Significance
We identify cellular senescence occurring in neural progenitor cells (NPCs) from primary progressive multiple sclerosis (PPMS). In this study, senescent progenitor cells were identified within demyelinated white matter lesions in progressive MS (PMS) autopsy tissue, and induced pluripotent stem-derived NPCs from patients with PPMS were found to express cellular senescence markers compared with age-matched control NPCs. Reversal of this cellular senescence phenotype, by treatment with rapamycin, restored PPMS NPC-mediated support for oligodendrocyte (OL) maturation. Proteomic and histological analyses identify senescent progenitor cells in PMS as a source of high-mobility group box-1, which limits maturation and promotes transcriptomic changes in OLs. These findings provide evidence that cellular senescence is an active process in PMS that may contribute to limited remyelination in disease.
Keywords: oligodendrocyte, aging, neural progenitor cell, rapamycin, proteomics
Abstract
Cellular senescence is a form of adaptive cellular physiology associated with aging. Cellular senescence causes a proinflammatory cellular phenotype that impairs tissue regeneration, has been linked to stress, and is implicated in several human neurodegenerative diseases. We had previously determined that neural progenitor cells (NPCs) derived from induced pluripotent stem cell (iPSC) lines from patients with primary progressive multiple sclerosis (PPMS) failed to promote oligodendrocyte progenitor cell (OPC) maturation, whereas NPCs from age-matched control cell lines did so efficiently. Herein, we report that expression of hallmarks of cellular senescence were identified in SOX2+ progenitor cells within white matter lesions of human progressive MS (PMS) autopsy brain tissues and iPS-derived NPCs from patients with PPMS. Expression of cellular senescence genes in PPMS NPCs was found to be reversible by treatment with rapamycin, which then enhanced PPMS NPC support for oligodendrocyte (OL) differentiation. A proteomic analysis of the PPMS NPC secretome identified high-mobility group box-1 (HMGB1), which was found to be a senescence-associated inhibitor of OL differentiation. Transcriptome analysis of OPCs revealed that senescent NPCs induced expression of epigenetic regulators mediated by extracellular HMGB1. Lastly, we determined that progenitor cells are a source of elevated HMGB1 in human white matter lesions. Based on these data, we conclude that cellular senescence contributes to altered progenitor cell functions in demyelinated lesions in MS. Moreover, these data implicate cellular aging and senescence as a process that contributes to remyelination failure in PMS, which may impact how this disease is modeled and inform development of future myelin regeneration strategies.
Cellular senescence is a component of the aging process that is increasingly recognized as an important pathophysiological mechanism associated with a variety of neurodegenerative human diseases (1, 2). Cellular senescence is distinct from cellular quiescence in that senescent cells develop a unique level of cellular activity that often exerts a proinflammatory paracrine effect on surrounding cells that can impair or alter tissue function, termed the senescence-associated secretory phenotype (SASP) (3, 4). Cellular senescence is linked with increased activity of mammalian target of rapamycin (mTOR) (5). Rapamycin has been found to decrease the production of the SASP through inhibition of mTORC1 (4), which has enabled studies on cellular plasticity under cellular senescence and the contribution of SASP factors to tissue and organ function. One notable influence of cellular senescence is on stem cells, where senescence has been shown to reduce trophic support and lessen regenerative capacity, which can contribute to impaired healing and tissue atrophy in aging and disease (6–8).
Progressive multiple sclerosis (PMS), which can develop either as primary (with no or minimal identified relapses) or secondary (with previous and sometimes ongoing relapses) progressive forms, represents the phase of MS where there is accrual of irreversible neurological disability. The unabating disability in PMS is attributed to chronic CNS demyelination and neurodegeneration with minimal remyelination (9). Promoting brain repair represents a potential strategy to restore neurological function in patients with PMS (10). However, the success of potential remyelinating therapies relies on endogenous oligodendrocyte progenitor cells (OPCs) to differentiate into myelinating oligodendrocytes (OLs) (11–15). Therefore, understanding what limits the potential of endogenous OPCs for remyelination will be required to realize remyelination as a therapeutic opportunity for regeneration.
Studies analyzing white matter lesion pathology in PMS brain tissues have identified immature, premyelinating oligodendroglia in association with progenitor cells (16). Endogenous and transplanted progenitor cells have been found to be capable of secreting factors that are antiinflammatory and support remyelination (17–20). Therefore, both of these cell types could be viewed as capable of promoting regeneration; however, OPC differentiation in vivo is limited and mostly arrested, resulting in chronic demyelination (21–24). We have recently identified that neural progenitor cells (NPCs) differentiated from induced pluripotent stem cells (iPSCs) derived from patients with primary PMS (PPMS) were less able to provide neuroprotection to myelin injury or support OPC differentiation in vitro (25). Importantly, OPCs differentiated from iPSC lines of patients with PPMS are equally capable of maturing into myelin-forming OLs (26), demonstrating that OPCs in this disease have potential that may be limited by a disease phenotype in NPCs.
In this study, we report evidence for cellular senescence in progenitor cells in PMS and provide functional evidence that this is a reversible process responsible for limiting OL maturation. These data suggest that while MS is not a disease of aging, at the cellular level, it may be an aging-related disease.
Materials and Methods
Human Tissue Immunohistochemistry.
Unfixed frozen postmortem tissue was provided by the UK Multiple Sclerosis Tissue Bank via a UK prospective donor scheme with full ethical approval (MREC/02/2/39) and the MRC Sudden Death Brain Bank (ethical approval LREC16/ES/0084) (SI Appendix, Table S1). Colorimetric staining was used to detect labeling in human tissue. Ten-micrometer sections were fixed in 4% paraformaldehyde (PFA), and antigen retrieval was performed by microwaving sections in Vector H-3300 antigen unmasking solution (Vector Laboratories). Slides were blocked with Bloxall blocking solution (Vector Laboratories), and further blocking was then performed with 2.5% horse serum (Vector Laboratories). Sections were incubated with the primary antibody high-mobility group box-1 (HMGB1; 1:500 mouse; BioLegend) or p16Ink4a (1:500 mouse; Thermo Fisher Scientific) in antibody diluent (Spring Bioscience, Abcam) overnight at 4 °C. Sections were incubated with Impress HRP anti-mouse and then colorized with 3,3′-diaminobenzidine (DAB; Vector Laboratories). Sections were washed and blocked in 2.5% horse serum and then incubated with antibodies against SOX2 (1:500 rabbit; Reprocell) overnight at 4 °C. Sections were incubated with Impress AP anti-rabbit, colorized with Vector Blue alkaline phosphatase, and then mounted in Fluoromount-G (Southern Biotech). Sections were imaged on a Zeiss Axio Scan.Z1 microscope and processed using Qupath software (27). Four areas measuring 0.37 mm2 each were counted within the demyelinated lesion, remyelinated lesion, and normal-appearing white matter (NAWM) in MS tissue and white matter of control tissue.
iPSC and NPC Culture Derivation.
The iPSCs and NPCs were generated as described previously (25), and iPSC lines from the laboratory of one of the authors (V.F.) were derived as described previously (26) (SI Appendix, Table S2). Cell colonies were passaged manually when needed, and NPC passage number was kept consistent between lines for all experiments described.
Isolation of Rat OPCs and Treatment with NPC Conditioned Media.
OPCs were obtained from the cerebral cortices of neonatal rat pups (postnatal day 0–2), and were plated on poly-l-ornithine–coated coverslips, as previously described (28). OPCs were cultured in varying conditioned media (CM) for 48 h. CM were taken from the NPCs after 48 h on 80% confluent cells, at least 4 d after passage, immediately spun down to remove debris, and then frozen at −80 °C. The CM experiments were performed in quadruplicate technical replicates and repeated in triplicate using separately developed cultures (i.e., unique biological replicates). For the HMGB1 blocking experiments, CM were collected and then incubated with 1 μg/mL function blocking α-HMGB1 antibody (BioLegend) or the appropriate isotype control (IgG) for 1 h at 37 °C on a shaker. The treated CM were then applied to OPCs for 48 h, and differentiation was assayed. For recombinant HMGB1 treatment, OPCs were grown in differentiation media [neurobasal media (Thermo Fisher Scientific), 2% B27, 2 mM l-glutamine, and 10 ng/mL T3] and treated with recombinant human HMGB1 (R&D Systems) every 24 h for 48 h, for a total of two doses. Doses consisting of 0.5, 5, 50, 500, and 5,000 ng/mL OPCs were then fixed, and differentiation was assayed. OLs on coverslips were fixed and stained for OLIG2 (1:500; Millipore) and myelin basic protein (MBP; 1:500; Millipore). For all experiments, five fields of view at a magnification of 20× using identical image capture settings were assessed by an experimenter blinded to treatments (Olympus IX71, CellSens Software; Olympus). For analysis of OPC differentiation, all OLIG2+ cells and MBP+ cells were counted and the percentage of MBP+ cells was calculated. Data are represented as the percentage of MBP+/OLIG2+ cells relative to the control condition set to 100%.
NPC Treatments.
NPCs were treated with rapamycin (12.5 nM, daily for 48 h) once cells became 80% confluent. After treatment, CM were collected, and cells were collected in TriReagent (Millipore Sigma) for RNA isolation or radioimmunoprecipitation assay (RIPA) buffer for protein isolation, or were fixed (4% PFA) for immunocytochemistry.
Immunocytochemistry.
NPCs were plated on laminin-coated coverslips (50 μg/mL; Sigma–Aldrich) and fixed using 4% PFA. Following fixation, they were permeabilized using Triton X-100 (0.5%; Millipore Sigma) and blocked with 10% normal goat serum (Thermo Fisher Scientific). Cells were then stained using the following primary antibodies overnight: p16Ink4a (1:500; Thermo Fisher Scientific) and SOX2 (1:250; Stemgent). Appropriate secondary antibodies were then applied.
Quantitative Real-Time PCR.
Total RNA was isolated from NPCs plated on laminin as described previously (29), and converted into cDNA via reverse transcription (iScript cDNA synthesis kit; BioRad), according to the manufacturer’s protocol. Synthesized cDNA samples were amplified for quantitative real-time PCR using primer pairs for human p16Ink4a [forward (For): 5′ GAAGGTCCCTCAGACATCCCC 3′, reverse (Rev): 5′ CCCTGTAGGACCTTCGGTGAC 3′], human p53 (For: 5′ CCCCTCCTGGCCCCTGTCATCTTC 3′, Rev: 5′ GCAGCGCCTCACAACCTCCGTCAT 3′), human HMGB1 (For: 5′ ACATCCAAAATCTTGATCAGTTA 3′, Rev: 5′ AGGACAGACTTTCAAAATGTTT 3′), and human β-actin (For: 5′ GGACTTCGAGCAAGAGATGG 3′, Rev: 5′ AGCACTGTGTTGGCGTACAG 3′) using SsoAdvanced Universal SYBR Green Supermix (BioRad) according to the manufacturer’s protocol. The PCR cycle conditions were one cycle at 95 °C for 2 min, 40 cycles of 95 °C for 5 s, and 40 cycles at 60 °C for 30 s run on a BioRad CFX96 Touch Real-Time PCR Detection System. Relative expression of mRNA was calculated relative to β-actin using comparative cycle threshold analysis.
Senescence β-Gal Staining.
Human NPCs, derived from iPSCs, were plated onto laminin-coated cover glass at equal densities (2 million cells per 35-mm well), and β-gal staining was performed according to the manufacturer’s instructions (Cell Signaling Technology). Images were captured at a magnification of 10× using identical phase contrast settings (Olympus IX71, CellSens Software; Olympus). The method for quantifying senescence-associated (SA)–β-gal is provided in SI Appendix, Fig. S5.
Immunoblots.
Cells were lysed in RIPA buffer, protein content was determined by bicinchoninic acid assay (BCA), and 20 μg of protein was separated by electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked in Tris-buffered saline (1% Tween-20) (TBST) + 5% BSA, incubated overnight with primary antibodies [phospho-mTOR (Ser2448; Cell Signaling Technology) and GAPDH (Cell Signaling)], washed in TBST, and incubated with the appropriate HRP-conjugated secondary antibody for an hour. Proteins were visualized by chemiluminescence.
Mass Spectrometry.
CM from iPS-derived NPCs from either nondisease control, PPMS, or PPMS lines treated with rapamycin were collected and sent to the Mass Spectrometry and Proteomics Keck Laboratory at Yale University, where liquid chromatography (LC)-tandem mass spectrometry analysis was performed on a Thermo Scientific Q Exactive Plus with a Waters nanoACQUITY UPLC system, using a Waters Symmetry C18 180-μm × 20-mm trap column and an ACQUITY UPLC Peptide Separation Technology [ethyline bridged hybrid (BEH)] C18 nanoACQUITY 75-μm × 250-mm (37 °C) 1.7-μm column for peptide separation. Trapping was done at 5 μL⋅min−1 with 97% buffer A (100% water, 0.1% formic acid) for 3 min. Peptide separation was performed at 330 nL⋅min−1 with buffer A (100% water, 0.1% formic acid) and buffer B (100% acetonitrile, 0.1% formic acid). A linear gradient (90 min) was run with 3% buffer B at initial conditions, 5% buffer B at 1 min, 35% buffer B at 50 min, 50% buffer B at 60 min, 90% buffer B at 65–70 min, and back to initial conditions at 71 min. Mass spectrometry was acquired in profile mode over the range of 300–1,700 m/z, using one microscan, a resolution of 70,000, an automatic gain control (AGC) target of 3E6, and a full maximum ion time of 45 ms. Tandem mass spectrometry was acquired in centroid mode using one microscan, a resolution of 17,500, an AGC target of 1E5, a full maximum ion time of 100 ms, an isolation window of 1.7 m/z, normalized collision energy of 28, and a scan range of 200–2,000 m/z. Up to 20 tandem mass spectra were collected per MS scan on species with an intensity threshold of 2 × 104, using charge states +2 through +6, where peptide match was preferred and dynamic exclusion was set to 20 s. Data were viewed using Scaffold Viewer (Proteome Software). Analysis of protein lists was performed using the DAVID (Database for Annotation, Visualization, and Integrated Discovery) bioinformatics database (30, 31), and the percentage of proteins from the varying media associated with disease was analyzed using the Genetic Association Database disease class. Percentages shown in the pie graph are the percentage of proteins involved in that disease category over total proteins (Fig. 3B). A proteomics dataset is provided (Dataset S1). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (32) partner repository with dataset ID PXD013060.
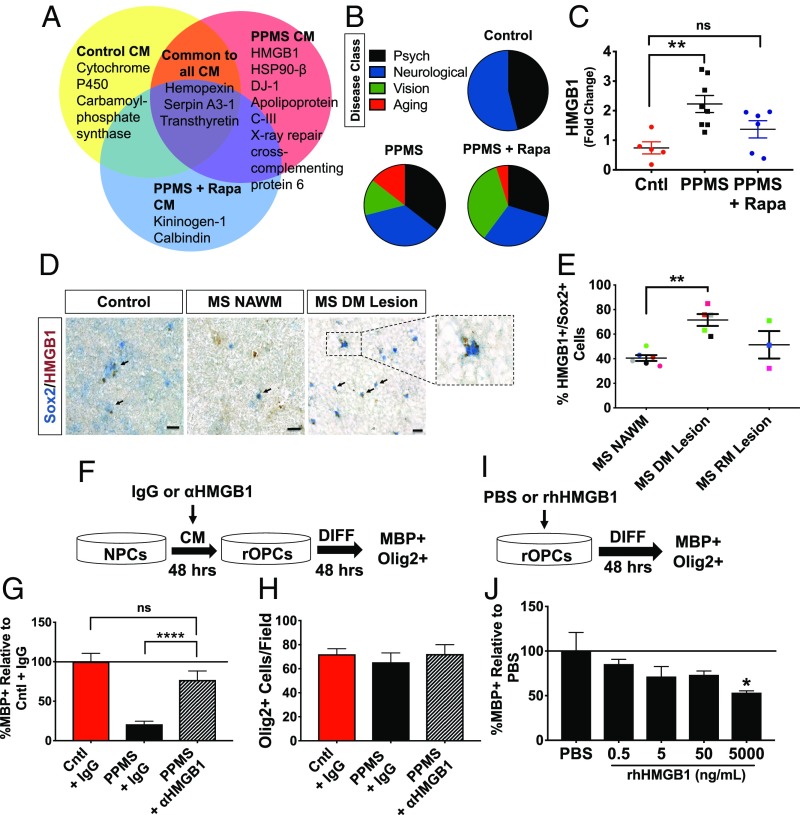
Fig. 3.
Proteomic analysis of the iPS-derived NPC secretome and characterization of HMGB1 as a directly acting inhibitor of OPC differentiation. (A) Venn diagram of comparison strategy for proteomic data analysis from control CM, PPMS CM, and PPMS + rapamycin (Rapa) treatment CM. Peptides were identified by LC-tandem mass spectrometry from each treatment group. The protein threshold was set at 95% confidence of identification. (B) Functional classification of secreted peptides from PPMS NPCs and nondisease control NPCs as determined by the DAVID bioinformatics database (https://david.ncifcrf.gov/). Pie graphs depict the proportions of peptides identified associated with disease classifications as determined by Genetic Association Database disease class. Psych, psychological. (C) Increased HMGB1 mRNA expression in PPMS NPCs was decreased with Rapa treatment [Control (Cntl) vs. PPMS (**P < 0.01), Cntl vs. PPMS + Rapa (P = 0.4182); ANOVA with Tukey’s multiple comparison test]. Data were normalized to Cntl NPC lines. Each data point represents an individual stem cell line and/or clone performed in triplicate. ns, not significant. (D) Human tissue staining for SOX2 (blue) and HMGB1 (red) in control and patients with PMS in the frontal lobe, subcortical white matter. The representative control image is CO 39 (SI Appendix, Table S1), and representative MS NAWM and MS demyelinated (DM) lesion images are from MS 640 (SI Appendix, Table S1). Arrows point to cells double positive for SOX2 and HMGB1. (Scale bars, 20 μm.) (E) Increase in percentage of SOX2+ cells expressing HMGB1 in a MS DM lesion compared with MS NAWM (**P < 0.01, two-tailed t test). Individual points are individual patients, and matching colors represent the same patient (SI Appendix, Table S1). Values are mean ± SEM. RM, remyelinated. (F) Experimental design to test whether functional blocking antisera against HMGB1 (1 μg/mL) in PPMS CM (or IgG control, 1 μg/mL) affected OPC maturation. rOPCs, rat OPCs. CM from NPCs were collected after 48 h, and α-HMGB1 or IgG control was incubated in CM for 1 h and applied to OPCs. Differentiation (DIFF) of OPCs was assayed after 48 h. (G) Quantification of OL maturation from experiment design in F. All conditions are relative to Cntl + IgG, set to 100% [PPMS + IgG vs. PPMS + α-HMGB1 (****P < 0.0001); ANOVA with Tukey’s multiple comparison test]. All conditions were performed in triplicate with the varying NPC lines. (H) No significant differences in the number of OLIG2+ cells were observed from the varying treatments on the OPCs (P = 0.7095; ANOVA). (I) Experimental design to test whether rhHMGB1 affected OPC maturation. Forty-eight hours after treatment of OPCs with varying concentrations of rhHMGB1, DIFF was assayed. (J) Treatment of OPCs with increasing doses of rhHMGB1 (ng/mL) decreases OL differentiation after 48 h in the presence of differentiation media. All conditions are relative to PBS, set to 100% [PBS vs. 5,000 ng/mL rhHMGB1 (*P < 0.05); ANOVA with Tukey’s multiple comparison test].
RNA Sequencing.
OPCs were plated out and treated with NPC CM from control, PPMS, and PPMS + α-HMGB1 CM as described above. After 48 h, the OPCs were collected in TRIzol, and RNA was isolated as described previously (29). RNA was given to The Jackson Laboratory, where 1 μg of total RNA was processed using the TruSeq RNA Library Preparation Kit v2 (RS-122-2001; Illumina) according to the manufacturer’s instructions. The protocol starts from using oligo-dT attached magnetic beads to purify the poly-A containing mRNA molecules. The purified mRNA was fragmented and reversed to first-strand cDNA. Then, second-strand cDNA was synthesized. After end repair and after a single “A” nucleotide was added to the 3′ ends, cDNA was ligated to its indexing adapter. Four cycles of PCR were used to enrich the adapter ligated DNA fragments. Following bead purification, libraries were quantified and equally pooled together. The pooled libraries were sequenced on an Illumina HiSeq 4000 platform using a 75-bp end protocol and sequenced to a depth of up to 40 million reads per library. The RNA-sequencing (RNA-seq) samples were processed using the in-house pipeline at The Jackson Laboratory. The reference for rat (version 6.0.91) was obtained from Ensemble. Alignment estimation of gene expression levels using the EM algorithm for single-ended and paired-end read data was performed using the RSEM package (version 1.3.0); default settings were used for alignment. Data quality control (QC) was performed using Picard (version 1.95) and qualimap (version 2.2.1). The qualimap output was utilized to examine the alignment data and to detect potential biases in mapping data; this was computed using two analysis types: (i) BAMQC and (ii) RNA-seq QC within the tool. BAMQC provided basic alignment statistics [e.g., coverage, guanine-cytosine (GC) content]. RNA-seq QC provided QC metrics, bias estimations, transcript coverage, and 5′–3′ bias computation. Analyses of aligned reads were performed using the edgeR (v3.30.7) and Biobase (2.38.0) packages in R (v3.4.3). Following normalization, read counts were filtered to include genes with counts per million greater than 1 in at least two samples. The quantile-adjusted conditional maximum likelihood method was used to determine differentially expressed genes between libraries as follows: (i) control and PPMS, (ii) control and PPMS + α-HMGB1, and (iii) PPMS and PPMS + α-HMGB1. Genes with false discovery rates (FDRs) < 0.05 were sorted by gene ontology using Gene Ontology Consortium and PANTHER analysis to get varying clusters, including myelin, oxidative stress, epigenetics, and senescence. All heatmaps were made in RStudio (V. 1.1.419) using the packages “gplots” and “RColorBrewer,” and were organized by gene log fold change. An RNA-seq dataset is provided (Dataset S2).
Statistical Analysis.
Data were analyzed by Student’s t test or one-way ANOVA with Tukey’s multiple comparison test, where appropriate and as indicated, using GraphPad Prism version 7 for Mac OS X (GraphPad Software; https://www.graphpad.com). Differences were considered significant when P < 0.05. Data are presented as mean ± SEM.
Results
Cellular Senescence Is Present in Progenitor Cells of PMS Brain Tissue and in iPS-Derived NPCs from Patients with PPMS.
Age is recognized as an irreversible process that limits tissue regeneration and impairs CNS remyelination (33, 34). We hypothesized that the process of cellular aging called cellular senescence may contribute to differences in support for myelination previously reported by NPCs derived from PPMS and nondisease control iPSC lines (25). Human autopsy brain tissue samples from patients confirmed to have PMS and age-matched controls were immunostained for the progenitor cell marker SOX2, along with p16Ink4a, a cyclin-dependent kinase inhibitor and an established marker of senescence (35). Within the demyelinated lesions of the PMS brain, we found there to be a significant increase in the number of SOX2+ progenitor cells in white matter lesions, compared with either NAWM or white matter of control brain samples, and a decrease in SOX2+ progenitor cells in the remyelinated lesions (Fig. 1 A and B and SI Appendix, Table S1). We also confirmed that the p16Ink4a detected was not an artifact of DAB staining (SI Appendix, Fig. S1) and established the presence of SOX2+ cells in both control white matter and demyelinated MS tissue using BaseScope mRNA detection (SI Appendix, Fig. S2). This increase in progenitor cells within demyelinated lesions was consistent with previously reported observations (16). However, when colabeled with p16Ink4a, we identified that the majority of SOX2+ progenitor cells within the demyelinated lesion area of the PMS brain expressed this senescence marker (Fig. 1 A and C). To further support these data, we performed confocal immunofluorescence and found cells expressing both SOX2+ and p16ink4a within MS lesion tissue [SI Appendix, Fig. S3 and accompanying Z-stack Graphics Interchange Format (GIF) (Movie S1)]. These data indicated the presence of elevated cellular senescence in progenitor cells in the PMS brain.
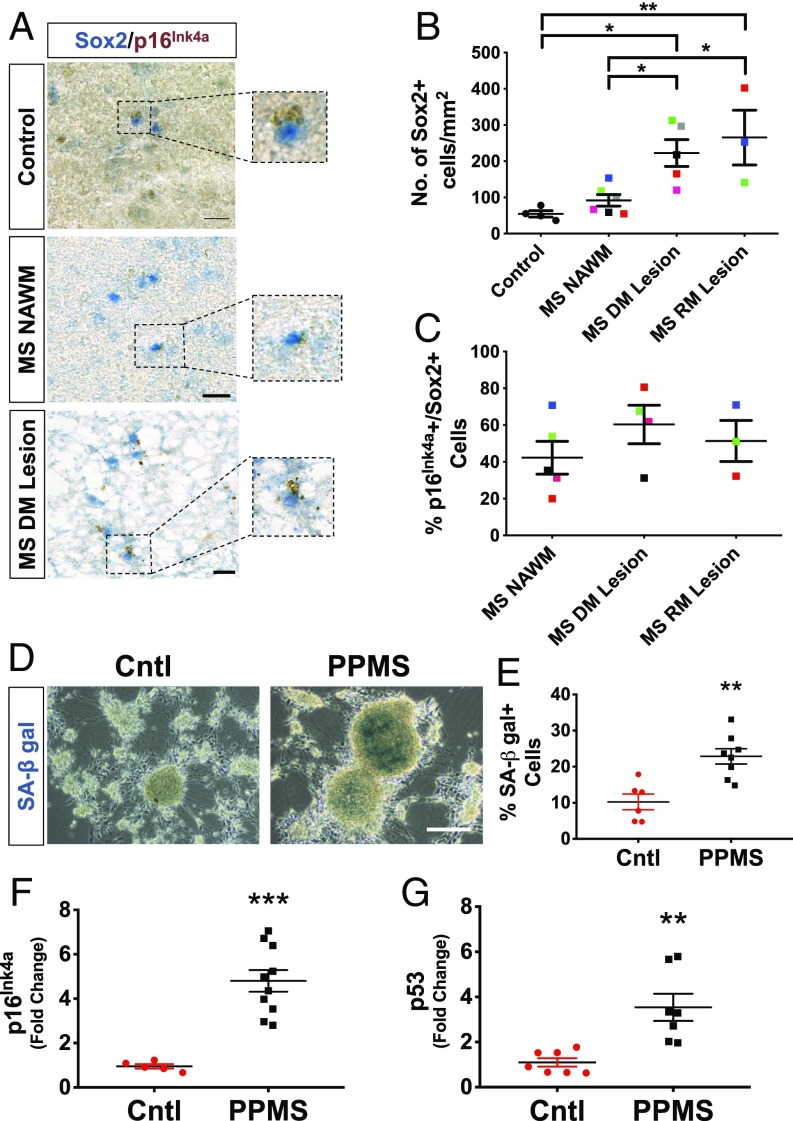
Fig. 1.
Senescent markers identify progenitor cells within demyelinated lesions of human PMS brain tissue and in iPS-derived NPCs from patients with PPMS. (A) Human brain tissue from the frontal lobe, subcortical white matter was stained for SOX2 (blue, progenitor marker) and p16Ink4a (red, senescence marker) in neurospecimens from controls and PMS patients. The representative control image is SD 22/16 (SI Appendix, Table S1), and representative MS NAWM and MS demyelinated (DM) lesion images are from MS 640 (SI Appendix, Table S1). (Scale bars, 20 μm.) (B) Increase in total number of SOX2+ cells in the MS DM lesion compared with control and MS NAWM area and control tissue (*P < 0.05, **P < 0.01; ANOVA with Tukey’s multiple comparison test). Individual points are individual patients, and matching colors between MS lesion samples represent the same patient. Color coding for patient samples is presented in SI Appendix, Table S1. RM, remyelinated. (C) Percentage of SOX2+ NPCs colabeling with p16Ink4a in MS NAWM, MS DM lesion, and MS RM lesion areas in the same patients identified by color coding (P = 0.4430, one-way ANOVA). (D) SA–β-gal staining in control (Cntl) NPC and PPMS iPS-derived NPC cultures in vitro revealed LacZ activity (shown in blue) in PPMS NPC cultures. Figures show representative lines 3.1 and 3 (SI Appendix, Table S2). (Scale bar, 400 μm.) (E) Quantification of SA–β-gal staining in Cntl and PPMS NPCs shows an increase in LacZ in PPMS cultures (**P < 0.01, t test). Staining was performed and quantified in triplicate in each NPC line. (F and G) p16Ink4a and p53 mRNA expression by qPCR in Cntl NPC and PPMS NPC cultures revealed a significant increase in p16Ink4a and p53 mRNA levels in PPMS NPCs (***P < 0.001, **P < 0.0022; unpaired t tests). All qPCR data were normalized to Cntl NPC lines. Each data point represents an individual stem cell line and/or clone performed in triplicate.
To further characterize cellular senescence in the PMS progenitor cells, and to interrogate a functional role for this aging process in human progenitor cells, we differentiated NPCs from iPSC lines of patients with PPMS and age-matched control donors (SI Appendix, Table S2). We found that control and PPMS iPS-derived NPCs exhibited equivalent differentiation into NPCs, and did not differ based on the iPSC of origin (SI Appendix, Fig. S4). Next, we determined whether expression of the lysosomal enzyme β-gal, also a marker of cellular senescence (36), was present in the iPS-derived NPCs. Elevated activity of SA–β-gal in PPMS NPCs was observed, while little to no activity was detected in any NPCs from nondisease control iPSC lines (Fig. 1 D and E and SI Appendix, Fig. S4, demonstration of quantification methodology). We then examined by quantitative PCR (qPCR) the expression of p16Ink4a and p53, which are both known markers of cellular senescence (37–39). Significantly higher mRNA expression of p16Ink4a and p53 was measured in PPMS NPCs compared with control NPCs (Fig. 1 F and G). Since cellular senescence is known to be associated with cell cycle arrest (40), we performed cell cycle analysis and examined the proportion of NPCs in G0/G1, G2/M, and S phases. We found no significant differences between control and PPMS NPCs in terms of cell cycle, indicating that evidence of senescence in PPMS NPCs was not accountable to replicative senescence (SI Appendix, Fig. S5B). Collectively, these independent markers of cellular senescence identified increased senescence in PPMS NPCs compared with the age-matched control NPCs, even though the chronological age of the human donors in both groups did not differ (SI Appendix, Table S2). We also considered whether these changes may relate to differentiation toward NPCs. To test this, we assayed p16Ink4a expression in the undifferentiated iPSC state. Interestingly, we found no differences in p16Ink4a mRNA expression among the different cell lines (SI Appendix, Fig. S6), suggesting that these early-passage iPSC lines adopted a cellular senescent phenotype only when differentiated to NPCs.
Reversal of Cellular Senescence in NPCs Promotes OPC Maturation.
The identification of cellular senescence markers in progenitor cells of PMS tissue and in PPMS NPCs led us to hypothesize that this intrinsic process may underlie the failure of PPMS NPCs to support OPC maturation as we had previously reported (25). Given that a senescent phenotype also contributes to a secretory phenotype, we selected to affect cellular senescence in PPMS NPCs using rapamycin, which would also block the SASP. We first confirmed that treatment of PPMS NPCs with rapamycin (12.5 nM, 48 h) resulted in decreased mTOR phosphorylation in PPMS NPCs, as determined by Western blotting (Fig. 2 A and B). We then assayed expression of senescence markers. This brief rapamycin treatment of PPMS NPCs decreased p16Ink4a mRNA and protein expression assayed by qPCR and immunocytochemistry, respectively (Fig. 2 C and D). SA–β-gal staining also decreased in intensity to that observed in nondiseased control NPCs following rapamycin treatment (Fig. 2E). These findings demonstrated that rapamycin effectively reduced expression of the key cellular senescence markers p16Ink4a and SA–β-gal in PPMS NPCs.
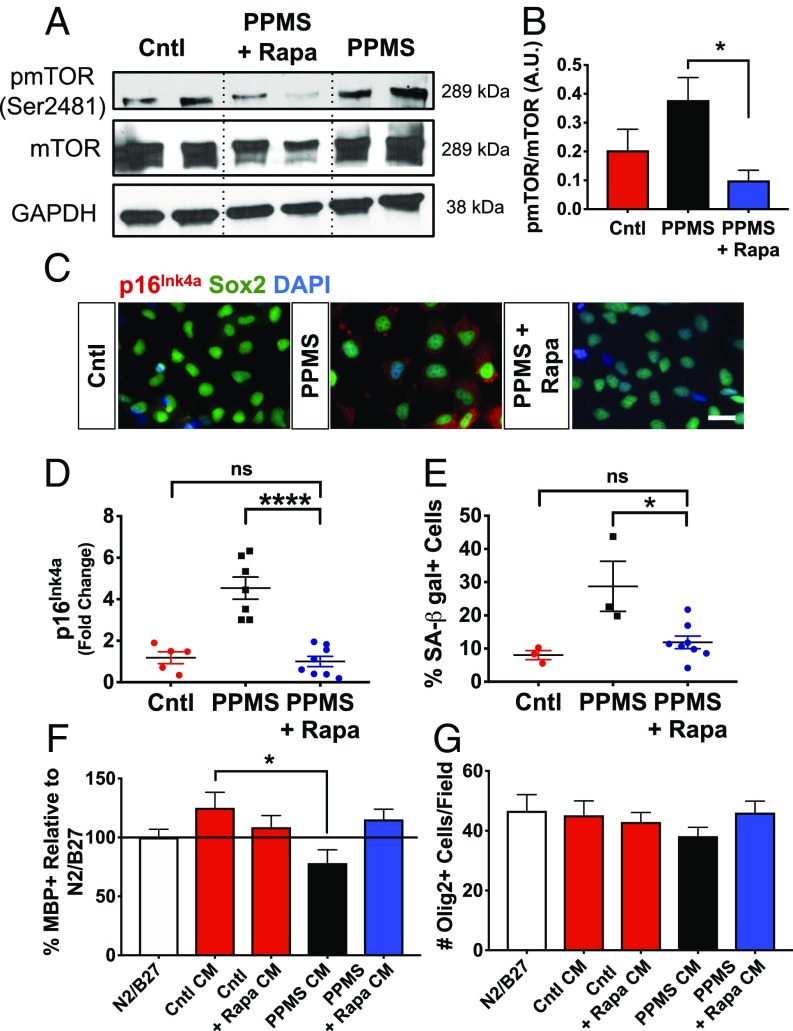
Fig. 2.
Cellular senescence phenotype in iPS-derived NPCs from patients with PPMS is reversed by treatment with rapamycin. (A) Western blot of protein lysates from control (Cntl) NPCs, PPMS NPCs, and PPMS NPCs treated with rapamycin (Rapa) for 48 h probed for phospho-mTOR (pmTOR; Ser2481), mTOR, and GAPDH. (B) Densitometry of pmTOR over mTOR shows a significant decrease in pmTOR after Rapa treatment (*P < 0.05, t test). (C) p16Ink4a and SOX2 staining in Cntl, PPMS, and PPMS NPCs treated with Rapa shows a decrease in p16Ink4a protein expression in the PPMS NPCs. The figure shows representative lines 1 and 1.1. (Scale bar, 50 μm.) (D) Decrease in PPMS NPC p16Ink4a mRNA expression after treatment with Rapa for 48 h [PPMS vs. PPMS + Rapa (****P < 0.0001), Cntl vs. PPMS (***P < 0.001), Cntl vs. PPMS + Rapa (P = 0.6418); ANOVA with Tukey’s multiple comparison test]. Data are normalized to Cntl NPC lines. Each data point represents an individual stem cell line and/or clone performed in triplicate. (E) Quantification of SA–β-gal staining in Cntl NPCs, PPMS NPCs, and Rapa-treated PPMS NPCs [PPMS vs. PPMS + Rapa (*P < 0.05), Cntl vs. PPMS (*P < 0.05), Cntl vs. PPMS + Rapa (P = 0.7483); ANOVA with Tukey’s multiple comparison test]. Staining was performed and quantified in triplicate in each NPC line and/or clone. ns, not significant. (F) Quantitative analysis of OL maturation (MBP+/OLIG2+) resulting from culture of OPCs in various CM, as indicated, after 48 h of treatment. CM were collected from Cntl NPCs, Cntl NPCs + Rapa, PPMS NPCs, and PPMS NPCs + Rapa. All conditions were quantified relative to differentiation under N2/B27 (standardized Cntl) non-CM set to 100% [Cntl CM vs. PPMS CM (*P < 0.05), Cntl CM vs. PPMS + Rapa CM (P = 0.9974)]; ANOVA with Tukey’s multiple comparison test]. All conditions were performed in triplicate with the varying NPC lines. (G) Quantification of total OLIG2+ cells under each treatment condition showing equivalent densities of cells under all conditions, indicating that differences in differentiation were not a result of cellular proliferation or cell death. Values are mean ± SEM (P = 0.5030; ANOVA).
We next determined whether rapamycin affected the potential for PPMS NPCs to support OPC differentiation. To test this, we collected CM from PPMS NPC cultures that had been treated with either rapamycin or vehicle, and these media were separately applied to OPC cultures. Consistent with our previous work (25), we found that CM from PPMS NPCs did not promote the differentiation of OPCs, as assayed by MBP expression, compared with control NPC CM, and did not alter OPC numbers (Fig. 2F). In contrast, OPCs grown in CM from rapamycin-treated PPMS NPCs were found to differentiate into MBP+ OLs at a rate comparable to OPCs grown under control NPC CM conditions (Fig. 2F). Treatment of the control NPCs with rapamycin did not negatively affect their ability to promote OL maturation via CM (Fig. 2F). This effect of rapamycin on human NPCs contrasts with previous studies examining the direct effects of mTOR signaling on OPCs (41, 42), indicating that rapamycin was not transferred to the OPCs in the CM. However, to verify that this was the case, we also tested whether carryover of rapamycin could have independently promoted OPC differentiation in OPCs grown in PPMS CM. OPCs were treated directly with the same concentration of rapamycin used to treat the NPCs (12.5 nM), and we found no increase in differentiation (SI Appendix, Fig. S7). This was consistent with previous work that had shown direct treatment of OPCs with rapamycin may actually inhibit OPC differentiation (43). Importantly, treatment of OPCs under any of these media conditions tested did not result in differences in cell death, as the number of OLIG2+ cells did not differ among any of the experimental treatment conditions (Fig. 2G). Hence, reversing senescence in PPMS NPCs using rapamycin was not only found to provide a change in senescence marker expression but to result in a significant improvement in PPMS NPC function reflected by enhanced OPC differentiation.
Proteomic Analysis of NPC Secretome Identified HMGB1 as a Potent Inhibitor of OL Differentiation.
To identify what factor(s) in the NPC CM were responsible for impaired OPC differentiation, we performed LC-tandem mass spectrometry on the CM from control NPCs, PPMS NPCs, and rapamycin-treated PPMS NPCs to identify and compare peptides from each of these conditions (Dataset S1). For this analysis, we exploited the heterogeneity of the human donors to provide higher stringency to the comparisons, where only factors found in all samples in a treatment group were considered valid targets. From these proteomic analyses, we identified several factors uniquely expressed by PPMS NPCs. Many of these peptides represented secreted proteins previously known to be associated with cellular senescence, including heat shock proteins 90 and 60, DJ-1, and HMGB1 (Fig. 3A). Importantly, these factors were not found to be produced by either the control NPCs or when PPMS NPCs had been treated with rapamycin (Fig. 3A). Ontology analysis of the identified peptides using the DAVID bioinformatics database revealed that 14.5% of proteins secreted by PPMS NPCs were associated with “aging.” This aging profile was reduced to only 4.8% of peptides identified from PPMS NPCs that had been treated with rapamycin, which represents a decrease of 67%. No aging ontology was identified from the secretome of control NPCs (Fig. 3B). These findings lend further support to cellular senescence as a feature of NPCs in PPMS and rapamycin as a pharmacological means to affect the SASP of NPCs.
Of the candidate proteins secreted by PPMS NPCs, we focused our attention on HMGB1 for several reasons: It is produced at higher levels in demyelinated lesions in MS brain tissues (39, 44), it is produced at higher levels by senescent cells (39), and it has been implicated as a mediator of neuroinflammation (45). We evaluated HMGB1 gene expression and found significantly increased mRNA expression in PPMS NPCs, which was also down-regulated by rapamycin treatment (Fig. 3C).
We determined whether progenitor cells could be a source of HMGB1 within demyelinated lesions in human MS brain tissues. Immunohistochemistry identified a significantly increased proportion of SOX2+/HMGB1+ progenitor cells in demyelinated white matter lesions compared with NAWM, which decreases in remyelinated lesions (Fig. 3 D and E). These progenitor cells, expressing both SOX2 and HMGB1, were further confirmed using confocal immunofluorescence within the MS lesion tissue [SI Appendix, Fig. S8 and accompanying Z-stack GIF (Movie S2)]. These data indicated the presence of elevated cellular senescence in progenitor cells in the PMS brain.
To address the contribution of extracellular HMGB1 to impaired OPC differentiation, we repeated the OPC differentiation experiment using CM from PPMS NPCs but added a well-characterized function blocking antibody to inhibit HMGB1 (α-HMGB1) or the matched IgG control (Fig. 3F). Maturation of OLs in the α-HMGB1–treated PPMS CM was significantly higher compared with OPCs grown in CM with serotype-matched IgG antisera (control) (Fig. 3G). Blocking HMGB1 was not found to affect either OLIG2+ cell proliferation or cell death (Fig. 3H). To test whether extracellular HMGB1 was acting directly as an inhibitor of OPC differentiation, we applied recombinant human HMGB1 (rhHMGB1) to cultures of differentiating OPCs and measured the proportion of OL maturation (Fig. 3I). Extracellular rhHMGB1 had a concentration-dependent effect on reducing OPC differentiation in vitro (Fig. 3J). These data provide direct evidence that extracellular HMGB1 produced by senescent NPCs can act to directly suppress OPC maturation.
HMGB1 Mediates Senescence-Related Transcriptomic Changes in OPCs.
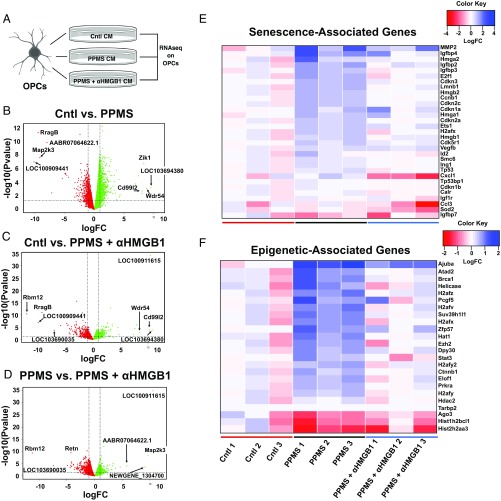
To better understand how cellular senescence of NPCs and their production of extracellular HMGB1 affects the potential for OPCs to mature into MBP+ OLs with myelinating potential, we performed transcriptomic analyses of OPCs (Dataset S2). Specifically, we collected cultures of primary OPCs that had been grown in CM from either PPMS NPCs or nondisease control NPCs or CM from PPMS NPCs that had been treated with α-HMGB1 antisera. OPCs were grown for 48 h and then collected for RNA-seq analyses (Fig. 4A). Genes were filtered for an FDR of <0.05. Substantial transcriptome changes were seen in the OPCs treated with either control CM, PPMS CM, or PPMS + α-HMGB1 (Fig. 4 B–D). Of all genes filtered, Cd99l2 and Wdr54 were found to be the most up-regulated genes in the OPCs treated with PPMS CM compared with the OPCs treated with control CM (Fig. 4B). Cd99l2 has been found to allow for the entrance of leukocytes into the CNS in the experimental autoimmune encephalomyelitis mouse model of MS. When knocked out in this disease model, leukocyte entry is inhibited and disease is ameliorated (46). Treatment of the PPMS CM with α-HMGB1 did not decrease Wdr54 and Cd99l2 to levels found in OPCs treated with control CM (Fig. 4C). Treatment of the OPCs with PPMS + α-HMGB1 CM did increase expression of Map2k3 (Fig. 4B), which was found to be significantly decreased in OPCs treated with PPMS CM (Fig. 4D). Map2k3, a dual-specificity kinase, has been found to be associated with a resolution in inflammation (47). Our data therefore implicate HMGB1 in perpetuating inflammation in OPCs. Raw sequencing files are available from the National Center for Biotechnology Information (NCBI) BioProject (accession no. PRJNA524718).
Fig. 4.
Transcriptomic analysis of OPCs treated with human NPC CM identifies marked differences in expression of cellular senescence genes (E) and epigenetic regulators (F) regulated by HMGB1. (A) OPC RNA was collected after 48 h of treatment with control (Cntl) CM, PPMS CM, or PPMS + α-HMGB1 CM, and RNA-seq was performed. The image was generated using BioRender. (B–D) Volcano plots of gene expression changes in the OPC transcriptome after treatment with Cntl CM, PPMS CM, or PPMS + α-HMGB1 CM. The x axis specifies the log of the fold change (logFC), and the y axis specifics the negative logarithm to the base 10 of the P values. Vertical lines reflect an FC of ±2, and the horizontal line reflects a P value of 0.05. (B) Gene expression changes in Cntl vs. PPMS CM-treated OPCs. (C) Gene expression changes in Cntl vs. PPMS + α-HMGB1 CM-treated OPCs. (D) Gene expression changes in PPMS vs. PPMS α-HMGB1 CM-treated OPCs. (E) Heatmap of expression differences in senescence-associated genes in OPCs after treatment with CM. (F) Heatmap of expression differences in epigenetic-associated genes in OPCs after treatment with CM. Three individual Cntl and PPMS lines were selected and are demonstrated individually in the heatmaps. RNA was aligned to the latest version of the rat genome (version 6.0.91). The heatmap for each class of genes is as indicated. Genes with an FDR < 0.05 were sorted by gene ontology using Gene Ontology Consortium and PANTHER (protein analysis through evolutionary relationships) analysis to get the varying clusters.
Our first step in curating these data was to identify differentially expressed genes, which defined several notable ontological differences between the treatment groups. Senescent cells also are known to induce senescence through secretion of the SASP as a paracrine effect, so we examined markers of senescence in the OPCs. Overall, we found an increase in senescence-associated genes, such as Mmp2, p16Ink4a (Cdkn2a), and Igfbp2, when OPCs were treated with PPMS CM, which was reduced close to normal in PPMS CM containing HMGB1 blocking antibody (Fig. 4E). We also investigated expression of epigenetic modifiers since epigenetics plays a pivotal role in determining the potential for OPC differentiation (48). We found that PPMS CM significantly induced expression of numerous epigenetic regulators in OPCs (Fig. 4F). Interestingly, when HMGB1 was functionally blocked from the PPMS NPC CM, most of these changes in epigenetic-associated genes were normalized to levels observed in control NPC CM (Fig. 4F). These RNA-seq data indicate that the expression and production of HMGB1 by NPCs play important roles, influencing the potential for OPCs to mature by modulating gene expression. Moreover, these differences in senescence-associated and epigenetic modifier genes in OPCs demonstrate that it is likely locally produced factors from progenitor cells within demyelinated lesions that contribute to the microenvironment that determines OPC fate. These observations provide a compelling link between the presence of senescent progenitor cells in demyelinated lesions in PMS and ascribe a deleterious role for cellular senescence in NPCs, which we have functionally characterized using iPSCs from patients with PPMS.
Discussion
Our data provide a perspective on the pathophysiology of demyelinating lesions in MS. We have identified cellular senescence in SOX2+ progenitor cells, which influence, through paracrine activity, the maturation of OPCs with myelinating potential. We have also determined that cellular senescence is potentially reversible or amenable to therapeutic intervention using rapamycin as a prototypic compound. We used proteomics to determine that senescent NPCs secrete extracellular HMGB1, which altered gene expression and impaired maturation of OPCs (Fig. 5). This was also found to be corroborated in human pathology, demonstrating progenitor cells expressing HMGB1 in PMS lesions. Age is recognized as a process limiting the myelinating potential of the CNS (33). Although MS is not typically considered a disease of aging, because it is generally diagnosed in early-to-middle adulthood, the functional impact of cellular senescence in progenitor cells we report here may indicate that premature cellular aging is an important component of this disease.
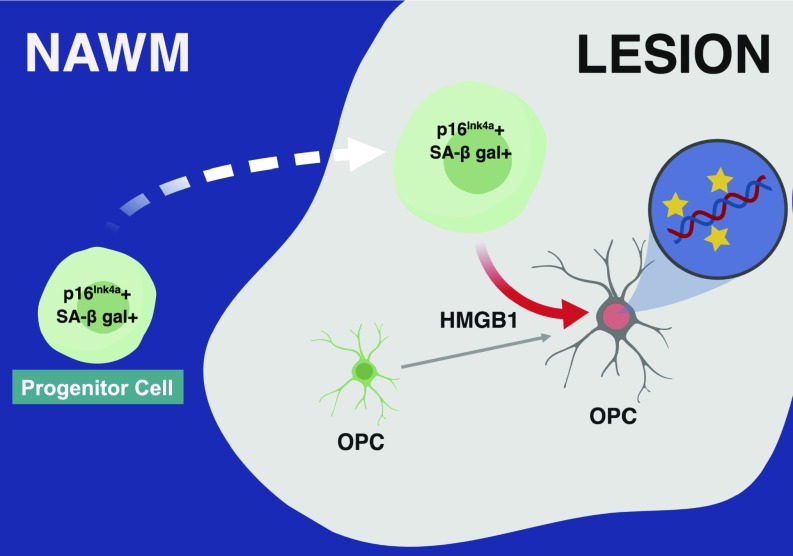
Fig. 5.
Schematic diagram outlining the hypothesized impact of senescent progenitor cells on remyelinating potential in the MS brain. The recruitment and presence of senescent progenitor cells in demyelinated white matter lesions exert a negative impact on the differentiation of OPCs, also within the demyelinated lesion, by modifying the transcriptional activity of OPCs via secretion of factors in the SASP, such as HMGB1. Collectively, this model suggests that cellular senescence in progenitor cells, either acquired as a result of disease or as a predisposing feature of individuals who develop MS, contributes to the limited potential of OPCs to foster remyelination in the MS brain. The image was generated using BioRender.
Progenitor cells are of particular interest because they have been found within demyelinated lesions in human tissue as well as in mouse models of MS (16, 49). Our data suggest that progenitor cells in the MS brain are actively contributing to the diminished regenerative capacity of OPCs within the lesion environment, yet NPCs are otherwise known to secrete regenerative, antiinflammatory, and promyelinating factors, or themselves differentiate into myelinating OLs (17, 50, 51). Currently, there are no markers that reliably distinguish human NPCs from other resident progenitor cell populations that could to be used to refine our identification of the senescent cell types in pathological analyses (52). Therefore, the senescent SOX2+ progenitor cells we identified in human brain tissues could represent a continuum of progenitor cells from NPCs to include OPC lineage cells (53, 54). As our data support, progenitor cells are found in greater numbers within the lesion area, compared with NAWM and control white matter, where they can contribute to the regulation of OPC maturation and myelin regeneration (16). However, our data would suggest that in PMS, endogenous progenitor cells exhibit higher levels of senescent markers, and, experimentally, this phenotype suppresses OPC maturation through the secretion of factors (the SASP), including HMGB1 (Fig. 5). These findings contribute to our understanding of the lesion microenvironment and may explain why immature oligodendroglia within lesions fail to remyelinate in PMS.
To model SOX2+ progenitor cells found within these lesion areas in vitro, NPCs were generated from iPSC lines derived from patients with PPMS (SI Appendix, Table S2). These cells were found to exhibit equivalent differentiation to an NPC fate, express identical markers to NPCs from nondisease age-matched donor iPSC lines (SI Appendix, Fig. S4), and differentiate toward the OPC lineage in vitro (26); yet, these PPMS NPCs are notable in their significantly reduced support for OL differentiation (25). We have now determined that PPMS NPCs display key hallmarks of cellular senescence, including SA–β-gal staining and increased p16Ink4a. p16Ink4a is a cyclin-dependent kinase inhibitor, a hallmark for identifying senescent cells (55), that plays a central role in the function of senescent cells (55–61). It acts through the retinoblastoma pathway, which inhibits the action of cyclin-dependent kinases (62). Up-regulation of p16Ink4a results in chromatin reorganization, which leads to changes in genes related to inflammation and oxidative damage, including COX1 and COX2 enzymes, as well as other inflammatory mediators (62, 63). The methylation pattern of p16Ink4a has been proposed to be an epigenetic risk factor in the development of MS (64). We found that the expression of p16Ink4a in PPMS NPCs was effectively reduced by rapamycin, and this was reflected in a functional change in the NPC secretome evidenced by enhanced OPC maturation after NPC treatment. Experimental deletion of p16Ink4a-positive cells in mice has been found to mitigate accelerated aging in models of disease, including progeria, and, more recently, in glia in a model of Alzheimer’s disease (57, 60).
Using both patient iPSCs and the study of human autopsy tissues, we implicate cellular senescence as a feature of progenitor cells in MS, offering potential for new approaches to treat this disease. Whether progenitor cells in PMS acquire senescence because of the disease, or a disease treatment (e.g., steroids), or are predisposed to developing senescence that then promotes the disease is presently unclear. It has been shown that the DNA damage response is strongly linked to senescence in other cell types where mitochondrial dysfunction has also been implicated (44). Cells with damaged mitochondria more easily acquire a senescent phenotype and secrete factors such as CCL27, TNF-α, and HMGB1 (44). We found HMGB1 only in CM from PPMS cells, and after treatment with rapamycin, HMGB1 secretion was abrogated. Extracellular HMGB1 acts as a cytokine and generates inflammatory responses, such as the production of additional cytokines and chemokines, continuing the cycle of chronic inflammation (45). HMGB1 can also be secreted by activated astrocytes and microglia in lesion areas; this has been proposed to help recruit NPCs to facilitate repair (65). However, as our data indicate, if NPCs become senescent and begin secreting HMGB1, this impairs OPC differentiation, and thus is deleterious. Our findings demonstrate HMGB1 is a potent suppressor of OL differentiation. The complexity of human subjects and the heterogeneity of donor lines in this study enabled us to refine the list of candidate factors emanating from PPMS NPCs. However, additional factors on this list, including XRCC6 and DJ-1, associated with oxidative stress and mitochondrial dysfunction, reflect the recapitulation of disease phenotype in these iPS-derived cells (66). While these factors were not addressed specifically in this report, future studies determining their influence on OPCs and myelination would be expected to provide additional translational understanding to the impact of the SASP on diminishing remyelination potential in MS.
Senescence can be induced from natural aging (termed replicative senescence) or can be induced by stressors, including inflammation and oxidative stress (44, 67–69). Persistent signs of oxidative stress and inflammation are associated with demyelination, and both are measurable in the brain and in the blood in PMS (70, 71). The negative impact of senescent cells on a tissue is mediated through release of factors (the SASP) that can promote inflammation and disrupt tissue microenvironments through paracrine activity on surrounding cells (40, 44, 72, 73). Senescent PPMS NPCs may be affecting other progenitor cells within the lesion microenvironment by preventing differentiation through secretion of the SASP. This concept has been termed the “senescence-stem lock model” (6).
To assess the impact of the PPMS NPC SASP on OPCs, we performed RNA-seq and identified induction of oxidative stress markers, epigenetic regulators, and senescence genes. Interestingly, genes up-regulated in OPCs treated with PPMS CM included VCAM1, STAT3, and MYB(AHI1), which had been previously identified by genome-wide association studies in MS (74). We identified extracellular HMGB1 as a putative mediator of transcriptional changes in OPCs linked to their differentiation potential. Our data show that HMGB1 had a significant negative impact on the expression of key epigenetic regulators in OPCs, such as helicase (75, 76) and the histones H2afz, H2afv, and H2afx (77). Previous research has implicated HMGB1 as a modifier of gene promoters and repressors involved in inflammation (78). These data support previous studies demonstrating that epigenetic modifiers are critical determinants of OPC differentiation potential and remyelination in mouse models (79–81).
The cause(s) of PMS is not known. Based on these data, we hypothesize that cellular senescence in progenitor cells impairs the regenerative potential of the lesion microenvironment. Current treatments for MS suppress inflammation and block access of immune cells into the CNS, yet this does not fully prevent demyelination and axonal degeneration (82). Additionally, current disease-modifying therapies for MS offer limited/no benefit to patients with PMS (83). The targeting and stimulating of resident OPCs to differentiate offers important therapeutic potential, but we speculate that the efficacy of any candidate drug also relies on overcoming the lesion microenvironment, which would include addressing the inhibitory influence of senescent progenitor cells. At present, it is unclear whether cellular senescence is present in other types of MS, such as relapsing-remitting multiple sclerosis (RRMS), or what existing therapies may affect cellular senescence. Future studies would be expected to extend the findings reported herein to address salient questions regarding the impact of cellular senescence in all forms of MS, including whether vulnerability to cellular senescence is a predisposing risk factor to developing this disease.
In summary, our identification of cellular senescence within the MS brain provides a mechanism by which aberrant cellular aging silently subverts natural aging by impairing OPCs and promoting chronic demyelination. Targeting cellular senescence in progenitor cells, as a means by which to unlock the differentiation potential of endogenous OPCs, may represent a novel therapeutic approach to promote remyelination in MS.
Supplementary Material
Acknowledgments
We acknowledge Dr. Matthieu Vermeren for assisting with confocal imaging and image processing, Lili Sun (The Jackson Laboratory for Genomic Medicine) for RNA-seq, Dr. Evan Jellison for help with the cell cycle analysis, and Olivia Heintz and Marwa Elamin for data collection. We thank the MS Society UK Tissue Bank and the MRC Sudden Death Brain Bank for human tissue donation and collection. This work was supported, in part, by a pilot grant from Connecticut Innovations (Grant 12-SCA-UCHC-06), the National Multiple Sclerosis Society (Grant RG-1802-30211), and the MS Society UK. We also acknowledge the WM Keck Foundation Biotechnology Resource Laboratory at Yale University for mass spectrometry and proteomics (NIH SIG, ODOD018034).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNAseq data have been deposited in the Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (BioProject accession no. PRJNA524718); Proteomics data have been deposited in the PRoteomics IDEntification (PRIDE) database, https://www.ebi.ac.uk/pride/ (accession no. PXD013060).
See Commentary on page 8646.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818348116/-/DCSupplemental.
References
- 1.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat Med. 2015;21:1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tacutu R, Budovsky A, Yanai H, Fraifeld VE. Molecular links between cellular senescence, longevity and age-related diseases–A systems biology perspective. Aging (Albany NY) 2011;3:1178–1191. doi: 10.18632/aging.100413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Y, Armstrong JL, Tchkonia T, Kirkland JL. Cellular senescence and the senescent secretory phenotype in age-related chronic diseases. Curr Opin Clin Nutr Metab Care. 2014;17:324–328. doi: 10.1097/MCO.0000000000000065. [DOI] [PubMed] [Google Scholar]
- 4.Laberge RM, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17:1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Keizer PL. The fountain of youth by targeting senescent cells? Trends Mol Med. 2017;23:6–17. doi: 10.1016/j.molmed.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Sousa-Victor P, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 8.van Wijngaarden P, Franklin RJ. Ageing stem and progenitor cells: Implications for rejuvenation of the central nervous system. Development. 2013;140:2562–2575. doi: 10.1242/dev.092262. [DOI] [PubMed] [Google Scholar]
- 9.Lassmann H, van Horssen J, Mahad D. Progressive multiple sclerosis: Pathology and pathogenesis. Nat Rev Neurol. 2012;8:647–656. doi: 10.1038/nrneurol.2012.168. [DOI] [PubMed] [Google Scholar]
- 10.Huang JK, et al. Myelin regeneration in multiple sclerosis: Targeting endogenous stem cells. Neurotherapeutics. 2011;8:650–658. doi: 10.1007/s13311-011-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Back SA, et al. Hyaluronan accumulates in demyelinated lesions and inhibits oligodendrocyte progenitor maturation. Nat Med. 2005;11:966–972. doi: 10.1038/nm1279. [DOI] [PubMed] [Google Scholar]
- 12.Sloane JA, et al. Hyaluronan blocks oligodendrocyte progenitor maturation and remyelination through TLR2. Proc Natl Acad Sci USA. 2010;107:11555–11560. doi: 10.1073/pnas.1006496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc Natl Acad Sci USA. 2009;106:19162–19167. doi: 10.1073/pnas.0902834106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferent J, Zimmer C, Durbec P, Ruat M, Traiffort E. Sonic Hedgehog signaling is a positive oligodendrocyte regulator during demyelination. J Neurosci. 2013;33:1759–1772. doi: 10.1523/JNEUROSCI.3334-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Paired related homeobox protein 1 regulates quiescence in human oligodendrocyte progenitors. Cell Rep. 2018;25:3435–3450.e6. doi: 10.1016/j.celrep.2018.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snethen H, Love S, Scolding N. Disease-responsive neural precursor cells are present in multiple sclerosis lesions. Regen Med. 2008;3:835–847. doi: 10.2217/17460751.3.6.835. [DOI] [PubMed] [Google Scholar]
- 17.Pluchino S, et al. Neurosphere-derived multipotent precursors promote neuroprotection by an immunomodulatory mechanism. Nature. 2005;436:266–271. doi: 10.1038/nature03889. [DOI] [PubMed] [Google Scholar]
- 18.Einstein O, Friedman-Levi Y, Grigoriadis N, Ben-Hur T. Transplanted neural precursors enhance host brain-derived myelin regeneration. J Neurosci. 2009;29:15694–15702. doi: 10.1523/JNEUROSCI.3364-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laterza C, et al. iPSC-derived neural precursors exert a neuroprotective role in immune-mediated demyelination via the secretion of LIF. Nat Commun. 2013;4:2597. doi: 10.1038/ncomms3597. [DOI] [PubMed] [Google Scholar]
- 20.Samanta J, et al. Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature. 2015;526:448–452. doi: 10.1038/nature14957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sim FJ, Zhao C, Penderis J, Franklin RJ. The age-related decrease in CNS remyelination efficiency is attributable to an impairment of both oligodendrocyte progenitor recruitment and differentiation. J Neurosci. 2002;22:2451–2459. doi: 10.1523/JNEUROSCI.22-07-02451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolswijk G. Chronic stage multiple sclerosis lesions contain a relatively quiescent population of oligodendrocyte precursor cells. J Neurosci. 1998;18:601–609. doi: 10.1523/JNEUROSCI.18-02-00601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- 24.Kuhlmann T, et al. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain. 2008;131:1749–1758, and erratum (2009) 132:1118. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- 25.Nicaise AM, et al. iPS-derived neural progenitor cells from PPMS patients reveal defect in myelin injury response. Exp Neurol. 2017;288:114–121. doi: 10.1016/j.expneurol.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 26.Douvaras P, et al. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports. 2014;3:250–259. doi: 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore CS, et al. Astrocytic tissue inhibitor of metalloproteinase-1 (TIMP-1) promotes oligodendrocyte differentiation and enhances CNS myelination. J Neurosci. 2011;31:6247–6254. doi: 10.1523/JNEUROSCI.5474-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Claycomb KI, et al. Aberrant production of tenascin-C in globoid cell leukodystrophy alters psychosine-induced microglial functions. J Neuropathol Exp Neurol. 2014;73:964–974. doi: 10.1097/NEN.0000000000000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruckh JM, et al. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell. 2012;10:96–103. doi: 10.1016/j.stem.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker DJ, Petersen RC. Cellular senescence in brain aging and neurodegenerative diseases: Evidence and perspectives. J Clin Invest. 2018;128:1208–1216. doi: 10.1172/JCI95145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campisi J, d’Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 36.Lee BY, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 37.Coppé JP, et al. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J Biol Chem. 2011;286:36396–36403. doi: 10.1074/jbc.M111.257071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campisi J, Robert L. Cell senescence: Role in aging and age-related diseases. Interdiscip Top Gerontol. 2014;39:45–61. doi: 10.1159/000358899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davalos AR, et al. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J Cell Biol. 2013;201:613–629. doi: 10.1083/jcb.201206006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai J, Bercury KK, Macklin WB. Interaction of mTOR and Erk1/2 signaling to regulate oligodendrocyte differentiation. Glia. 2014;62:2096–2109. doi: 10.1002/glia.22729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahl SE, McLane LE, Bercury KK, Macklin WB, Wood TL. Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination. J Neurosci. 2014;34:4453–4465. doi: 10.1523/JNEUROSCI.4311-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyler WA, et al. Activation of the mammalian target of rapamycin (mTOR) is essential for oligodendrocyte differentiation. J Neurosci. 2009;29:6367–6378. doi: 10.1523/JNEUROSCI.0234-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiley CD, et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 2016;23:303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erlandsson Harris H, Andersson U. Mini-review: The nuclear protein HMGB1 as a proinflammatory mediator. Eur J Immunol. 2004;34:1503–1512. doi: 10.1002/eji.200424916. [DOI] [PubMed] [Google Scholar]
- 46.Samus M, Seelige R, Schäfer K, Sorokin L, Vestweber D. CD99L2 deficiency inhibits leukocyte entry into the central nervous system and ameliorates neuroinflammation. J Leukoc Biol. 2018;104:787–797. doi: 10.1002/JLB.1A0617-228R. [DOI] [PubMed] [Google Scholar]
- 47.Swaroop S, Sengupta N, Suryawanshi AR, Adlakha YK, Basu A. HSP60 plays a regulatory role in IL-1β-induced microglial inflammation via TLR4-p38 MAPK axis. J Neuroinflammation. 2016;13:27. doi: 10.1186/s12974-016-0486-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Moyon S, Hernandez M, Casaccia P. Epigenetic control of oligodendrocyte development: Adding new players to old keepers. Curr Opin Neurobiol. 2016;39:133–138. doi: 10.1016/j.conb.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picard-Riera N, et al. Experimental autoimmune encephalomyelitis mobilizes neural progenitors from the subventricular zone to undergo oligodendrogenesis in adult mice. Proc Natl Acad Sci USA. 2002;99:13211–13216. doi: 10.1073/pnas.192314199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michailidou I, de Vries HE, Hol EM, van Strien ME. Activation of endogenous neural stem cells for multiple sclerosis therapy. Front Neurosci. 2015;8:454. doi: 10.3389/fnins.2014.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peruzzotti-Jametti L, et al. Macrophage-derived extracellular succinate licenses neural stem cells to suppress chronic neuroinflammation. Cell Stem Cell. 2018;22:355–368.e3. doi: 10.1016/j.stem.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliver-De La Cruz J, et al. SOX2+ cell population from normal human brain white matter is able to generate mature oligodendrocytes. PLoS One. 2014;9:e99253. doi: 10.1371/journal.pone.0099253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellis P, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 54.Suh H, et al. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 57.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhat R, et al. Astrocyte senescence as a component of Alzheimer’s disease. PLoS One. 2012;7:e45069. doi: 10.1371/journal.pone.0045069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baar MP, et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169:132–147.e16. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bussian TJ, et al. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature. 2018;562:578–582. doi: 10.1038/s41586-018-0543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu M, et al. Senolytics improve physical function and increase lifespan in old age. Nat Med. 2018;24:1246–1256. doi: 10.1038/s41591-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130:1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HJ, Kim KW, Yu BP, Chung HY. The effect of age on cyclooxygenase-2 gene expression: NF-kappaB activation and IkappaBalpha degradation. Free Radic Biol Med. 2000;28:683–692. doi: 10.1016/s0891-5849(99)00274-9. [DOI] [PubMed] [Google Scholar]
- 64.Sokratous M, et al. CpG island methylation patterns in relapsing-remitting multiple sclerosis. J Mol Neurosci. 2018;64:478–484. doi: 10.1007/s12031-018-1046-x. [DOI] [PubMed] [Google Scholar]
- 65.Sun Y, et al. HMGB1 expression patterns during the progression of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2015;280:29–35. doi: 10.1016/j.jneuroim.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 66.van Horssen J, et al. Nrf2 and DJ1 are consistently upregulated in inflammatory multiple sclerosis lesions. Free Radic Biol Med. 2010;49:1283–1289. doi: 10.1016/j.freeradbiomed.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 67.Ren JL, Pan JS, Lu YP, Sun P, Han J. Inflammatory signaling and cellular senescence. Cell Signal. 2009;21:378–383. doi: 10.1016/j.cellsig.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang C, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349:aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lasry A, Ben-Neriah Y. Senescence-associated inflammatory responses: Aging and cancer perspectives. Trends Immunol. 2015;36:217–228. doi: 10.1016/j.it.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 70.Haider L, et al. Oxidative damage in multiple sclerosis lesions. Brain. 2011;134:1914–1924. doi: 10.1093/brain/awr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasquali L, et al. Plasmatic oxidative stress biomarkers in multiple sclerosis: Relation with clinical and demographic characteristics. Clin Biochem. 2015;48:19–23. doi: 10.1016/j.clinbiochem.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 72.Coppé JP, Kauser K, Campisi J, Beauséjour CM. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem. 2006;281:29568–29574. doi: 10.1074/jbc.M603307200. [DOI] [PubMed] [Google Scholar]
- 73.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawcer S, et al. International Multiple Sclerosis Genetics Consortium; Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhan R, et al. A DEAD-box RNA helicase Ddx54 protein in oligodendrocytes is indispensable for myelination in the central nervous system. J Neurosci Res. 2013;91:335–348. doi: 10.1002/jnr.23162. [DOI] [PubMed] [Google Scholar]
- 76.Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: Past and future. Epigenetics. 2015;10:103–121. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Henikoff S, Smith MM. Histone variants and epigenetics. Cold Spring Harb Perspect Biol. 2015;7:a019364. doi: 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.El Gazzar M, et al. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen S, et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conway GD, O’Bara MA, Vedia BH, Pol SU, Sim FJ. Histone deacetylase activity is required for human oligodendrocyte progenitor differentiation. Glia. 2012;60:1944–1953. doi: 10.1002/glia.22410. [DOI] [PubMed] [Google Scholar]
- 82.Brück W, et al. Therapeutic decisions in multiple sclerosis: Moving beyond efficacy. JAMA Neurol. 2013;70:1315–1324. doi: 10.1001/jamaneurol.2013.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Comi G. Disease-modifying treatments for progressive multiple sclerosis. Mult Scler. 2013;19:1428–1436. doi: 10.1177/1352458513502572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.