHY-1734030. Work on this project at the Aspen Center for Physics was supported by NSF Grant PHY-1607611.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812810116/-/DCSupplemental.
References
- 1.Perkins TJ, Swain PS. Strategies for cellular decision-making. Mol Syst Biol. 2009;5:326. doi: 10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi TJ. Implementation of dynamic Bayesian decision making by intracellular kinetics. Phys Rev Lett. 2010;104:228104. doi: 10.1103/PhysRevLett.104.228104. [DOI] [PubMed] [Google Scholar]
- 3.Siggia ED, Vergassola M. Decisions on the fly in cellular sensory systems. Proc Natl Acad Sci USA. 2013;110:E3704–E3712. doi: 10.1073/pnas.1314081110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivak DA, Thomson M. Environmental statistics and optimal regulation. PLoS Comput Biol. 2014;10:e1003826. doi: 10.1371/journal.pcbi.1003826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeWeese MR, Zador AM. Asymmetric dynamics in optimal variance adaptation. Neural Comput. 1998;10:1179–1202. [Google Scholar]
- 6.Beck JM, et al. Probabilistic population codes for Bayesian decision making. Neuron. 2008;60:1142–1152. doi: 10.1016/j.neuron.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deneve S. Bayesian spiking neurons I: Inference. Neural Comput. 2008;20:91–117. doi: 10.1162/neco.2008.20.1.91. [DOI] [PubMed] [Google Scholar]
- 8.Wark B, Fairhall A, Rieke F. Timescales of inference in visual adaptation. Neuron. 2009;61:750–761. doi: 10.1016/j.neuron.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy K, Travers P, Walport M. Janeway’s Immunobiology. 7th Ed. Vol 2 Garland Science; Abingdon, UK: 2001. [Google Scholar]
- 10.Farmer JD, Packard NH, Perelson AS. The immune system, adaptation, and machine learning. Phys D. 1986;22:187–204. [Google Scholar]
- 11.Ahmed R, Gray D. Immunological memory and protective immunity: Understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 12.Perelson AS, Weisbuch G. Immunology for physicists. Rev Mod Phys. 1997;69:1219–1268. [Google Scholar]
- 13.Farber DL, Yudanin NA, Restifo NP. Human memory T cells: Generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchholz VR, Schumacher TNM, Busch DH. T cell fate at the single-cell level. Annu Rev Immunol. 2016;34:65–92. doi: 10.1146/annurev-immunol-032414-112014. [DOI] [PubMed] [Google Scholar]
- 15.Mayer A, Balasubramanian V, Mora T, Walczak AMA. How a well-adapted immune system is organized. Proc Natl Acad Sci USA. 2015;112:5950–5955. doi: 10.1073/pnas.1421827112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen ZHE. Bayesian filtering: From Kalman filters to particle filters, and beyond. Statistics. 2003;182:1–69. [Google Scholar]
- 17.Deem MW, Lee HY. Sequence space localization in the immune system response to vaccination and disease. Phys Rev Lett. 2003;91:068101, and erratum (2003) 91:229902. doi: 10.1103/PhysRevLett.91.068101. [DOI] [PubMed] [Google Scholar]
- 18.Antia R, Ganusov VV, Ahmed R. The role of models in understanding CD8+ T-cell memory. Nat Rev Immunol. 2005;5:101–111. doi: 10.1038/nri1550. [DOI] [PubMed] [Google Scholar]
- 19.François P, Altan-Bonnet G. The case for absolute ligand discrimination: Modeling information processing and decision by immune T cells. J Stat Phys. 2016;162:1130–1152. [Google Scholar]
- 20.George JT, Kessler DA, Levine H. Effects of thymic selection on T cell recognition of foreign and tumor antigenic peptides. Proc Natl Acad Sci USA. 2017;114:E7875–E7881. doi: 10.1073/pnas.1708573114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty AK. A perspective on the role of computational models in immunology. Annu Rev Immunol. 2017;35:403–439. doi: 10.1146/annurev-immunol-041015-055325. [DOI] [PubMed] [Google Scholar]
- 22.Mayer A, Zhang Y, Perelson AS, Wingreen NS. Regulation of T cell expansion by antigen presentation dynamics. Proc Natl Acad Sci USA. 2019;116:5914–5919. doi: 10.1073/pnas.1812800116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Percus JK, Percus OE, Perelson AS. Predicting the size of the T-cell receptor and antibody combining region from consideration of efficient self-nonself discrimination. Immunology. 1993;90:1691–1695. doi: 10.1073/pnas.90.5.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mora T, Walcak A. Systems Immunology: An Introduction to Modeling Methods for Scientists. Taylor & Francis; Boca Raton, FL: 2019. Quantifying lymphocyte receptor diversity. [Google Scholar]
- 25.Kimura M. Diffusion models in population genetics. J Appl Probab. 1964;1:177–232. [Google Scholar]
- 26.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 27.Zarnitsyna VI, Evavold BD, Schoettle LN, Blattman JN, Antia R. Estimating the diversity, completeness, and cross-reactivity of the T cell repertoire. Front Immunol. 2013;4:485. doi: 10.3389/fimmu.2013.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antia R, Pilyugin SS, Ahmed R. Models of immune memory: On the role of cross-reactive stimulation, competition, and homeostasis in maintaining immune memory. Proc Natl Acad Sci USA. 1998;95:14926–14931. doi: 10.1073/pnas.95.25.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- 30.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macallan DC, Borghans JA, Asquith B. Human T cell memory: A dynamic view. Vaccines. 2017;5:E5. doi: 10.3390/vaccines5010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker NB, Mugler A, Ten Wolde PR. Optimal prediction by cellular signaling networks. Phys Rev Lett. 2015;115:258103. doi: 10.1103/PhysRevLett.115.258103. [DOI] [PubMed] [Google Scholar]
- 33.De Boer RJ, Perelson AS. Antigen-stimulated CD4 T cell expansion can be limited by their grazing of peptide-MHC complexes. J Immunol. 2013;190:5454–5458. doi: 10.4049/jimmunol.1203569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellebedy A, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci USA. 2014;111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, et al. Metabolic phenotypes of response to vaccination in humans. Cell. 2017;169:862–877.e17. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bocharov G, et al. Feedback regulation of proliferation vs. differentiation rates explains the dependence of CD4 T-cell expansion on precursor number. Proc Natl Acad Sci USA. 2011;108:3318–3323. doi: 10.1073/pnas.1019706108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarnitsyna VI, Lavine J, Ellebedy A, Ahmed R, Antia R. Multi-epitope models explain how pre-existing antibodies affect the generation of broadly protective responses to influenza. PLoS Pathog. 2016;12:e1005692. doi: 10.1371/journal.ppat.1005692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quiel J, et al. Antigen-stimulated CD4 T-cell expansion is inversely and log-linearly related to precursor number. Proc Natl Acad Sci USA. 2011;108:3312–3317. doi: 10.1073/pnas.1018525108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saule P, et al. Accumulation of memory T cells from childhood to old age: Central and effector memory cells in CD4+ versus effector memory and terminally differentiated memory cells in CD8+ compartment. Mech Ageing Dev. 2006;127:274–281. doi: 10.1016/j.mad.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Shearer WT, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: The Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112:973–980. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Perelson AS, Oster GF. Theoretical studies of clonal selection: Minimal antibody repertoire size and reliability of self-non-self discrimination. J Theor Biol. 1979;81:645–670. doi: 10.1016/0022-5193(79)90275-3. [DOI] [PubMed] [Google Scholar]
- 42.Britanova OV, et al. Dynamics of individual T cell repertoires: From cord blood to centenarians. J Immunol. 2016;196:5005–5013. doi: 10.4049/jimmunol.1600005. [DOI] [PubMed] [Google Scholar]
- 43.Netea MG, et al. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaleyat-Maurel M, Genon-Catalot V. 2007, July 4. Filtering the Wright-Fisher diffusion. arXiv:0707.0537.
- 45.Johnstone IM, Titterington DM. Statistical challenges of high-dimensional data. Philos Trans A Math Phys Eng Sci. 2009;367:4237–4253. doi: 10.1098/rsta.2009.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Advani M, Ganguli S. Statistical mechanics of optimal convex inference in high dimensions. Phys Rev X. 2016;6:031034. [Google Scholar]
- 47.Glaze CM, Kable JW, Gold JI. Normative evidence accumulation in unpredictable environments. eLIFE. 2015;4:e08825. doi: 10.7554/eLife.08825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glaze CM, Filipowicz ALS, Kable JW, Balasubramanian V, Gold JI. A bias-variance trade-off governs individual differences in on-line learning in an unpredictable environment. Nat Hum Behav. 2018;2:213. [Google Scholar]
- 49.Piet AT, El Hady A, Brody CD. Rats adopt the optimal timescale for evidence integration in a dynamic environment. Nat Commun. 2018;9:4265. doi: 10.1038/s41467-018-06561-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vergassola M, Villermaux E, Shraiman BI. ‘Infotaxis’ as a strategy for searching without gradients. Nature. 2007;445:406–409. doi: 10.1038/nature05464. [DOI] [PubMed] [Google Scholar]
- 51.Łuksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- 52.Luksza M, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551:517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Etheridge A. 2012. Some Mathematical Models from Population Genetics, Lecture Notes in Mathematics (Springer, Berlin)
- 54.Griffiths RC, Spano D. 2010, March 24. Diffusion processes and coalescent trees. arXiv:1003.4650.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.