Significance
IgE antibody interaction with allergen proteins is the main driver for life-threating allergic responses. Using a nanoparticle-based method, we identified the most crucial binding sites for IgEs on peanut proteins by testing a small population of patient serum with a clinical history of severe allergies to peanuts. We then synthesized inhibitors, we call covalent heterobivalent inhibitors, which specifically and permanently prevent IgEs from binding to peanut proteins and triggering allergic responses. This is significant as it seems that only a few binding sites on peanut allergen proteins drive allergic responses. Also, this is a functional example of selective IgE inhibition to a relevant food allergen and establishes cHBIs as a class of allergy therapeutics.
Keywords: IgE, allergy, epitope, selective inhibition, peanut
Abstract
Allergies are a result of allergen proteins cross-linking allergen-specific IgE (sIgE) on the surface of mast cells and basophils. The diversity and complexity of allergen epitopes, and high-affinity of the sIgE–allergen interaction have impaired the development of allergen-specific inhibitors of allergic responses. This study presents a design of food allergen-specific sIgE inhibitors named covalent heterobivalent inhibitors (cHBIs) that selectively form covalent bonds to only sIgEs, thereby permanently inhibiting them. Using screening reagents termed nanoallergens, we identified two immunodominant epitopes in peanuts that were common in a population of 16 allergic patients. Two cHBIs designed to inhibit only these two epitopes completely abrogated the allergic response in 14 of the 16 patients in an in vitro assay and inhibited basophil activation in an allergic patient ex vivo analysis. The efficacy of the cHBI design has valuable clinical implications for many allergen-specific responses and more broadly for any antibody-based disease.
Allergy symptoms can vary from a rash to a potentially fatal anaphylactic reaction. Food allergies, especially peanut allergy, fall into the latter. These types of allergic reactions (IgE-dependent type-I hypersensitivity reactions) can lead to life-threatening systemic inflammatory responses such as anaphylactic shock (1, 2). The critical step in a peanut-allergy reaction is the binding of peanut allergens to allergen-specific IgE antibodies (sIgEs) attached to the high-affinity Fc epsilon receptors (FcεRIs) on mast cell surfaces that results in their cross-linking. This cross-linking triggers intracellular signaling that culminates in cellular degranulation that releases preformed mediators stored in cytoplasmic granules, including vasoactive amines, neutral proteases, proteoglycans, cytokines, and chemokines (3).
Currently, in the clinic, there are no medications that can prevent the degranulation response from mast cells, aside from a nonspecific IgE depletion therapy, wherein omalizumab, a monoclonal antibody, binds and inhibits all IgE from binding the surface receptor. Although it has shown effectiveness in clinical treatment of asthma, omalizumab has not been approved for the treatment of peanut allergies (4, 5). Furthermore, the inhibition of all IgE can lead to nonspecific immune suppression, leading to increases in parasitic infection and cancer (6, 7). A targeted and specific sIgE inhibitor would alleviate these concerns. One strategy for inhibition of peanut-allergy reactions (and any other food allergy) is to prevent the binding of allergen epitopes to sIgEs, but there are currently no functional examples of specific sIgE inhibitory compounds in the clinic or scientific literature.
The major challenge in developing targeted sIgE inhibitors is to identify the critical immunogenic epitopes from the large number of potential epitopes on allergen proteins (8, 9). Literature reports of several sIgE binding epitopes of the two most immunogenic peanut proteins, Ara h 2 and Ara h 6, exist (10, 11). These studies, however, did not assess the relative importance of those epitopes for IgE-dependent activation, instead only determining that they bind sIgEs. Here, we developed a physiologically relevant, nanoparticle-based assay system we termed nanoallergens, to identify the most critical immunogenic epitopes of Ara h 2 and Ara h 6. Next, we used this information to rationally design a class of sIgEs inhibitors, named covalent heterobivalent inhibitors (cHBIs), to irreversibly bind to and inhibit peanut allergen/sIgE interactions, thereby selectively and exhaustively inhibiting peanut-allergy reactions (SI Appendix, Fig. S1). We demonstrated, by using peanut allergic patient sera, that cHBIs specifically and irreversibly target the most crucial sIgEs for allergic responses and yield almost complete inhibition of degranulation responses to peanuts in cellular assays with patient serum. This study presents compelling preclinical data for further development and assessment of the cHBI strategy as inhibitors of peanut allergies and other food allergies to improve patient outcomes.
Results
Identification of Immunodominant Ara h 2 and Ara h 6 Epitopes Using the Nanoallergen Assay.
To facilitate design of cHBIs, we first identified the most critical immunogenic epitopes from Ara h 2 and Ara h 6 using the nanoallergen assay system developed in our laboratory (12). Nanoallergens provide multivalent display for allergen epitopes on liposomal surfaces, which are then used to evaluate their immunogenicity via in vitro mast cell degranulation responses (Fig. 1A). Currently used microarray techniques only determine if IgE binds a particular epitope but lack proper epitope display and valency to trigger cellular degranulation (13). Nanoallergens overcome these shortcomings and provide a physiologically relevant assay system to evaluate immunogenicity of epitopes by directly comparing cellular responses not just relative binding.
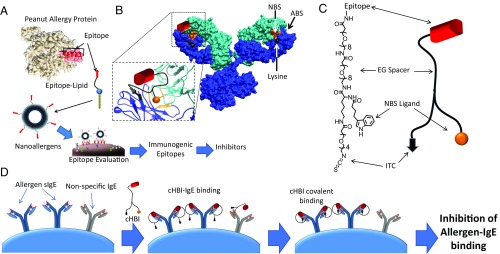
Fig. 1.
cHBIs are designed to selectively and irreversibly bind sIgEs and inhibit cellular degranulation. (A) Cartoon demonstration of the nanoallergen assay system for evaluating potential immunogenic epitopes. Potential epitopes of allergen proteins are synthesized as linear peptides conjugated to lipid tails (epitope-lipid). Epitope-lipids are then incorporated during liposome synthesis to produce nanoallergens. Nanoallergens are used to screen for epitope immunogenicity in cellular degranulation assays. (B) cHBI design. cHBIs were synthesized to display an immunogenic epitope that binds to the ABS of sIgEs, a NBS ligand that binds to the NBS of sIgEs, and an amine/lysine-reactive moiety (ITC) to facilitate irreversible binding to sIgEs. Representative crystal structure of an Ig with the ABS, NBS (orange), and a lysine proximal to NBS (black) is shown. The zoomed-in Fab region describes cHBI’s bivalent NBS and ABS binding, which promotes covalent conjugation to the proximal lysine. (C) Chemical structure of cHBI. The immunodominant epitope, NBS ligand, and ITC moiety are linked using flexible EG linkers to facilitate bivalent binding and covalent bond formation to sIgEs. (D) Schematic illustration of allergy inhibition by cHBIs. After initial bivalent binding to ABS and NBS, cHBI forms a covalent bond with sIgEs via ITC cross-linking of the nearby lysine, thereby inhibiting cellular degranulation.
Nanoallergen preparation required synthesis of epitope-lipids, for which potential epitopes (synthesized as linear peptides) are conjugated to palmitic acid tails via ethylene glycol (EG). The epitope-lipid was purified, mixed at precise ratios with phospholipids [1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC)] and extruded through membranes to form 100-nm nanoparticles (SI Appendix, Fig. S2). These nanoallergens multivalently present epitopes on their surfaces with high-precision and controlled stoichiometry (12, 14). Nanoallergens were used to trigger degranulation response in RBL-SX38 cells [model mast cell line which expresses human IgE receptor (FcεRI)] (15) primed with a specific patient serum (Fig. 1A). This assay enables patient-specific assessment of epitope immunogenicity, rather than only assessing binding interactions, and hence accurately identifies the most allergenic epitopes.
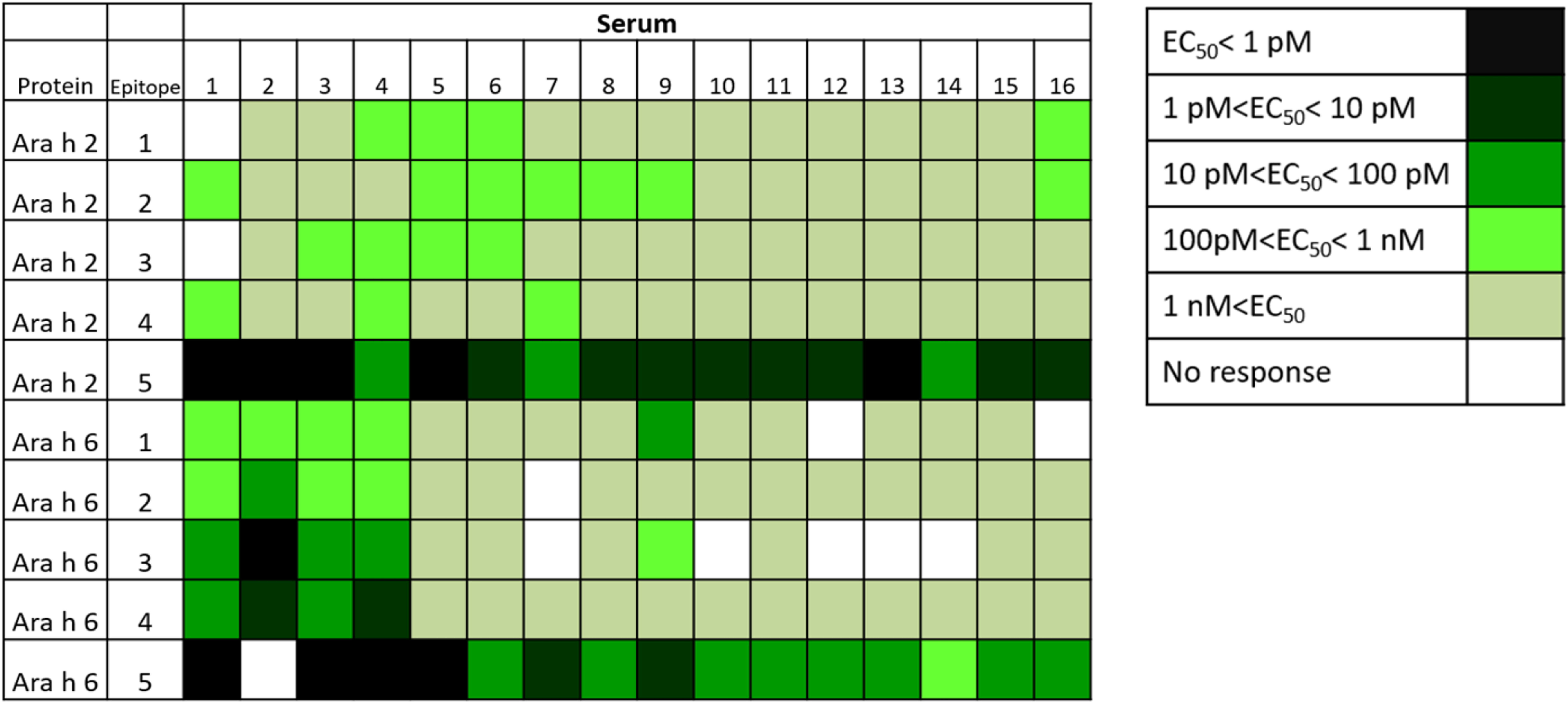
We synthesized nanoallergens which present epitopes for 10 commonly cited peanut protein epitopes in the literature, epitopes 1–5 of Ara h 2, and epitopes 1–5 of Ara h 6 (SI Appendix, Table S1) (10, 11, 16–18). To evaluate these epitopes, we used human serum from 16 peanut-allergic patients obtained from PlasmaLabs and Massachusetts General Hospital (SI Appendix, Table S2). Serum samples were selected based on clinical history of peanut-allergic responses (as verified by commercial source or by food challenge) and a peanut sIgE >50 kU/L as determined by ImmunoCAP (SI Appendix, Table S2). Serum from each patient was used to prime RBL-SX38 cells before incubation with nanoallergens (0–5 nM) presenting each of the epitopes (SI Appendix, Fig. S3). Degranulation was monitored via beta-hexosaminidase assay and EC50 values were determined (Table 1 and SI Appendix, Table S3). Our results identified Ara h 2 epitope 5, and Ara h 6 epitope 5 to be immunodominant, given their low EC50 (EC50 < 10 pM for Ara h 2 epitope 5 and EC50 < 100 pM for Ara h 6 epitope 5) for most patient samples. Note that as demonstrated previously (12), degranulation intensity does not correlate with epitope dominance due to inhibitory cascades [such as Src homology 2 (SH2) domain containing inositol polyphosphate 5-phosphatase 1 (SHIP)] that attenuate degranulation (19). Hence, EC50 values were used as the primary metric for epitope dominance.
Table 1.
Identification of the immunogenic epitopes from Ara h 2 and Ara h 6 by nanoallergen analysis
 |
Identification of Potentially Cross-Reactive Peanut Protein Epitopes via Analysis of Epitope Sequence Homology.
The major challenge of preventing allergic reactions with sIgE inhibitors is the large number of epitopes on allergy proteins that could bind to a diverse set of sIgEs (20). In peanuts, among the 13 known immunogenic proteins, Ara h 2 and Ara h 6 are the immunogenic proteins most closely associated with severe allergic reactions (21). Other proteins such as Ara h 1 and Ara h 3, however, are also immunogenic, albeit triggering less severe reactions (21). While it is not practical to develop inhibitors for each epitope/sIgE interaction, it is known that allergens often have homologous epitopes between two different proteins, and sIgEs can be potentially designed to have cross-reactivity for several allergenic proteins simultaneously (17, 22). We noticed a high level (>50%) of homology between the most immunogenic epitopes identified in our nanoallergen analysis, Ara h 2 epitope 5 and Ara h 6 epitope 5, and epitopes from Ara h 1, Ara h 2, Ara h 3, and Ara h 6 (SI Appendix, Table S4). Hence, we hypothesized that by developing inhibitors of immunodominant epitope/sIgE interactions, namely Ara h 2 epitope 5 and Ara h 6 epitope 5, we could inhibit epitope/sIgE interactions for a broad range of immunogenic epitopes because of homology and possible cross-reactivity with sIgEs and improve the therapeutic potential of cHBI.
Rational Design and Synthesis of cHBIs to Inhibit Binding of sIgEs to Immunodominant Allergy Epitopes.
Next, we engineered cHBIs that irreversibly inhibit binding of sIgEs to these two immunodominant epitopes. The cHBI design required three moieties linked by ethylene-glycol spacers: (i) the immunodominant epitopes from Ara h 2 and Ara h 6 to target the antigen binding site (ABS), (ii) a ligand to target the nucleotide binding site (NBS), and (iii) a reactive functional group (RFG) to form a covalent bond with target sIgEs (Fig. 1 B and C). NBS is a conserved binding site located proximal to the ABS between the heavy and light chains of all immunoglobulins (Fig. 1B) (23, 24). We identified a molecule, indole-3 butyric acid (IBA) that specifically binds to NBS with moderate affinity (Kd = 2–10 µM) (23, 25, 26). Targeting two independent sites on sIgEs simultaneously, ABS (with a peptide epitope) and NBS (with IBA), enables cHBIs to have both increased avidity and selectivity for target sIgEs. The moderate affinity of IBA minimizes its binding to off-target antibodies and requires an ABS ligand for effective inhibition, while the bivalent binding to both sites improves overall avidity and selectivity for sIgEs (23). The third critical design factor for cHBIs is the RFG which facilitates covalent bond formation after a cHBI binds bivalently to target sIgEs, thereby permanently inhibiting sIgE/allergen interaction. For this purpose, we chose an amine-reactive moiety, isothiocynate (ITC), as the RFG in the design of cHBIs (Fig. 1C). ITC compounds are common in nature and are nontoxic (27). ITC groups form thiourea bonds with primary amines, such as lysines, in elevated pH solutions (>9) but have limited reactivity at pH 7.4 (t1/2 > 50 h) (28). Consequently, under physiological conditions, ITC has negligible reactivity with random lysines, reducing off-target conjugations with other serum proteins. Furthermore, this limited reactivity means the preassociation of cHBIs with an ABS/NBS is necessary to enhance the kinetics of covalent bond formation on sIgEs via increased ITC effective molarity, thereby providing selectivity. We previously confirmed that the NBS pocket is highly conserved in antibodies by analyzing over 260 antibody crystal structures (29) and have identified a lysine within 2 nm of the NBS pocket lip on several antibody crystal structures (SI Appendix, Fig. S4). This lysine provided the potential reactive site on sIgE Fab for the irreversible conjugation of cHBIs, thus enabling selective and irreversible inhibition of allergic reactions (Fig. 1D).
We synthesized two versions of cHBIs displaying Ara h 2 epitope 5 (cHBIArah2/ep5) or Ara h 6 epitope 5 (cHBIArah6/ep5) using Fmoc solid-phase peptide synthesis (SI Appendix, Fig. S5). ITC on cHBI was stable at pH 7.4, but became reactive at pH 10.0, indicating that cHBIs will have negligible off-target conjugation at pH 7.4 (SI Appendix, Fig. S6). We also synthesized fluorescein-tagged cHBIs which were used to detect cHBI binding to sIgEs (Table 2). Note that a 1:1 combination of cHBIArah2/ep5:cHBIArah6/ep5 (which will be referred to as cHBIArah2:Arah6) was used for all following experiments unless otherwise noted.
Table 2.
Chemical structures of cHBI inhibitors used in this study
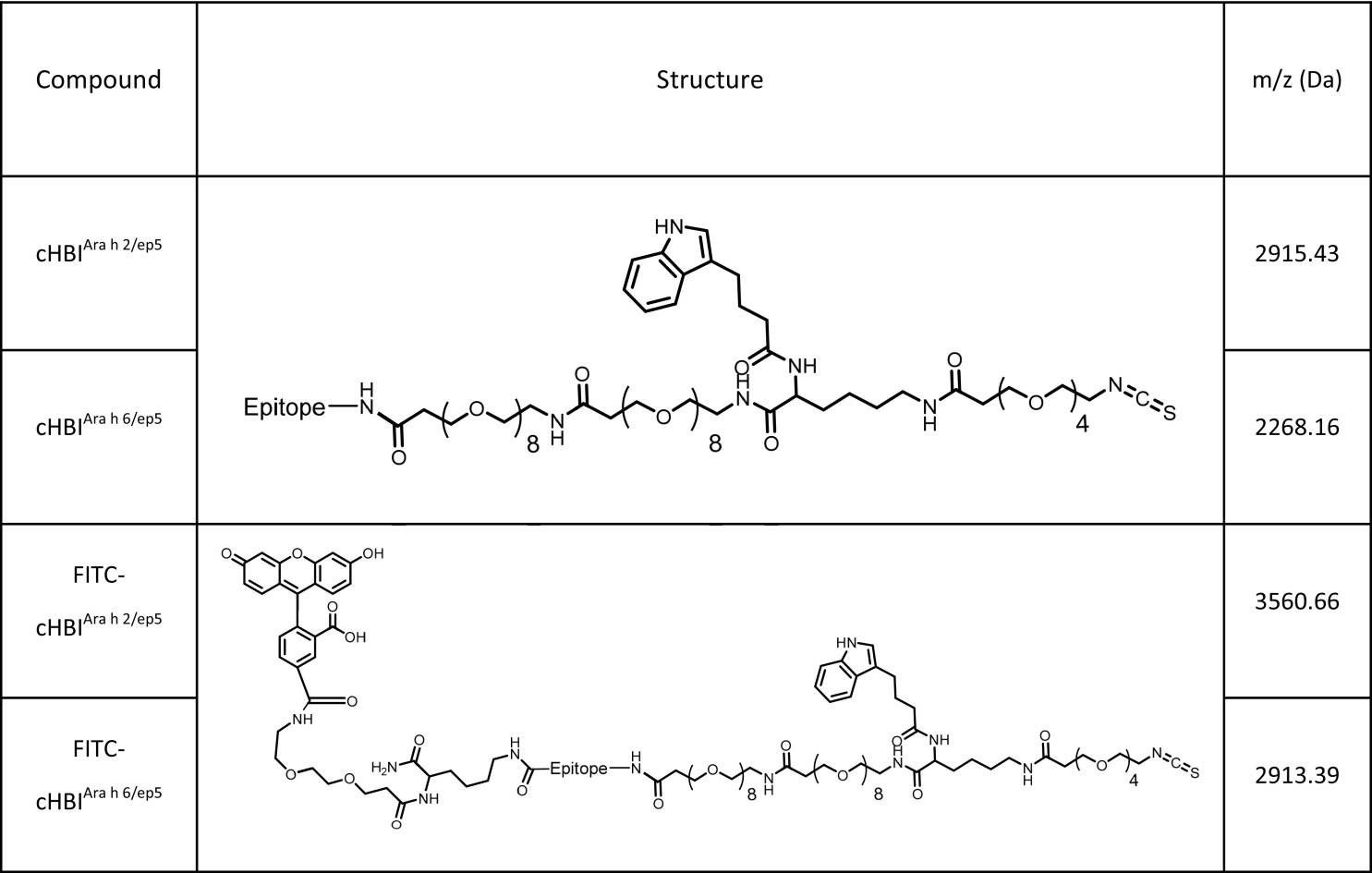 |
cHBIs Selectively Bind to Peanut sIgEs in Clinically Relevant Patient Serum Samples.
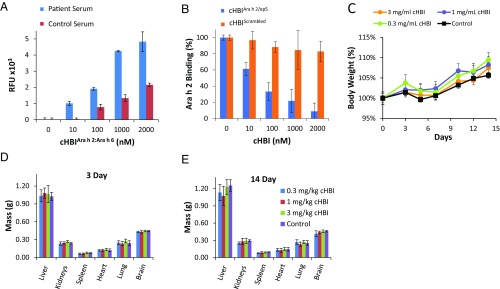
To determine if cHBIArah2:Arah6 binds specifically to sIgEs in clinically relevant patient serum samples, fluorescein-tagged cHBIArah2:Arah6 was incubated with serum obtained from either a peanut-allergic patient (serum 4) or a healthy volunteer (control). IgE-cHBI binding was determined using ELISA by incubating cHBI-treated serum on anti-human IgE-coated plates. The fluorescein-tagged cHBIArah2:Arah6 showed significantly higher conjugation to IgE in peanut-allergic patient serum than to control serum (Fig. 2A, P < 0.05). In a separate ELISA experiment, we incubated cHBIArah 2/ep5 or a cHBI presenting a Scrambled peptide (cHBIScrambled) with patient serum, incubated on anti-human IgE-coated plates and performed an ELISA to observe IgE binding to Ara h 2. Our results demonstrated a significant drop in Ara h 2 binding when serum was incubated with cHBIArah2/ep5, but not when incubated with cHBIScrambled (Fig. 2B). Furthermore, we performed an ELISA to demonstrate that cHBIs do not nonspecifically label BSA protein and that the cHBI design is highly antigen specific (SI Appendix, Fig. S7). These results demonstrate that cHBIArah2/ep5 targeted sIgEs for Ara h 2 in an epitope-specific fashion.
Fig. 2.
cHBIs form covalent bonds selectively with target sIgEs. (A) Fluorescein-tagged cHBIArah2:Arah6 binds to sIgEs in patient serum (serum 4) as detected by ELISA. Serum from healthy volunteers was used as a control. RFU, relative fluorescence unit. (B) cHBIArah2/epi5 inhibits the binding of Ara h 2 to IgE in serum 4. ELISA was used to monitor binding of Ara h 2 to IgE from serum 4 after incubation with cHBIArah2/epi5. cHBIScrambled (cHBI presenting a nonspecific peptide) was used as control. (C) Assessment of cHBI toxicity in vivo. Body weight of C57BL/6 mice injected (i.v.) with a single injection of 0.3, 1, or 3 mg/kg cHBIArah2:Arah6 was assessed as a measure of systemic toxicity. PBS was used as control. (D) Organ weight of mice killed 3 d after receiving a single i.v. injection of 0.3, 1, or 3 mg/kg cHBIArah2:Arah6 was assessed. (E) Organ weight from D, 14 d postinjection. Data represent the mean ± SEM of six mice per group.
Assessment of cHBI Toxicity in Vivo.
To evaluate the therapeutic potential of cHBI, next, we assessed cHBI toxicity in vivo using C57BL/6J mice. Acute and chronic systemic toxicity of cHBIs was assessed 3 d and 14 d post single i.v. injection of cHBIArah2:Arah6 at 0.1, 1, and 3 mg/kg. The doses of cHBIArah2:Arah6 were selected based on our earlier studies (23). PBS injection was used as control. Body weight was monitored every 2–3 d for 14 d and did not significantly vary from control (Fig. 2C). Mice were killed at 3 and 14 d after cHBIArah2:Arah6 injection, and organ weight and blood counts were analyzed. There was no change in liver, kidney, spleen, heart, lung, or brain organ weights at 3 (Fig. 2D) or 14 d (Fig. 2E) postinjection. The WBC, RBC, and platelet counts were also unchanged (SI Appendix, Fig. S8). Based on these results, cHBIs did not demonstrate any significant toxicity in vivo, and therefore is likely to be well tolerated in a clinical setting.
cHBIs Prevent Cellular Degranulation Induced by Crude Peanut Extract in Cellular Assays Performed with Patient Serum.
To further evaluate the therapeutic potential of cHBIs, we tested if cHBIs could inhibit degranulation induced by peanut allergen using cells primed with allergic patient serum. In this experiment, we primed RBL-SX38 cells with patient serum, incubated them with cHBIArah2/ep5 or cHBIArah2:Arah6, and then challenged cells with (i) nanoallergens, (ii) Ara h proteins, or (iii) crude peanut extract.
For nanoallergen challenge, primed RBL-SX38 cells were incubated with varying concentrations (0–1,000 nM) of cHBIArah2/ep5 overnight, washed, and challenged with 1–1,000 nM of nanoallergens presenting Ara h 2 epitope 5 (NanoallergenArah2/ep5). We observed >95% inhibition for all cHBIArah2/ep5 concentrations tested (SI Appendix, Fig. S9A), confirming that cHBIArah2/ep5 effectively inhibit degranulation triggered by Ara h 2 epitope 5. We also observed that cHBIArah2/ep5 inhibited degranulation triggered by a nanoallergen loaded with a homologous epitope from Ara h 6 (NanoallergenAra h 6/YDSYDIR, see SI Appendix, Fig. S9B).
For the purified component allergen challenge experiments, primed RBL-SX38 cells were incubated overnight with varying concentrations (0–2,000 nM) of cHBIArah2:Arah6, washed, and challenged with one of the major peanut allergen proteins: Ara h 2 (10 nM), Ara h 6 (10 nM), Ara h 1 (100 nM), or Ara h 3 (100 nM) (SI Appendix, Fig. S10). cHBIArah2:Arah6 inhibited >90% of the degranulation triggered by Ara h 2 or Ara h 6 for all sera tested. In addition, cHBIArah2:Arah6 inhibited between 40% and 80% of the degranulation triggered by Ara h 1 and Ara h 3, suggesting that cHBIArah2:Arah6 inhibits sIgEs for Ara h 1 and Ara h 3 via epitope cross-reactivity. In an additional control experiment, we also tested the selectivity of cHBIArah2:Arah6 for peanut sIgEs by using the RBL-2H3 cell line. RBL-2H3 were primed with antidinitrophenol (DNP)-IgE, and challenged with DNP-conjugated (∼17 DNP conjugations per BSA) BSA (DNP-BSA) in the presence or absence of cHBIArah2:Arah6. As expected, cHBIArah2:Arah6 did not inhibit degranulation induced by DNP-BSA, demonstrating that cHBIs are allergen-selective inhibitors (SI Appendix, Fig. S11).
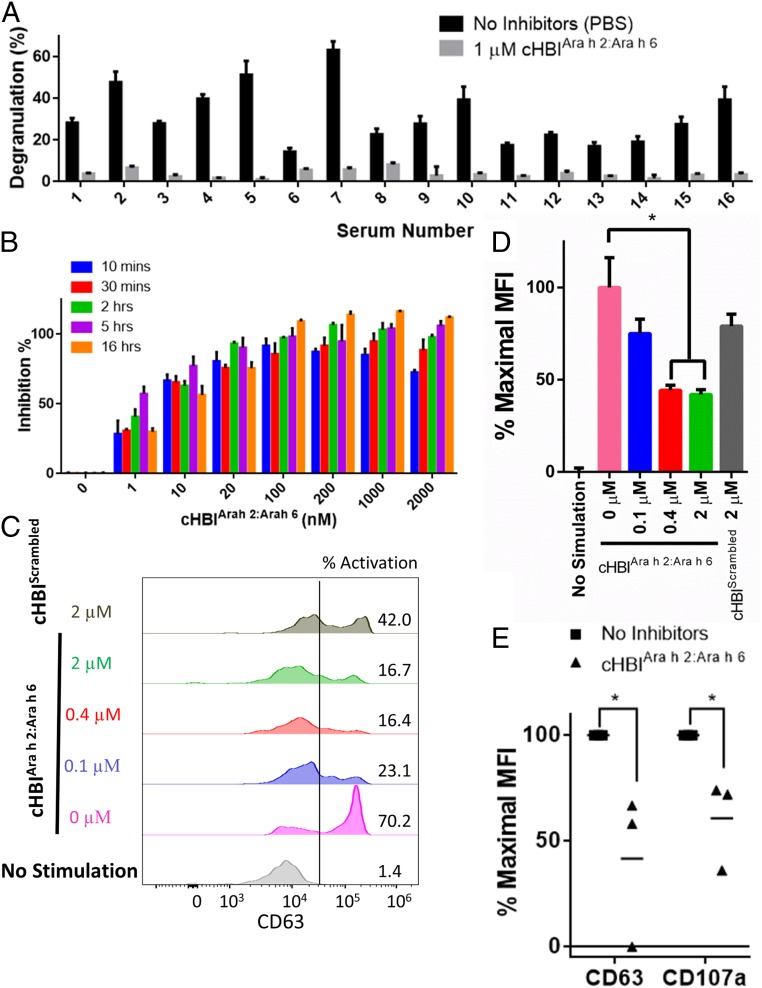
For the crude peanut-extract challenge, primed RBL-SX38 cells were incubated with 1 µM cHBIArah2:Arah6 overnight, washed, and then challenged with 1–1,000 ng/mL of crude peanut extract. cHBIArah2:Arah6 inhibited >80% of degranulation in 14 of 16 sera for at least one of the peanut-extract concentrations tested. Note that due to varying levels of sensitivity of the individual subjects, some patients had lower inhibition at higher allergen concentrations (Fig. 3A and SI Appendix, Figs. S12 and S13). In control experiments performed using either cHBIArah2/ep5 or cHBIArah6/ep5 alone, we observed <30% inhibition, demonstrating that effective inhibition required a combination of more than one immunodominant epitope (SI Appendix, Fig. S14A). Western blot analysis revealed that cHBIArah2:Arah6 suppressed crude peanut extract-induced phosphorylation of intracellular proteins associated with degranulation response (SI Appendix, Fig. S14B). This inhibition was also long lasting as primed RBL-SX38 cells incubated with cHBIArah2:Arah6 retained resistance to crude peanut extract-induced degranulation even when challenged 3 d after cHBIArah2:Arah6 was washed away (SI Appendix, Fig. S15). Additionally, inhibition via cHBIArah2:Arah6 was concentration and time dependent; more inhibition occurred at increased concentrations and incubation times (Fig. 3B). Taken together, these results demonstrate a combination of just two engineered epitope inhibitors, cHBIArah2:Arah6 deliver potent inhibition of immune cell and peanut allergen interactions.
Fig. 3.
cHBIs inhibit degranulation induced by crude peanut extract in clinical patient samples. (A) cHBIArah2:Arah6 inhibits degranulation induced by crude peanut extract. A total of 1 µM cHBIArah2:Arah6 inhibited degranulation induced by crude peanut extract in RBL-SX38 cells primed with 16 different patient sera. Note that due to differences in patient sensitivity to crude peanut extract, patient samples were challenged with varying amount of crude peanut extract after cHBI incubation. Patients 1, 2, and 4 were challenged with 1 μg/mL peanut extract, patients 3 and 7 with 100 ng/mL, patient 5 with 10 ng/mL, and all others with 1 ng/mL PBS, used as a control. (B) RBL-SX38 cells primed with patient serum (serum 4) were incubated with varying concentration of cHBIArah2:Arah6 for varying time points (10 min, 30 min, 2 h, 5 h, and 16 h), washed, and then challenged with 100 ng/mL of crude peanut extract. (C) cHBIArah2:Arah6 inhibits degranulation in BAT. BAT data for whole blood incubated with 0 μM (pink), 0.1 µM (blue), 0.4 µM (red), or 2 µM (green) cHBIArah2:Arah6 and then challenged with 100 ng/mL crude peanut extract are shown. Flow cytometry plots are shown with fluorescence intensity on the x axis and number of basophils on the y axis. Controls include blood incubated without crude peanut extract (no treatment), and blood incubated with 2 µM of a 1:1 mixture of cHBIs presenting Scrambled sequences of Ara h 2 epitope 5 and Ara h 6 epitope 5, respectively (cHBIScrambled). Basophils were considered CD63 positive if fluorescence intensity was greater than the threshold of 2 × 104 (black line). Percent activated basophils is shown on Right side of plot. (D) Percent maximal MFI of flow cytometry plots from D is shown, calculated as percentage between the “0 µM” and “no stimulation” MFI values. Error bars indicate ± SD of triplicate experiments. (E) Three additional patients were assessed using BAT assay. Whole blood from three patients was incubated with (triangles) or without (squares) 1 μM cHBIArah2:Arah6 and then challenged with 100 ng/mL of crude peanut extract. Percent maximal MFI was similarly calculated as in D. Each point represents an average of three technical replicates of each patient; horizontal lines represent mean of all three patients. *P < 0.05.
cHBIs Prevent Cellular Degranulation Induced by Crude Peanut Extract in a Basophil Activation Test.
Finally, to test the potential for a clinical evaluation of the inhibitors, we evaluated cHBIs by using a basophil activation test (BAT). The BAT is a physiologically relevant assay in which whole blood is taken from a patient, incubated briefly with an allergen, and the allergic response is monitored via flow cytometry by measuring cellular markers of activation and degranulation, including CD63, CD107a, or CD203c (30). Whole blood drawn in heparin vacutainer tubes was taken from a single patient with a history of severe allergic reactions to peanuts, incubated with cHBIArah2:Arah6, challenged with 100 ng/mL crude peanut extract, and analyzed with BAT. The percent of cells expressing CD63 above a threshold value [CD63+, threshold shown via black line at 2 × 104 mean fluorescence intensity (MFI)] increased from 1.4% to 70% when blood was challenged with crude peanut extract in the absence of cHBIArah2:Arah6 inhibitors (Fig. 3C). When the blood was treated with cHBIArah2:Arah6, however, CD63+ cells decreased from 70% to 16–23% (Fig. 3C). In parallel, the percent maximal MFI of CD63 decreased when blood was treated with 0.4 and 2 µM of cHBIArah2:Arah6 (to 44% and 40% of maximal response respectively, Fig. 3D, P < 0.05). As a control, we took the epitope sequences in cHBIArah2:Arah6, synthesized cHBIs presenting Scrambled epitopes of Ara h 2 epitope 5 and Ara h 6 epitope 5, and mixed them at a 1:1 ratio (cHBIScrambled). cHBIScrambled did not reduce the levels of CD63 expression on basophils after crude peanut extract challenge, demonstrating that cHBIs have high selectivity (Fig. 3 C and D). In addition to CD63, cHBIArah2:Arah6 treatment reduced the levels of other markers of degranulation, such as CD107a and CD203c (SI Appendix, Fig. S16). Among these markers, CD63 expression in particular has been shown to be a predictive marker of physiological severity of an allergic response (31). To further validate these inhibitors, we performed the BAT assay on three additional allergic patients and two nonallergic control patients (Fig. 3E and SI Appendix, Fig. S17). There was a significant drop in CD63 and CD107a expression when patient samples were treated with 1 μM cHBIArah2:Arah6, while the nonallergic patients had no significant change in CD63 or CD107a expression. In particular, the significant reduction in the number of CD63+ cells in allergen-challenged cells when treated with cHBIArah2:Arah6 demonstrate the clinical potential for cHBIs to prevent severe allergic reactions to peanuts. Combined, these results demonstrate that cHBIs have promising clinical potential to prevent allergic reactions to peanuts and improve patient outcomes; nevertheless, studies with larger patient populations are required to further validate these claims.
Discussion
Here, we described the design and evaluation of cHBIs, a selective sIgE inhibitor, to target and permanently inhibit the antigen binding site of allergen-specific IgE antibodies that recognize specific allergen epitope sequences. The most critical component of cHBI design required the identification of the immunodominant epitopes that are critical mediators during the initiation of degranulation responses. Using the nanoallergen platform, we identified these immunodominant peanut-allergen epitopes for a diverse set of serum samples in a functional cellular degranulation assay. The results showed that the immunodominant epitopes were commonly shared by all of the 16 subjects. Therefore, two cHBI molecules to mimic these two epitopes were synthesized and evaluated.
One of the most significant and surprising results was that two cHBIs, targeting only two of the peanut epitopes, was sufficient to inhibit degranulation responses to crude peanut extract with over 80% efficiency in 14 of 16 patient samples, although this effect was dependent upon the concentration of allergen challenge. Given that patients generate IgE to multiple proteins and multiple epitopes within each protein, the inhibition of responses by only two epitopes highlights that we still know very little about engagement of IgE by complex allergens. In our previous work we demonstrated that low-affinity epitopes can cooperate with high-affinity epitopes in IgE engagement and degranulation responses (32). Moreover, we previously observed that certain peanut epitopes trigger degranulation at significantly lower concentrations of nanoallergens, but when used in combination with an epitope of lower potency, there is a synergistic enhancement of degranulation (12). This combinatorial aspect is important and contrasts with many studies that use “allergens” with high numbers of multimerized epitopes. This work suggests several things that are important in considering allergen reactivity. First, we suggest that there are public epitopes, reactivity that is shared across a broad portion of the peanut-allergic human population, in IgE recognition of peanut proteins. Second, we suggest that these public allergen epitopes are required for allergen responses and can be therapeutically targeted to inhibit responses. This finding potentially eliminates a major obstacle in designing allergen-specific inhibitors, suggesting that inhibitors would not need to be personalized and that common reagents could be used by a broader segment of the peanut-allergic population than initially perceived. This will be a critical concept to explore in other allergens to determine if our approach to inhibitor generation can be broadly applied to allergy treatment.
The second most significant result is increase in efficacy and the lack of complications or off-target effects due to incorporation of ITC in the cHBI design. From our experience with allergen model systems, we predicted that without a permanent covalent linkage, the epitope mimics would be unlikely to sufficiently inhibit sIgEs due to the short half-life of peptide-based therapeutics and the eventual dissociation of the inhibitor from sIgEs (33). One major concern is that cHBIs will indiscriminately conjugate proteins and trigger nonspecific inflammatory responses or side effects. While we did not observe any acute toxicity in our mouse experiments and have demonstrated that these inhibitors are very selective in vitro, further studies concerning the chronic toxicology of these inhibitors on immune responses is necessary. Based on the data from this study, we expect cHBIs to be nontoxic and the irreversible action of cHBI inhibitors could potentially yield long-lasting inhibition, since IgE bound to mast cells and basophils can remain in the body for months (34).
The cHBI design would be a significant improvement over current allergy therapeutics. Drugs that are currently available, such as epinephrine or antihistamines, only transiently attenuate the intensity of the degranulation response and allergic symptoms. This type of approach to immune inhibition is nonspecific and results in blocking immune components that could be important for antipathogen or antitumor immunity. Attempts at developing specific allergy inhibitors have not been successful due to the complexity of the immune response and the challenges arising from the multivalent nature of the interactions. This study describes the cHBI design, which provides a solution to these issues by using a lesser-known binding site on the Fab arm of antibodies with a multivalent design, and presents a successful example of a specific allergy inhibitor to be used on a clinically relevant protein-based allergen.
Materials and Methods
Materials.
NovaPEG Rink amide resin, 5(6)-carboxy-fluorescein, 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluroniumhexafluorophosphate (HBTU), Fmoc-Lys(IvDde)-OH, all Fmoc protected amino acids, and 10 kDa 0.5 mL centrifugal filters were purchased from EMD Millipore.
N,N-dimethylformamide (DMF) (>99.8%), dichloromethane (DCM) (>99.8%), N,N-diisopropylethylamine (DIEA), methanol, hydrazine, piperidine, trifluoroacetic acid (TFA), triisopropylsilane (TIS), tryptamine, 2-naphthaleneacetic acid, ethylene diamine, biotin, di-tert-butyl carbonate (BOC2O), 4-(dimethylamino)pyridine (DMAP), succinic anhydride, carbon disulfide (CS2), diisopropylazocarboxylate (DIAD), 2,4-dinitro-1-fluorobenzene (DNFB), acetonitrile, acetic acid, methanol, carbonate-bicarbonate buffer, Tween 20, indole-3-butyric acid (IBA), biotin, PBS, streptavidin conjugated to HRP (Step- HRP), defattened peanut meal (crude peanut extract), carboxyfluorescein (CF), and cytochalasin D were purchased from Sigma-Aldrich. IBA-Boc is a Boc protected variant of IBA, which was synthesized separately in house.
Ara h 1, Ara h 2, biotin-Ara h 2, Ara h 3, and Ara h 6 proteins were purchased from Indoors Biotechnologies.
DSPC, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-mPEG2000) (ammonium salt), membranes, and all mini extruder components were purchased from Avanti Polar Lipids (Alabaster).
High-binding and non-binding 96-well plates were purchased from Corning. RPMI media, minimum essential media, penicillin-strep solution, l-glutamine, and Amplex Red ELISA kits were purchased from Life Technologies. BSA, G418 disulfide salt, and FBS were purchased from Gemini Biosciences. The 96-well tissue culture plates were purchased from Falcon.
Fmoc-N-amido-dPEG2-acid (EG2), Fmoc-N-amido-dPEG4-acid (EG4) and Fmoc-N-amido-dPEG8-acid (EG8) were purchased from Quanta Biodesign. Fluorescein isothiocyanate (FITC) was purchased from Toronto Research Chemistry. Anti-human cyclinA IgEs (clone BF683) were purchased from BD Biosciences. Mouse IgGPenicillin (monoclonal antibody clone P2B9) anti-human IgE (clone XTE4) was purchased from Abcam anti-DNP IgE (clone SPE-7) was purchased from Sigma-Aldrich. Anti-FITC mouse IgG-HRP conjugate was purchased from Jackson Immunoresearch. All antibodies used for BAT (visibility dye, anti-HLA-DR, anti-CD123, anti- CD63, anti-CD107a and anti-CD203c) were purchased from Fisher. ACK lysis buffer was purchased from Lonza.
Human serum was obtained from two different sources. Peanut-allergic patient serum (sera 1, 2, 3, and 4) and healthy nonallergic control patient serum were purchased from Plasma Lab International. Sera 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, and 16 were taken from patients enrolled in an oral peanut immunotherapy (OIT) clinical trial administered at Massachusetts General Hospital under the direction of Wayne Shreffler (NCT01324401). Samples were obtained from patients before OIT and selected from patients having sIgEs for peanuts >50 kU/L as determined by ImmunoCAP.
Blood from human subjects was obtained by venipuncture from patients with a clinical history of severe allergic reactions to peanuts or from healthy volunteers. All volunteers consented and all studies were approved by either the Massachusetts General Hospital Institutional Review Board or the Indiana University Institutional Review Board.
Methods.
Nanoallergen synthesis and characterization.
We utilized the epitope-lipid synthesis strategy as seen in previous publications (12). We synthesized epitope-lipids of all five Ara h 2 epitopes and five Ara h 6 epitopes using standard Fmoc solid-phase peptide synthesis (SPPS) chemistry on NovaPEG Rink amide resin. Residues were activated with HBTU and DIEA in DMF for 5 min and allowed to couple to resin for 30–45 min. Coupling efficiency was monitored using a Kaiser test. The Fmoc-protected residues were deprotected with a 3-min application of piperidine in DMF three times. After synthesis of epitope as a linear peptide, an EG2 spacer, three lysines, three additional EG6 spacers, a tryptophan residue, an Fmoc-Lys(Fmoc)-OH and palmitic acid (C16) tails were added in that order to the resin. Molecules were cleaved from the resin using a 95/2.5/2.5 TFA/water/TIS mixture for two cycles for 45 min each. We purified the molecules using RP-HPLC on an Agilent 1200 series system with a semipreparative Zorbax C18 column or Zorbax C3 column with either acetonitrile or 2-propanol gradients in the mobile phase. We monitored the column eluent with a diode array detector allowing a spectrum from 200 to 400 nm to be analyzed. The purified product was characterized using a Bruker Autoflex III Smartbeam matrix-assisted laser desorption ionization time-of-flight mass spectrometer. Liposomal nanoallergens were prepared using a procedure as previously described (35). Epitope lipids were then incorporated into liposomes using dry film hydration and extrusion. The liposomes were composed of 2% epitope lipid, 5% polyethylene glycol 2000(mPEG2000)–DSPE, 5% cholesterol, and 93% DSPC. The lipid mixture was dissolved in chloroform, dried with nitrogen lyophilized for 30 min, rehydrated in PBS at 60 °C, and then extruded through a 100-nm polycarbonate filter (Avanti).
The liposomes were characterized by DLS analysis via a 90Plus nanoparticle size analyzer (Brookhaven Instruments Corp.), using 658 nm light observed at a fixed angle of 90° at 20 °C. All nanoallergens had particle sizing between 115 and 125 nm, depending on the epitope lipid (Fig. 2C and SI Appendix).
cHBI synthesis.
All inhibitors (cHBI and FITC-cHBI) were synthesized using SPPS (23), with several modifications. First, the epitope was synthesized as a linear peptide using Fmoc-amino acids activated with a 3.6-fold excess of HBTU with 20-fold DIEA for 5 min before addition. Note that for FITC-cHBI, before epitope synthesis, Fmoc-Lys-IvDdE-OH was added, then deprotected and Fmoc-EG2-OH was added. The Fmoc was deprotected and a HBTU-activated CF was added. Then the IvDdE group was deprotected with 2% hydrazine in DMF and epitope synthesis was performed.
Following epitope synthesis for all cHBIs, a Fmoc-EG8-OH spacer was added, followed by conjugation of Fmoc-Lys(IvDdE)-OH. To the Fmoc-protected amine, IBA-BOC was added. Following IBA-Boc addition, the IvDdE group of lysine was deprotected using 2% hydrazine in DMF in the same fashion. Fmoc-EG4-OH spacer was added to the ε-amine of this lysine.
After deprotection of the Fmoc group of the EG4 spacer, ITC domain was added, just before cleavage from resin. The deprotected amines were chemically modified into ITC moieties using a modified procedure from Munch et al. (36). Briefly, resins with deprotected primary amines were washed in anhydrous DMF three times. A 10-fold excess CS2 with a 20-fold excess of DIEA was added in DMF and allowed to react for 30 min. Resin was then drained and washed once with anhydrous DMF. One milliliter of DMF with a 20-fold excess of DIEA was added to resin and cooled to ∼0 °C in a −20 °C freezer. Then, a 2-fold excess of Boc2O and 0.2-fold of DMAP was added to the vessel and allowed to react for 20 min at −20 °C. The vessel was removed, allowed to warm to room temperature for 30 min, and then washed with DMF, DCM, and diethyl ether and allowed to dry in a vacuum chamber. See SI Appendix, Fig. S5 for further details.
Molecules were cleaved from the resin using a 95/2.5/2.5 TFA/water/TIS mixture for two cycles for 45 min each. The resulting solution was evaporated using a Buchi rotor-evaporator to remove TFA, rehydrated in 50/50 ACN/water, and purified by RP-HPLC using an Agilent 1200 series HPLC with a Zorbax C18 semiprep column using an ACN/water gradient between 20% and 60% ACN in 10 min with a flow rate of 4 mL/min. Product was collected, evaporated, lyophilized, and redissolved in DMSO. Concentration was determined by absorbance at 280 nm or 335 nm. All molecules were characterized using high-resolution MicroTOF MS analysis. Purity was determined by analytical RP-HPLC using Zorbax Eclipse XBD-C18 with a 20–60% ACN gradient; all molecules were assessed to have >95% purity through analysis of a 220-nm signal.
The following molecules used in this study were analyzed using high-resolution mass spectroscopy using a Bruker microTOF II mass spectrometer. Note that all molecular weights have +1 mass increase from the addition of a proton. cHBIArah2/ep5 (C126H207N27O49S) was determined to be 2916.45 m/z (2915.43 m/z expected); cHBIArah6/ep5 (C99H170N18O39S) was determined to be 2269.17 m/z (2268.16 m/z expected); FITC-cHBIArah2/ep5 (C160H242N30O59S) was determined to be 3561.67 m/z (3560.66 m/z expected); and FITC-FITC-cHBIArah6/ep5 (C133H205N21O49S) was determined to be 2914.40 m/z (2913.39 m/z expected).
Cell culture.
RBL-SX38 cells (cells expressing human FcεRI) were a generous gift from Jean-Pierre Kinet, Harvard University, Cambridge, MA. RBL-SX38 were cultured in MEM (Gibco) with 10% FBS (Gemini BioProducts), l-glutamine, penicillin-streptomycin, and 1.2 mg/mL of G418 salt (Sigma) as previously described (37). RBL-2H3 cells (ATCC) were cultured in a similar manner but without the addition of G418.
Degranulation assays.
Degranulation assays were performed as previously described using nanoallergens, Ara h proteins, or crude peanut extract as the allergen with some modifications (12). RBL-SX38 cells were plated (100 µL each, 50,000 cells per well) into 96-well dishes for 24 h and then incubated with 10% of patient sera (sera 1–16 or a serum taken from healthy volunteer as control) in cell culture media for an additional 24 h before the degranulation assay. Cells were then washed and incubated in RBL media for 1 h. Cells were then washed, incubated in tyrodes buffer with 0.5 mg/mL BSA and 1 µM Cytochalasin D, and challenged with allergens or lysed with Triton X (as a positive control). After allergen challenge, the supernatant was incubated with pNAG solution for 45 min, the reaction halted with a stopping solution (1.5 mg/mL glycine, pH 10.7), and the degranulation measured by reading the absorbance at 405 nm after subtracting the background signal at 630 nm.
To evaluate the cHBI inhibitors, RBL-SX38 cells were similarly plated and primed with patient serum. However, before incubation with allergen, cells were incubated with cHBIs (15 min–24 h), and similarly challenged with allergens as described above. Note that concentrations of crude peanut extract used for inhibition assays shown in Fig. 3A varied, given the sensitization of the patient to crude peanut extract. A total of 1 ng/mL was used for serum 6 and sera 8–16, 100 ng/mL was used for sera 3 and 7, and the remaining sera 1, 2, and 4 were challenged with 1,000 ng/mL.
For DNP-BSA degranulation assays, a similar procedure was used with the following exceptions: RBL-2H3 cells were used instead of RBL-SX38 cells, and cells were incubated with a 25%/75% mixture of anti-DNP IgE and anti-cyclin A IgE for 24 h instead of patient serum, and DNP-BSA was used as the allergen.
Percent degranulation was calculated as previously decribed (32). Briefly, experimental signals were calculated as a percentage between positive control (100%) and negative control (0%). Degranulation maximum (Dmax) and EC50 values were calculated using Origin 7 software using Hill’s curve fit for sigmoidal curves using percent degranulation values. Note that if nanoallergen did not induce significant degranulation above error (determined as SD of triplicate experiments, P < 0.05), it was deemed to have “no response.” If a significant signal was present but there was insufficient data to determine an EC50 value, the EC50 value was estimated to be greater than an EC50 value calculated assuming that the highest signal reached was Dmax.
ELISA assay.
Binding interactions between cHBI and antibodies were detected using a sandwich ELISA. Before the ELISA test, patient serum was incubated at various concentrations with FITC-cHBIs at 37 °C for 5 h. Then, a high-binding 96-well ELISA plate was coated with 100 μL of 2 nM of anti-IgE antibody for 2 h in bicarbonate buffer (pH = 10). Plates were washed to remove unbound antibody. Wells were blocked with a blocking buffer (5% BSA, 0.2% Tween-20 solution in PBS) for 1 h, washed, and incubated with serum-cHBI mixture mixed 1:1 with the blocking buffer for 16 h. The plate was washed again and incubated with an anti-FITC mouse IgG-HRP conjugate (diluted 1:5,000 in blocking buffer) for 1 h. The plate was washed again and an Amplex Red Kit was used to quantify the ELISA signal using a SpectraMax M5 spectrophotometer according to the manufacturer’s instructions.
For the Ara h 2 competitive binding inhibition, a similar ELISA was performed with some modifications. Before the ELISA test, patient serum was incubated in various concentrations with unlabeled cHBIs at 37 °C for 5 h. Then, a high-binding 96-well ELISA plate was coated with 100 μL of 2 nM of anti-IgE antibody for 2 h in bicarbonate buffer. Plates were washed with PBS to remove unbound antibody. Wells were blocked with the blocking buffer for 1 h, washed, and incubated with serum-cHBI mixture mixed 1:1 with the blocking buffer for 16 h. The plate was washed again and incubated with biotin-tagged HRP (diluted 1:5,000 in blocking buffer) for 1 h. The plate was washed again, incubated with streptavidin-HRP conjugate, washed, and an Amplex Red Kit was used to quantify the ELISA signal using a SpectraMax M5 spectrophotometer according to the manufacturer’s instructions.
Animal studies.
C57BL/6 female mice (7- to 8-wk old) were obtained from Harlan Biosciences. Mice were maintained in pathogen-free conditions, and studies were approved by the Indiana University Institutional Animal Care and Use Committee. Toxicology studies were carried out in a similar manner as previously published in the University of Indiana In Vivo Therapeutics Core (32).
Basophil activation test.
The BAT assay protocol was used as previously described (38). Briefly, blood collected in vacutainer tubes containing heparin was processed within 24 h of collection. For basophil activation: 100 µL of whole blood was diluted 1:1 in serum-free RPMI containing cHBIs for 4 h at 37 °C. Cells were activated with 100 ng/mL of crude peanut extract or media (blank) for 30 min at 37 °C. RBs were lysed using ACK lysis buffer and remaining cells were stained and fixed for flow cytometry. Basophils were selected as viability dye negative, SSClow, HLA-DR−, and CD123+. Each sample was analyzed in triplicate with a cutoff of 1,000 basophils per sample. Basophil activation was assessed by measuring fluorescence of CD63, CD107a, and CD203c. Percentage maximal CD63 MFI was found by calculating the percentage between the blank (negative control) and the no inhibitor (positive control) MFI for CD63.
Supplementary Material
Acknowledgments
We thank Dr. Jean-Pierre Kinet for his kind gift of the RBL-SX38 cells; Mr. Douglas Zych and Mr. and Mrs. Jim and Annette Lecinski for funding; the NIH for Grants R01 AI108884 (to B.B. and M.H.K.), R01 AI129241 (to M.H.K.), U19 AI095261 (to W.G.S.), and T32 DK007519 (to A.A.Q.); the University of Notre Dame Proteomics Facility for use of their mass spectroscopy equipment; and Indiana University (IU)’s In Vivo Therapeutics Core for performing our toxicology experiments. M.J.T. was supported by VA CDA2 (IK2 CX001019). Core facility usage was supported by IU Simon Cancer Center Support Grants P30 CA082709 and U54 DK106846. A pilot grant and additional support provided by the Herman B Wells Center was in part from the Riley Children’s Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.J.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820417116/-/DCSupplemental.
References
- 1.Cromheecke JL, Nguyen KT, Huston DP. Emerging role of human basophil biology in health and disease. Curr Allergy Asthma Rep. 2014;14:408. doi: 10.1007/s11882-013-0408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iweala OI, Burks AW. Food allergy: Our evolving understanding of its pathogenesis, prevention, and treatment. Curr Allergy Asthma Rep. 2016;16:37. doi: 10.1007/s11882-016-0616-7. [DOI] [PubMed] [Google Scholar]
- 3.Blank U, Rivera J. Assays for regulated exocytosis of mast cell granules. Curr Protoc Cell Biol. 2006;Chapter 15:Unit 15.11. doi: 10.1002/0471143030.cb1511s32. [DOI] [PubMed] [Google Scholar]
- 4.Sampson HA, et al. A phase II, randomized, double-blind, parallel-group, placebo-controlled oral food challenge trial of Xolair (omalizumab) in peanut allergy. J Allergy Clin Immunol. 2011;127:1309–10.e1. doi: 10.1016/j.jaci.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 5.Lafeuille MH, et al. Association between consistent omalizumab treatment and asthma control. J Allergy Clin Immunol Pract. 2013;1:51–57. doi: 10.1016/j.jaip.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Cooper PJ, et al. Geohelminth infections: A review of the role of IgE and assessment of potential risks of anti-IgE treatment. Allergy. 2008;63:409–417. doi: 10.1111/j.1398-9995.2007.01601.x. [DOI] [PubMed] [Google Scholar]
- 7.Busse W, et al. Omalizumab and the risk of malignancy: Results from a pooled analysis. J Allergy Clin Immunol. 2012;129:983–989.e6. doi: 10.1016/j.jaci.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Handlogten MW, Serezani AP, Kaplan MH, Bilgicer B. What potential do heterobivalent inhibitors have for the treatment of severe allergic reactions? Immunotherapy. 2014;6:223–225. doi: 10.2217/imt.14.1. [DOI] [PubMed] [Google Scholar]
- 9.Kühne Y, et al. A novel multipeptide microarray for the specific and sensitive mapping of linear IgE-binding epitopes of food allergens. Int Arch Allergy Immunol. 2015;166:213–224. doi: 10.1159/000381344. [DOI] [PubMed] [Google Scholar]
- 10.Otsu K, Guo R, Dreskin SC. Epitope analysis of ara h 2 and ara h 6: Characteristic patterns of IgE-binding fingerprints among individuals with similar clinical histories. Clin Exp Allergy. 2015;45:417–484. doi: 10.1111/cea.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller GA, et al. Ara h 2: Crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011;66:878–885. doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deak PE, Vrabel MR, Kiziltepe T, Bilgicer B. Determination of crucial immunogenic epitopes in major peanut allergy protein, ara h2, via novel nanoallergen platform. Sci Rep. 2017;7:3981. doi: 10.1038/s41598-017-04268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, et al. A bioinformatics approach to identify patients with symptomatic peanut allergy using peptide microarray immunoassay. J Allergy Clin Immunol. 2012;129:1321–1328.e5. doi: 10.1016/j.jaci.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deak PE, Vrabel MR, Pizzuti VJ, Kiziltepe T, Bilgicer B. Nanoallergens: A multivalent platform for studying and evaluating potency of allergen epitopes in cellular degranulation. Exp Biol Med (Maywood) 2016;241:996–1006. doi: 10.1177/1535370216644533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ladics GS, et al. Assessment of three human FcepsilonRI-transfected RBL cell-lines for identifying IgE induced degranulation utilizing peanut-allergic patient sera and peanut protein extract. Regul Toxicol Pharmacol. 2008;51:288–294. doi: 10.1016/j.yrtph.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Albrecht M, et al. Relevance of IgE binding to short peptides for the allergenic activity of food allergens. J Allergy Clin Immunol. 2009;124:328–336, 336.e1-6. doi: 10.1016/j.jaci.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 17.Bublin M, et al. IgE cross-reactivity between the major peanut allergen Ara h 2 and the nonhomologous allergens Ara h 1 and Ara h 3. J Allergy Clin Immunol. 2013;132:118–124. doi: 10.1016/j.jaci.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Mishra A, Jain A, Arora N. Mapping B-cell epitopes of major and minor peanut allergens and identifying residues contributing to IgE binding. J Sci Food Agric. 2016;96:539–547. doi: 10.1002/jsfa.7121. [DOI] [PubMed] [Google Scholar]
- 19.Huber M. Activation/Inhibition of mast cells by supra-optimal antigen concentrations. Cell Commun Signal. 2013;11:7. doi: 10.1186/1478-811X-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanabe S. Epitope peptides and immunotherapy. Curr Protein Pept Sci. 2007;8:109–118. doi: 10.2174/138920307779941569. [DOI] [PubMed] [Google Scholar]
- 21.Koppelman SJ, Wensing M, Ertmann M, Knulst AC, Knol EF. Relevance of Ara h1, Ara h2 and Ara h3 in peanut-allergic patients, as determined by immunoglobulin E Western blotting, basophil-histamine release and intracutaneous testing: Ara h2 is the most important peanut allergen. Clin Exp Allergy. 2004;34:583–590. doi: 10.1111/j.1365-2222.2004.1923.x. [DOI] [PubMed] [Google Scholar]
- 22.Aalberse RC, Akkerdaas J, van Ree R. Cross-reactivity of IgE antibodies to allergens. Allergy. 2001;56:478–490. doi: 10.1034/j.1398-9995.2001.056006478.x. [DOI] [PubMed] [Google Scholar]
- 23.Handlogten MW, et al. A heterobivalent ligand inhibits mast cell degranulation via selective inhibition of allergen-IgE interactions in vivo. J Immunol. 2014;192:2035–2041. doi: 10.4049/jimmunol.1301371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handlogten MW, Kiziltepe T, Serezani AP, Kaplan MH, Bilgicer B. Inhibition of weak-affinity epitope-IgE interactions prevents mast cell degranulation. Nat Chem Biol. 2013;9:789–795. doi: 10.1038/nchembio.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mustafaoglu N, Kiziltepe T, Bilgicer B. Site-specific conjugation of an antibody on a gold nanoparticle surface for one-step diagnosis of prostate specific antigen with dynamic light scattering. Nanoscale. 2017;9:8684–8694. doi: 10.1039/c7nr03096g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mustafaoglu N, Kiziltepe T, Bilgicer B. Antibody purification via affinity membrane chromatography method utilizing nucleotide binding site targeting with a small molecule. Analyst (Lond) 2016;141:6571–6582. doi: 10.1039/c6an02145j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hecht SS. Inhibition of carcinogenesis by isothiocyanates. Drug Metab Rev. 2000;32:395–411. doi: 10.1081/dmr-100102342. [DOI] [PubMed] [Google Scholar]
- 28.Ji Y, Kuo Y, Morris ME. Pharmacokinetics of dietary phenethyl isothiocyanate in rats. Pharm Res. 2005;22:1658–1666. doi: 10.1007/s11095-005-7097-z. [DOI] [PubMed] [Google Scholar]
- 29.Handlogten MW, Kiziltepe T, Moustakas DT, Bilgiçer B. Design of a heterobivalent ligand to inhibit IgE clustering on mast cells. Chem Biol. 2011;18:1179–1188. doi: 10.1016/j.chembiol.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 30.Santos AF, et al. Basophil activation test discriminates between allergy and tolerance in peanut-sensitized children. J Allergy Clin Immunol. 2014;134:645–652. doi: 10.1016/j.jaci.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos AF, et al. Distinct parameters of the basophil activation test reflect the severity and threshold of allergic reactions to peanut. J Allergy Clin Immunol. 2015;135:179–186. doi: 10.1016/j.jaci.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handlogten MW, Deak PE, Bilgicer B. Two-allergen model reveals complex relationship between IgE crosslinking and degranulation. Chem Biol. 2014;21:1445–1451. doi: 10.1016/j.chembiol.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penchala SC, et al. A biomimetic approach for enhancing the in vivo half-life of peptides. Nat Chem Biol. 2015;11:793–798. doi: 10.1038/nchembio.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsuka A, Kabashima K. Mast cells and basophils in cutaneous immune responses. Allergy. 2015;70:131–140. doi: 10.1111/all.12526. [DOI] [PubMed] [Google Scholar]
- 35.Stefanick JF, Ashley JD, Bilgicer B. Enhanced cellular uptake of peptide-targeted nanoparticles through increased peptide hydrophilicity and optimized ethylene glycol peptide-linker length. ACS Nano. 2013;7:8115–8127. doi: 10.1021/nn4033954. [DOI] [PubMed] [Google Scholar]
- 36.Munch H, Hansen JS, Pittelkow M, Christensen JB, Boas U. A new efficient synthesis of isothiocyanates from amines using di-tert-butyl dicarbonate. Tetrahedron Lett. 2008;49:3117–3119. [Google Scholar]
- 37.Dibbern DA, Jr, Palmer GW, Williams PB, Bock SA, Dreskin SC. RBL cells expressing human Fc epsilon RI are a sensitive tool for exploring functional IgE-allergen interactions: Studies with sera from peanut-sensitive patients. J Immunol Methods. 2003;274:37–45. doi: 10.1016/s0022-1759(02)00369-1. [DOI] [PubMed] [Google Scholar]
- 38.Patil SU, Shreffler WG. BATting above average: Basophil activation testing for peanut allergy. J Allergy Clin Immunol. 2014;134:653–654. doi: 10.1016/j.jaci.2014.07.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.