Significance
A growing body of literature has shown that, aside from carrying genetic information, both nuclear and mitochondrial DNA can be released by innate immune cells and promote inflammatory responses. Here we show that when CD4+ T lymphocytes, key orchestrators of adaptive immunity, are activated, they form a complex extracellular architecture composed of oxidized threads of DNA that provide autocrine costimulatory signals to T cells. We named these DNA extrusions “T helper-released extracellular DNA” (THREDs). We suggest that inhibiting this inflammatory process might represent a new therapeutic modality for CD4+ T cell-mediated autoimmune disorders, such as experimental autoimmune encephalomyelitis, the varied forms of which provide animal models for multiple sclerosis and neuromyelitis optica.
Keywords: CD4+ T cells, DNA threads, autoimmunity, EAE, multiple sclerosis
Abstract
The extrusion of DNA traps contributes to a key mechanism in which innate immune cells clear pathogens or induce sterile inflammation. Here we provide evidence that CD4+ T cells, a critical regulator of adaptive immunity, release extracellular threads of DNA on activation. These DNA extrusions convey autocrine costimulatory signals to T lymphocytes and can be detected in lymph nodes isolated during the priming phase of experimental autoimmune encephalomyelitis (EAE), a CD4+ T cell-driven mouse model of multiple sclerosis. Pharmacologic inhibition of mitochondrial reactive oxygen species (mtROS) abolishes the extrusion of DNA by CD4+ T cells, reducing cytokine production in vitro and T cell priming against myelin in vivo. Moreover, mtROS blockade during established EAE markedly ameliorates disease severity, dampening autoimmune inflammation of the central nervous system. Taken together, these experimental results elucidate a mechanism of intrinsic immune costimulation mediated by DNA threads released by activated T helper cells, and identify a potential therapeutic target for such disorders as multiple sclerosis, neuromyelitis optica, and CD4+ T cell-mediated disorders.
Cells belonging to the innate arm of the immune system release fibers of extracellular DNA, which support pathogen clearance or promote sterile inflammation in autoimmune disorders, such as lupus and vasculitis (1–4). The formation of neutrophil extracellular traps (NETs) has been described as a peculiar form of neutrophilic death, called suicidal NETosis, and is characterized by the extrusion of decondensed chromatin and granule proteins (5). An early and vital release of NETs, composed mainly of mitochondrial DNA (mtDNA), has also been reported (5, 6). This latter modality resembles the catapult-like release of mtDNA, which has been originally identified in eosinophils and is independent of cell death (2).
More recently, B cells have been shown to extrude mtDNA webs in vitro following exposure to synthetic CpG; however, there is no clear indication of the occurrence of this process in vivo or of its physiological significance (7). In this work, we report that CD4+ T lymphocytes, key regulator cells of adaptive immunity, release extracellular threads of DNA and investigated their specific biologic activity in vitro and in vivo in experimental autoimmune encephalomyelitis (EAE), mouse model of CD4+ T cell-mediated autoimmunity against the central nervous system (CNS) mimicking aspects of multiple sclerosis (MS) and neuromyelitis optica (NMO).
Results
CD4+ T Cells Release Extracellular Threads of DNA on Activation.
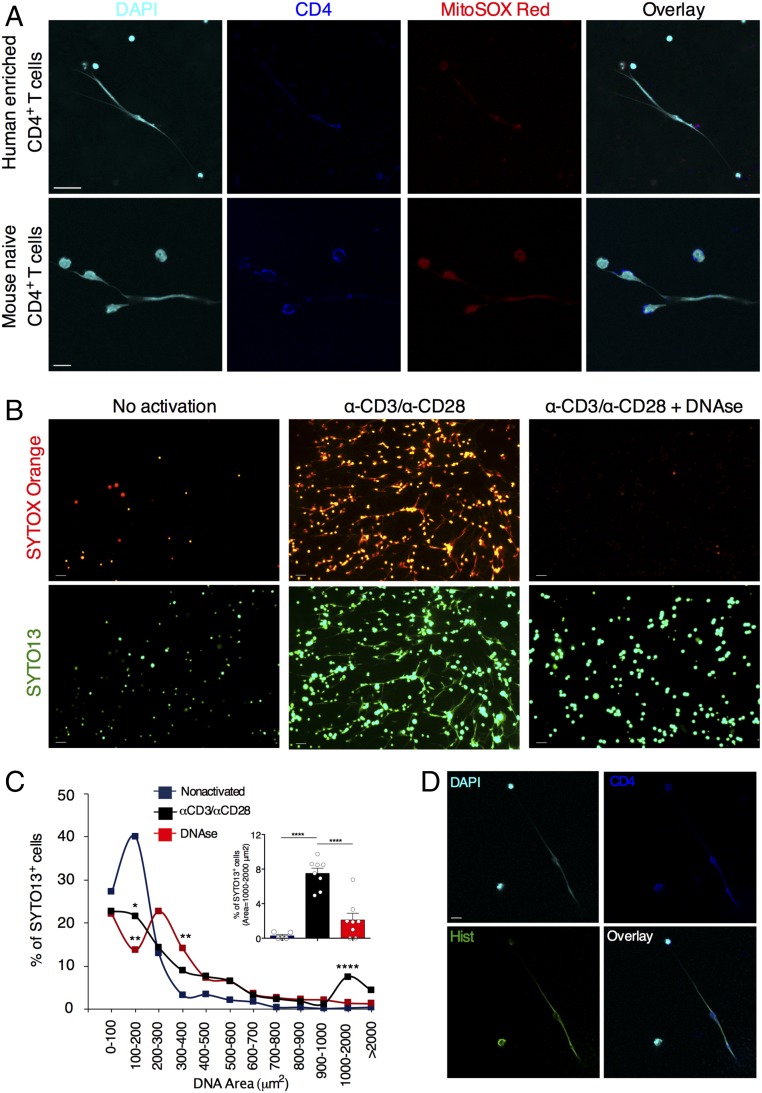
To evaluate whether T lymphocytes extrude DNA fibers, we seeded human CD4+ T cells enriched from buffy coats suspensions or highly purified mouse naïve CD4+ T cells on poly-d-lysine–coated glass and activated them with anti-CD3/anti-CD28 antibodies (Abs) for 24 h. Confocal microscopy analysis revealed the presence of numerous filaments of DNA ejected by CD4+ cells with a disrupted cell membrane and connected to other CD4+ T lymphocytes with an intact morphology (Fig. 1A). These DNA extrusions also costained for MitoSOX Red, a mitochondria-targeted probe that emits red fluorescence when oxidized by reactive oxygen species (ROS), suggesting an oxidized status of these nucleic acid filaments (Fig. 1A). We could observe both narrow threads and thicker fibers of DNA, dotted with the CD4+ marker (Fig. 1A). We did not observe extracellular threads of DNA in cultures of nonactivated CD4+ T lymphocytes or in samples activated and exposed to DNase (SI Appendix, Fig. S1 A and B).
Fig. 1.
CD4+ T cells release extracellular threads of DNA on activation. (A) Representative confocal immunofluorescence microscopy images of human CD4+ T cells enriched from buffy coats or mouse naïve CD4+ T cells activated with anti-CD3 (α-CD3) or α-CD28 Ab for 24 h. Cells were fixed with 4% PFA and stained for DNA (DAPI; cyan), CD4 (blue), and MitoSOX (red). (Scale bars: Upper, 25 μm; Lower, 10 μm.) (B) Fluorescence micrographs of mouse naïve CD4+ T cells left nonactivated or activated (24 h) with α-CD3/α-CD28 Abs in the absence or presence of DNase. Nucleic acids were labeled with SYTOX Orange and SYTO13 (green). (Scale bars: 20 μm.) (C) Quantification of DNA threads released in B. *P < 0.05; **P < 0.005; ****P < 0.0001 versus nonactivated. P values were determined by one-way ANOVA with Tukey’s multiple comparisons test (D) Representative confocal immunofluorescence microscopy images of mouse naïve CD4+ T cells activated with α-CD3/α-CD28 for 24 h. Cells were fixed with 4% PFA and stained for DNA (DAPI; cyan), CD4 (blue), and histones (green). (Scale bar: 10 μm.) The data in A–D are representative of three independent experiments.
In another set of experiments, mouse naïve CD4+ T cells were cultured with the vital nucleic acid dyes Sytox Orange and SYTO13, which are live cell-impermeable and live cell-permeant, respectively, and analyzed them by fluorescence microscopy. We found numerous Sytox Orange+/SYTO13+ filaments of nucleic acids with an extracellular localization on activation (Fig. 1B). Of note, DNase treatment dissolved most Sytox Orange staining but not SYTO13 staining, suggesting that activated CD4+ T cells were enwrapped into a tangle of extracellular Sytox Orange+ DNA (Fig. 1B). A few activated lymphocytes were labeled with Sytox Orange after exposure to DNase, probably due to cell death. Extrusions of DNA were quantified with software-assisted analysis (3, 8), by calculating the percentages of SYTO13+ cells within a given DNA area (Fig. 1C). Nonactivated T cells exhibited small and narrowly distributed SYTO13+ nuclear areas (Fig. 1C and SI Appendix, Fig. S1C). Conversely, activated T lymphocytes displayed a wide range of nuclear areas, with a SYTO13+ peak between 1 and 2 × 103 μm2, typical of extruded DNA, which was abolished by DNase digestion (Fig. 1C). The fragmentation of the released nucleic acids mediated by DNase resulted in a SYTO13+ peak between 3 and 4 × 102 μm2 (Fig. 1C and SI Appendix, Fig. S1C). We then evaluated whether DNA extrusion was associated with cell death processes in mouse CD4+ T cell cultures. Interestingly, we found that thicker DNA fibers were coated with histones (Fig. 1D), indicating that the activation of naïve CD4+ T cells can also trigger the release of DNA resembling suicidal NETosis. No extracellular staining of histones or DNA was observed in nonactivated samples (SI Appendix, Fig. S1D). Overall, our analysis shows that on activation, both human and mouse CD4+ T cells extrude extracellular threads of DNA, which we term T helper-released extracellular DNAs (THREDs).
THREDs Provide Autocrine Costimulation to CD4+ T Cells.
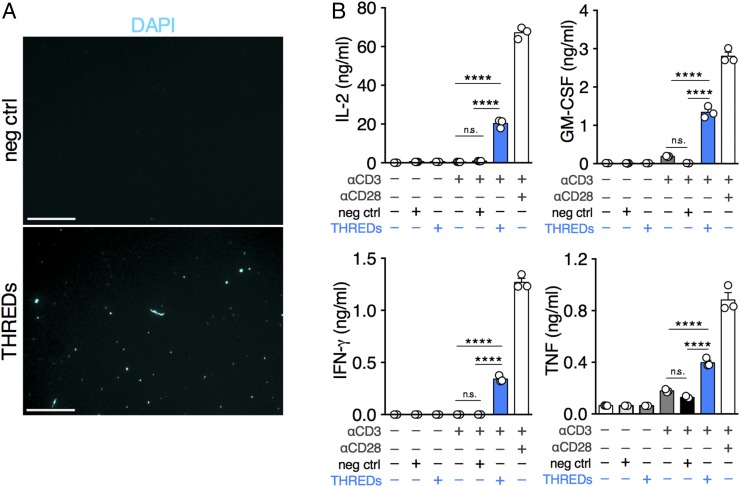
We next sought to understand whether the release of extracellular DNA fibers has any functional significance for CD4+ T cell activity. Synthetic CpG and non-CpG oligodeoxynucleotides have been shown to induce costimulation of T cells (9, 10); therefore, we speculated that self-DNA extruded by activated CD4+ T lymphocytes might support the full expression of T cell effector functions by conveying costimulatory signals in an autocrine fashion. To explore this possibility, extracellular DNA was isolated as previously described for NETs (11), by applying the same purification procedure to supernatants from both activated and resting mouse CD4+ T cells. The extraction products were then seeded overnight at 37 °C on poly-d-lysine–coated glass and analyzed by staining with DAPI. In the extract from stimulated T cells (Fig. 2A, Lower, THREDs), we detected numerous spots of DNA with occasionally unrolled fibers, even though we could not visualize the same structural organization shown in Fig. 1B where, directly released by T lymphocytes, DNA formed a narrow-mesh web among the cells. Virtually no DNA spots were detected in extracts from nonactivated cells (Fig. 2A, Upper, negative control). In parallel cultures, mouse naïve CD4+ T cells were stimulated with anti-CD3 Abs alone or in combination with negative control, THREDs, or anti-CD28 Abs (as a positive control). We found that THREDs were able to induce significantly increased production of IL-2, GM-CSF, IFN-γ, and TNF-α compared with cells activated with anti-CD3 alone or in combination with negative control (Fig. 2B). Treatment of CD4+ T cells with THREDs or negative control in the absence of anti-CD3 Abs did not result in the secretion of any cytokine. These results suggest that THREDs provide a second signal to CD4+ T lymphocytes.
Fig. 2.
THREDs provide autocrine costimulation to CD4+ T cells. (A) Immunofluorescence for extracellular DNA (DAPI; cyan) isolated from resting (negative control [neg ctrl]) or activated (THREDs) mouse naïve CD4+ T cells and spotted on poly-d-lysine–coated plates. (Scale bars: 25 μm.) (B) Cytokine production by mouse naïve CD4+ T cells left unstimulated or activated with α-CD3 for 96 h in combination with negative control, THREDs, or α-CD28 Abs. Data are mean ± SEM of triplicate wells pooled and tested in triplicate. ****P < 0.0001. P values were determined by one-way ANOVA with Tukey’s multiple comparisons test. The data in A and B are representative of three independent experiments.
Release of THREDs Is Reliant on ROS Production.
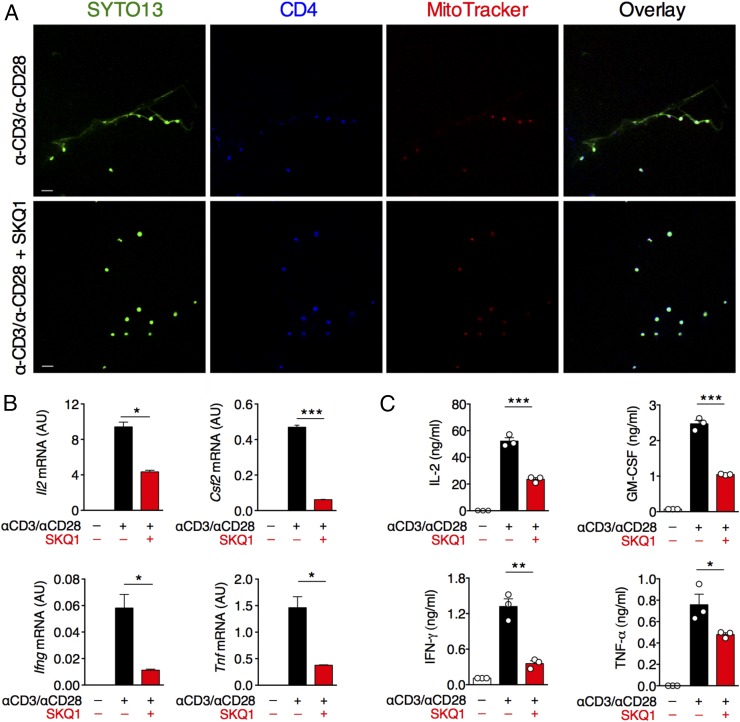
The extrusion of NETs has been shown to depend on so-called “burst” of ROS, catalyzed mainly in neutrophils by NADPH oxidase (6). Because T cells express a phagocyte-type NADPH oxidase (12), we evaluated whether the generation of THREDs depends on this enzyme. Treatment of activated CD4+ T cells with diphenyleneiodonium (DPI), an NADPH inhibitor, reduced the T cell release of DNA filaments (SI Appendix, Fig. S2). Mitochondria have been recently suggested as a crucial site for ROS production, and mitochondrial ROS (mtROS) also have been implicated in the formation of NETs (6). THREDs stained with both MitoSOX Red (Fig. 1A) and MitoTracker, a dye that labels mitochondria (Fig. 3A). Therefore, we assessed the specific involvement of mtROS in the extrusion of DNA by CD4+ T lymphocytes, by exposing activated CD4+ T cells to a nontoxic dose of SKQ1 (SI Appendix, Fig. S3B), a potent mitochondria-targeted ROS scavenger (13). We observed that mtROS inhibition hindered the release of THREDs (Fig. 3A). Inhibition of THREDs extrusion by SKQ1 was not associated with any modification of either T cell proliferation (SI Appendix, Fig. S3A) or expression of the activation markers CD25 and CD69 (SI Appendix, Fig. S3 C and D), but resulted in a significant reduction in the secretion of cytokines, such as IL-2, GM-CSF, IFN-γ, and TNF-α at both transcript and protein levels (Fig. 3 B and C). We did not detect IL-17A, IL-4, IL-10, or IL-1β in supernatants of cultured naïve T cells (SI Appendix, Fig. S3A).
Fig. 3.
The release of THREDs depends on mtROS, and their inhibition is associated with reduced production of inflammatory cytokines. (A) Mouse naïve CD4+ T cells were left nonactivated or activated as depicted with or without mtROS inhibitor SKQ1. After 24 h, cells were analyzed by confocal microscopy with staining for DNA (SYTO13, green), CD4 (blue), and MitoTracker (red). (Scale bars: 20 μm.) (B and C) Parallel cultures were analyzed for gene expression (B) and cytokine release (C) after 72 h of stimulation. In B, expression of target genes is presented in arbitrary units (AU), as a percentage of the housekeeping gene Gapdh ± SD. In C, cytokine production is expressed as mean ± SEM of quintuplicate wells pooled and tested in triplicate. *P < 0.05; **P < 0.005; ***P ≤ 0.001. P values were determined by the two-tailed unpaired t test. The data in A–C are representative of three independent experiments.
THREDs Are Detectable in the Lymph Nodes of EAE Mice and Can Be Inhibited in Vivo by mtROS Blockade.
To explore the occurrence of this phenomenon in vivo, we investigated the formation of THREDs and their dependence on mtROS in the priming phase of chronic EAE (14). In this model, immunization of C57BL/6 mice with myelin oligodendrocyte glycoprotein peptide 35–55 (MOG35–55) and adjuvants results in the expansion of MOG35–55-reactive CD4+ Th1/Th17 cells (priming phase), which orchestrate an autoimmune reaction against myelin in the CNS, culminating in paralysis (14–16).
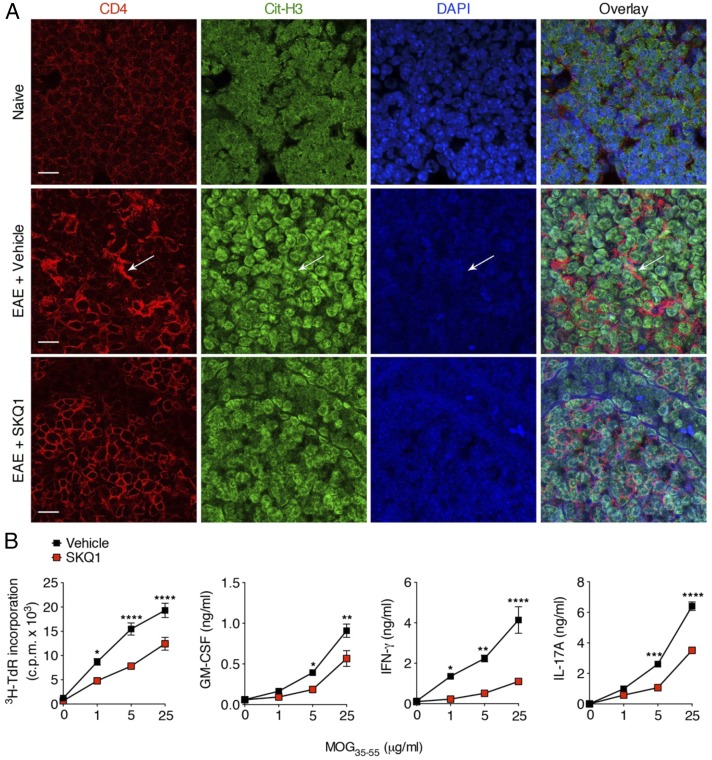
We induced EAE and injected mice intraperitoneally with vehicle or SKQ1 from day 0 (time of disease induction) to 7 d postimmunization (dpi). In lymph nodes from vehicle-treated EAE mice at 7 dpi, several areas of extracellular deposition of DNA were detected, often in colocalization with extracellular staining for citrullinated-histone H3 and in close proximity to CD4+ cells (Fig. 4A). Of note, marked reductions in extracellular DNA and citrullinated-histone H3 staining were found in the lymph nodes of SKQ1-treated EAE mice, and no extracellular staining for DNA or citrullinated histone H3 was observed in the lymph nodes of naïve mice (Fig. 4A). Interestingly, the decrease in extracellular DNA deposits in the lymph nodes of SKQ1-treated mice was paralleled by a reduced proliferative response and reduced secretion of proinflammatory cytokines (i.e., GM-CSF, IFN-γ, and IL-17A) by lymph node cells (LNCs) on ex vivo restimulation with MOG35–55 compared with LNCs from vehicle-treated mice (Fig. 4B).
Fig. 4.
Inhibition of mtROS decreases the deposition of extracellular DNA in the lymph nodes of EAE mice and reduces T cell priming against MOG35–55. (A) Representative confocal immunofluorescence microscopy images of inguinal lymph node sections derived from naïve mice or MOG35–55-immunized mice injected i.p. with vehicle or SKQ1 on day 0 to day 7 dpi (n = 3 mice/group). Sections were stained for CD4 (red), citrullinated histone H3 (Cit-H3; green), and DNA (DAPI; blue). The arrow indicates extracellular deposition of DNA and citrullinated histones in nearby CD4+ cells. (Scale bars: 20 μm.) (B) LNCs obtained from the contralateral inguinal lymph nodes of EAE mice in A were pooled and restimulated with decreasing concentrations of MOG35–55 and then analyzed for proliferation rate by a 3H-thymidine (3H-TdR) incorporation assay and for cytokine release by ELISA. Data represent the mean ± SEM cpm of quadruplicate wells. For cytokine production, the data represent mean ± SEM of quintuplicate wells pooled and tested in duplicate. *P < 0.05; **P < 0.005; ***P < 0.001; ****P < 0.0001. P values were determined by two-way ANOVA with Sidak’s multiple comparisons test. Data in A and B are representative of three independent experiments.
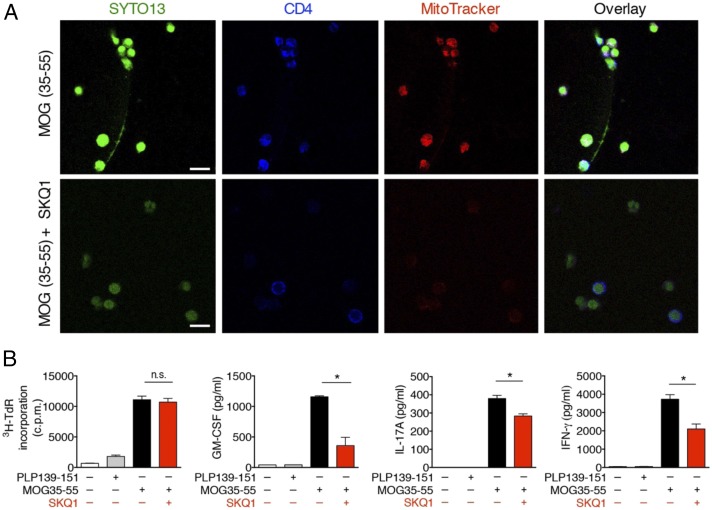
To confirm that THREDs detected in vivo could indeed originate from T lymphocytes, CD4+ T cells purified from the lymph nodes during the priming phase of EAE were restimulated with MOG35–55 peptide (H-2b–restricted) or proteolipid protein peptide 139–151 (PLP139–151), an encephalitogenic H-2s–restricted myelin peptide inducing EAE in SJL/J mouse strain (15), as negative control, in the presence of mitomycin-treated antigen-presenting cells (APCs). Restimulation of CD4+ T cells with MOG35–55 (Fig. 5A), but not with PLP139–151 (SI Appendix, Fig. S4), resulted in the release of THREDs. Treatment with DNase, but not with RNase, dissolved nucleic acids released by T cells exposed to MOG35–55 peptide (SI Appendix, Fig. S4). These data confirm that DNA extrusions observed in the lymph nodes of EAE mice were derived from CD4+ T cells, and also suggest that the activation of CD4+ T cells with a more physiological stimulus such as an antigen, rather than α-CD3/α-CD28 Abs, can trigger the release of extracellular DNA.
Fig. 5.
THREDs extrusion by MOG35–55-primed T cells is reliant on mtROS. (A) CD4+ T cells purified from chronic EAE mice at 7 dpi (n = 5) were restimulated with mytomicin C-treated splenocytes and MOG35–55 in the absence or presence of SKQ1. After 72 h, cells were analyzed by confocal microscopy for SYTO13 (green), CD4 (blue), and MitoTracker (red). (Scale bars: 10 μm.) (B) Parallel cultures were tested as depicted for proliferation by 3H-TdR incorporation and cytokine production by ELISA. Data represent mean ± SEM of quadruplicate wells for proliferation and quintuplicate wells pooled and tested in duplicate for cytokine production. *P < 0.05, two-tailed unpaired t test. n.s., not significant.
Along with supporting the occurrence of THREDs in vivo, these experiments provide evidence that not only naïve, but also primed CD4+ T cells extrude extracellular DNA on restimulation. To assess whether the release of THREDs by primed cells also is dependent on mtROS, we restimulated parallel cultures of CD4+ T cells purified from EAE mice with MOG35–55 in the presence of SKQ1. In line with the data obtained with naïve T cells, treatment of primed CD4+ T cells isolated from EAE mice with SKQ1 inhibited extrusion of THREDs in response to MOG35–55 restimulation (Fig. 5A) and promoted a significant reduction in the production of proinflammatory cytokines (i.e., GM-CSF, IFN-γ, and IL-17A) without affecting the proliferation rate, compared with untreated cells (Fig. 5B).
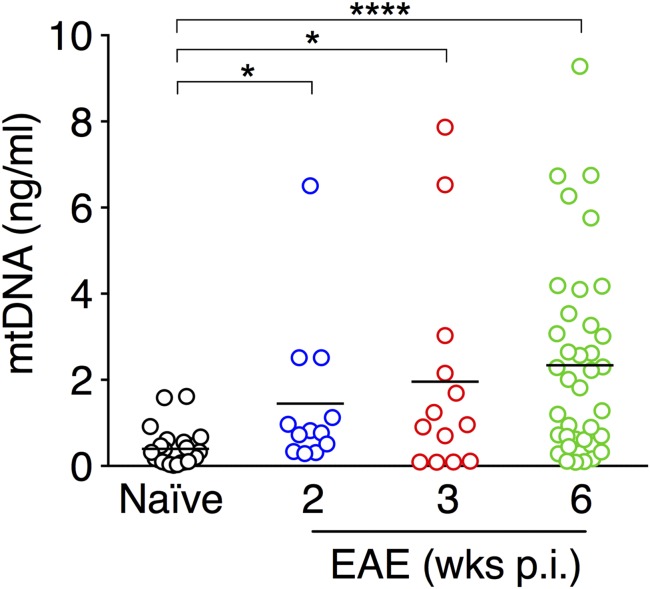
The colocalization of mitochondrial markers such as MitoSOX and MitoTracker on THREDs in vitro supports the hypothesis that mtDNA might be an important component of these DNA filaments. We assessed whether cell-free mtDNA could be detected in serum during the course of chronic EAE. Interestingly, we found that mtDNA concentrations were significantly increased during EAE in both acute (n = 12 mice, P < 0.05) and chronic (3 wk postimmunization, n = 13 mice, P < 0.05; 6 wk postimmunization, n = 38 mice, P < 0.001) phases of the disease compared with naïve mice (n = 24), suggesting that mtDNA serum levels are persistently up-regulated during EAE (Fig. 6).
Fig. 6.
Serum levels of mtDNA are significantly increased during EAE. Sera were collected from naïve C57BL/6 mice (n = 24) or C57BL/6 mice with chronic EAE at different weeks after disease induction (2 wk, n = 12; 3 wk, n = 13; 6 wk, n = 38). Data represent serum mtDNA concentration (ng/mL) of individual mice tested in duplicate. Bars indicate mean values. *P < 0.05; ****P < 0.0001, Kruskal–Wallis test with Dunn’s multiple comparisons test.
Pharmacologic Inhibition of mtROS During Established EAE Dampens Clinical Severity and Autoimmune Inflammation.
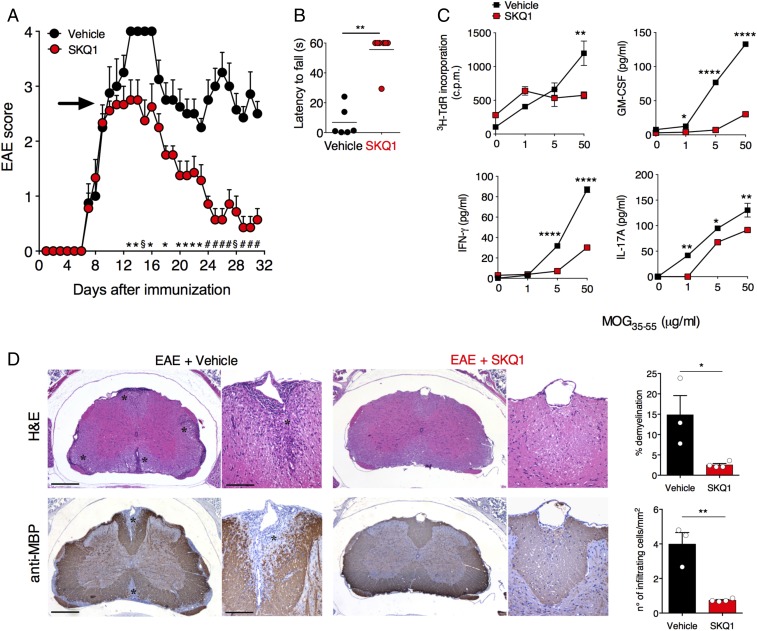
Given that THREDs provide support to full expression of T cell functions and that encephalitogenic CD4+ T cells rely on mtROS for THREDs secretion, we investigated whether mtROS blockade could be exploited for therapeutic purposes in a CD4+-dependent CNS autoimmune pathology such as EAE, widely used as an animal model for multiple sclerosis (MS). To test this hypothesis, mice with chronic EAE were treated daily with either vehicle or SKQ1 from the onset of clinical signs of disease until 31 dpi. We observed that SKQ1 drastically dampened EAE severity (Fig. 7A and SI Appendix, Table S1) and significantly improved motor performance, as measured by the wire-hang test (17) (Fig. 7B and Movies S1–S3), compared with control vehicle-treated mice. Splenocytes from SKQ1-treated mice exhibited a slightly but significantly reduced proliferative response to MOG35–55 peptide and a markedly decreased secretion of the proinflammatory cytokines GM-CSF, IFN-γ, and IL-17A compared with vehicle-treated group (Fig. 7C). Histopathological analysis corroborated the clinical findings by showing that the number of immune cell infiltrates and the extent of demyelination in the spinal cord of EAE mice were significantly reduced by SKQ1 treatment (Fig. 7D).
Fig. 7.
Therapeutic treatment of chronic EAE with mtROS inhibitor lessens clinical severity and dampens CNS autoimmune inflammation. (A) C57BL/6 mice with chronic EAE were i.p. injected from 9 dpi (arrow) to 31 dpi with vehicle or SKQ1 (n = 8–9/group) and monitored for clinical signs of disease (mean ± SEM). Data represent one of two independent experiments that yielded similar results. (B) Motor function was evaluated by the wire-hang test at 31 dpi. (C) Proliferation and cytokine production of splenocytes isolated from EAE mice (n = 5/group) at 31 dpi, pooled, and restimulated with MOG35–55. Data represent mean ± SEM of duplicate wells. (D) Neuropathological analysis of spinal cord sections from EAE mice (31 dpi) stained with H&E and antimyelin basic protein (MBP) (n = 3–4/group). (Scale bars: 500 μm and 100 μm.) *Immune cell infiltration and demyelinated areas. Data in plots are mean ± SEM. *P < 0.05; § or **P < 0.01; # or ***P < 0.001; ****P < 0.0001. P values were determined by the Mann–Whitney U test in A and B, the two-tailed unpaired t test with Holm–Sidak correction in C, and the two-tailed unpaired t test in D.
Discussion
Early studies in the 1970s detected DNA in supernatants of peripheral blood mononuclear cells stimulated with phytohemagglutinin (18, 19). More recently, B cells have been shown to extrude DNA after exposure to synthetic CpG ligands. Our results show that both human and mouse CD4+ T cells release a web of extracellular threads of DNA on activation. The concurrent staining of THREDs with mitochondrial probes, such as MitoSOX and MitoTracker, and occasionally even histones suggests that both mitochondrial and nuclear DNA might be implicated in the formation of these DNA structures. Whether the formation of THREDs is more reminiscent of vital or suicidal NETosis needs further investigation; however, it is possible that both these modalities are involved in the release of DNA by CD4+ T cells.
Our data suggest that DNA extruded from mouse cells is endowed with autocrine costimulatory properties for T lymphocytes. Additional work is needed to unravel the intracellular machinery underlying the actions exerted by THREDs. Owing to its ancestral origin from prokaryotes, mtDNA contains typical motifs of bacterial DNA such as hypomethylated CpG sequences and has been shown to convey “danger signals” to the innate immune system through at least three different routes, including Toll-like receptor (TLR)-9, NLRP3 inflammasome, and STING cytosolic DNA-sensing pathways (20). To date, the intracellular signaling driving the costimulatory effects of synthetic CpG sequences on CD4+ T cells is still obscure. Indeed, both genetic and pharmacologic approaches have shown that CpG activity is independent of both TLR-9–mediated and Myd88-mediated cascades (10). In addition, non-CpG oligonucleotides and sequences that are effective in inhibiting TLR-9 signaling in other immune cell types have been shown to activate T cells, suggesting that these cells are particularly sensitive to oligonucleotide exposure (10). Interestingly, the colocalization of DNA extrusions from T cells with MitoSOX suggests an oxidized status of DNA. The extent of DNA oxidation, rather than differences in the composition between mitochondrial and nuclear DNA, has been identified as responsible for the immunogenic potential of DNA forming NETs (6). Nonetheless, probably due to its proximity to key ROS sources, such as mitochondrial respiratory complexes, mtDNA seems more prone to undergo oxidative damage (21) and NETs are particularly enriched in oxidized mtDNA (6). Interestingly, oxidized mtDNA has been demonstrated to be required for activation of the NLRP3 inflammasome complex in macrophages (22), and NLRP3 inflammasome has been recently implicated in the control of CD4+ Th1 responses (23).
As has been observed for innate immune cells, the release of THREDs also appears to depend on ROS. We specifically tested the contribution of mtROS to this process using the recently-developed mtROS-scavenger SKQ1, which is effective in vitro at lower doses than those required for other available mtROS inhibitors (13). Of note, treatment of naïve CD4+ T cells with SKQ1 did not alter cell activation or cell proliferation (SI Appendix, Fig. S3 A, C, and D), but more specifically hindered the release of THREDs and reduced cytokine production, thus suggesting that SKQ1-mediated effects are not the result of a general inhibition of cell energy machinery. In line with our data, a recent study showed that mtROS are required for optimal T cell function by promoting translocation of nuclear factor of activated T cells (NFAT) to the nucleus and production of IL-2 (24).
We found that both naïve and in vivo-primed CD4+ T cells can extrude THREDs on activation, and that this process occurs both after strong stimulation with α-CD3/α-CD28 Abs and with a more physiological activation, such as that with an antigen such as MOG35–55. The formation of THREDs was detected in vivo in the lymph nodes analyzed during the priming phase of chronic EAE and was inhibited by mtROS blockade with SKQ1 given to mice in vivo. Furthermore, serum mtDNA concentrations increased progressively throughout the course of EAE. It is possible to hypothesize that at least a fraction of the mtDNA detected in sera might derive from activated T cells, even though the mtDNA cell of origin cannot be unambiguously identified and might also include damaged cells within the CNS or other cell types undergoing inflammatory cell death. Three recent reports have shown that cell-free mtDNA is elevated in the cerebrospinal fluid of patients with both relapsing remitting and progressive MS (25–27). Notably, significantly lower levels of mtDNA were found in MS subjects treated with fingolimod (25), an immunomodulatory drug that sequestering lymphocytes in lymph nodes impairs T cell migration to the CNS (14). Currently no data are available on the oxidative status of circulating mtDNA in MS or any other pathological condition; however, autoantibodies against oxidized mtDNA have been detected in a subset of patients with systemic lupus erythematosus (SLE) (28). This phenomenon has been causally linked to the peculiar ability of neutrophils from SLE patients to extrude oxidized nucleoids of mtDNA with potent interferogenic properties without undergoing cell death (28). Understanding the oxidative status of circulating mtDNA in mice with EAE remains an interesting topic for future investigation.
Given our observation that mtROS inhibition was effective in blocking the release of THREDs from MOG-primed CD4+ T cells and in reducing their production of proinflammatory cytokines, we exploited mtROS inhibition for therapeutic purposes in a chronic EAE animal model for MS. We observed that SKQ1 promoted a robust clinical improvement in the disease. It must be noted that SKQ1 was administered systemically in EAE mice, and thus cells and mechanisms other than DNA extrusion by T cells might have been affected by mtROS inhibition during the disease. However, CNS inflammation in MOG35–55-induced EAE is considered to be driven by an adaptive immune response lead by CD4+ T cells, with B cells and CD8+ T cells playing redundant and immunoregulatory roles, respectively (29, 30). In our setting, the significant reduction of the proinflammatory responses that we observed in CD4+ T cells from SKQ1-treated mice suggest that at least part of the SKQ1-mediated effects in dampening EAE could be associated with an effect of mtROS inhibition in CD4+ T cells and THREDs formation.
It is noteworthy that in an animal model of NMO type I IFN stimulates neutrophils to release neutrophil elastase through NET formation. Administration of type 1 IFN exacerbates NMO, a Th17-mediated disease with a prominent neutrophilic infiltrate at the site of disease (31). Neutrophils are also prominent in the early phase of MS, and neutrophil-related markers are present in MS (32). Thus, NET formation has been associated with the pathogenesis of NMO and MS and in its animal models. Our finding that THREDs produced by CD4+ T cells are prominent in the pathogenesis of experimental inflammation may have implications for new therapeutic approaches in MS and in NMO, targeting extrusions from T cells. Overall, our results suggest that the inhibition of THREDs formation by mtROS blockade might represent a novel target of therapy for CD4+ T cell-mediated disorders.
Materials and Methods
Animals.
C57BL/6 mice were all female 8- to 12-wk-old (Charles River Laboratories). All procedures involving animals were approved by the Ethical Committee of the Carlo Besta Neurological Institute and by the Italian General Direction for Animal Health at the Ministry of Health. All animal studies were performed in accordance with the institutional guidelines and national law (DL116/92) and carried out according to the Principles of Laboratory Animal Care (European Communities Council Directive 2010/63/EU), to minimize discomfort for animals.
Human Samples.
Peripheral blood was obtained from healthy individuals at the National Tumor Institute after provision of informed consent, according to institutional guidelines. Samples were deidentified before processing in the laboratory.
Cell Isolation.
Human CD4+ T cells were enriched from suspensions of buffy coats from healthy donors collected at the National Tumor Institute with the Naive CD4+ T Cell Isolation Kit II, human (Miltenyi Biotec). The preparation contained approximately 30% CD4+ CD45RA+ T cells and 60% CD3+ T cells by flow cytometry. Untouched naïve CD4+ T helper cells were magnetically purified from suspensions of spleen cells and LNCs isolated from naïve C57BL/6 mice with the Naive CD4+ T Cell Isolation Kit, mouse (Miltenyi Biotec). The purity of bead-isolated T naïve lymphocytes (CD3+CD4+CD62L+CD44−) was >95% by flow cytometry. Untouched CD4+ T cells were magnetically purified by negative selection (CD4+ T cell Isolation Kit; Miltenyi Biotec) from suspensions of LNCs and splenocytes isolated from chronic EAE mice at 7 dpi. CD3+CD4+ cell purity >95% was confirmed by flow cytometry.
Mouse T Cell Cultures.
Mouse naïve CD4+ T cells (1 × 105/well) were cultured in 96-well U-bottom plates in 200 μL of enriched RPMI, composed of RPMI 1640 (EuroClone) supplemented with l-glutamine (2 mM), sodium pyruvate (1 mM), nonessential amino acids (0.1 mM), penicillin (100 U/mL), streptomycin (0.1 mg/mL), Hepes buffer (0.01 M), and 10% Nu-Serum (355500; Corning). T cells were activated for 72 h with plate-bound anti-CD3 mAb (5 μg/mL, 145–2C11; BD Biosciences) and soluble anti-CD28 mAb (2 μg/mL, 37.51; BD Biosciences) in the absence or presence of SKQ1 (150 nM; AdooQ Bioscience). In costimulation experiments, mouse naïve CD4+ T cells (1 × 106/well) were cultured in 24-well plates in enriched RPMI and left unstimulated or stimulated with plate-bound anti-CD3 mAb (5 μg/mL, clone 145–2C11; BD Biosciences) in the presence or absence of THREDs, negative control, or anti-CD28 mAb (1 μg/mL, clone 37.51; BD Biosciences) for 96 h. CD4+ T cells purified from EAE mice were cultured in enriched RPMI (1 × 105/well) with mitomycin-treated splenocytes (2 × 105/well) isolated from naïve C57BL/6 mice (as a source of APCs), and stimulated with PLP139–151 (100 μg/mL) or MOG35–55 (100 μg/mL) in the absence or presence of SKQ1 (50 nM). LNCs and splenocytes were isolated from chronic EAE mice at 7 and 31 dpi and then restimulated with MOG35–55 (1–50 μg/mL) or concanavalinA (2 μg/mL) in 96-well U-bottom plates at a density of 2 × 105 cells/well in 200 μL of RPMI 1640 (EuroClone) supplemented with l-glutamine (2 mM), sodium pyruvate (1 mM), nonessential amino acids (0.1 mM), penicillin (100 U/mL), streptomycin (0.1 mg/mL), Hepes buffer (0.01 M), and 10% FBS. To measure proliferation rate, cells were incubated for 48 h at 37 °C with 5% CO2 and then pulsed with 0.5 μCi [3H]-thymidine per well for 18 h, and proliferation was measured from triplicate cultures on a beta counter (PerkinElmer). Cytokine release was evaluated in supernatants of parallel cultures by ELISA (Mouse DuoSet; R&D Systems).
THREDs Detection and Quantification.
Human enriched CD4+ T cells and mouse naïve CD4+ T cells were seeded on poly-d-lysine–coated glass (3 × 105/well) in enriched RPMI and left unstimulated or stimulated for 24 h with plate-bound anti-CD3 mAbs (5 μg/mL; for human cells, clone OKT3, eBioscience; for mouse cells, clone 145–2C11, BD Biosciences) and soluble anti-CD28 mAbs (2 μg/mL) (for human cells, clone CD28.2, eBioscience; for mouse cells, clone 37.51, BD Biosciences). For confocal microcopy analyses, cells were treated as indicated above and incubated with live-cell dyes MitoSOX Red (0.5 μM; Thermo Fisher Scientific) or MitoTracker Red (100 nM; Thermo Fisher Scientific) in the last 30 min or 45 min of culture, respectively. After 24 h, T cells were fixed with 4% paraformaldehyde (PFA) and stained with an mAb to CD4 (clone OKT4, APC-conjugated for human samples or clone GK-1.5, APC-conjugated for mouse samples; both from eBioscience), DAPI (ProLong Gold Antifade Mountant with DAPI; Thermo Fisher Scientific), and, where indicated, a polyclonal Ab to histones (pan histone, clone H11-4; EMD Millipore) conjugated with Alexa Fluor 488 dye (labeling kit for mouse IgG) or SYTO13 (1 μM). Alternatively, after 24 h of culture, mouse naïve CD4+ T cells were stained for 15 min with SYTOX Orange (0.5 μM), SYTO13 (1 μM), and DAPI (ProLong Gold Antifade Mountant with DAPI) (all from Thermo Fisher Scientific) and then imaged immediately for the presence of THREDs with a Leica DM4 B optical microscope and a Leica DFC450 digital camera.
THREDs quantification was performed as described for NETs by Metzler et al. (3). Untouched CD4+ T cells from EAE mice (3 × 105/well) were cultured with mytomicin-treated splenocytes (6 × 105/well) from naïve C57BL/6 mice (as a source of APCs) in the absence or presence of PLP139–151 (100 μg/mL) or MOG35–55 (100 μg/mL) in enriched RPMI, with the addition of SYTO13 (1 μM) and MitoTracker Red (100 nM) in the last 45 min of culture. After 72 h, T cells were fixed with 4% PFA and stained with an mAb to CD4 (APC-conjugated; eBioscience). Where indicated, T cells were treated with 150 nM SKQ1 (AdooQ Bioscience), 10 μM DPI (Sigma-Aldrich), 10 μg/mL DNase I (Sigma-Aldrich), or 5 μg/mL RNase (Roche).
To assess THREDs formation in vivo, we isolated inguinal lymph nodes from naïve and EAE mice at 7 dpi. Confocal microscopy analysis of lymph node sections was performed by staining with mAb to CD4 (4SM95; e-Bioscience) and a polyclonal Ab to histone H3 (citrulline R2 + R8 + R17; Abcam, ab5103). In detail, tissue sections (4 μm) were deparaffinized and rehydrated. An antigen unmasking technique was performed using Novocastra Epitope Retrieval Solution (pH 9) at 98 °C for 30 min. Subsequently, the sections were brought to room temperature and washed in PBS. After Fc-blocking, the samples were incubated overnight at 4 °C with primary antibodies. Primary antibody binding was amplified and visualized using Alexa Fluor 488-conjugated goat anti-rat IgG (H+L) (Invitrogen), Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) (Invitrogen), or Alexa Fluor 633-conjugated goat anti-rat IgG (H+L) (A-21094; Invitrogen). The slides were counterstained with DAPI Nucleic Acid Stain (Invitrogen Molecular Probes). All confocal microscopy analyses were performed using a Leica TCS-SP8-X confocal laser scanning microscope.
THREDs Preparation.
To evaluate the costimulatory properties of THREDs, we used a protocol similar to that previously described for NETs isolation (11). In brief, untouched naïve CD4+ T cells (1 × 106/well) were left unstimulated or stimulated with plate-bound anti-CD3 mAb (5 μg/mL, 145-2C11; BD Biosciences) and soluble anti-CD28 mAb (2 μg/mL, 37.51; BD Biosciences). After 24 h, cells were centrifuged at 450 × g for 10 min at 4 °C. The cell-free supernatant was recovered and centrifuged at 18,000 × g for 10 min at 4 °C, after which the pellet was resuspended in 50 μL of PBS. The extraction products from activated cells (THREDs) or nonactivated cells (negative control) were pooled from three different experiments and then used for costimulation.
To visualize DNA production from cultured T cells, the extraction products were incubated on poly-d-lysine–coated glass overnight at 37 °C, stained with DAPI, fixed with PFA 4%, and analyzed by fluorescence microscopy (Leica).
Flow Cytometry.
Single-cell suspensions were incubated with blocking mAb anti-CD16/CD32 (2.4G2; 1:100) and then stained with FITC anti-CD3, BV510 anti-CD45RA, and APC anti-CD4 (for human samples; all from eBioscience except anti-CD45RA, from BD Biosciences) or FITC anti-CD3, PE anti-CD11c, PE-Cy7 anti-CD62L, APC anti-CD44, and BV510 anti-CD4 (for mouse samples, all from eBioscience except anti-CD4, from BD Bioscience) for cell purity control. For activation and survival analysis, T cells were stained with 7AAD, PE-Cy7 anti-CD4 (GK1.5), APC anti-CD25 (PC61 5.3), and PE anti-CD69 (H1.2F3) (all from eBioscience). Cells were analyzed by flow cytometry on a BD LSRFortessa (BD Biosciences) or Attune NxT (Thermo Fisher Scientific) flow cytometer, and analyses were performed using FlowJo software (Treestar).
Gene Expression Analysis.
Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and then reverse-transcribed with the QuantiTect Reverse Transcription Kit (Qiagen), which includes a genomic DNA removal step. Real-time PCR was performed on 7500 Fast Real-Time PCR system (Applied Biosystems). PCR reactions were performed with Fast Universal Master Mix (Applied Biosystems) and the following primer/probe sets (Applied Biosystems): Csf2, Mm01290062_m1; Ifng, Mm01168134_m1; Il2, Mm00434256_m1; Tnf, Mm00443258_m1; and Gapdh, Mm99999915_g1. Expression of target genes was quantified by the comparative threshold cycle method, and Gapdh was used as housekeeping gene. Data are presented as percentage of the housekeeping gene Gapdh ± SD from duplicate wells.
MtDNA Quantification.
DNA was purified from 25 μL of mouse serum with the Qiagen QIAamp DNA Mini and Blood Mini Kit. For absolute quantification, mtDNA was purified from the livers of naïve C57BL/6 mice as described previously (33), quantified with a NanoDrop 2000 spectrophotometer, and used to create a standard curve. Samples and standards were tested in duplicate by real-time PCR for the mitochondrial gene Atp8 (Mm04225236_g1) on an Applied Biosystems 7500 Fast Real-Time PCR system (Thermo Fisher Scientific).
Statistical Analysis.
Statistical analyses were performed with GraphPad Prism software. Data are presented as mean ± SEM or ± SD as indicated in the figure legends and compared using a two-tailed unpaired t test or one-way or two-way ANOVA with the Tukey or Sidak multiple-comparison test, as appropriate. For EAE clinical data, the Mann–Whitney U test was used to compare results between two groups. In all tests, *P < 0.05 was considered statistically significant.
Information on animals, EAE studies, and histopathological analysis of CNS inflammation during EAE is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of the Cell Imaging Facility at the National Tumor Institute for confocal microscopy image acquisition. This work was supported by the Italian Multiple Sclerosis Foundation (Grant 2015/R19, to R.P.) and the Italian Ministry of Health (Grant GR-2013-02355637, to S.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822013116/-/DCSupplemental.
References
- 1.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 2.Yousefi S, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 3.Sangaletti S, et al. Neutrophil extracellular traps mediate transfer of cytoplasmic neutrophil antigens to myeloid dendritic cells toward ANCA induction and associated autoimmunity. Blood. 2012;120:3007–3018. doi: 10.1182/blood-2012-03-416156. [DOI] [PubMed] [Google Scholar]
- 4.Okubo K, et al. Macrophage extracellular trap formation promoted by platelet activation is a key mediator of rhabdomyolysis-induced acute kidney injury. Nat Med. 2018;24:232–238. doi: 10.1038/nm.4462. [DOI] [PubMed] [Google Scholar]
- 5.Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med. 2017;23:279–287. doi: 10.1038/nm.4294. [DOI] [PubMed] [Google Scholar]
- 6.Lood C, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22:146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingelsson B, et al. Lymphocytes eject interferogenic mitochondrial DNA webs in response to CpG and non-CpG oligodeoxynucleotides of class C. Proc Natl Acad Sci USA. 2018;115:E478–E487. doi: 10.1073/pnas.1711950115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripodo C, et al. Persistent immune stimulation exacerbates genetically driven myeloproliferative disorders via stromal remodeling. Cancer Res. 2017;77:3685–3699. doi: 10.1158/0008-5472.CAN-17-1098. [DOI] [PubMed] [Google Scholar]
- 9.Gelman AE, et al. The adaptor molecule MyD88 activates PI-3 kinase signaling in CD4+ T cells and enables CpG oligodeoxynucleotide-mediated costimulation. Immunity. 2006;25:783–793. doi: 10.1016/j.immuni.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrigan A, Wong MT, Utz PJ. CpG and non-CpG oligodeoxynucleotides directly costimulate mouse and human CD4+ T cells through a TLR9- and MyD88-independent mechanism. J Immunol. 2011;187:3033–3043. doi: 10.4049/jimmunol.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franck G, et al. Roles of PAD4 and NETosis in experimental atherosclerosis and arterial injury: Implications for superficial erosion. Circ Res. 2018;123:33–42. doi: 10.1161/CIRCRESAHA.117.312494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson SH, Devadas S, Kwon J, Pinto LA, Williams MS. T cells express a phagocyte-type NADPH oxidase that is activated after T cell receptor stimulation. Nat Immunol. 2004;5:818–827. doi: 10.1038/ni1096. [DOI] [PubMed] [Google Scholar]
- 13.Plotnikov EY, et al. Protective effect of mitochondria-targeted antioxidants in an acute bacterial infection. Proc Natl Acad Sci USA. 2013;110:E3100–E3108. doi: 10.1073/pnas.1307096110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinman L. Immunology of relapse and remission in multiple sclerosis. Annu Rev Immunol. 2014;32:257–281. doi: 10.1146/annurev-immunol-032713-120227. [DOI] [PubMed] [Google Scholar]
- 15.Pedotti R, et al. An unexpected version of horror autotoxicus: Anaphylactic shock to a self-peptide. Nat Immunol. 2001;2:216–222. doi: 10.1038/85266. [DOI] [PubMed] [Google Scholar]
- 16.Costanza M, Musio S, Abou-Hamdan M, Binart N, Pedotti R. Prolactin is not required for the development of severe chronic experimental autoimmune encephalomyelitis. J Immunol. 2013;191:2082–2088. doi: 10.4049/jimmunol.1301128. [DOI] [PubMed] [Google Scholar]
- 17.Lewis KE, et al. Microglia and motor neurons during disease progression in the SOD1G93A mouse model of amyotrophic lateral sclerosis: Changes in arginase1 and inducible nitric oxide synthase. J Neuroinflammation. 2014;11:55. doi: 10.1186/1742-2094-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers JC, Boldt D, Kornfeld S, Skinner A, Valeri CR. Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen. Proc Natl Acad Sci USA. 1972;69:1685–1689. doi: 10.1073/pnas.69.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers JC. Identification of an intracellular precursor to DNA excreted by human lymphocytes. Proc Natl Acad Sci USA. 1976;73:3211–3215. doi: 10.1073/pnas.73.9.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West AP, Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol. 2017;17:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama H, Otsu K. Mitochondrial DNA as an inflammatory mediator in cardiovascular diseases. Biochem J. 2018;475:839–852. doi: 10.1042/BCJ20170714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Z, et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560:198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arbore G, et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science. 2016;352:aad1210. doi: 10.1126/science.aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sena LA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leurs CE, et al. Cerebrospinal fluid mtDNA concentration is elevated in multiple sclerosis disease and responds to treatment. Mult Scler. 2018;24:472–480. doi: 10.1177/1352458517699874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varhaug KN, et al. Increased levels of cell-free mitochondrial DNA in the cerebrospinal fluid of patients with multiple sclerosis. Mitochondrion. 2017;34:32–35. doi: 10.1016/j.mito.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Fissolo N, et al. July 1, 2018. Cerebrospinal fluid mitochondrial DNA levels in patients with multiple sclerosis. Mult Scler. doi.org/10.1177/1352458518786055.
- 28.Caielli S, et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyons JA, San M, Happ MP, Cross AH. B cells are critical to induction of experimental allergic encephalomyelitis by protein but not by a short encephalitogenic peptide. Eur J Immunol. 1999;29:3432–3439. doi: 10.1002/(SICI)1521-4141(199911)29:11<3432::AID-IMMU3432>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Ortega SB, et al. The disease-ameliorating function of autoregulatory CD8 T cells is mediated by targeting of encephalitogenic CD4 T cells in experimental autoimmune encephalomyelitis. J Immunol. 2013;191:117–126. doi: 10.4049/jimmunol.1300452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herges K, et al. Protective effect of an elastase inhibitor in a neuromyelitis optica-like disease driven by a peptide of myelin oligodendroglial glycoprotein. Mult Scler. 2012;18:398–408. doi: 10.1177/1352458512440060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rumble JM, et al. Neutrophil-related factors as biomarkers in EAE and MS. J Exp Med. 2015;212:23–35. doi: 10.1084/jem.20141015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathew A, et al. Degraded mitochondrial DNA is a newly identified subtype of the damage associated molecular pattern (DAMP) family and possible trigger of neurodegeneration. J Alzheimers Dis. 2012;30:617–627. doi: 10.3233/JAD-2012-120145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.