Significance
TRPV5 (transient receptor potential vanilloid 5) is a unique calcium-selective ion channel in the TRP family that functions as a gatekeeper of active calcium reabsorption in the kidney. Despite increasing knowledge of TRP channel structures, little is known about the structural arrangements controlling TRPV5 function. The most notable feature of TRPV5 is its calcium-dependent channel inactivation, which relies on binding of the calcium-sensing protein, calmodulin. In this work, we present TRPV5 structures in open and closed states, as well as in complex with calmodulin. These findings contribute to a more complete understanding of the structural mechanisms underlying TRPV5 function, providing important fundamental knowledge about the maintenance of calcium homeostasis.
Keywords: TRP channel, calcium, calmodulin, cryo-EM
Abstract
TRPV5 (transient receptor potential vanilloid 5) is a unique calcium-selective TRP channel essential for calcium homeostasis. Unlike other TRPV channels, TRPV5 and its close homolog, TRPV6, do not exhibit thermosensitivity or ligand-dependent activation but are constitutively open at physiological membrane potentials and modulated by calmodulin (CaM) in a calcium-dependent manner. Here we report high-resolution electron cryomicroscopy structures of truncated and full-length TRPV5 in lipid nanodiscs, as well as of a TRPV5 W583A mutant and TRPV5 in complex with CaM. These structures highlight the mechanism of calcium regulation and reveal a flexible stoichiometry of CaM binding to TRPV5.
The transient receptor potential (TRP) family of ion channels is large and functionally diverse, as exemplified by a wide range of physiological and pharmacological modulators, differences in ion selectivity, and tissue and subcellular distributions. Transient receptor potential vanilloid 5 (TRPV5) and its close homolog TRPV6 are unique among all TRP channels because of their high selectivity for calcium over monovalent cations (PCa/PNa >100:1) (1). They are also distinct from other members of the vanilloid subfamily (TRPV1–4) in that they are not known to be thermosensitive or ligand-activated (2, 3). Physiologically, TRPV5 and TRPV6 serve as apical entryways into epithelial cells that line segments of the intestine and kidney, initiating transcellular calcium transport that helps fine-tune serum calcium levels (4). Disturbances in the body’s calcium balance can cause major health problems, including neurologic and cardiac aberrations, stone formation, and bone disorders (5–7). Indeed, genetic variants of TRPV5 are associated with altered renal calcium handling and increased risk of recurrent kidney stones (8–10).
In addition to calcium selectivity, another functional hallmark of TRPV5 is a pronounced inward rectification of the current-voltage relationship, in contrast to an outward rectification observed for most other TRPV channels. This allows TRPV5 to move calcium ions into the cell at negative membrane potential, resulting in significant calcium permeation at physiological membrane potentials. Furthermore, TRPV5 channels exhibit calcium-dependent feedback regulation that includes fast inactivation and slow current decay (11). This inhibition is likely controlled by the concentration of calcium locally near the cytoplasmic side of the channel, preventing excessive calcium influx. Part of the calcium-mediated negative feedback mechanism also involves calcium-dependent binding of calmodulin (CaM), a ubiquitous calcium-binding protein (12, 13).
To understand how TRPV5 conveys extraordinary calcium selectivity and calcium-dependent channel inactivation, we set out to determine its structure using single particle electron cryomicroscopy (cryo-EM). Here we report structures of truncated and full-length rabbit TRPV5 channels at nominal resolutions of 2.9 Å and 3.0 Å, respectively. In addition, we determined the structure of the TRPV5-CaM complex in LMNG detergent at a nominal resolution of 3.0 Å, as well as the TRPV5 W583A mutant as an open-channel structure at 2.8 Å.
Results
Structure of TRPV5.
We expressed and purified rabbit TRPV5 as a full-length protein (1–730) or bearing a C-terminal truncation (1–660). Proteins purified from these constructs were stable and monodispersed (SI Appendix, Fig. S1 A and B). The truncated TRPV5 retains most of its functional domains but lacks one of the postulated CaM-binding sites (701–730) that has been shown to regulate TRPV5 in a calcium-dependent manner (12, 13). The truncated channel shows significant but slightly less robust TRPV5-mediated calcium uptake compared with the full-length protein (SI Appendix, Fig. S1C).
We solubilized and purified both full-length and truncated TRPV5 in the detergent n-Dodecyl-β-d-maltoside (DDM), followed by exchange into lauryl maltose neopentyl glycol (LMNG) or reconstitution into lipid nanodiscs for single-particle cryo-EM studies. EM of frozen hydrated proteins in LMNG and nanodiscs showed monodispersed particles without obvious preferred orientations (SI Appendix, Fig. S2 A and B). Two-dimensional class averages calculated from selected particles of both samples showed clear features of both transmembrane and intracellular domains (SI Appendix, Fig. S2C), as well as a density corresponding to the nanodisc surrounding the transmembrane core of the channel (Fig. 1 A and B). We determined a total of four 3D reconstructions, including truncated and full-length TRPV5 in lipid nanodisc at overall resolutions of 2.9 Å (Fig. 1 and SI Appendix, Fig. S2) and 3.0 Å, respectively, as well as TRPV5-CaM complex in LMNG detergent and full-length protein with a W583A point mutation in lipid nanodisc to resolutions of 3.0 Å and 2.8 Å, respectively (SI Appendix, Figs. S3 and S4). The resolutions and qualities of all maps were sufficient to trace protein backbone and model side chains of most residues unambiguously (residues 27–638) (Fig. 1 and SI Appendix, Fig. S5). Overall, the structures are similar to other recently published TRPV5 structures (14, 15).
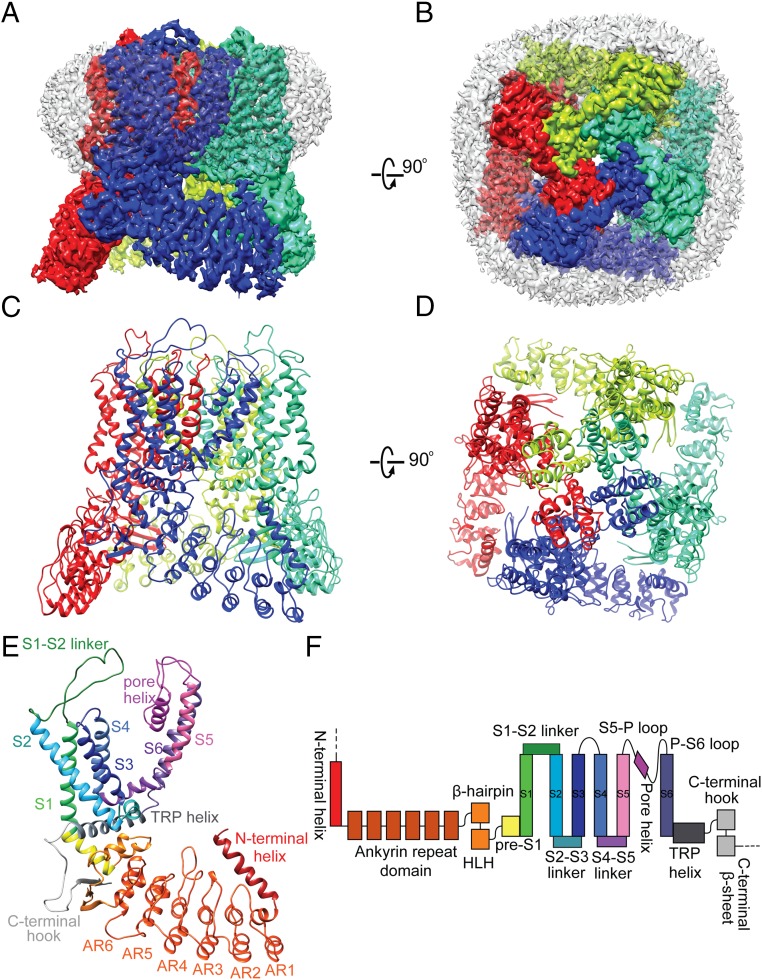
Fig. 1.
Cryo-EM structure of TRPV5. (A and B) Side (A) and top (B) views of nanodisc-reconstituted TRPV5 1–660 at 2.9-Å resolution, with the subunits colored individually. The nanodisc density surrounds the transmembrane region of the channel (white). (C and D) Side (C) and top (D) views of the TRPV5 tetrameric complex, using the same subunit colors as in A and B. (E) Side view of a TRPV5 monomer with the functional domains colored individually. Structural elements are depicted as ribbons for α-helices, arrows for β-sheets, and ropes for unordered loops. (F) Domain organization of TRPV5, with domains colored as in E. The dashed line denotes a region that was not resolved in the structure but is present in the full-length channel.
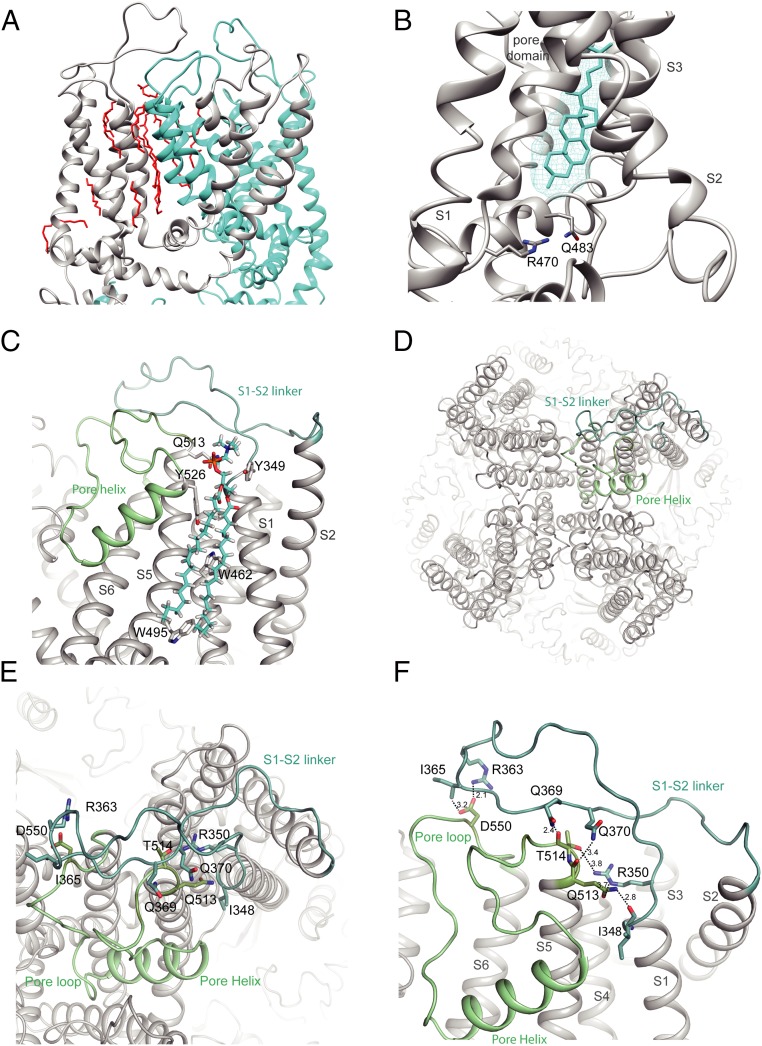
In all of our structures, many lipid-like densities were well resolved, occupying crevasses between two neighboring units (Fig. 2A and SI Appendix, Fig. S6 A and B). One lipid density was found within the lower segment of the S1–S4 domain with location and shape resembling a lipid found in TRPV1 that was interpreted as phosphatidylcholine (16). Another lipid is seen in a pocket akin to the vanilloid-binding site in TRPV1, which is occupied by a phosphatidylinositol lipid in apo TRPV1. The binding of a vanilloid compound in this pocket replaces the phosphatidylinositol and activates TRPV1 (16). In our TRPV5 structures, the exact identity of this lipid is unclear; however, its shape is clearly not consistent with a phosphatidylinositol but may fit nicely with cholesterol (Fig. 2B), as observed in TRPV6 (17). If so, this cholesterol molecule must be endogenously bound, since we observed the same density in all of our structures even though we did not add cholesterol or its mimetic, cholesterol-hemisuccinate, during protein purification. A similar density interpreted as bound econazole or annular lipid was also seen in recently published TRPV5 structures (14, 15).
Fig. 2.
S1-S2 linker position in TRPV5. (A) Expanded view of the lipid densities observed within the pore domain. Two monomers are shown and individually colored. Lipids are in red. (B) Zoom of the resident lipid in the vanilloid pocket. The density of the lipid is visualized with cyan mesh, and the fitted acyl chain is in cyan. The side chains of residues important in phosphatidylinositol binding in TRPV1 are shown. (C) Zoom-in view of the interaction formed by the S1-S2 linker, annular lipid, and pore domain. The side chains of residues interacting with the lipid are shown. (D) Top view of TRPV5 tetramer showing the positions of the S1-S2 linker (light teal) and the S5-P-S6 domain (pale green). (E and F) Zoom-in view of the intersubunit interface formed by the S1-S2 linker and the pore helix. Putative hydrogen bonds and electrostatic interactions are shown as dashed lines. Side chains of interacting residues are shown as sticks for both, and interatomic distances (F) between side chains are depicted.
Overall, TRPV5 closely resembles the structures of other TRPV subfamily members with its six transmembrane helices arranged in a domain-swapped architecture and its N terminus dominated by an ankyrin repeat domain (ARD) (Fig. 1 C–F) (14, 15, 17–21). Similar to TRPV6 (17), but not observed in TRPV1–4, is a helix preceding the first ankyrin repeat that forms a major interface between neighboring subunits at each corner of the channel. Moreover, an extended extracellular linker between S1 and S2 is present in TRPV5 (and TRPV6) but lacking in other TRPV members (SI Appendix, Figs. S6 C and D and S7). This linker interacts with the pore region of the neighboring subunit and contains the N-glycosylation site N358 (Fig. 2D and SI Appendix, Fig. S6 C and D) (22). It is stabilized by forming rather tight interactions through hydrogen bonds between the carboxyl groups of D550 in the pore loop and R363 and I365 in the S1-S2 loop (Fig. 2 E and F), as well as a web of other hydrogen bonds and salt bridges between I348, R350, Q369, and Q370 in the S1-S2 loop and Q513 and T514 in the S5-pore loop, plus some main chain interactions. Q513 and T514 are located at the top of the pore domain and seem to play major roles in interacting with the S1-S2 loop via numerous hydrogen bonds and salt bridges (Fig. 2 E and F). As one of the few TRPV5/TRPV6-specific regions, such interactions may constrain the movement of the selectivity filter. Another interesting structural feature for TRPV5 and TRPV6 is a C-terminal helix uniquely observed in our CaM-bound structure, that we discuss in more detail below.
Ion Permeation Pathway.
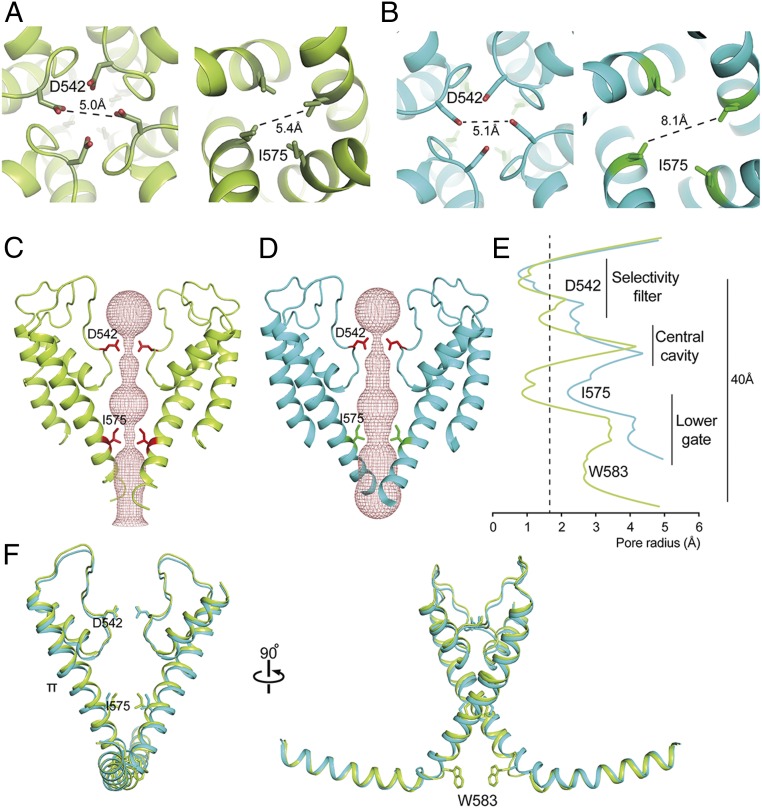
Similar to other TRPV ion channels, the ion permeation pathway of TRPV5 has two main constriction points. The upper constriction is formed by D542, the main residue of the selectivity filter, and is located in the lower part (but not the bottom) of the pore loop. Above the selectivity filter is a large cavity formed by the upper segment of the pore loop, which is stabilized by the S1-S2 linker loop. This conformation is somewhat different from both the funnel-shaped configuration of the outer pore in the apo structure of TRPV1, where the upper gate is in a closed conformation, and the more parallel configuration observed in an open-gate conformation when the gating modifier spider toxin, DkTx, is bound to the outer pore region (23). In our TRPV5 structure, a density is located in the middle of the pore between the side chains of D542 (SI Appendix, Fig. S8A), where similar densities are found in the same location in both half maps. The distance between side chains of opposite D542 residues is ∼5 Å (Fig. 3A), sufficient to coordinate a cation. Thus, this density likely corresponds to a coordinated cation, consistent with D542 serving as a cation selectivity filter.
Fig. 3.
Pore domain characteristics of TRPV5. (A and B) Top (Left) and bottom (Right) view of TRPV5 full-length (A) and TRPV5 W583A (B). Side chains are shown for two constricting residues in the selectivity filter and lower part of the pore, D542 and I575. (C and D) The ion permeation pathway of closed TRPV5 (C; yellow) and open TRPV5 W583A (D; cyan) is shown as a ribbon diagram with the solvent-accessible space depicted as a pink mesh. Only two subunits are shown for clarity, with side chains of the restricting residues D542 and I575. (E) Diagram of the pore radius calculated with HOLE shown for TRPV5 (yellow) and TRPV5 W583A (cyan). The dotted line indicates the radius of a hydrated calcium ion. (F) Comparison of the TRPV5 (yellow) and TRPV5 W583A (cyan) pore domains with the TRP helix attached. Key residues in the pore domain are depicted.
In contrast to what is seen in the crystal structure of TRPV6 (24), there are no significant densities in our structure coordinated by the side chains of T539 or M570 within the lower gate. Instead, we see a weaker density positioned between the four G579 residues and a second density between the W583 side chains (SI Appendix, Fig. S8 C and E). Both densities are found in the same locations in both half maps and are present when the structure is determined without symmetry, suggesting that they are indeed coordinated cations. W583 is the last residue in the S6 helix, located just before the TRP helix. It is conserved in TRPV6 but not in the other TRPV channels and is essential for TRPV5 function (25) (SI Appendix, Fig. S7). To explore the influence of W583, we determined a structure of full-length TRPV5 with a W583A point mutation in nanodisc to 2.8-Å resolution (SI Appendix, Fig. S3). Consistent with a previously reported gain-of-function phenotype for the W583 mutant (25), the lower gate of the TRPV5 W583A structure has an open conformation (Fig. 3 and SI Appendix, Fig. S8 D and F).
Comparing ion permeation profiles in both TRPV5 structures (Fig. 3 C–E), the side chains of D542 form the tightest constriction in the selectivity region, just like the corresponding residue D541 in TRPV6 (17). They project into and partially occlude the permeation pathway, creating a minimum interatomic distance of 5.0 Å (Fig. 3A). The W583A mutation does not affect the conformation at the upper restriction site, where the distance between the D542 side chains is 5.1 Å (Fig. 3B). Analysis of solvent accessibility with HOLE revealed a pore diameter of ∼1.4 Å at this position (Fig. 3E). Further down, the pore of WT TRPV5 is also tightly constricted at I575, illustrated by a solvent-accessible pore radius of 1.0 Å (Fig. 3E). The interatomic distance between these lower gate side chains is 5.4 Å, approximating the interatomic distances found for lower gate residues of other TRPV channel structures (Fig. 3A). In contrast, the lower gate of TRPV5 W583A is clearly open for conductance of hydrated calcium ions (Fig. 3 B and E).
The S6 helix in both WT and mutant TRPV5 adopts a π-helical conformation (Fig. 3F), which typically results from a single amino acid insertion that produces a bulge in the helix. From other recent structural studies of TRP channels, such a π-helical conformation is suggested to be involved in channel gating (17, 19, 26–28). Of note, this does not appear to be dependent on sequence, since the residue that marks the register shift is different in TRPV1 (Y671) than in TRPV5 (T567), but similar in TRPV5 and TRPV6 (SI Appendix, Fig. S7).
TRPV5-CaM Complex Structure.
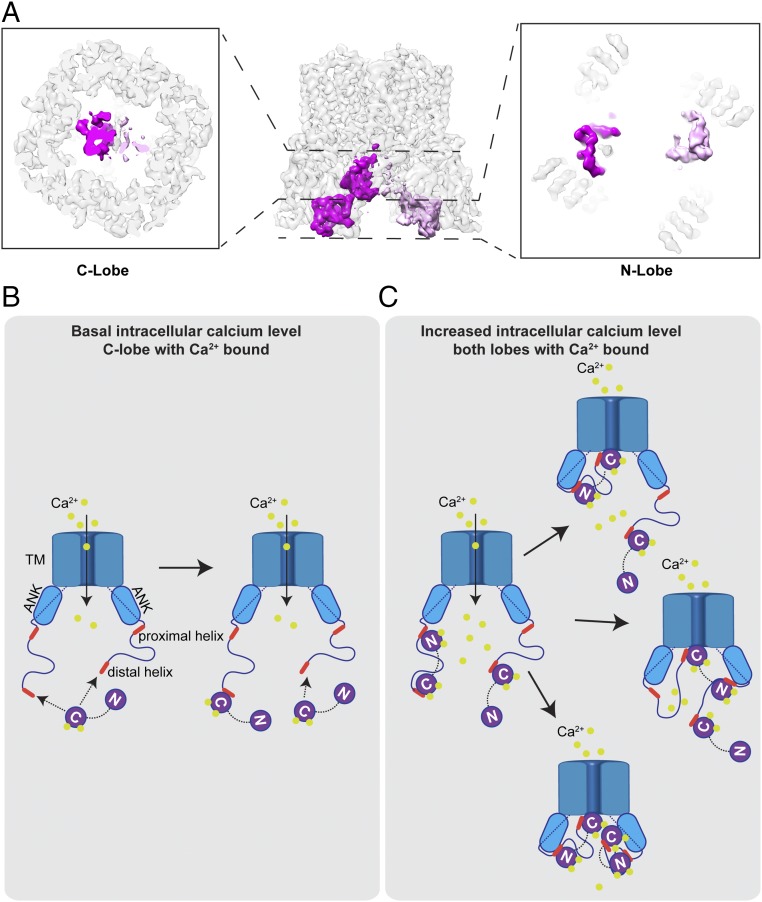
Previous studies suggest that TRPV5 is inactivated in a calcium-dependent manner through the action of CaM (12, 13). CaM consists of an N-lobe and a C-lobe, each of which contains two calcium-binding sites. Generally, CaM adopts a closed conformation in the calcium-free state and an open, extended conformation in the calcium-bound state. Both states are able to bind various target proteins, including TRP channels, and consequently transduce relevant signals on calcium binding (29). In the case of modulating TRPV5, the calcium influx induced by channel opening presumably generates calcium-bound CaM that binds to and somehow inactivates the channel.
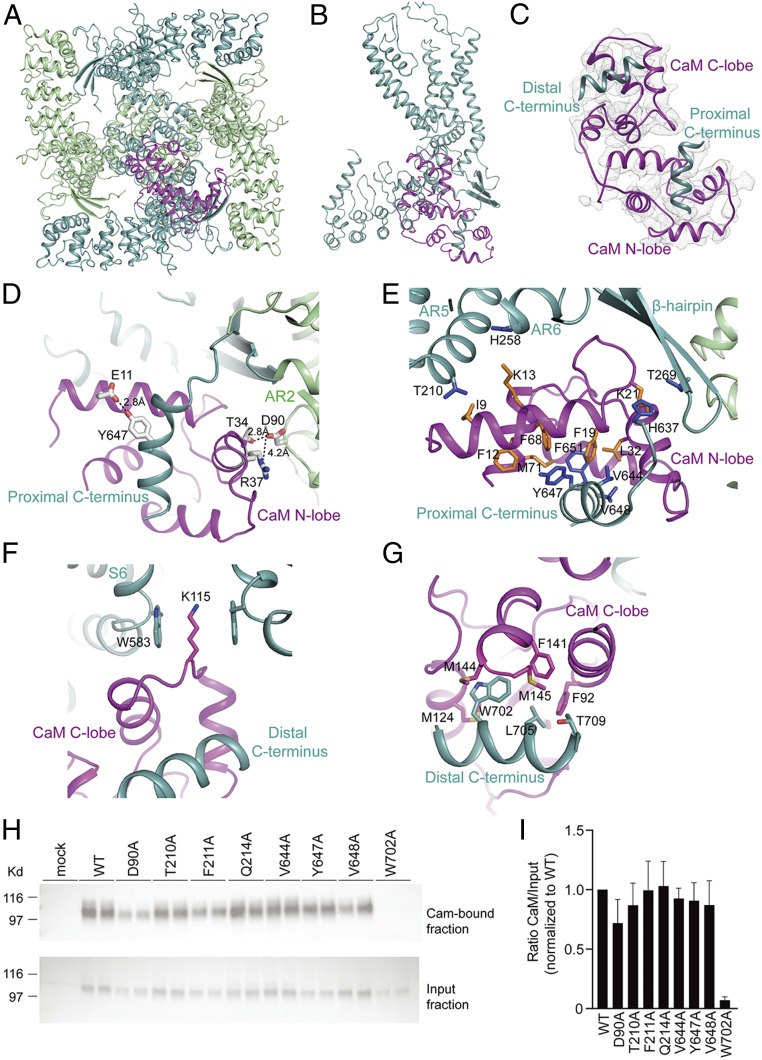
To gain further insight into this regulatory mechanism, we added bovine CaM to detergent (LMNG)-solubilized full-length TRPV5 in the presence of 5 mM CaCl2 and determined the structure of the complex (SI Appendix, Fig. S4). We tested determining the structure under C4, C2, or no symmetry (C1). In all these reconstructions, we observed major extra densities at the cytosolic face of the channel that are contributed by the bound CaM (SI Appendix, Fig. S4E). Within an asymmetric unit, one density is located directly under the lower gate and in the symmetry axis, and another is located next to the β-sheet and the ankyrin repeats of the two neighboring subunits. Even in the C1 reconstruction, along with the strongest CaM density are significant other densities likely corresponding to other CaMs, suggesting the possibility of more than one CaM bound to one channel.
To determine the stoichiometry of CaM binding to TRPV5, we performed 3D classification on the C1 reconstruction (SI Appendix, Fig. S9) and found three different classes. In one class, which contains 35% of particles, one CaM binds to TRPV5 such that its C-lobe interacts with the channel directly below the lower gate and the N-lobe binds to the β-sheet from inside of the tetramer (SI Appendix, Fig. S9A). After further refinement, the resolution is sufficient to model the bound CaM (SI Appendix, Fig. S5B) and to reveal the interactions between bound CaM and TRPV5 (Fig. 4). In two other classes, however, there are densities corresponding to additional CaMs. In these two classes, one CaM binds to the channel exactly as described above, while another, slightly weaker CaM density binds to either the opposite or a neighboring subunit (SI Appendix, Fig. S9 B and C), suggesting that there may be a second CaM bound to the channel.
Fig. 4.
Structure of the TRPV5-CaM complex. (A) Bottom view of TRPV5-CaM with the two subunits including the CaM-interacting one (light teal) and the other two (pale green). CaM is in magenta. (B) Side view of a TRPV5 monomer (light teal) with CaM (magenta) interaction at the C terminus. (C) Overview of CaM (magenta) interacting with two C-terminal helices of TRPV5 (light teal). (D–H) Close-up views of the CaM N-lobe (D and E) and CaM C-lobe (F and G) interacting with the TRPV5 N and C termini. Side chains of hydrophobic interactions are shown as sticks for both CaM (orange) and TRPV5 (blue). In D, interatomic distances between side chains are depicted. (H) CaM-binding assay of HEK293 cells transfected with WT TRPV5 and the indicated mutants. Samples were analyzed by immunoblotting with GFP antibody. The CaM fraction represents the TRPV5 bound to the CaM agarose beads (Top), and input demonstrates TRPV5 expression in total cell lysates (Bottom). A representative immunoblot of three independent experiments is shown. (I) Quantification of the immunoblots is depicted as percentage of WT, which represents the relative CaM binding compared with input.
We carried out further alignment and classification (Methods) to rule out the possibility that the second CaM density is generated by misaligning some single CaM-bound particles that are rotated by 90° or 180° around the symmetry axis. We confirmed that >20% of TRPV5 particles have two CaMs bound to two opposite subunits. In most cases, the two CaMs bind opposite to each another, but in a small number of cases, they bind to adjacent sites. Thus, there is a flexible stoichiometry of CaM binding to TRPV5 at either 1:1 or 2:1. Interestingly, in our reconstruction of full-length TRPV5 in nanodiscs (to which no CaM was added), we also found extra densities that are similar but significantly weaker, corresponding to the class of one CaM bound to the channel. Most likely, these channels have bound endogenous CaM from HEK293 cells.
The space within the channel encircled by four ARDs is sufficient to accommodate up to four CaM N-lobes bound rather symmetrically. However, the space in which the CaM C-lobe binds is limited. Based on our classification, two C-lobes do not bind the channel symmetrically; rather, the first C-lobe binds the channel in a manner similar to that seen in the single CaM-bound class, while the second C-lobe binds slightly away from the symmetry axis and with a density weaker than that of the first bound C-lobe (Fig. 5A).
Fig. 5.
Inactivation of TRPV5 by CaM in a calcium-dependent manner. (A) Cryo-EM density map showing two CaM molecules bound to one TRPV5 channel. (B) Under a basal intracellular calcium level, the C-lobe of CaM with calcium bound is able to interact with the C-terminal distal helix of TRPV5, while the N-lobe is in a calcium-free state. (C) Influx of calcium from TRPV5 increases the intracellular calcium concentration, which allows calcium binding to the N-lobe of CaM and further interaction with the C-terminal proximal helix of TRPV5. This interaction positions the C-lobe of CaM to block the lower gate of the channel. Binding of CaMs to TRPV5 presents a flexible stoichiometry.
Analysis of the binding interface of TRPV5 and CaM shows that both the CaM N-lobe and C-lobe can bind tightly to TRPV5 at different regions (Fig. 4 C–G). The CaM N-lobe is sandwiched between the β-sheet and a C-terminal helix (residues 639–653, termed the proximal C-terminal helix) (Fig. 4 D and E). Binding of the CaM N-lobe to this helix is in line with a previous biochemical finding that the same region in TRPV6, which is highly homologous in TRPV5, can pull down CaM (30). In the TRPV5-CaM complex, this helix—which is not resolved in the other TRPV5 structures—is stabilized by CaM binding, consistent with its important contribution to channel–CaM intermolecular interactions (Fig. 4 D and E). For example, residue Y647 in this helix can form a hydrogen bond with E11 in CaM (Fig. 4D). In addition, four hydrophobic residues in the TRPV5 helix—V644, Y647, V648, and F651—are surrounded by hydrophobic residues from CaM, including F12, F19, K21, L32, F68, and M71 (Fig. 4E). This “V”-shaped hydrophobic groove not only stabilizes the helix of TRPV5, but also increases the interaction between TRPV5 and the N-lobe of CaM (Fig. 4E). There is another interaction interface mediated by hydrogen bonds between D90 from the neighboring TRPV5 subunit and T34 in CaM; this interaction is further strengthened by a salt bridge between D90 and R37 in CaM (Fig. 4D). Indeed, our CaM agarose pull-down assay showed that the D90A mutation slightly reduces binding between CaM and TRPV5 (Fig. 4 H and I).
Besides the interface described above, there are three hydrophobic interaction pairs between TRPV5 and CaM: residues T210 and H258 of the TRPV5 ARD can form hydrophobic interactions with I9 and K13 in CaM, respectively, and T269 in the β-hairpin can hydrophobically interact with K21 in CaM (Fig. 4E). Together, these interactions sandwich the CaM N-lobe into a specific stable orientation.
The distal C-terminal helix of TRPV5, composed of residues 698–710, is responsible for CaM C-lobe binding (Fig. 4 F and G). The three hydrophobic residues in this helix (W702, L705, and T709) are surrounded by several hydrophobic residues from the CaM C-lobe, including F92, M124, F141, M144, and M145 (Fig. 4G). Of note, our CaM agarose pull-down confirmed the described interaction of W702 with CaM (Fig. 4 H and I) (13). A loop of 44 residues (654–697) that connects the proximal and distal C-terminal helices is unresolved and presumably remains flexible. Therefore, it is likely that the CaM C-lobe bound to the TRPV5 distal C-terminal helix is placed in this specific position by the stably bound CaM N-lobe via a short linker between two lobes. Similar to TRPV6 (31), the C-lobe of CaM occupies the lumen close to the lower gate such that K115 of CaM can insert deeply into the lower pore region surrounded by W583 from four subunits, thereby blocking the channel (Fig. 4F). The steric hindrance will preclude binding of a second CaM lobe to the same site. Interestingly, in the case in which two CaMs are bound to TRPV5, the second CaM C-lobe presumably binds to the same distal C-terminal helix of another TRPV5 subunit, but its K115 residue is located away from the pore, nevertheless enhancing the local CaM concentration near the channel gate.
Discussion
While generally similar to other TRPV channels, TRPV5 has a number of interesting structural features that are either unique or shared with TRPV6 (17, 24). Both TRPV5 and TRPV6 have an extensive S1-S2 linker that covers a large portion of the S5-pore and pore-S6 linkers, possibly stabilizing the upper pore. In the TRPV1 apo structure, the vanilloid-binding pocket is occupied by a phosphatidylinositol lipid. TRPV5 and TRPV6 both have an analogous pocket occupied by a lipid that is clearly not a phosphatidylinositol, but more likely a cholesterol (17). A density of almost identical shape at this same location is observed in all our structures, including closed and open channels (W583A), suggesting that it likely does not have a role in activating TRPV5, but more likely has a role in stabilizing the channel.
An important feature of TRPV5 and TRPV6 is CaM-dependent regulation. Indeed, without CaM binding, the C-terminal region after D638 is completely unresolved in all our cryo-EM structures, presumably reflecting its inherent flexibility. Similar to recent CaM-bound structures of TRPV5 and TRPV6 (15, 31), we resolved two key helices within the C terminus of TRPV5 that participate in CaM binding, including a proximal helix (639–653) immediately after D638 that interacts with the CaM N-lobe and a second distal helix (698–710) that binds the CaM C-lobe. A loop of >40 amino acids that links the two helices remains unresolved.
Early studies established that the CaM C-lobe has greater calcium-binding affinity than the N-lobe (32). Thus, in the case of TRPV5, we speculate that the C-lobes can bind to the C-terminal distal helix under basal calcium levels (Fig. 5B). Without constraint, each channel can accommodate more than one bound CaM C-lobe, but this is likely insufficient to inhibit the channel, as the proximal C-terminal helix is still very flexible. Channel opening and calcium influx increase intracellular local calcium concentrations and recruit a calcium-bound N-lobe that subsequently binds to the proximal C-terminal helix. Binding of the N-lobe then stabilizes the conformation of the proximal C-terminal helix and rearranges the entire CaM into a specific conformation that brings the C-lobe to the bottom of the pore to block the channel (Fig. 5C).
Many ion channels use CaM, either as a constitutively bound subunit or as a dissociable partner, to regulate channel activity in a calcium-dependent manner. In this regard, a comparison of TRPV5 and well-studied potassium channels reveals some interesting similarities and differences. For KCNQ1 and SK (small-conductance calcium activated potassium) channels, CaMs bind either constitutively (33) or as an obligate subunit (34), even before calcium is bound (35). Binding of calcium to CaM induces conformational changes that activate the channel either directly (33) or by releasing them from inhibition (34). In contrast, TRPV5 is constitutively open for calcium influx, and it uses calcium-bound CaM as a dissociable factor to inhibit the channel conductance by sensing the local calcium concentration. Inhibition of TRPV5 requires binding of C- and N-lobes from only one CaM, while activation of SK channel requires stoichiometric binding of four CaMs per tetramer. In contrast to previous studies on TRPV5 and TRPV6 (15, 31), we also found that some TRPV5 particles have two bound CaMs, with the second C-lobe positioned off the lower gate. This configuration may result in more effective calcium-dependent channel inhibition by increasing local concentration of CaM in close proximity to the lower gate.
As illustrated in Fig. 5, we speculate that CaM binds to TRPV5 in a stepwise manner such that the C-lobe binds first to the C-terminal distal helix at a relative low calcium concentration, followed by N-lobe binding with increased local calcium concentration. Only after the N-lobe binds to the C-terminal proximal helix is the C-lobe positioned to block the lower gate of the channel. Given the unresolved long flexible loop of 44 residues between the proximal and distal helices, it is possible that the C- and N-lobes of CaM are bound to distal and proximal helices within the same C terminus or on adjacent channel subunits. Further studies are needed to resolve these possibilities and appreciate any functional consequences.
Materials and Methods
Cloning, Expression, and Purification.
Using standard PCR cloning methods, rabbit TRPV5 (rbTRPV5) full-length and residues 1–660 were cloned into a modified BacMam pFastBac1 vector containing an N-terminal MBP tag separated by a tobacco etch virus (TEV) protease cleavage site. Recombinant bacmid DNA and baculovirus of rbTRPV5 were produced as described previously (36). The plasmid for the membrane scaffold protein MSP2N2 was obtained from Addgene (29520) and was expressed in Escherichia coli. It was purified using the N-terminally tagged hexahistidine (His) as described previously (37). Afterward, the tag was cleaved with TEV protease, and MSP2N2 was snap-frozen and stored at −80 °C.
TRPV5 was purified and reconstituted into nanodiscs in similar fashion as described previously for TRPV1 (16, 38), with minor alterations to the protocol. TRPV5 expression was carried out in HEK293s GnTi− or HEK293F cells, grown in Freestyle 293 Expression medium (Invitrogen) supplemented with 2% FBS at 37 °C and 8% CO2 on an orbital shaker. Cells were transduced when cell density reached 2–2.5 × 106 cells/mL, and sodium butyrate was added at 24 h after transduction to a final concentration of 10 mM. Cells were collected by centrifugation at 48 h after transduction and then resuspended in hypotonic buffer (50 mM Tris, 36.5 mM sucrose, and 2 mM TCEP, pH 8.0) with freshly added protease inhibitor mixture (EDTA-Free SigmaFAST tablet; Sigma-Aldrich). Resuspended cells were broken by passing them through an emulsifier (Avestin Emulsiflex) five times, and the lysate was cleared by low-speed centrifugation at 8,000 × g for 20 min at 4 °C. Subsequently, the membrane fraction was collected by ultracentrifugation at 200,000 × g for 1 h, and resuspended in buffer A [150 mM NaCl, 2 mM Tris(2-carboxyethyl)phosphine, 10% glycerol, and 50 mM Hepes, pH 8.0]. Membranes were solubilized in buffer A supplemented with 20 mM DDM (Anatrace) under mild agitation for 2 h at 4 °C. Detergent-insoluble material was removed by centrifugation at 30,000 × g for 30 min, and the supernatant was mixed with amylose resin for 1–2 h to bind the MBP-tagged TRPV5. The resin was washed in a gravity-flow column with five column volumes of buffer A supplemented with a 10 μg/mL polar soybean lipid mixture and 0.5 mM DDM. The same buffer, with 20 mM maltose added, was used for elution of MBP-TRPV5. The eluted fraction was concentrated to ∼1 mg/mL using a 100-kDa Amicon Ultra-4 filter unit.
Sample Preparation.
Soybean polar lipids were prepared from a chloroform-dissolved stock solution (Avanti Polar Lipids). In short, chloroform was removed from the lipid solution by nitrogen gas drying and subsequent vacuum desiccation for 12 h. Lipids were rehydrated in buffer A (supplemented with 14 mM DDM) and sonicated, yielding a clear lipid stock at 10 mM concentration.
Reconstitution was performed at two different ratios (TRPV5 monomer:MSP2N2:lipids): 1:1.5:150 for rbTRPV5 1–660 and 1:2.5:100 for rbTRPV5 full-length and rbTRPV5 W583A. The concentrated TRPV5 sample was mixed with soybean polar lipids for 1 h at 4 °C. Next, MSP2N2 was added, followed by incubation on ice for 5 min and then the addition of 100 μL of Bio-Beads SM2 resin (Bio-Rad) to remove detergents and initiate nanodisc reconstitution. After rotation for 1 h at 4 °C, a second batch of Bio-Beads (same volume), together with 150 μg of TEV protease was added, and the mixture was rotated overnight at 4 °C. The mixture was filtered to remove all Bio-Beads and injected onto a Superdex 200 increase column (10/300 GL; GE Healthcare Life Sciences) in buffer B (150 mM NaCl, 2 mM TCEP, and 50 mM Hepes, pH 8.0).
For the TRPV5-CaM complex, the detergent-solubilized TRPV5 was concentrated to ∼1 mg/mL and incubated with bovine CaM (1:2 molar ratio of TRPV5 monomer:CaM; Sigma-Aldrich) and 150 μg of TEV protease at 4 °C overnight. The sample was separated on a Superdex 200 increase column (10/300 GL; GE Healthcare Life Sciences) in buffer B supplemented with 0.02% LMNG. Of note, 5 mM CaCl2 was added to all buffers.
In all instances, the fractions corresponding to tetrameric TRPV5 were collected and pooled after assessment by SDS/PAGE and negative-stain EM. They were then concentrated and used for single-particle cryo-EM.
Radioactive Calcium Uptake Assay.
To assess the activity of rbTRPV5 full-length and 1–660, a radioactive 45Ca isotope uptake assay was performed as described previously (39). In brief, HEK293 cells were seeded onto six-well plates at ∼1.4 × 106 cells/well and transiently transfected 6–8 h later with the corresponding TRPV5-containing pcDNA-GFP-FRT/TO plasmids using polyethylenimine (PEI) at a 1:6 ratio. The next day, cells were trypsinized and reseeded onto poly-l-lysine–coated 24-well plates (300 μg of poly-l-lysine and 3.0 × 105 cells per well). Before the start of the experiment, cells were pretreated with 25 μM BAPTA-AM [1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester] for 30 min. The cells were then washed once with warm KHB buffer (110 mM NaCl, 5 mM KCl, 1.2 mM MgCl2, 0.1 mM CaCl2, 10 mM Na acetate, 2 mM NaHPO4, and 20 mM Hepes, pH 7.4, with added inhibitors of voltage-gated calcium channels 10 μM felodipine and 10 μM verapamil) and then incubated with KHB buffer supplemented with 1 μCi/mL 45Ca for 10 min at 37 °C. Subsequently, the assay was stopped with three washes of ice-cold stop buffer (110 mM NaCl, 5 mM KCl, 1.2 mM MgCl2, 10 mM Na acetate, 0.5 mM CaCl2, 1.5 mM LaCl2, and 20 mM Hepes, pH 7.4). Cell were lysed with 0.05% SDS, and 45Ca uptake was then quantified by liquid scintillation counting.
CaM-Binding Assay.
CaM binding was assessed as described previously (25). In short, HEK293 cells were transiently transfected at 6–8 h after seeding with the corresponding mutant TRPV5 pcDNA-GFP-FRT/TO plasmids using PEI and then lysed at 36 h after transfection. The lysis buffer [50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% (vol/vol) Triton X-100, 1 mM sodium orthovanadate, 10 mM Na-β-glycerophosphate, 50 mM Na fluoride, 10 mM Na+ pyrophosphate, 0.27 M sucrose, and the protease inhibitors 1 mM PMSF, 10 μg/mL leupeptin, 10 μg/mL pepstatin A, and 5 μg/mL aprotinin] for cell lysis also contained 5 mM CaCl2. Subsequently, 1 mg of clarified cell lysate was incubated with CaM agarose beads (Sigma-Aldrich) for 2 h at 4 °C under gentle agitation. Subsequently, the immunoprecipitates were washed three times with lysis buffer and proteins were eluted in 30 μL of 2× Laemmli sample buffer [5× stock buffer contained 250 mM Tris⋅HCl pH 6.8, 10% (wt/vol) SDS, 30% (vol/vol) glycerol, 5% (vol/vol) β-mercaptoethanol, and 0.02% (wt/vol) bromophenol blue]. Samples were subjected to 8% (wt/vol) SDS/PAGE, transferred to polyvinylidene fluoride membranes that were blocked for 30 min with 5% (wt/vol) nonfat dry milk in Tris-buffered saline, 0.1% Tween 20 (TBS-T) and immunoblotted overnight at 4 °C using anti-GFP antibody (1:5,000; Sigma-Aldrich). The blots were then washed with TBS-T and incubated with secondary peroxidase-labeled goat anti-rabbit IgG (1:10,000; Chemie Brunschwig) for 1 h at room temperature. After subsequent washes, they were visualized with an enhanced chemiluminescence reagent using the Bio-Rad ChemiDoc XRS imaging system.
EM Data Acquisition.
For negative staining, 3 μL of the purified TRPV5 sample (∼0.01–0.1 mg/mL) was applied to glow-discharged EM grids covered by a thin layer of continuous carbon film (Ted Pella) and stained with 0.75% (wt/vol) uranyl formate solution as described previously (40). EM grids were imaged on a FEI Tecnai T12 microscope (Thermo Fisher Scientific) operated at 120 kV with a TVIPS TemCaM F816 (8K × 8K) scintillator-based CMOS camera (TVIPS). Images were recorded at a magnification of 52,000×, which resulted in a 2.21-Å pixel size on the specimen. Defocus was set to 1.5 μm.
For cryo-EM, 3 μL of purified TRPV5 sample (∼1.0 mg/mL) was applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, 400 copper mesh). The grids were blotted with Whatman no. 1 filter paper and then plunge-frozen in liquid ethane using a Mark III Vitrobot (FEI) with a blotting time of 5–8 s at room temperature and 100% humidity. Cryo-EM data collection of TRPV5 was done with a FEI Titan Krios electron microscope (Thermo Fisher Scientific) equipped with a high-brightness field emission gun operated at 300 kV at the Howard Hughes Medical Institute’s cryo-EM facility at Janelia Research Campus (rbTRPV5 1–660), and the University of California San Francisco’s cryo-EM facility (rbTRPV5 full-length, W583A, and CaM complex). Images were collected with a K2 Summit direct electron detector (Gatan) using SerialEM in superresolution mode at a calibrated magnification of 130,000× (0.84 Å physical pixel size) for TRPV5 1–660 or 22,500× (1.059 Å physical pixel size) for TRPV5 full-length, TRPV5 W583A, and TRPV5-CaM. Dose rate, total dose, and defocus range used for data collection are summarized in SI Appendix, Table S1.
Data Processing.
For negative-stain data, RELION was used for particle picking and 2D classification. For cryo-EM data, the 60-frame image stacks were drift-corrected and 2 × 2 binned by Fourier cropping using MotionCor2 (41). All micrographs were converted to PNG image format and manually inspected. Motion-corrected sums without dose-weighting were used for contrast transfer function estimation with GCTF (42). Motion-corrected sums with dose-weighting were used for all other image processing.
All datasets were processed using a similar strategy (SI Appendix, Fig. S10). In brief, particles picked automatically by Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) were extracted by RELION and then imported into cryoSPARC (43) for 3D classification and refinement procedures. The initial model was generated using ab initio reconstruction. Particles from good classes by ab initio reconstruction were further sorted by iterative 2D classification and heterogenous refinement. Well-sorted particles were finally subjected to homogenous refinement to generate the final maps, including two half maps, an unsharpened map, and a sharpened map. Resolution was estimated using the Fourier shell correlation 0.143 criterion. Local resolution estimates were calculated with the unsharpened raw density map using ResMap.
Additional CaM densities might arise either from bona fide 1:2 TRPV5-CaM complexes or from misalignment of 1:1 complexes with 90° or 180° rotations around the symmetry axis. Because CaM represents only ∼5% of the total complex mass and has a concomitantly small effect on particle alignment probabilities (cross-correlation), such misalignment may persist even after many rounds of refinement have led to a high-resolution reconstruction. To rule out the possibility that the second CaM density is generated by misaligning single CaM-bound particles, we applied further refinement and classification approach within the cisTEM image processing suite (44) (SI Appendix, Fig. S11). First, we aligned TRPV5-CaM particles from all the foregoing classes according to the pose of TRPV5 only, imposing C4 symmetry and using a mask that excludes CaM. The resulting structure has 2.8-Å resolution and equal CaM densities at each of the four binding sites.
Next, we conducted a global angular search of rotations around the symmetry axis (ϕ angle). The search was made sensitive to CaM by applying a mask focused on a single CaM density and using a relatively fine step size of 2°. So as not to compromise alignment of the whole particle given the small size of CaM, information outside the mask was low-pass-filtered to 10 Å and down-weighted to the finite value of 0.3 rather than 0. This resulted in an asymmetric complex structure with strong CaM density at a single site and reduced resolution of 3.5 Å. Weak CaM density remained at the opposite binding site, with weaker density still at an adjacent site (counterclockwise from the primary site when viewed from the cytosolic side).
Finally, we performed focused classification without alignment, now using a mask corresponding to two bound CaM molecules (either opposite or adjacent configuration). Information outside the mask was not discarded but rather was again down-weighted to 0.3 and low-pass-filtered at 10 Å. This classification led to structures of TRPV5-CaM (34,606 particles; 51.%), and TRPV5-CaM2 in an opposite binding site (13,633 particles; 20.6%) configuration. The remaining particles may be from partially disordered TRPV5-CaM complexes with only the C-lobe bound and/or TRPV5-CaM2 complexes with CaM in adjacent binding sites. However, unambiguous density assignment in these classes is difficult owing to the low number of particles and the resulting decrease in resolution.
Model Building.
Ab initio model building was carried out in Coot (45) and PHENIX (46). The initial model was generated with the SWISS-MODEL server on the basis of sequence alignment of TRPV5 with the crystal structure of TRPV6 (Protein Data Bank ID code 5IWK). The resulting model was refined in real space with Phenix.real_space_refine and subsequently adjusted manually in Coot. This process was repeated until the Ramachandran validation was satisfied. Further validation was carried out using EMRinger and MolProbity in PHENIX. The HOLE program (47) was used to calculate the pore profile of TRPV5. Chimera (48) and PyMOL were used to prepare images.
Data Availability.
Cryo-EM density maps of TRPV5 have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-0593, EMD-0594, EMD-0605, and EMD-0607). Particle image stacks after motion correction related to TRPV5 have been deposited in the Electron Microscopy Public Image Archive (www.ebi.ac.uk/pdbe/emdb/empiar/; accession nos. EMPIAR-10255 for TRPV5 1–660 reconstituted in nanodiscs, EMPIAR-10254 for full-length TRPV5 reconstituted in nanodiscs, EMPIAR-10256 for complex of TRPV5 and CaM in LMNG, and EMPIAR-10253 for TRPV5 with W583A mutant reconstituted in nanodiscs). Atomic coordinates for TRPV5 have been deposited in the Protein Data Bank (ID codes 6O1N, 6O1P, 6O1U, and 6O20). All functional data generated or analyzed during this study are included in the published paper.
Supplementary Material
Acknowledgments
We thank N. Thijssen for technical support; C. Hong, R. Huang, and Z. Yu (Howard Hughes Medical Institute’s Janelia cryo-EM facility) for help with data acquisition; and D. Bulkley, A. Myasnikov, and M. Braunfeld (University of California San Francisco’s cryo-EM facility) for help with data acquisition. J.W. is supported by the European Union’s Horizon 2020 Marie Skłodowska Curie Actions (Grant Agreement 748058) and the Netherlands Organisation for Health Research and Development (Off Road Grant 451001 004). S.D. is supported by a Human Frontier Science Program Postdoctoral Fellowship. M.K.v.G. was supported by a Dutch Kidney Foundation student abroad fellowship and by the Nora Baart Foundation of the Netherlands Foundation for the Advancement of Biochemistry. This work was supported in part by grants from the National Institutes of Health (R01 GM098672, S10 OD020054, and S10 OD021741, to Y.C. and R35 NS105038, to D.J.). Y.C. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-0593, EMD-0594, EMD-0605, and EMD-0607); Electron Microscopy Public Image Archive (accession nos. EMPIAR-10255 for TRPV5 1–660 reconstituted in nanodiscs, EMPIAR-10254 for full-length TRPV5 reconstituted in nanodiscs, EMPIAR-10256 for the complex of TRPV5 and CaM in LMNG, and EMPIAR-10253); and the Protein Data Bank www.wwpdb.org (PDB ID codes 6O1N, 6O1P, 6O1U, and 6O20).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820323116/-/DCSupplemental.
References
- 1.Nilius B, et al. The single pore residue Asp542 determines Ca2+ permeation and Mg2+ block of the epithelial Ca2+ channel. J Biol Chem. 2001;276:1020–1025. doi: 10.1074/jbc.M006184200. [DOI] [PubMed] [Google Scholar]
- 2.Vennekens R, et al. Permeation and gating properties of the novel epithelial Ca(2+) channel. J Biol Chem. 2000;275:3963–3969. doi: 10.1074/jbc.275.6.3963. [DOI] [PubMed] [Google Scholar]
- 3.Nilius B, et al. Whole-cell and single channel monovalent cation currents through the novel rabbit epithelial Ca2+ channel ECaC. J Physiol. 2000;527:239–248. doi: 10.1111/j.1469-7793.2000.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Goor MKC, Hoenderop JGJ, van der Wijst J. TRP channels in calcium homeostasis: From hormonal control to structure-function relationship of TRPV5 and TRPV6. Biochim Biophys Acta. 2017;1864:883–893. doi: 10.1016/j.bbamcr.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 5.Ryan LE, Ing SW. Idiopathic hypercalciuria: Can we prevent stones and protect bones? Cleve Clin J Med. 2018;85:47–54. doi: 10.3949/ccjm.85a.16090. [DOI] [PubMed] [Google Scholar]
- 6.Reid IR, Birstow SM, Bolland MJ. Calcium and cardiovascular disease. Endocrinol Metab (Seoul) 2017;32:339–349. doi: 10.3803/EnM.2017.32.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peacock M. Calcium metabolism in health and disease. Clin J Am Soc Nephrol. 2010;5(Suppl 1):S23–S30. doi: 10.2215/CJN.05910809. [DOI] [PubMed] [Google Scholar]
- 8.Oddsson A, et al. Common and rare variants associated with kidney stones and biochemical traits. Nat Commun. 2015;6:7975. doi: 10.1038/ncomms8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaleel A, et al. A single nucleotide polymorphism (rs4236480) in TRPV5 calcium channel gene is associated with stone multiplicity in calcium nephrolithiasis patients. Mediators Inflamm. 2015;2015:375427. doi: 10.1155/2015/375427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Na T, et al. The A563T variation of the renal epithelial calcium channel TRPV5 among African Americans enhances calcium influx. Am J Physiol Renal Physiol. 2009;296:F1042–F1051. doi: 10.1152/ajprenal.90771.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilius B, et al. Modulation of the epithelial calcium channel, ECaC, by intracellular Ca2+ Cell Calcium. 2001;29:417–428. doi: 10.1054/ceca.2001.0201. [DOI] [PubMed] [Google Scholar]
- 12.Nilius B, et al. The carboxyl terminus of the epithelial Ca(2+) channel ECaC1 is involved in Ca(2+)-dependent inactivation. Pflugers Arch. 2003;445:584–588. doi: 10.1007/s00424-002-0923-9. [DOI] [PubMed] [Google Scholar]
- 13.de Groot T, et al. Molecular mechanisms of calmodulin action on TRPV5 and modulation by parathyroid hormone. Mol Cell Biol. 2011;31:2845–2853. doi: 10.1128/MCB.01319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes TET, et al. Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM. Nat Struct Mol Biol. 2018;25:53–60. doi: 10.1038/s41594-017-0009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes TET, et al. Structural insights on TRPV5 gating by endogenous modulators. Nat Commun. 2018;9:4198. doi: 10.1038/s41467-018-06753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGoldrick LL, et al. Opening of the human epithelial calcium channel TRPV6. Nature. 2018;553:233–237. doi: 10.1038/nature25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zubcevic L, et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat Struct Mol Biol. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huynh KW, et al. Structure of the full-length TRPV2 channel by cryo-EM. Nat Commun. 2016;7:11130. doi: 10.1038/ncomms11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Z, et al. Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat Struct Mol Biol. 2018;25:252–260. doi: 10.1038/s41594-018-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu P, Boros S, Chang Q, Bindels RJ, Hoenderop JG. The beta-glucuronidase klotho exclusively activates the epithelial Ca2+ channels TRPV5 and TRPV6. Nephrol Dial Transplant. 2008;23:3397–3402. doi: 10.1093/ndt/gfn291. [DOI] [PubMed] [Google Scholar]
- 23.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saotome K, Singh AK, Yelshanskaya MV, Sobolevsky AI. Crystal structure of the epithelial calcium channel TRPV6. Nature. 2016;534:506–511. doi: 10.1038/nature17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Wijst J, et al. A gate hinge controls the epithelial calcium channel TRPV5. Sci Rep. 2017;7:45489. doi: 10.1038/srep45489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palovcak E, Delemotte L, Klein ML, Carnevale V. Comparative sequence analysis suggests a conserved gating mechanism for TRP channels. J Gen Physiol. 2015;146:37–50. doi: 10.1085/jgp.201411329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh AK, McGoldrick LL, Sobolevsky AI. Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat Struct Mol Biol. 2018;25:805–813. doi: 10.1038/s41594-018-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zubcevic L, et al. Conformational ensemble of the human TRPV3 ion channel. Nat Commun. 2018;9:4773. doi: 10.1038/s41467-018-07117-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu MX. Multiple roles of calmodulin and other Ca(2+)-binding proteins in the functional regulation of TRP channels. Pflugers Arch. 2005;451:105–115. doi: 10.1007/s00424-005-1427-1. [DOI] [PubMed] [Google Scholar]
- 30.Lambers TT, Weidema AF, Nilius B, Hoenderop JGJ, Bindels RJM. Regulation of the mouse epithelial Ca2(+) channel TRPV6 by the Ca(2+)-sensor calmodulin. J Biol Chem. 2004;279:28855–28861. doi: 10.1074/jbc.M313637200. [DOI] [PubMed] [Google Scholar]
- 31.Singh AK, McGoldrick LL, Twomey EC, Sobolevsky AI. Mechanism of calmodulin inactivation of the calcium-selective TRP channel TRPV6. Sci Adv. 2018;4:eaau6088. doi: 10.1126/sciadv.aau6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James P, Vorherr T, Carafoli E. Calmodulin-binding domains: Just two faced or multi-faceted? Trends Biochem Sci. 1995;20:38–42. doi: 10.1016/s0968-0004(00)88949-5. [DOI] [PubMed] [Google Scholar]
- 33.Lee C-H, MacKinnon R. Activation mechanism of a human SK-calmodulin channel complex elucidated by cryo-EM structures. Science. 2018;360:508–513. doi: 10.1126/science.aas9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J, MacKinnon R. Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell. 2017;169:1042–1050.e9. doi: 10.1016/j.cell.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shamgar L, et al. Calmodulin is essential for cardiac IKS channel gating and assembly: Impaired function in long-QT mutations. Circ Res. 2006;98:1055–1063. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
- 36.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie TK, et al. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao E, Cordero-Morales JF, Liu B, Qin F, Julius D. TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron. 2013;77:667–679. doi: 10.1016/j.neuron.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Goor MK, Verkaart S, van Dam TJ, Huynen MA, van der Wijst J. Interspecies differences in PTH-mediated PKA phosphorylation of the epithelial calcium channel TRPV5. Pflugers Arch. 2017;469:1301–1311. doi: 10.1007/s00424-017-1996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—Powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 44.Grant T, Rohou A, Grigorieff N. cisTEM, user-friendly software for single-particle image processing. eLife. 2018;7:e14874. doi: 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, et al. Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems. Annu Rev Biophys. 2013;42:265–287. doi: 10.1146/annurev-biophys-083012-130253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps of TRPV5 have been deposited in the Electron Microscopy Data Bank (accession nos. EMD-0593, EMD-0594, EMD-0605, and EMD-0607). Particle image stacks after motion correction related to TRPV5 have been deposited in the Electron Microscopy Public Image Archive (www.ebi.ac.uk/pdbe/emdb/empiar/; accession nos. EMPIAR-10255 for TRPV5 1–660 reconstituted in nanodiscs, EMPIAR-10254 for full-length TRPV5 reconstituted in nanodiscs, EMPIAR-10256 for complex of TRPV5 and CaM in LMNG, and EMPIAR-10253 for TRPV5 with W583A mutant reconstituted in nanodiscs). Atomic coordinates for TRPV5 have been deposited in the Protein Data Bank (ID codes 6O1N, 6O1P, 6O1U, and 6O20). All functional data generated or analyzed during this study are included in the published paper.