Significance
Chronic CNS hypoxia is a characteristic of diverse vascular disorders and environmental conditions. There is strong physiologic evidence that neurons and glia mount a protective response in the face of hypoxic stress. We used single-cell RNA sequencing and metabolic profiling to study the hypoxia responses of neurons and glia in the retina in a mouse model of familial exudative vitreoretinopathy, a developmental disorder in which retinal vascularization is incomplete. These responses were compared with those induced in the mouse cerebral cortex by a 1-week exposure to low atmospheric oxygen. These experiments reveal a distinctive set of genomic and metabolic responses in the hypoxic retina and a related set of genomic responses in the hypoxic cerebral cortex.
Keywords: Norrie disease, familial exudative vitreoretinopathy, single-cell RNA-seq, serine synthesis, metabolomics
Abstract
The mammalian CNS is capable of tolerating chronic hypoxia, but cell type-specific responses to this stress have not been systematically characterized. In the Norrin KO (NdpKO) mouse, a model of familial exudative vitreoretinopathy (FEVR), developmental hypovascularization of the retina produces chronic hypoxia of inner nuclear-layer (INL) neurons and Muller glia. We used single-cell RNA sequencing, untargeted metabolomics, and metabolite labeling from 13C-glucose to compare WT and NdpKO retinas. In NdpKO retinas, we observe gene expression responses consistent with hypoxia in Muller glia and retinal neurons, and we find a metabolic shift that combines reduced flux through the TCA cycle with increased synthesis of serine, glycine, and glutathione. We also used single-cell RNA sequencing to compare the responses of individual cell types in NdpKO retinas with those in the hypoxic cerebral cortex of mice that were housed for 1 week in a reduced oxygen environment (7.5% oxygen). In the hypoxic cerebral cortex, glial transcriptome responses most closely resemble the response of Muller glia in the NdpKO retina. In both retina and brain, vascular endothelial cells activate a previously dormant tip cell gene expression program, which likely underlies the adaptive neoangiogenic response to chronic hypoxia. These analyses of retina and brain transcriptomes at single-cell resolution reveal both shared and cell type-specific changes in gene expression in response to chronic hypoxia, implying both shared and distinct cell type-specific physiologic responses.
The mammalian CNS is exquisitely sensitive to hypoxia. When combined with nutrient deprivation, hypoxia in the setting of acute ischemia causes rapid and irreversible cell loss in the brain and retina (1, 2). In contrast, mild to moderate hypoxia with continued nutrient delivery—as occurs at high altitude—can be tolerated for days to years (3, 4). In the laboratory, rodents can tolerate weeks to months in a hypoxia chamber with minimal long-term effects on brain morphology or neuronal function (5, 6). In the short term, hypoxia tolerance is likely mediated by metabolic suppression and reduced ion channel activity; in the longer term, it is likely mediated by changes in gene expression driven predominantly by hypoxia-inducible factors (HIFs) (7). These gene expression changes also protect against subsequent ischemia, a phenomenon known as hypoxic preconditioning (8). Previous work has hinted at CNS cell type-specific responses to hypoxia based, for example, on differences between cultured neurons and glia in their gene expression response to hypoxia (9) and on the activation of a latent angiogenic sprouting response by vascular endothelial cells (ECs) in the chronically hypoxic mouse brain (10). However, CNS cell type-specific responses to chronic hypoxia have not been examined in vivo on a genome-wide basis.
Hypoxia tolerance in the retina is likely to be a feature of familial exudative vitreoretinopathy (FEVR), an inherited disorder in which retinal blood vessels fail to develop normally (11). Approximately 50% of FEVR patients carry a loss-of-function mutation in one of the genes coding for the components of a canonical beta-catenin signaling pathway (11). In this system, the ligand Norrin [the protein product of the X-linked Norrie disease protein (Ndp) gene] is secreted by Muller glia and activates a complex of receptor, coreceptor, and coactivator (Frizzled4, LRP5, and TSPAN12, respectively) on the surface of retinal ECs, leading to the accumulation of the intracellular effector beta-catenin (CTNNB1) (12–14). We refer hereafter to this signaling system as Norrin/Frizzled4 signaling. The virtually identical cross-sectional anatomies of the human and mouse vasculatures made it possible to accurately model FEVR in mice with Ndp, Fz4, Lrp5, and Tspan12 loss-of-function mutations (13–16). In both species, the choroidal circulation supplies the metabolic requirements of the cells in the outer nuclear layer, the rod and cone photoreceptors, whereas vessels on the vitreal surface of the retina together with the intraretinal vasculature, comprising two tiers of capillaries that flank the inner nuclear layer (INL), supply the metabolic requirements of the inner two layers of retinal neurons and glia. In FEVR patients (17) and mouse models (18, 19), reduced or absent Norrin/Frizzled4 signaling results in reduced or aborted development of the intraretinal and peripheral retinal blood vessels.
Despite the absence of intraretinal capillaries in murine retinas lacking Norrin/Frizzled4 signaling and the resulting hypoxia of the INL, the overall structure and cellularity of the mutant retina appear to be largely preserved, with minimal loss of INL cells over the first several months of life (20). It is likely that INL hypoxia in these retinas is substantially mitigated by loss of the blood–retina barrier (BRB), which allows small molecules, such as glucose, to diffuse into the INL (19). However, the function of the inner retina is severely affected: the b-wave of the electroretinogram, which arises from the summed responses of INL neurons to a flash of light, is greatly reduced, and there is no detectable transmission of visual information from rod and cone photoreceptors to retinal ganglion cells (RGCs) as measured by the optokinetic response (14, 15). Remarkably, when these retinas are placed in oxygenated Ringer’s buffer, neurotransmission from the photoreceptors to the RGCs is restored within tens of minutes as evidenced by light-dependent RGC action potentials recorded with a multielectrode array (14). These observations hint at the existence of a reversible metabolic state that silences the hypoxic inner retina in vivo, and they suggest that studying the retinas of Norrin/Frizzled4-deficient mice could provide general insights into hypoxia tolerance in the CNS.
In this paper, we have explored the adaptations of murine Ndp KO (NdpKO) retinas to chronic hypoxia by (i) comparing the transcriptomes of WT and NdpKO retinas at single-cell resolution, (ii) comparing the metabolomes of WT and NdpKO retinas, and (iii) correlating these “omic” observations with histologic analyses. We have also analyzed, at single-cell resolution, the transcriptomes of the cerebral cortex from mice under normoxic conditions vs. mice exposed to 7 d of hypoxia (7.5% oxygen). These experiments reveal a distinctive set of genomic and metabolic responses in the hypoxic NdpKO retina and a related set of genomic responses in the hypoxic cerebral cortex.
Results
Minimal Morphologic Change in the NdpKO Retina.
The absence of intraretinal capillaries in NdpKO retinas leads to chronic hypoxia that predominantly affects interneurons and glia that have their cell bodies (horizontal, bipolar, Muller, and nondisplaced amacrine cells) in the INL as demonstrated by the accumulation of pimonidazole (Hypoxyprobe) (Fig. 1A). The predominance of pimonidazole accumulation in the INL is consistent with the observations, in NdpKO mice, that (i) the pupillary light reflex (i.e., pupil constriction in response to light) is nearly normal, indicating that intrinsically photoreceptive RGCs are functional, and that (ii) the a-wave of the electroretinogram is unaffected, indicating that photoreceptors are functional (14, 15).
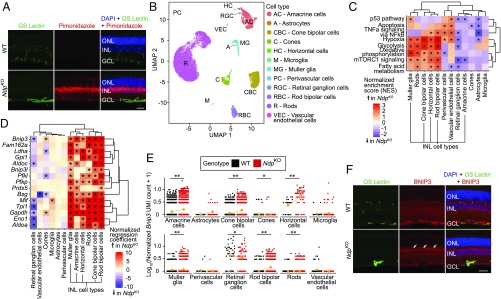
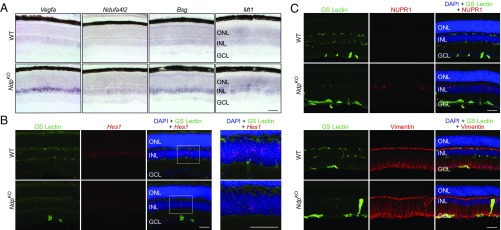
Fig. 1.
scRNA-seq analysis of NdpKO retinas reveals transcriptional changes in INL cell types. (A) Immunofluorescence of adult WT and NdpKO retina cross-sections showing blood vessels (GS Lectin; green), hypoxic tissue (antipimonidazole; red), and nuclei (DAPI; blue). In this and other retina cross-sections, ganglion cell layer (GCL), INL, and outer nuclear layer (ONL) are shown. (Scale bar: 50 μm.) (B) UMAP plot showing different cell-type clusters in a merged dataset from duplicate samples of WT and NdpKO retinas. (C) Statistically significant MSigDB Hallmark pathways in three or more cell types on GSEA reveal pathways enriched in NdpKO over WT retinas across inner retinal cell types. *P value of <0.05. (D) Known hypoxia-inducible genes that are up-regulated in NdpKO compared with WT retinas in three or more cell types. *q value of <0.05. (E) Dot plots showing the log10[number of Bnip3 unique molecular identifiers (UMIs) + 1] for each cell for WT and NdpKO retinas plotted by cell type. Each orange dot and its associated vertical line represent the mean UMI count and its nonparametric bootstrap confidence interval, respectively, for each cell type of each genotype. The line connecting two oranges dots represents the trend in mean expression between the two genotypes for each cell type. *q value of <0.05 on differential gene expression testing; **q value of <0.01 on differential gene expression testing. (F) Immunofluorescence of adult WT and NdpKO retina cross-sections showing blood vessels (GS Lectin; green), BNIP3 (red), and nuclei (DAPI; blue). BNIP3 immunofluorescence is more intense in the NdpKO INL (indicated by white arrows) compared with the WT INL. (Scale bar: 50 μm.)
To assess the structure of the NdpKO retina in greater detail, we compared 2-mo-old NdpKO and WT retinas by transmission EM (TEM) and by immunostaining. By TEM, the presence of ribbon synapses and the density and overall structure of the neuropil in the outer plexiform layer (OPL) and inner plexiform layer (IPL) were indistinguishable between NdpKO and WT retinas, as was the appearance of nuclei in the INL (SI Appendix, Fig. S1A). Immunostaining for markers in the INL and its adjacent plexiform layers generally showed indistinguishable patterns of staining in NdpKO and WT retinas. These include vesicular glutamate transporter-1, a presynaptic marker of glutamatergic synapses (SI Appendix, Fig. S1B); postsynaptic density protein 95, a postsynaptic marker (SI Appendix, Fig. S1B); and markers for horizontal cells (Neurofilament-H and Calbindin), RGC axons (Neurofilament-H), cholinergic amacrine cells, and various amacrine and RGC subsets (Calretinin) (SI Appendix, Fig. S1C). A marker for rod bipolar cells (PKC-alpha) showed the same pattern but was less intense in NdpKO compared with WT retinas (SI Appendix, Fig. S1C). Vascular markers (GS-lectin for ECs and Aquaporin-4 for EC-associated Muller processes) showed the expected absence of intraretinal capillaries in NdpKO retinas (SI Appendix, Fig. S1C).
The essentially normal neuronal structure in the NdpKO retina, as seen by TEM and immunostaining, is consistent with the earlier observation of a rapid recovery of neurotransmission across the inner retina when NdpKO retinas are provided with oxygen and glucose ex vivo (14). Taken together, the data suggest that the dormancy of INL neurons in NdpKO retinas likely arises from a reversible change in metabolic state.
Single-Cell RNA-seq of NdpKO Retinas Reveals both Shared and Cell Type-Specific Gene Expression Changes.
Using a droplet-based single-cell RNA-seq (scRNA-seq) platform (10× Genomics), we characterized 32,825 dissociated retinal cells from 8-wk-old NdpKO mice and WT littermate controls. The data were derived from two independent samples per genotype. All 12 major retinal cell types were readily identified based on known markers (SI Appendix, Fig. S2 B and C) and showed distinct clusters on a Uniform Manifold Approximation and Projection (UMAP) (21) plot (Fig. 1B); 1,133 presumed multiplets were excluded from subsequent analyses. When cells from NdpKO and WT retinas were visualized separately, they showed very similar clustering behavior (SI Appendix, Fig. S2A). Cross-sample normalization was performed via mean scaling of the raw transcript copies per cell in each cell type independently. Although a previous study suggested inflammation as a consequence of the loss of BRB integrity in NdpKO retinas (22), we did not observe infiltrating immune cells by scRNA-seq.
Using Monocle2 (23, 24), we next generated a negative binomial regression model that included parameters for genotype, cell type, sequencing depth, and batch effects. This permitted the calculation of a z-scored genotype regression coefficient for each gene and the identification of differentially expressed genes on a per cell type basis. To identify gene sets with differential representation in WT vs. NdpKO datasets, genes were ranked based on the z-scored genotype regression coefficient for each gene in each cell type, and this ranking was used for a preranked gene set enrichment analysis (GSEA) analysis (25). This preranked GSEA performed with the “Hallmark” gene sets curated by the Molecular Signatures DataBase (MSigDB) (26) identified gene sets, including the “Hypoxia” gene set, that were positively enriched in NdpKO retinas in multiple INL cell types, including Muller glia, rod bipolar, cone bipolar cells, and horizontal cells (Fig. 1C).
The cell type-specific gene expression changes (Monocle2 test; q value ≤ 0.05) observed by scRNA-seq are consistent with the hypothesis that the INL is the site of the greatest hypoxic stress in the NdpKO retina. A subset of known hypoxia-induced genes from the Hypoxia gene set (MSigDB) was significantly up-regulated in most INL cell types (Fig. 1D and SI Appendix, Fig. S2D), consistent with the pimonidazole labeling experiment. Interestingly, many of the same transcripts were also enriched in rods but not enriched in cones. One possible explanation for the different responses of rods and cones is that cones are intrinsically less energy consuming than rods. Another more speculative possibility is that, based on the steep decline in oxygen concentration between retinal pigment epithelium (RPE; ∼70 mmHg) and the OPL (∼10 mmHg) (27), the loss of intraretinal vasculature might produce a greater hypoxic stress in those rods with nuclei closer to the INL compared with cones, which have their nuclei closer to the RPE where the oxygen concentration is higher.
Many of the differentially expressed genes in INL cells in the hypoxic NdpKO retina are targets of HIFs. Two examples shown in detail are Bnip3 and Bsg. Bnip3 is up-regulated approximately twofold in all INL cell types by scRNA-seq (Fig. 1E), consistent with the modest elevation in BNIP3 immunostaining intensity in the INL in NdpKO retinas (Fig. 1F). Bnip3 up-regulation has been shown to mitigate oxidative stress in the context of hypoxia by mediating mitophagy (28). Bsg, another HIF-regulated gene, is up-regulated approximately twofold in INL cells in NdpKO retinas but is paradoxically down-regulated >10-fold in ECs (SI Appendix, Fig. S2 E and F). Bsg codes for Basigin (CD147), a widely expressed multifunctional transmembrane protein that associates with and directs the trafficking of integral membrane monocarboxylate transporters to promote glycolysis in hypoxia (29, 30). However, the presence of T cell factor/lymphocyte enhancer-binding factor (TCF/LEF) binding motifs in accessible chromatin regions upstream of the Bsg promoter in CNS ECs (31) suggests that Bsg may also be a beta-catenin target gene required for the maturation of cell–cell junctions in CNS ECs (32). Its down-regulation in NdpKO ECs is consistent with a loss of beta-catenin signaling in ECs in the absence of Ndp.
In addition to Bnip3 and Bsg (described above), the glycolytic enzyme genes Aldoa, Aldoc, Ldha, Pfkp, Pfkl, Gapdh, Tpi1, Eno1, and Gpi1 are all targets of HIF regulation (33) and are significantly up-regulated in INL cells in NdpKO retinas (Fig. 1D). Prdx5, which is up-regulated in a subset of INL cells, is a known hypoxia-inducible gene (34) that codes for Peroxiredoxin-5, an enzyme that confers antioxidant protection by reducing hydrogen peroxide (35). Taken together, these gene expression changes are consistent with a model in which transcriptional changes during hypoxia, driven primarily by HIFs, promote glycolysis and/or inhibit the mitochondrial respiratory chain, which together minimize the production of reactive oxygen species (ROS) (36). Additional defenses against ROS are provided by up-regulation of genes, such as Prdx5.
Metabolic Changes in the NdpKO Retina Are Centered Around Glutathione and One-Carbon Metabolism.
As many of the transcripts that are enriched in NdpKO retinas are linked to metabolism, we performed an untargeted survey of the metabolome using capillary electrophoresis (CE)–TOF MS on ionic metabolites isolated from whole NdpKO and WT retinas (Dataset S1). Based on six independent samples per genotype, CE-TOF MS identified dozens of metabolites that showed consistent changes in abundances (as measured by the CE-TOF MS relative peak areas) between NdpKO and WT retinas (Fig. 2 A and C and SI Appendix, Fig. S3 A and E and Table S1). By principal component analysis (PCA) of the untransformed CE-TOF MS relative peak areas across metabolites, the NdpKO and WT retina metabolomes are clearly distinct (SI Appendix, Fig. S3B).
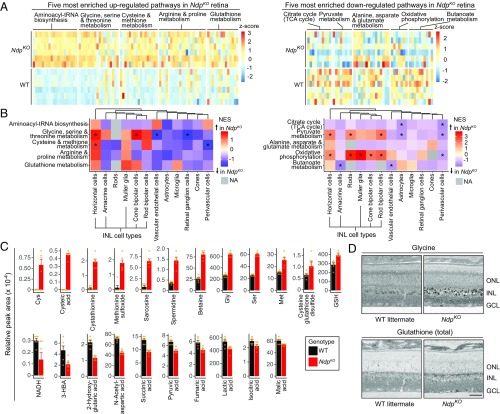
Fig. 2.
Untargeted metabolic profiling of WT and NdpKO retinas reveals metabolic changes centered around glutathione and one-carbon metabolism. (A) Heat maps showing individual metabolites in the top five up-regulated in NdpKO (Left) and top five down-regulated in NdpKO (Right) metabolic pathways as determined by overrepresentation analysis. Each row represents an independent retina sample, and each column represents a metabolite. (B) Heat maps showing enrichment scores for each cell type in GSEA for the same 10 NdpKO-enriched pathways shown in A. NA, nonapplicable (gray), indicating that GSEA could not be performed due to the low signal for the relevant pathway genes in a particular cell type; NES, normalized enrichment score. *P value of <0.05. (C) Bar plots showing relative MS peak areas for representative metabolites in the NdpKO-enriched metabolic pathways listed in A. Error bars represent the SEM. Orange dots represent individual samples. (D) Immunogold staining with antibodies raised against metabolite-hapten conjugates followed by silver intensification. Elevated levels of glycine and total glutathione in NdpKO retinas were mainly localized to the INL. GCL, ganglion cell layer; ONL, outer nuclear layer. (Scale bar: 50 μm.)
The hypoxic state of the INL and the marked reduction in inner retinal neurotransmission might reasonably suggest a reduction in ATP levels and a reduction in the ATP/ADP ratio in the INL. Surprisingly, these values for the entire retina were essentially identical in NdpKO vs. WT retinas (SI Appendix, Fig. S3C and Table S1). Classical microchemical studies of ATP and ADP content in different retinal layers found that the ATP content of the INL represents ∼28% of total retina ATP (37). Thus, even a relatively modest (e.g., 30%) reduction in the INL ATP level or in the INL ATP/ADP ratio should have produced a detectable reduction (∼10%) in the whole-retina analysis. In contrast, NADH levels and the NADH/NAD+ ratio were roughly halved in the NdpKO retina (SI Appendix, Fig. S3C), consistent with oxidative stress in the NdpKO INL and/or decreased entry of pyruvate into the TCA cycle via acetyl-CoA.
An overrepresentation analysis, implemented with the MetaboAnalystR software (38), identified statistically significant up- and down-regulated metabolic pathways in NdpKO vs. WT retinas. Pathways that were most significantly up-regulated are centered around glutathione and one-carbon metabolism, including glycine, serine, threonine, cysteine, and methionine metabolism (Fig. 2 A, Left and C and SI Appendix, Fig. S3D, Left). There were marked increases in cystathionine and cysteine in the NdpKO retina (Fig. 2C), consistent with increased activity in the transsulfuration pathway. Pathways that were significantly down-regulated include the TCA cycle and the mitochondrial electron transport chain, which are known to be inhibited in chronic hypoxia (Fig. 2 A, Right and C and SI Appendix, Fig. S3D, Right) (36). These metabolite changes correlate broadly with gene expression changes as determined by GSEA of the scRNA-seq data from NdpKO vs. WT INL cell types (Fig. 2B), especially in glycine, serine, and threonine metabolism. However, some genes involved in oxidative phosphorylation are up-regulated in NdpKO retinas (Fig. 2B, Right and SI Appendix, Table S2), despite some metabolic intermediates in oxidative phosphorylation being down-regulated (Fig. 2C and SI Appendix, Table S1). This apparent discrepancy most likely reflects hypoxia-induced changes in the subunit composition of the mitochondrial respiratory chain complexes (39–41). More generally, apparent discrepancies between transcriptome and metabolome changes in NdpKO vs. WT retinas could represent homeostatic gene regulatory responses to altered levels of particular metabolites.
To localize and independently assess changes in metabolite levels, we performed computational molecular phenotyping (CMP) (42, 43). CMP utilizes immunogold staining of resin-embedded tissues with antibodies raised against hapten-metabolite conjugates followed by silver intensification. In general agreement with the metabolomics data (Fig. 2C), CMP showed increases in glycine and glutathione in the INL (Fig. 2D).
13C Metabolite Labeling Reveals the Requirement for de Novo Serine Synthesis in the NdpKO Retina.
Since the NdpKO vs. WT retina metabolomics analysis described above revealed differential signals in pathways linked to glucose metabolism, we performed in vivo metabolite labeling with uniformly labeled 13C-glucose by injecting NdpKO and WT mice i.p. with 2 g/kg 13C-glucose and harvesting retinas 45 min later (four groups of mice per genotype) (Dataset S2). In Fig. 3 and SI Appendix, Fig. S4, we refer to chemical species observed on MS as “M + X” to indicate X additional mass units beyond the nonlabeled species. Since the natural isotope of carbon is 12C, each 13C adds one mass unit.
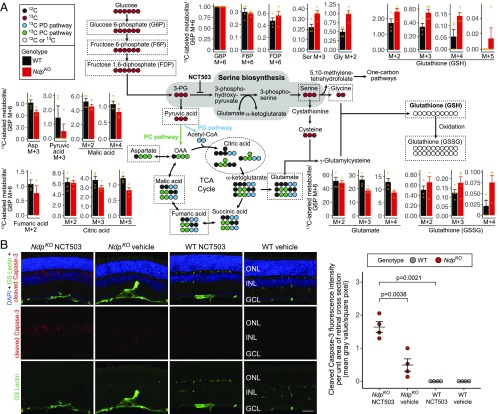
Fig. 3.
Metabolic flux analysis and pharmacological inhibition reveal enhanced de novo serine biosynthesis in NdpKO retinas in vivo. (A) Schematic showing the fates of 13C atoms from uniformly labeled 13C-glucose. For metabolites enclosed in dashed lines in the schematic, the surrounding bar plots show the mean ratios of the measured abundances of 13C-labeled metabolites to the abundance of 13C-glucose 6-phosphate (M + 6) for WT and NdpKO retinas. Orange dots represent individual samples. PC pathway, pyruvate carboxylase pathway; PD pathway, pyruvate dehydrogenase pathway. (B) The effect on apoptotic pathway activation of systemic treatment with NCT503 (an inhibitor of PHGDH; 40 mg/kg vs. vehicle, daily i.p. injections for 12 d). (Left) Representative images of immunofluorescence of retina cross-sections showing blood vessels (GS Lectin; green), cleaved Caspase-3 (red), and nuclei (DAPI; blue). (Right) Quantification of cleaved Capase-3 immunofluorescence of two random whole-retina cross-sections per eye. Horizontal bars represent the mean and the SEM. GCL, ganglion cell layer; ONL, outer nuclear layer.
As a preliminary experiment, we injected NdpKO and WT mice i.p. with 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG), a fluorescent glucose analog, and found that, despite the hypovascularization of the NdpKO retina, 2-NBDG was present in the INL of the NdpKO retina at a higher level compared with the WT retina (SI Appendix, Fig. S4A). This apparent paradox is likely explained by the loss of vascular barrier function in the NdpKO retina, which allows rapid equilibration of low-molecular weight compounds between serum and the extravascular space (18). Interestingly, the scRNA-seq data showed that the genes coding for hexokinase (Hk1 and Hk2), the enzyme responsible for the first enzymatic transformation of glucose (to glucose 6-phosphate) after cellular uptake, are down-regulated in photoreceptors but up-regulated in INL cell types in the NdpKO retina (SI Appendix, Fig. S4B). Overall, however, the uptake efficiency of 13C-glucose into retinal cells seems to be similar for WT and NdpKO mice as judged by the levels of glucose 6-phosphate M + 6 (SI Appendix, Fig. S4C), which constituted 16–26% of total glucose 6-phosphate in the samples (SI Appendix, Fig. S4D). For each sample in the 13C experiment, the abundances of labeled metabolites were divided by the level of glucose 6-phosphate (M + 6) to account for differences in 13C-glucose uptake (Dataset S3). The overall pattern of 13C-labeled metabolites showed a clear distinction between NdpKO and WT samples by PCA (SI Appendix, Fig. S4E).
Despite the dilution of 13C-glucose throughout the body, the small amount of retinal tissue, and the relatively brief labeling period, there was detectable 13C incorporation into multiple metabolites across glycolysis, the TCA cycle, the pentose phosphate pathway, and glutathione metabolism (Fig. 3A and SI Appendix, Fig. S4F). NdpKO retinas showed decreases in glutamate and in multiple 13C-labeled TCA cycle metabolites (Fig. 3A and SI Appendix, Table S3). In contrast, serine M + 3 and glycine M + 2 were increased in the NdpKO samples, suggesting an increase in glucose flux into de novo serine synthesis and serine metabolism (Fig. 3A). Although transcripts coding for the rate-limiting enzyme in de novo serine synthesis, phosphoglycerate dehydrogenase (PHGDH), were only detected at low abundance on scRNA-seq and show no significant difference in INL cell types, immunofluorescence showed an apparent increase in PHGDH protein in Muller glia in NdpKO retinas (compare staining intensity of radial processes in SI Appendix, Fig. S4H). The increase in 13C-labeled serine and glycine was accompanied by an increase in 13C-labeled reduced and oxidized glutathione (GSH and GSSG, respectively) (Fig. 3A), which is consistent with the increase in steady-state serine, glycine, and glutathione seen in the nonisotopic metabolomics data (Fig. 2 C and D). Increased de novo serine synthesis is likely to contribute to increased glutathione synthesis through the transsulfuration pathway as evidenced by the marked increases in cystathionine and cysteine in the NdpKO retina (Fig. 2C).
De novo serine synthesis is an important source of glutathione (Fig. 3A), a major cellular antioxidant. In cultured Muller glia, disruption of de novo serine synthesis through inhibition of PHGDH reduces glutathione levels and increases cellular damage under mild oxidative stress (44). We, therefore, hypothesized that inhibiting de novo serine synthesis could exacerbate cellular oxidative stress specifically in the hypoxic NdpKO inner retina. To test this hypothesis, we injected NdpKO mice and their WT littermates i.p. with 40 mg/kg NCT503 or vehicle control daily for 12 consecutive days and then analyzed their retinas for cleaved Caspase-3, a marker of apoptosis. NCT503 is a well-characterized small molecule inhibitor of PHGDH (45). As seen in Fig. 3B, WT retinas show undetectable levels of cleaved Caspase-3, irrespective of whether they have been treated with vehicle or NCT503, whereas NdpKO retinas show low levels of cleaved Caspase-3 after vehicle injection and high levels of cleaved Caspase-3 after NCT503 injection. In both cases, the cleaved Caspase-3 was confined to the INL, and it was enriched in regions of cellular disorganization that resemble the cystoid lesions that develop in the INL in older NdpKO retinas (22), suggesting that NCT503 may accelerate what would otherwise be a slower degenerative process. These data are consistent with a model in which a combination of hypoxia and reduced de novo serine synthesis produces a level of cellular stress that is sufficient to activate Caspase-3 cleavage.
Muller Glia in the NdpKO Retina Exhibit Distinct Transcriptional Changes.
In the NdpKO retina, Vegfa transcripts are selectively up-regulated in Muller glia (18). To verify additional transcriptome changes in NdpKO Muller glia identified with scRNA-seq, a Muller glia-specific CreER transgene (GLAST-CreER) (46) was crossed to RiboTag mice (47) to activate production of an epitope-tagged ribosomal protein in a Cre recombinase-dependent manner in Muller glia. This allowed us to immunoaffinity purify polyribosomes from Muller glia for RNA-seq. We will refer to the resulting RNA-seq datasets as “Muller glia-RiboTag” datasets.
Duplicate NdpKO and WT RNA-seq datasets from whole retina and from Muller glia-RiboTag–purified transcripts were appropriately clustered (SI Appendix, Fig. S5A). To identify patterns in the Muller glia-enriched and whole-retina RNA-seq datasets in an unbiased fashion, we used the Coordinated Gene Activity in Pattern Sets (CoGAPS) algorithm (48). CoGAPS decomposes gene expression into a set of patterns that quantify the association of each sample with a vector of relative gene expression changes. Unlike clustering methods, CoGAPS allows a gene to contribute to more than one pattern, which more effectively captures the complexity of gene expression patterns associated with specific cell types and physiologic perturbations.
CoGAPS identified a pattern with higher weights in Muller glia-RiboTag samples compared with the whole-retina samples (Fig. 4A, Left). When projected into the scRNA-seq dataset using the projectR software (49), this Muller glia-RiboTag–enriched CoGAPS pattern showed the highest projection scores in the Muller glia cluster for both NdpKO and WT scRNA-seq datasets, consistent with the expectation that it reflects a Muller glial transcriptional signature. Indeed, known Muller glia markers (50) were found to specifically have high gene weights for this pattern and also showed significant enrichment in both Muller glia-Ribotag samples and Muller glia scRNA-seq data for NdpKO and WT samples (Fig. 4B). In addition, this pattern showed higher weights in the NdpKO compared with the WT Muller glia-RiboTag samples (Fig. 4A) and higher projection scores for NdpKO than WT Muller glia in the scRNA-seq data (Fig. 4C), implying that this CoGAPS pattern also includes a hypoxia response signature.
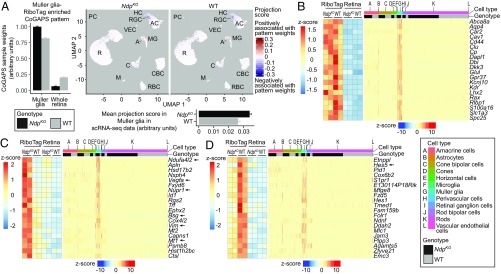
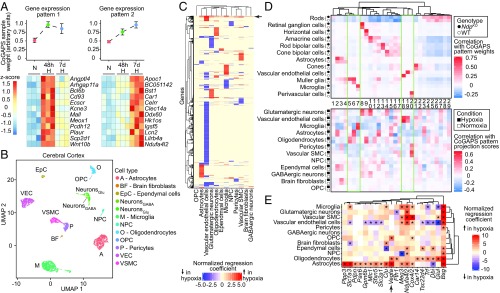
Fig. 4.
RiboTag and scRNA-seq analysis of Muller glia-enriched transcripts in WT and NdpKO retinas converge on a common set of differentially expressed genes. (A, Left) CoGAPS sample weights of the Muller glia-RiboTag–enriched pattern for Muller glia-RiboTag and whole-retina samples. (A, Upper Right) Projections of the Muller glia-RiboTag–enriched pattern into WT and NdpKO scRNA-seq datasets visualized on UMAP plots. (A, Lower Right) Mean projection scores of the Muller glia-RiboTag–enriched pattern in NdpKO and WT Muller glia clusters of the scRNA-seq datasets. Error bars represent the SEM. (B) Expression of Muller glia-enriched transcripts in Muller glia-RiboTag vs. whole-retina RNA-seq (Left) and in scRNA-seq (Right). In B–D, each cell in the scRNA-seq dataset is represented by a point along the horizontal axis, and the regions of the plot corresponding to different cell types and to WT vs. NdpKO are color coded across the top. (C) Analysis as in B, except that C shows Muller glia-enriched transcripts that were also up-regulated in NdpKO retinas in the Muller glia-RiboTag RNA-seq (Left) and scRNA-seq datasets (Right). (D) Analysis as in B, except that D shows Muller glia-enriched transcripts that were also down-regulated in NdpKO retinas in the Muller glia-RiboTag RNA-seq (Left) and scRNA-seq datasets (Right). As seen in B–D, among retinal cells, retinal astrocytes bear the closest resemblance to Muller glia as judged by their patterns of transcript abundances. A, astrocyte; AC, amacrine cell; C, cone; CBC, cone bipolar cell; HC, horizontal cell; M, microglia; MG, Muller glia; PC, perivascular cell; R, rod; RBC, rod bipolar cell; VEC, vascular endothelial cell.
We next intersected statistically significant (q value ≤ 0.05) gene expression changes in Muller glia from the RiboTag and scRNA-seq datasets. Using as thresholds a specificity score of ≥0.4 (Monocle2) for Muller glia in the scRNA-seq dataset and twofold enrichment for the Muller glia-RiboTag–enriched dataset, there were 27 up- and 65 down-regulated Muller glia-enriched genes in the intersection of the two datasets (SI Appendix, Fig. S5 B and C and Table S4); 24 of 27 up-regulated Muller enriched genes are also specific to the Muller glia CoGAPS pattern described above. Twenty examples each from the up- and down-regulated Muller glia-enriched genes are shown in Fig. 4 C and D. For five differentially regulated genes—Vegfa, Ndufa4l2, Bsg, Mt1, and Hes1—in situ hybridization to sections of NdpKO and WT retinas confirmed the changes in transcript abundance in the INL in NdpKO retinas (Fig. 5 A and B and SI Appendix, Fig. S2 E and F for Bsg). Immunostaining confirmed the increased abundances of the proteins encoded by the up-regulated genes Nupr1 and Vim (Fig. 5C). Gfap was identified as a Muller-enriched up-regulated gene in the scRNA-seq analysis (SI Appendix, Fig. S5E) but did not pass the enrichment threshold for RiboTag analysis. Immunostaining confirmed the increased abundance of GFAP in Muller glia (SI Appendix, Fig. S5F), suggesting that the scRNA-seq analysis may be able to better discern cell type-specific gene expression changes compared with the RiboTag analysis.
Fig. 5.
Histochemical assessment of transcripts and proteins validates differentially regulated Muller glia genes in WT and NdpKO retinas. (A) Abundance changes for four transcripts that are up-regulated in NdpKO compared with WT retinas. In situ hybridization shows increased abundance in the INL in the NdpKO retina. (Scale bar: 50 μm.) (B) Fluorescent Hes1 in situ hybridization shows decreased abundance in the INL in the NdpKO retina. The boxed region encompassing the INL is enlarged in Right. (Scale bars: 50 μm.) (C, Upper) Immunofluorescence of retina cross-sections showing blood vessels (GS Lectin; green), Nupr1 (red), and nuclei (DAPI; blue). NUPR1 is present in scattered Muller glial cell bodies in the NdpKO retina but is undetectable in WT retinas. (C, Lower) Immunofluorescence of retina cross-sections showing blood vessels (GS Lectin; green), Vimentin (Vim; red), and nuclei (DAPI; blue). Vimentin is modestly elevated in the NdpKO retina compared with the WT retina. GCL, ganglion cell layer; ONL, outer nuclear layer. (Scale bars: 50 μm.)
The Muller glia genes that are up-regulated in NdpKO retinas code for a functionally diverse set of proteins. Ndufa4l2 codes for a hypoxia-induced mitochondrial protein that inhibits Complex I and reduces oxygen consumption (41). Mt1 codes for Metallothionien-1, an intracellular metal chelator, that is induced by diverse cellular stresses, including hypoxia (51). Nupr1 codes for a nuclear protein that protects against DNA damage-induced cell death in hypoxia (52). Vim and Gfap code for Vimentin and Glial Fibrillary Acidic Protein, respectively, which are intermediate filaments up-regulated in Muller glia in response to a variety of retinal insults, and they are postulated to protect against mechanical stress (53). Hes1 is downstream of Notch signaling, and it is down-regulated in the INL in NdpKO retinas. Notch signaling has been implicated in generating and maintaining glial identity in postmitotic Muller glia (54).
A Comparison of Transcriptional Changes in NdpKO Retinas and in Chronic Brain Hypoxia.
Since the retina is part of the CNS, it was of interest to compare the gene expression changes identified in the hypoxic NdpKO INL with those induced by a similar stress in the brain. For this comparison, chronic global hypoxia in the brain—induced by housing mice for up to 7 d in an atmosphere of 7.5% oxygen—is presumed to be similar to the hypoxic NdpKO INL, since the former does not reduce the exchange of molecules other than oxygen.
In an initial set of experiments, cages housing 8- to 10-wk-old C57BL/6J male mice were maintained in room air (∼21% oxygen) or were placed in 7.5% oxygen for either 48 h or 7 d before harvesting their brains for bulk RNA-seq. CoGAPS analysis revealed two distinct patterns of gene expression responses to hypoxia (Fig. 6A and Dataset S4). The first pattern was characterized by an increase in transcript abundance between 0 and 48 h of hypoxia followed by a plateau or a decrease between 48 h and 7 d of hypoxia. The second pattern was characterized by a continuous increase in transcript abundance from 0 h through 7 d of hypoxia. Gene expression changes in response to 7 d of hypoxia that were not seen with 48 h of hypoxia likely represent the response to chronic hypoxia, suggesting that 7 d was sufficient to induce a chronically hypoxic state in the brain.
Fig. 6.
Single-cell RNA-seq reveals correlated transcriptional changes between chronically hypoxic mouse cerebral cortex and NdpKO INL cell types. (A, Upper) Plots showing CoGAPS sample weights of whole-brain RNA-seq from mice exposed to normoxia (N), 48-h hypoxia (48 h H), and 7-d hypoxia (7 d H) for two distinct expression patterns determined by CoGAPS. Error bars represent the SEM. (A, Lower) Heat maps for examples of genes specific to the two CoGAPS patterns, showing gene expression (z score) across triplicate samples for the three experimental conditions listed above the heat maps. Each column represents a sample. (B) UMAP plot of scRNA-seq data from cerebral cortex from mice exposed to normoxia (control) and 7-d hypoxia. NeuronsGABA, GABAergic neurons; NeuronsGlu, glutamatergic neurons; NPC, neural progenitor cells; OPC, oligodendrocyte precursor cells; VEC, vascular endothelial cells; VSMC, vascular smooth muscle cells. (C) Heat map showing normalized regression coefficients for transcripts with statistically significantly differences on scRNA-seq differential gene expression analysis in cortical cell types exposed to 7-d hypoxia vs. normoxia. The arrow points to a cluster of known hypoxia-indicible genes. (D, Upper) Heat map showing correlation of single-cell CoGAPS pattern weights with cell type and genotype in the retina scRNA-seq dataset. (D, Lower) Heat map showing correlation of cortical cell type and condition with projection scores of the retinal patterns above projected into the cortical scRNA-seq dataset. Patterns are numbered sequentially according to their order in the associated dendrogram. Each pattern in the heat map D, Upper is aligned with its corresponding projection in the heat map in D, Lower. (E) Heat map showing, for each cortical cell type, the normalized regression coefficients for transcripts enriched in the Muller glia-associated single-cell CoGAPS patterns that are also up-regulated in astrocytes and/or oligodendrocytes from 7-d hypoxia cerebral cortices. *q value of <0.05.
To characterize cell type-specific transcriptional changes in the chronically hypoxic brain, we performed scRNA-seq on 7,925 dissociated cells from cerebral cortices from 8- to 10-wk-old C57BL/6J male mice housed in room air or in 7.5% oxygen for 7 d, conditions that are referred to hereafter as normoxia and hypoxia, respectively. Based on known markers, we identified all of the major cortical cell types with similar yields in the normoxic and hypoxic samples (Fig. 6B and SI Appendix, Fig. S6 A, C, and D). SI Appendix, Fig. S6B shows that, in contrast to the interspersion of GABAergic and glutamatergic neurons in the UMAP plot of the entire dataset (Fig. 6B), these two cell types resolve into separate clusters when the input for the UMAP plot is restricted to neurons; 135 presumed multiplets were excluded from subsequent analyses.
Each cell type within the hypoxic cerebral cortex exhibits a distinctive set of up- and down-regulated transcripts as shown in the heat map in Fig. 6C. There are a small number of known hypoxia-inducible genes that exhibit common responses across multiple cell types (indicated by the horizontal arrow in Fig. 6C and shown in SI Appendix, Fig. S6E).
To compare transcriptome changes between the hypoxic retina and cortex at a cell type-specific level, we first defined a set of gene expression patterns in the NdpKO retina scRNA-seq dataset using CoGAPS (49) and projected these patterns into the hypoxic cortical scRNA-seq dataset (Fig. 6D). Several patterns that had high weights in specific cell types in the retina dataset had high projection weights in analogous cell types in the cortical dataset (enclosed by green lines in Fig. 6D): pattern 7 in microglia, patterns 18 and 19 in ECs, and pattern 20 in vascular smooth muscle cells and pericytes. As Muller glia exhibit distinctive transcriptome changes in the NdpKO retina (Fig. 4), it was of interest to assess the extent to which this response was shared with one or more hypoxic cortical cell types. By CoGAPS analysis, patterns 5 and 6 most closely resembled the Muller glia-specific hypoxic response (enclosed by green lines in Fig. 6D). When projected onto the cortical dataset, these patterns showed the highest weights in astrocytes and oligodendrocytes.
Additional evidence for a functional relationship between Muller glia and cortical astrocytes and oligodendrocytes is seen in the heat map in Fig. 6E, which shows individual transcripts that are most enriched in patterns 5 and 6 (Fig. 6D) and that are also up-regulated by hypoxia in either cortical astrocytes or oligodendrocytes. Only a few of these genes are up-regulated in cortical neurons (Fig. 6E). Interestingly, Vegfa induction is more significant in astrocytes, oligodendrocytes, and oligodendrocyte precursor cells than in neurons (arrow in Fig. 6E), reminiscent of its Muller glia-specific induction in the NdpKO retina (Fig. 4C).
Brain Vascular EC Responses to Hypoxia.
The scRNA-seq analysis of normoxic vs. hypoxic cerebral cortex presents an opportunity to explore the CNS EC-specific transcriptional changes in response to chronic tissue hypoxia. For a global assessment of transcript changes, we computed transcripts per million (TPM) from aggregated transcript abundances in either normoxic or hypoxic cortical ECs. For the identification of subtle and/or heterogenous gene expression changes, this approach is less sensitive than the differential gene expression test in Monocle2: when up- and down-regulated EC transcripts identified by Monocle2 are plotted on a TPM scatterplot, many show only small changes in TPM (SI Appendix, Fig. S7A). CNS EC-enriched transcripts (SI Appendix, Fig. S7A) [more than twofold enriched in CNS ECs relative to non-ECs (31)] and blood–brain barrier (BBB) transcripts (Fig. 7A) [more than twofold enriched in brain ECs relative to non-CNS ECs (31)] also show minimal differences in computed TPM between normoxic and hypoxic cortical ECs, consistent with previous observations that the BBB in cortical ECs is not disrupted in chronic hypoxia (10). To examine CNS EC subtypes, we combined published data and a recent scRNA-seq analysis of postnatal day 7 mouse brain ECs (31) to define subsets of transcripts specifically enriched in arterial ECs, venous ECs, and tip cells. (Tip cells are the highly motile ECs at the growing front of an angiogenic vascular plexus.) Arterial and venous transcripts show little or no change in abundance between normoxic and hypoxic conditions (Fig. 7A, panels 2 and 3). In contrast, markers for tip cells show a clear trend toward increased abundance in hypoxic ECs relative to normoxic ECs (Fig. 7A, panel 4).
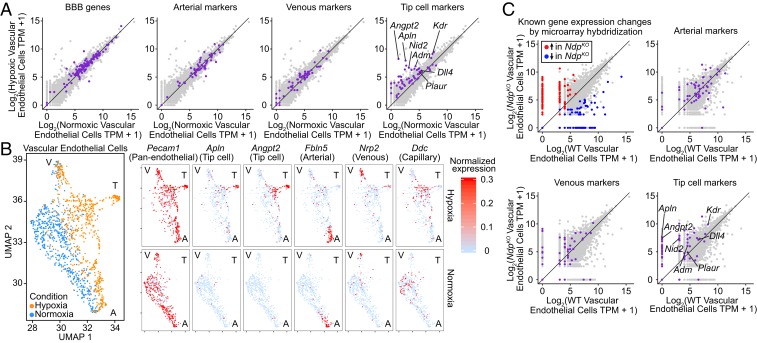
Fig. 7.
Brain and retinal vascular ECs activate the tip cell transcriptional program in response to chronic tissue hypoxia. (A) Scatterplots showing TPM for all transcripts expressed in normoxic and/or hypoxic cortical ECs (light gray) obtained by pooling transcript counts in the cortical scRNA-seq dataset. Transcripts previously defined as enriched in BBB-type, arterial, venous, or tip cell ECs are colored purple on individual plots. A 45° line is shown in each plot. (B, Left) UMAP plot for hypoxic and normoxic cortical ECs. (B, Right) Separate plots for each transcript showing hypoxic vs. normoxic cortical ECs [based on the combined UMAP plot (B, Left)] color coded by the level of normalized expression for a panendothelial marker (Pecam1), two tip cell markers (Apln and Angpt2), an arterial marker (Fbln5), a venous marker (Nrp2), and a capillary marker (Ddc). A, artery; T, tip cell; V, vein. (C) Scatterplots as in A showing TPM for all transcripts expressed in WT and NdpKO retinal ECs (light gray) obtained by pooling transcript counts in the retina scRNA-seq dataset. (C, Upper Left) Transcripts previously shown by microarray hybridization of immunoaffinity-purified ECs to be up- or down-regulated in NdpKO compared with WT ECs are highlighted (14). (C, Upper Right and Lower Right) Transcripts previously defined as enriched in arterial, venous, or tip cell ECs are colored purple on individual plots. A 45° line is shown in each plot.
We next visualized the cell by cell abundances of individual arterial, venous, and tip cell transcripts in normoxic and hypoxic ECs on a UMAP plot generated using transcripts specific to each of the EC subtypes previously defined (31) (Fig. 7B and SI Appendix, Fig. S7B). These plots reveal the clustering of venous and arterial EC subtypes at the top and bottom, respectively, with capillary ECs in the central region. Strikingly, at the right side of the UMAP plots, there is a cluster of ECs that consistently expresses tip cell markers and is only present in the hypoxic sample. Tip cell transcripts are also more abundant among ECs in the central region of the UMAP plot (corresponding to capillary ECs) in the hypoxic sample compared with the normoxic control.
We next asked whether an up-regulation of tip cell transcripts occurs in the hypoxic retina in the context of the NdpKO retinal vasculature. As a preliminary quality control step, we first determined that an earlier analysis of NdpKO vs. WT retinal EC transcriptomes—obtained by immunoaffinity purifying retinal ECs followed by microarray hybridization (14)—had converged on the same set of differentially expressed genes defined in the scRNA-seq analysis (Fig. 7C, Upper Left). Although ECs in the hypoxic cerebral cortex and ECs in the NdpKO retina are both in close proximity to hypoxic parenchymal cells, they differ with respect to (i) canonical beta-catenin signaling, which is active in cortical ECs and greatly reduced in NdpKO retinal ECs, and (ii) the luminal blood oxygen level, which is presumably lower in the hypoxic cortical vasculature than in the normoxic NdpKO retinal vasculature. Despite these differences, the basic pattern of up-regulation of tip cell transcripts with essentially no alteration in arterial and venous transcripts is conserved between these two models of CNS hypoxia (Fig. 7C). The greater noise in the retina EC dataset compared with the cortical EC dataset likely reflects the smaller number of ECs in the retinal scRNA-seq samples.
Taken together, these analyses show that, with 1 wk of chronic hypoxia, a substantial fraction of cortical ECs is mobilized to turn on tip cell genes, presumably as part of an adaptive neoangiogenic program to increase vascular density and cerebral blood flow (5, 10). A similar conversion characterizes the NdpKO retinal vasculature and is presumably responsible for the hyperdense capillary network that develops on the vitreal face of the NdpKO retina (18), implying that the tip cell genes analyzed here can be activated in the absence of canonical Wnt signaling. It is plausible that the tip cell transcriptional program is activated in CNS ECs in response to VEGFA (55) produced by hypoxic glia in both the brain and retina.
Discussion
In this study, we have combined scRNA-seq, RiboTag sequencing, metabolic profiling and flux analysis, pharmacologic inhibition, and ultrastructural and immunohistochemical analyses to (i) define the response of the NdpKO retina to chronic hypoxia and (ii) compare the retina’s hypoxia response with that of the cerebral cortex subjected to 7 d of hypoxia. For both retina and cortex, determining gene expression changes at single-cell resolution presented a critical advantage over whole-tissue analysis, since many of the cell type-specific responses would have been difficult or impossible to detect if gene expression had been analyzed in bulk.
Metabolic Responses of the NdpKO Retina.
The preservation of a nearly normal ATP level and ATP/ADP ratio in NdpKO retinas is striking, and it implies the existence of powerful homeostatic mechanisms that conserve ATP in chronic hypoxia. Since the plasma membrane Na/K-ATPase is responsible for ∼50% of ATP hydrolysis in the CNS (56), we speculate that such a reduction in ATP consumption could be brought about through a reduction in Na/K-ATPase activity mediated by hypoxia-related oxidative stress-induced nonenzymatic glutathionylation and/or PKC phosphorylation (57). Such posttranslational regulation of Na/K-ATPase activity and its rapid reversal would also dynamically affect neuronal membrane potential and account for the rapidly reversible reduction in inner retinal neurotransmission in NdpKO retinas (14).
Global metabolic profiling of NdpKO retinas shows prominent metabolic changes centered around glutathione and one-carbon metabolism. Immunostaining for selected metabolites shows that some of these changes are localized to the hypoxic INL of the NdpKO retina. Similar metabolic changes have been described in hypoxic cancer cells (58). These metabolic changes mirror recent findings in mitochondrial disorders, where mitochondrial respiratory chain dysfunction leads to changes in glutathione and one-carbon metabolic pathways (59, 60). We speculate that hypoxia-induced defects in mitochondrial respiration and induction of oxidative stress in the NdpKO INL may be analogous to the metabolic adaptations in primary mitochondrial disorders.
The role of glucose-driven de novo serine synthesis in glutathione and one-carbon metabolism has been described recently (45, 59, 60). Here, we provide in vivo evidence for increased flux of glucose to serine in the NdpKO retina. This finding is especially intriguing, as it implies that cells in the NdpKO retina—despite being under energy stress due to hypoxia—divert glucose away from pyruvate production and toward the synthesis of serine, a nonessential amino acid. The importance of this pathway for cell survival in the context of chronic hypoxia is implied by our observation that inhibition of de novo serine synthesis by the PHGDH inhibitor NCT503 resulted in increased levels of cleaved Caspase-3 in the NdpKO INL but not in the WT INL.
Muller Glia, Cortical Astrocytes, and Oligodendrocytes.
Muller glia, the major macroglia in the retina, exhibit a distinct gene expression profile in health (50, 54) and in disease (61). Here, we combined single-cell transcriptomics and RiboTag technology to further characterize the gene expression changes in Muller glia in NdpKO retinas. Notably, transcripts coding for NDUFA4L2, which inhibits mitochondrial Complex I, were up-regulated and enriched in NdpKO Muller glia, but they were unchanged in neurons. We also found evidence for lower Notch signaling in Muller glia as evidenced by a reduction in transcripts coding for HES1 and HES5. It is unclear if reduced Notch signaling is a specific response to hypoxia or a more generic response to cellular stress. In avian (62) and zebrafish (63) retinas, a reduction in Notch signaling in Muller glia has been linked to induction of a stem cell-like state through dedifferentiation and reentry into the cell cycle. Mammalian Muller glia seem to have a more limited capacity for this conversion (64).
We also compared the single-cell transcriptomes of the NdpKO retina and the hypoxic cerebral cortex. Similar to the NdpKO INL, glia and neurons in the hypoxic cortex displayed distinct changes in gene expression. Compared with other cell types in the cerebral cortex, astrocytes and oligodendrocytes exhibit a transcriptional response to hypoxia that most closely resembles the response of NdpKO Muller glia. This similarity suggests similar roles for cortical and retinal glia in mitigating the hypoxic stress of surrounding neurons.
ECs in the Hypoxic CNS.
The single-cell transcriptome analysis of hypoxic vs. normoxic cerebral cortex has provided a global view of the changes in EC gene expression that are associated with CNS hypoxia. This response is striking in its simplicity: capillary ECs in the hypoxic cerebral cortex activate an adaptive and previously dormant tip cell program of gene expression. We presume that this represents the gene expression correlate of the microvascular sprouting and formation of new capillaries observed by two-photon imaging in the hypoxic mouse cerebral cortex during an extended hypoxic exposure (5). In the NdpKO retina, ECs proliferate locally to form glomeruloids (18) but fail to form new intraretinal capillaries, despite the induction of the tip cell transcriptional program. Whether this failure reflects an additional requirement for beta-catenin signaling (which is absent in NdpKO retinal ECs), a nonpermissive environment for angiogenesis within the mature retina, or some combination of the two is unknown.
Clinical Relevance.
This work has implications beyond the pathophysiology of Norrie disease. The observation that NdpKO retinas exhibit an increase in de novo serine biosynthesis and that INL cells in NdpKO retinas are uniquely sensitive to inhibition of that pathway may be of relevance to the pathophysiology of macular telangiectasia (MacTel). MacTel is characterized by abnormal blood vessels in the fovea or perifoveal region, reduced capillary density in the surrounding retina (65), and loss of perifoveal Muller glia (66, 67). The disease phenotype has been modeled by genetic ablation of Muller glia in the mouse retina (68). A recent genome-wide association study of MacTel patients implicated genes involved in de novo serine biosynthesis and its associated one-carbon pathways, and it also demonstrated that serum levels of serine and glycine are decreased in MacTel cases compared with controls (69). These observations are consistent with a model in which a reduction in de novo serine synthesis and/or one-carbon metabolism renders the retina more susceptible to oxidative stress and/or hypoxia. Because complete disruption of de novo serine synthesis in Phgdh−/− mice leads to severe defects in CNS development (70), it would be interesting to determine whether heterozygosity for Phgdh deficiency alters neuronal function or survival in models of MacTel or other CNS diseases.
This work also has implications for hypoxia tolerance in the CNS more generally. First, the reversible inactivation of neurotransmission in the NdpKO inner retina implies that, with a reduced blood supply, neurons can remain alive for months in a state of suspended animation and still recover their excitability. This natural history might account for some of the functional recovery that accompanies revascularization in the penumbra of a stroke (1). Second, the oxidative stress that accompanies hypoxia in the NdpKO INL—as inferred from the accumulation of pimonidazole and the enhanced synthesis of glutathione—supports the counterintuitive idea that reduced oxygen availability is associated with increased oxidative stress, most likely due to reduced efficiency of the electron transport chain (36). The coupling of hypoxia and oxidative stress suggests that dietary supplements that support antioxidant defenses might be broadly efficacious in patients with diverse diseases involving CNS hypoxia, including FEVR, chronic obstructive pulmonary disease, diabetic retinopathy, and cerebrovascular disease. Finally, identifying the cell type-specific changes in gene expression that are associated with chronic CNS hypoxia suggests the possibility that selective pharmacological enhancement of these gene expression programs could promote cell survival and recovery.
Materials and Methods
All animal experiments were conducted in accordance with the approved Institutional Animal Care and Use Committee protocol MO16M367 of the Johns Hopkins Medical Institutions. Mouse husbandry, single-cell isolation, scRNA-seq, RNA-seq, RiboTag experiments, in situ hybridization, immunostaining, metabolomics, in vivo 13C-glucose labeling, and in vivo NCT503 experiments are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Robert E. Marc for advice on CMP; Laura Shelton and Takushi Oga (Human Metabolome Technologies) for their assistance with metabolomics; Kakali Sarkar and Melissa Olson [Johns Hopkins Medical Institutions (JHMI) Genetics Research Core Facility] for constructing 10X libraries; David Mohr (JHMI Genetics Research Core Facility), Haiping Hao (JHMI Deep Sequencing Core Facility), and Linda Orzolek (JHMI Deep Sequencing Core Facility) for NextGen sequencing; Sean Hackett and Peter Campochiaro for assistance with the brain hypoxia experiments; Mike Delannoy (JHMI Microscope Core Facility) for his assistance with EM; Mark Sabbagh and Yanshu Wang for their advice and assistance; and Gregg Semenza, Bindu Paul, and Michael Pacold for helpful discussions. This work was supported by a Genetics Research Core Facility Core Coins Grant from the Johns Hopkins School of Medicine (to J.S.H. and J.N.); the Thomas J. Kelly and Mary L. Kelly Young Scholar Award (to J.S.H.); Chan–Zuckerberg Initiative Donor Advised Fund Grant 2018-183445 (to G.L.S.-O. and L.A.G.); National Eye Institute (NIH) Grants EY015128 (to B.W.J.), EY014800 Vision Core (to B.W.J.), and R01EY018637 (to J.N.); an unrestricted grant from Research to Prevent Blindness, Inc. [to the Department of Ophthalmology & Visual Sciences, University of Utah (to B.W.J.)]; Johns Hopkins University Catalyst and Synergy awards (to L.A.G.); NSF Grant IOS-1665692 (to L.A.G.); the Howard Hughes Medical Institute (J.N.); the Arnold and Mabel Beckman Foundation (J.N.); and Mr. David Labovitz (J.N.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE125708). The annotated datasets can be viewed at https://jacobheng.shinyapps.io/cnshypoxia/ and loom.gofflab.org. Supplementary code for processing and visualizing the scRNA-seq data can be found in an R package, cellwrangler (https://github.com/jacobheng/cellwrangler).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821122116/-/DCSupplemental.
References
- 1.Seitz RJ, Donnan GA. Recovery potential after acute stroke. Front Neurol. 2015;6:238. doi: 10.3389/fneur.2015.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tobalem S, Schutz JS, Chronopoulos A. Central retinal artery occlusion–Rethinking retinal survival time. BMC Ophthalmol. 2018;18:101. doi: 10.1186/s12886-018-0768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrido E, et al. Are Himalayan Sherpas better protected against brain damage associated with extreme altitude climbs? Clin Sci (Lond) 1996;90:81–85. doi: 10.1042/cs0900081. [DOI] [PubMed] [Google Scholar]
- 4.Fan C, et al. Reversible brain abnormalities in people without signs of mountain sickness during high-altitude exposure. Sci Rep. 2016;6:33596. doi: 10.1038/srep33596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takuwa H, et al. Long-term adaptation of cerebral hemodynamic response to somatosensory stimulation during chronic hypoxia in awake mice. J Cereb Blood Flow Metab. 2013;33:774–779. doi: 10.1038/jcbfm.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari M, et al. Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc Natl Acad Sci USA. 2017;114:E4241–E4250. doi: 10.1073/pnas.1621511114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickler PE, Donohoe PH. Adaptive responses of vertebrate neurons to hypoxia. J Exp Biol. 2002;205:3579–3586. doi: 10.1242/jeb.205.23.3579. [DOI] [PubMed] [Google Scholar]
- 8.Gidday JM. Cerebral preconditioning and ischaemic tolerance. Nat Rev Neurosci. 2006;7:437–448. doi: 10.1038/nrn1927. [DOI] [PubMed] [Google Scholar]
- 9.Bernaudin M, Tang Y, Reilly M, Petit E, Sharp FR. Brain genomic response following hypoxia and re-oxygenation in the neonatal rat. Identification of genes that might contribute to hypoxia-induced ischemic tolerance. J Biol Chem. 2002;277:39728–39738. doi: 10.1074/jbc.M204619200. [DOI] [PubMed] [Google Scholar]
- 10.Masamoto K, et al. Microvascular sprouting, extension, and creation of new capillary connections with adaptation of the neighboring astrocytes in adult mouse cortex under chronic hypoxia. J Cereb Blood Flow Metab. 2014;34:325–331. doi: 10.1038/jcbfm.2013.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmour DF. Familial exudative vitreoretinopathy and related retinopathies. Eye (Lond) 2015;29:1–14. doi: 10.1038/eye.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Q, et al. Vascular development in the retina and inner ear: Control by Norrin and frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 13.Junge HJ, et al. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/β-catenin signaling. Cell. 2009;139:299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 14.Ye X, et al. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell. 2009;139:285–298. doi: 10.1016/j.cell.2009.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luhmann UFO, et al. Role of the Norrie disease pseudoglioma gene in sprouting angiogenesis during development of the retinal vasculature. Invest Ophthalmol Vis Sci. 2005;46:3372–3382. doi: 10.1167/iovs.05-0174. [DOI] [PubMed] [Google Scholar]
- 16.Xia C-H, et al. A model for familial exudative vitreoretinopathy caused by LPR5 mutations. Hum Mol Genet. 2008;17:1605–1612. doi: 10.1093/hmg/ddn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, et al. Optical coherence tomography angiography in familial exudative vitreoretinopathy: Clinical features and phenotype-genotype correlation. Invest Ophthalmol Vis Sci. 2018;59:5726–5734. doi: 10.1167/iovs.18-25377. [DOI] [PubMed] [Google Scholar]
- 18.Rattner A, Wang Y, Zhou Y, Williams J, Nathans J. The role of the hypoxia response in shaping retinal vascular development in the absence of Norrin/Frizzled4 signaling. Invest Ophthalmol Vis Sci. 2014;55:8614–8625. doi: 10.1167/iovs.14-15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, et al. Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell. 2012;151:1332–1344. doi: 10.1016/j.cell.2012.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck SC, et al. Long-term consequences of developmental vascular defects on retinal vessel homeostasis and function in a mouse model of Norrie disease. PLoS One. 2017;12:e0178753. doi: 10.1371/journal.pone.0178753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018;37:38–44. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 22.Beck SC, et al. Cystoid edema, neovascularization and inflammatory processes in the murine Norrin-deficient retina. Sci Rep. 2018;8:5970. doi: 10.1038/s41598-018-24476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu X, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberzon A, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wangsa-Wirawan ND, Linsenmeier RA. Retinal oxygen: Fundamental and clinical aspects. Arch Ophthalmol. 2003;121:547–557. doi: 10.1001/archopht.121.4.547. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Philp NJ, Ochrietor JD, Rudoy C, Muramatsu T, Linser PJ. Loss of MCT1, MCT3, and MCT4 expression in the retinal pigment epithelium and neural retina of the 5A11/basigin-null mouse. Invest Ophthalmol Vis Sci. 2003;44:1305–1311. doi: 10.1167/iovs.02-0552. [DOI] [PubMed] [Google Scholar]
- 30.Ke X, et al. Hypoxia upregulates CD147 through a combined effect of HIF-1α and Sp1 to promote glycolysis and tumor progression in epithelial solid tumors. Carcinogenesis. 2012;33:1598–1607. doi: 10.1093/carcin/bgs196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabbagh MF, et al. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. eLife. 2018;7:e36187. doi: 10.7554/eLife.36187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno V, et al. An EMMPRIN-γ-catenin-Nm23 complex drives ATP production and actomyosin contractility at endothelial junctions. J Cell Sci. 2014;127:3768–3781. doi: 10.1242/jcs.149518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benita Y, et al. An integrative genomics approach identifies hypoxia inducible factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37:4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiota M, et al. Ets regulates peroxiredoxin1 and 5 expressions through their interaction with the high-mobility group protein B1. Cancer Sci. 2008;99:1950–1959. doi: 10.1111/j.1349-7006.2008.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabharwal SS, Waypa GB, Marks JD, Schumacker PT. Peroxiredoxin-5 targeted to the mitochondrial intermembrane space attenuates hypoxia-induced reactive oxygen species signalling. Biochem J. 2013;456:337–346. doi: 10.1042/BJ20130740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger SJ, et al. The distribution of the components of the cyclic GMP cycle in retina. J Biol Chem. 1980;255:3128–3133. [PubMed] [Google Scholar]
- 38.Chong J, et al. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 40.Hwang HJ, et al. Hypoxia inducible factors modulate mitochondrial oxygen consumption and transcriptional regulation of nuclear-encoded electron transport chain genes. Biochemistry. 2015;54:3739–3748. doi: 10.1021/bi5012892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tello D, et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1α decreases oxygen consumption by inhibiting complex I activity. Cell Metab. 2011;14:768–779. doi: 10.1016/j.cmet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Marc RE, Murry RF, Basinger SF. Pattern recognition of amino acid signatures in retinal neurons. J Neurosci. 1995;15:5106–5129. doi: 10.1523/JNEUROSCI.15-07-05106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marc RE, Jones BW. Molecular phenotyping of retinal ganglion cells. J Neurosci. 2002;22:413–427. doi: 10.1523/JNEUROSCI.22-02-00413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang T, et al. Disruption of de novo serine synthesis in müller cells induced mitochondrial dysfunction and aggravated oxidative damage. Mol Neurobiol. 2018;55:7025–7037. doi: 10.1007/s12035-017-0840-8. [DOI] [PubMed] [Google Scholar]
- 45.Pacold ME, et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol. 2016;12:452–458. doi: 10.1038/nchembio.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Melo J, et al. Injury-independent induction of reactive gliosis in retina by loss of function of the LIM homeodomain transcription factor Lhx2. Proc Natl Acad Sci USA. 2012;109:4657–4662. doi: 10.1073/pnas.1107488109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fertig EJ, Ding J, Favorov AV, Parmigiani G, Ochs MF. CoGAPS: An R/C++ package to identify patterns and biological process activity in transcriptomic data. Bioinformatics. 2010;26:2792–2793. doi: 10.1093/bioinformatics/btq503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein-O’Brien GL, et al. 2018. Decomposing cell identity for transfer learning across cellular measurements, platforms, tissues, and species. bioRxiv:10.1101/395004.
- 50.Roesch K, et al. The transcriptome of retinal Müller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miles AT, Hawksworth GM, Beattie JH, Rodilla V. Induction, regulation, degradation, and biological significance of mammalian metallothioneins. Crit Rev Biochem Mol Biol. 2000;35:35–70. doi: 10.1080/10409230091169168. [DOI] [PubMed] [Google Scholar]
- 52.Hamidi T, et al. Nupr1-aurora kinase A pathway provides protection against metabolic stress-mediated autophagic-associated cell death. Clin Cancer Res. 2012;18:5234–5246. doi: 10.1158/1078-0432.CCR-12-0026. [DOI] [PubMed] [Google Scholar]
- 53.Verardo MR, et al. Abnormal reactivity of muller cells after retinal detachment in mice deficient in GFAP and vimentin. Invest Ophthalmol Vis Sci. 2008;49:3659–3665. doi: 10.1167/iovs.07-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson BR, et al. Genome-wide analysis of Müller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS One. 2011;6:e22817. doi: 10.1371/journal.pone.0022817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blanco R, Gerhardt H. VEGF and Notch in tip and stalk cell selection. Cold Spring Harb Perspect Med. 2013;3:a006569. doi: 10.1101/cshperspect.a006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howarth C, Gleeson P, Attwell D. Updated energy budgets for neural computation in the neocortex and cerebellum. J Cereb Blood Flow Metab. 2012;32:1222–1232. doi: 10.1038/jcbfm.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fuller W, et al. Regulation of the cardiac sodium pump. Cell Mol Life Sci. 2013;70:1357–1380. doi: 10.1007/s00018-012-1134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samanta D, et al. PHGDH expression is required for mitochondrial redox homeostasis, breast cancer stem cell maintenance, and lung metastasis. Cancer Res. 2016;76:4430–4442. doi: 10.1158/0008-5472.CAN-16-0530. [DOI] [PubMed] [Google Scholar]
- 59.Nikkanen J, et al. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab. 2016;23:635–648. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 60.Bao XR, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife. 2016;5:e10575. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roesch K, Stadler MB, Cepko CL. Gene expression changes within Müller glial cells in retinitis pigmentosa. Mol Vis. 2012;18:1197–1214. [PMC free article] [PubMed] [Google Scholar]
- 62.Hayes S, Nelson BR, Buckingham B, Reh TA. Notch signaling regulates regeneration in the avian retina. Dev Biol. 2007;312:300–311. doi: 10.1016/j.ydbio.2007.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sifuentes CJ, Kim J-W, Swaroop A, Raymond PA. Rapid, dynamic activation of Müller glial stem cell responses in zebrafish. Invest Ophthalmol Vis Sci. 2016;57:5148–5160. doi: 10.1167/iovs.16-19973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao K, et al. Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature. 2018;560:484–488. doi: 10.1038/s41586-018-0425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chidambara L, et al. Characteristics and quantification of vascular changes in macular telangiectasia type 2 on optical coherence tomography angiography. Br J Ophthalmol. 2016;100:1482–1488. doi: 10.1136/bjophthalmol-2015-307941. [DOI] [PubMed] [Google Scholar]
- 66.Powner MB, et al. Perifoveal müller cell depletion in a case of macular telangiectasia type 2. Ophthalmology. 2010;117:2407–2416. doi: 10.1016/j.ophtha.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Len ACL, et al. Pilot application of iTRAQ to the retinal disease macular telangiectasia. J Proteome Res. 2012;11:537–553. doi: 10.1021/pr200889t. [DOI] [PubMed] [Google Scholar]
- 68.Chung SH, et al. Differential gene expression profiling after conditional Müller-cell ablation in a novel transgenic model. Invest Ophthalmol Vis Sci. 2013;54:2142–2152. doi: 10.1167/iovs.12-11559. [DOI] [PubMed] [Google Scholar]
- 69.Scerri TS, et al. MacTel Project Consortium Genome-wide analyses identify common variants associated with macular telangiectasia type 2. Nat Genet. 2017;49:559–567. doi: 10.1038/ng.3799. [DOI] [PubMed] [Google Scholar]
- 70.Yoshida K, et al. Targeted disruption of the mouse 3-phosphoglycerate dehydrogenase gene causes severe neurodevelopmental defects and results in embryonic lethality. J Biol Chem. 2004;279:3573–3577. doi: 10.1074/jbc.C300507200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.