Significance
The vesicular membrane protein synaptobrevin-2 (syb-2) plays an important role in vesicular membrane fusion at the neuronal synapse by participating in the dynamic formation of the SNARE complex. Here, by a combination of solution-state NMR and fluorescence spectroscopy, we find that syb-2 in its prefusion form, before forming the SNARE complex, shows high internal flexibility, characteristic for an intrinsically disordered protein (IDP). But it also reveals an increasing rigidity from the N to C terminus that correlates with an observed increase in lipid binding affinity as well as the known increased rate for C-terminal compared with N-terminal SNARE zippering. This provides a mechanistic perspective on how an IDP and its lipid interactions can lower the energy barrier for membrane fusion.
Keywords: neurotransmitter release, SNARE proteins, membrane fusion, NMR, dynamics
Abstract
Intrinsically disordered proteins (IDPs) and their conformational transitions play an important role in neurotransmitter release at the neuronal synapse. Here, the SNARE proteins are essential by forming the SNARE complex that drives vesicular membrane fusion. While it is widely accepted that the SNARE proteins are intrinsically disordered in their monomeric prefusion form, important mechanistic aspects of this prefusion conformation and its lipid interactions, before forming the SNARE complex, are not fully understood at the molecular level and remain controversial. Here, by a combination of NMR and fluorescence spectroscopy methods, we find that vesicular synaptobrevin-2 (syb-2) in its monomeric prefusion conformation shows high flexibility, characteristic for an IDP, but also a high dynamic range and increasing rigidity from the N to C terminus. The gradual increase in rigidity correlates with an increase in lipid binding affinity from the N to C terminus. It could also explain the increased rate for C-terminal SNARE zippering, known to be faster than N-terminal SNARE zippering. Also, the syb-2 SNARE motif and, in particular, the linker domain show transient and weak membrane binding, characterized by a high off-rate and low (millimolar) affinity. The transient membrane binding of syb-2 may compensate for the repulsive forces between the two membranes and/or the SNARE motifs and the membranes, helping to destabilize the hydrophilic-hydrophobic boundary in the bilayer. Therefore, we propose that optimum flexibility and membrane binding of syb-2 regulate SNARE assembly and minimize repulsive forces during membrane fusion.
At the neuronal synapse, neurotransmitter release to the synaptic cleft requires exocytotic fusion of synaptic vesicles with the presynaptic plasma membrane. Fusion is elicited by the SNARE (soluble N-ethylmaleimide–sensitive factor attachment receptor) proteins synaptobrevin-2 (syb-2), residing in the synaptic vesicle membrane (also referred to as VAMP2), and SNAP-25 and syntaxin-1 (syx-1), residing in the plasma membrane (1–3).
While the SNARE proteins are, to a large extent, intrinsically disordered in their monomeric prefusion form (4–7), during exocytosis, they spontaneously assemble into a highly stable complex containing four α-helices. One each of the helices is contributed by the SNARE motif of syx-1 and syb-2, respectively, and two are contributed by the two SNARE motifs of SNAP-25 (8, 9). The coiled-coil assembly of the SNARE complex is thought to progress from the N terminus to the C terminus in a process called zippering, which provides the energy required to overcome the energy barrier for membrane fusion (1, 3). Additionally, several accessory proteins, such as Munc13, Munc18, complexin, and synaptotagmin, support assembly of the SNARE complex (10).
Although assembly of the SNARE complex has been intensely studied for many years and its role in fusion has been firmly established (1, 3, 10), the exact structural mechanism by which SNAREs remodel membranes to induce lipid mixing and the formation of the fusion pores is not well understood (3, 10), and different alternative models have been proposed (11–16). A key open question is how SNAREs interact with membranes in their prefusion and fusion states, and how these interactions might influence the SNARE zippering and membrane fusion (17). For example, it is generally accepted that a cluster of basic residues in the linker domain (LD) of SNAREs interacts with the anionic head groups of the membrane lipids (18–20). These interactions are thought to tilt the LD in the membrane interface, positioning the SNARE motif closer to the membrane surface (21–23). In contrast, the membrane interactions of the SNARE motifs and the N termini are still unclear. Using NMR and fluorescence interference contrast, it has recently been shown that the SNARE motif of syx-1 interacts with lipids, suggesting the formation of two well-ordered α-helices (7). However, similar experiments on syb-2 arrived at controversial conclusions (15, 16, 24). For instance, an NMR study on syb-2 reconstituted in dodecyl phosphocholine detergent micelles showed that the SNARE motif binds to lipids, forming an additional α-helical region (15). In contrast, NMR experiments on full-length syb-2 reconstituted in lipid nanodiscs as well as in liposomes, showed that the SNARE motif remains intrinsically disordered and does not interact with lipids (24). A more recent study, using a combination of NMR and EPR spectroscopy to study syb-2 and lipid interactions in dodecyl phosphocholine, as well as lipid bicelles and the lipid bilayer, has suggested exchange between a lipid-bound conformation of the SNARE motif and an intrinsically disordered unbound conformation, with the population of the bound conformation decreasing for a more planar lipid environment (16).
To resolve these discrepancies and to investigate the intrinsically disordered nature of syb-2 in its monomeric prefusion form, we performed solution-state NMR relaxation measurements and lipid binding studies on syb-2, supported by fluorescence steady-state and stopped-flow experiments, to obtain independent information on lipid binding and also binding kinetics. Our results indicate high internal flexibility of syb-2, characteristic for an intrinsically disordered protein (IDP), but also reveal a high dynamic range and an increasing stiffness from the N to C terminus that correlates with lipid binding affinity and may favor SNARE complex assembly and close membrane juxtaposition during SNARE complex zippering. We find by two independent methods, NMR and fluorescence, that the SNARE motif of syb-2, in addition to its LD, exhibits transient membrane binding, which likely facilitates membrane fusion.
Results
Soluble Syb-2 (1–96) Secondary Structure and Internal Dynamics.
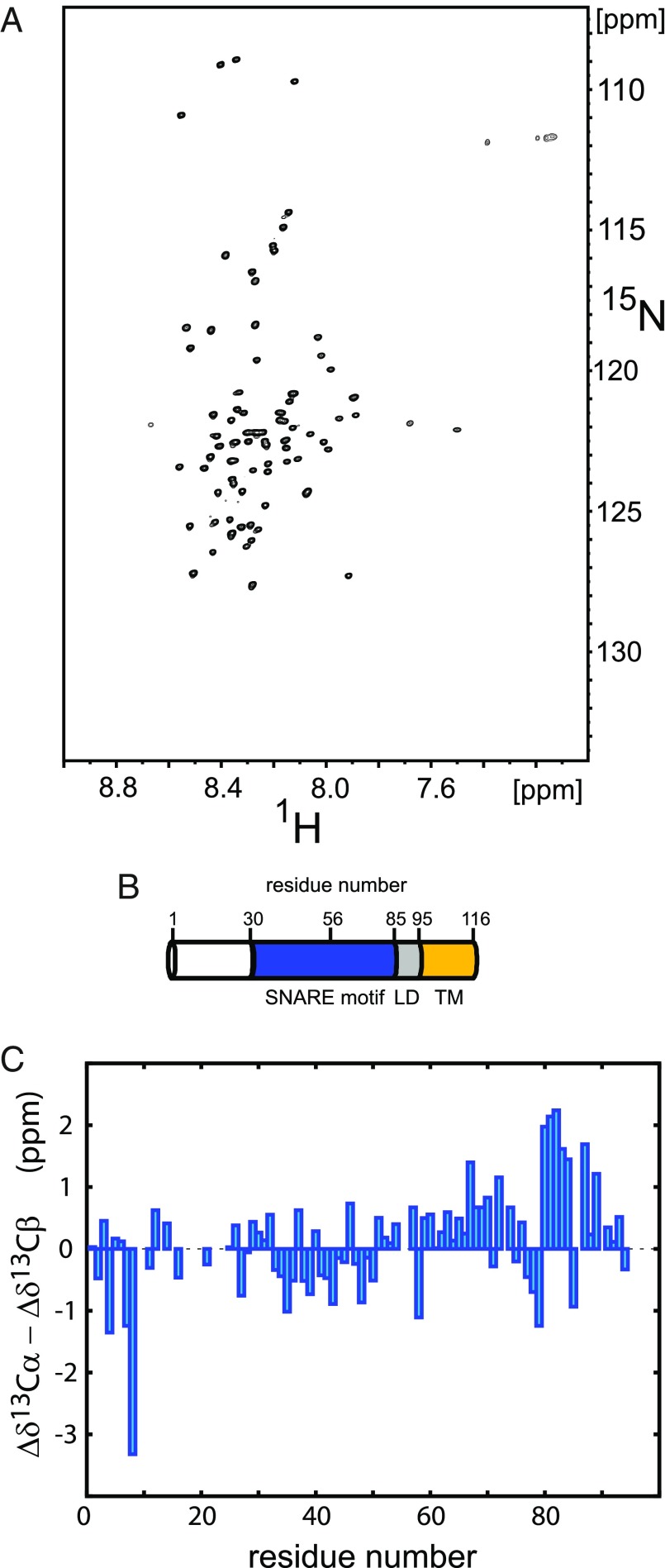
To investigate secondary structure propensities and internal dynamics of syb-2, NMR backbone assignment experiments and NMR relaxation measurements were performed on a soluble syb-2 [1–96, without transmembrane region (TMR)] construct (Materials and Methods). Syb-2 (1–96) showed well-resolved resonances with narrow line widths but low dispersion in the proton dimension (Fig. 1A), and random-coil–like Cα − Cβ secondary chemical shifts, Δδ13Cα − Δδ13Cβ (Fig. 1C), characteristic for an IDP (25–27). However, we observed increased Cα − Cβ secondary chemical shifts for the C terminus of the SNARE motif and N terminus of the adjacent LD, indicative of increased α-helical propensity, as reported previously (6). A spectrum that includes backbone assignments is shown in SI Appendix, Fig. S1; backbone assignments for syb-2 (1–96) were also reported previously (6).
Fig. 1.
NMR analysis of syb-2. (A) Representative 2D 1H-15N TROSY-HSQC spectrum of 200 μM uniformly 2H,13C,15N-enriched syb-2 (1–96) dissolved in 20 mM MES buffer (pH 6.1), containing 250 mM NaCl, 1 mM EDTA, and 0.1 μM TCEP, recorded at 800-MHz magnetic field strength and 5 °C. (B) Schematic view of the domain structure of syb-2 (1–116). Residue numbers above the cylinder indicate the boundaries of the SNARE motif, the zero layer, the LD, and the TMR, respectively. (C) Cα − Cβ secondary chemical shifts (Δδ13Cα − Δδ13Cβ), plotted as a function of amino acid sequence number [values around zero (between −1 and 1) indicate random coil-like behavior, and positive values (>1) indicate α-helical propensity (around 3–4 for fully α-helical)]. Data reveal increased α-helical propensity for the C terminus of the SNARE motif and the N terminus of the adjacent LD. The strongly negative outlier for Val-8 can be explained by nearest neighbor effects due to Pro-9 (48).
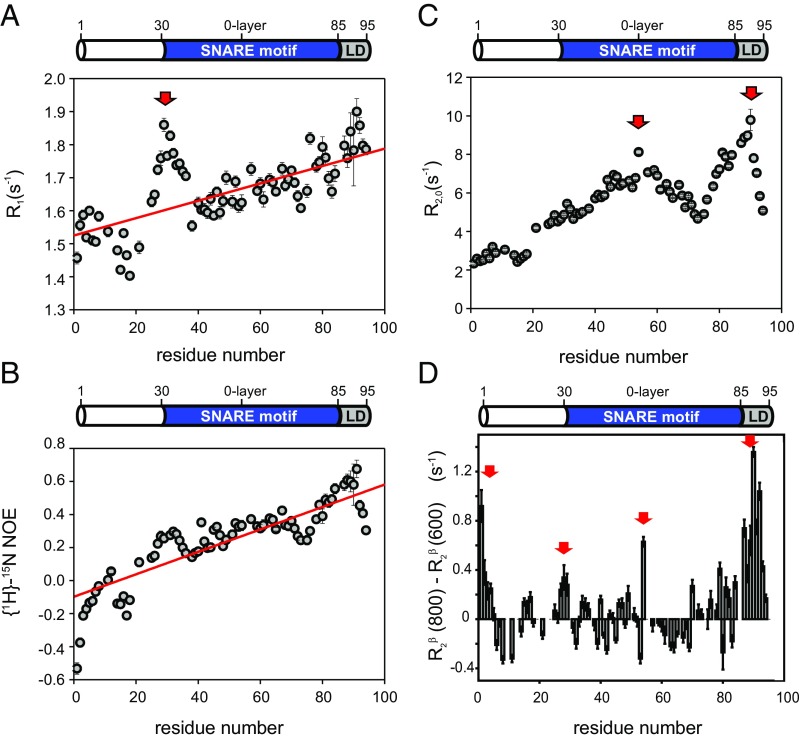
Next, 15N R1, R2 (derived from R1ρ) and 1H-15N NOE experiments were performed to investigate internal dynamics on the fast picosecond-nanosecond time scale (Materials and Methods). Those internal dynamics of the protein can be considered as flexibility of the protein backbone. The 15N R2 rate constants and {1H}-15N NOE values (Fig. 2 and SI Appendix, Table S2) show very low values overall, indicating high internal flexibility, characteristic for an IDP. However, both 15N R2 and {1H}-15N NOE values consistently reveal a high dynamic range, with a fivefold increase in 15N R2 rate constants from the N to C terminus, and negative {1H}-15N NOE values for the N terminus but values around 0.6 for the C-terminal LD. This points to a highly flexible and intrinsically disordered N terminus, particularly the N-terminal 20 residues that are not part of the SNARE motif, but a more ordered and secondary structure-like behavior of the LD, which is in line with the increased α-helical propensity of the LD, as observed by secondary chemical shifts (discussed above). Both 1H-15N NOE and 15N R2 rate constants are direct measures of internal dynamics. Low 1H-15N NOE and 15N R2 values correspond to high internal dynamics (high internal flexibility). While 1H-15N NOE values show little dependence on the size of the system (after their asymptotic steady-state value has been reached), 15N R2 rate constants scale with the weight-dependent overall tumbling correlation time of the protein. However, their relative values depend on the local internal flexibility. This relation holds true also for IDPs in the absence of an overall tumbling correlation time. For syb-2 (1–96), 1H-15N NOE and 15N R2 values increase linearly from the N terminus to the C terminus of syb-2 (1–96), indicating gradually decreasing flexibility (increasing rigidity) from the N terminus toward the C-terminal LD. However, 15N R2 rate constants show an additional local maximum (Fig. 2C and SI Appendix, Table S1) around L54, adjacent to the zero layer of the SNARE motif (centered at R56). This indicates increased secondary structure propensity for the zero layer and the C-terminal SNARE motif and LD, which is in line with increased Cα − Cβ secondary chemical shifts for S80–L84 and K87. The 15N R1 rate constants follow a very similar trend as the 1H-15N NOE values and the 15N R2 rate constants, but show unexpectedly high values (and thereby a higher rigidity) for residues A21 to Q36, with a maximum at R31, the N terminus of the SNARE motif. We speculate that R31 may act as a nucleation point for fast α-helix formation during SNARE complex formation. We also performed a model-free analysis of the measured NMR relaxation data to obtain additional insights into the time scale and amplitudes of motion (SI Appendix). Amplitudes of motion decrease and the residue-specific correlation time becomes longer from the N to C terminus (SI Appendix, Fig. S2), confirming the notion above that in syb-2 (1–96), flexibility decreases from the N to C terminus.
Fig. 2.
Backbone dynamics of syb-2. The 15N relaxation data recorded for syb-2 (1–96) are shown. (A) 15N R1 relaxation rate constants recorded at 600 MHz versus residue. The red arrow highlights the unusually high values of 15N R1. The red line corresponds to the linear fit indicating the increase of rigidity of the protein backbone versus residue. (B) {1H}-15N NOE values versus residue increase linearly from the N terminus to the C terminus. The red line corresponds to the linear fitting indicating the increase of rigidity of the protein backbone versus residue. (C) 15N R2,0 relaxation rate constants recorded at 600 MHz (derived from 15N R1ρ with a 2-kHz RF field; R1 contribution-corrected) plotted versus residue. Red arrows highlight the two maxima in the dataset. (D) Hahn-echo transverse relaxation rates, R2β, at 800 MHz minus those measured at 600 MHz. Positive values indicate Rex contributions due to conformational dynamics on a microsecond-millisecond time scale. Regions subject to such conformational exchange processes are highlighted by red arrows.
Conformational changes on the microsecond to millisecond time scale give rise to an exchange (Rex) contribution to R2. To probe the presence of such Rex contributions, we measured transverse relaxation rate constants of the slowly relaxing transverse relaxation optimized spectroscopy (TROSY) line of the NH doublet, R2β, using a Hahn-echo R2 experiment at two different magnetic field strengths (600 MHz and 800 MHz). Different from the R1ρ experiment and also different from a Carr–Purcell–Meiboom–Gill (CPMG) experiment, which both refocus exchange contributions of processes slower than inverse of the spin-lock or CPMG field strength, respectively, conformational exchange effect (R2ex) from the entire microsecond-millisecond time window (<10 ms) contributes to R2β. For most residues, R2β rate constants at 800 MHz are equal to or slightly lower than at 600 MHz due to absence of Rex contributions and more efficient cancellation of R2 autorelaxation and transverse NH-dipole, 15N chemical shift anisotropy cross-correlated relaxation (TROSY effect). Rex effects scale with the square of the magnetic field. Thus, elevated R2β rate constants, when the rate constants at 800 MHz exceed those measured at 600 MHz, are a clear indication for conformational exchange processes. By taking the difference between R2β rate constants measured at 800 MHz minus those measured at 600 MHz, we highlight residues with Rex contributions, indicated by positive values of the difference R2β (800 MHz) − R2β (600 MHz). We observed those conformational exchange processes at the N terminus (residues M1–T4), the N terminus of the SNARE motif (residues S28 and N29), adjacent to the zero layer (L54), and at the C terminus of the SNARE motif (residues A81 and A82) as well as the LD (residues K87 to K94). Taking into account that S80–L84 and K87 showed increased α-helical propensity (Fig. 1C), our findings suggest a conformational exchange process between an unstructured conformation and an α-helical conformation for the C terminus of the SNARE motif. The conformational exchange process for the N terminus appears refocused in the 15N R1ρ experiments, using a 2-kHz spin-lock, indicating conformational dynamics on a time scale slower than 80 μs. Together, our results reveal that syb-2, albeit mainly unstructured in solution, exhibits conformational dynamics over a wide range of time scales, as observed also for other IDPs recently (28, 29).
To probe for the presence of transient and long-range contacts in soluble syb-2 (1–96), we performed NMR paramagnetic relaxation enhancement (PRE) measurements (30) [SI Appendix, SI Results; transient and long-range contacts in syb-2 (1–96) are shown in SI Appendix, Fig. S4]. We observe a slight reduction of normalized intensities, paramagnetic spin-labeled sample intensities (Ipara)/diamagnetic reference sample intensities (Idia), for the N terminus of the SNARE motif (residues around R30) as well as for the C terminus of the soluble syb-2 (1–96) construct, but little interaction otherwise. In contrast, the mutants with the spin-label attached at the C-terminal end of the SNARE motif (S79C; SI Appendix, Fig. S4B) or in the LD (K91C; SI Appendix, Fig. S4C) show a significant reduction of Ipara/Idia for the C-terminal SNARE motif and complete bleaching (Ipara = 0) of the LD, indicating transient proximity between the C-terminal SNARE motif and LD in soluble syb-2 (1–96).
Syb-2 (1–116) Reconstituted in Liposomes Remains Intrinsically Disordered for the N Terminus and N-Terminal SNARE Motif.
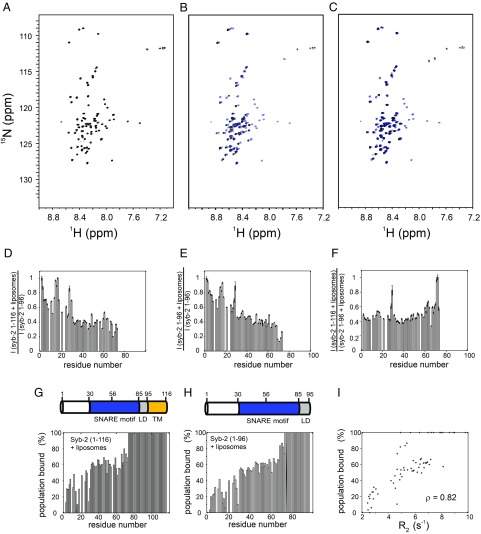
Next, to investigate lipid interactions, monomeric full-length syb-2 (1–116, with TMR) was reconstituted into liposomes composed of a lipid mix of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), and cholesterol (Chol) at a molar ratio of 5:2:2:1 DOPC/DOPS/DOPE/Chol, a lipid composition similar to the one in synaptic vesicles (31) (Materials and Methods). The spectra of syb-2 (1–116) reconstituted in liposomes superimpose well to those of soluble syb-2 (1–96), with identical chemical shifts for most of the resonances (Fig. 3 A and B). Residues M1–G73 remain visible in the spectra, pointing to intrinsic disorder, while residues A74–M96, comprising the C-terminal SNARE motif and LD, become invisible in the presence of lipids. Due to the high molecular mass of liposomes (about MDa), the liposome overall tumbling rotational correlation time is in the range of hundreds of nanoseconds. Therefore, anything that is attached to or embedded in the liposome adopts the relaxation properties of the liposome and shows severe line broadening resulting in resonance intensities below the detection threshold, and therefore invisible. This fact implies that a major population of the region M1–G73 remains intrinsically disordered in the presence of lipids and retains its high internal flexibility. In contrast, residues spanning from A74 to M96 either bind to liposomes or adopt a rigid helical structure such that the relaxation properties of the TMR propagate into the LD and C-terminal SNARE motif. To discriminate those two effects, we turned back to syb-2 (1–96) and investigated its spectra in the presence of liposomes of identical composition.
Fig. 3.
Lipid interactions of syb-2 monitored by NMR. (A) Reference 1H-15N TROSY-HSQC spectrum of 200 μM 2H,13C,15N syb-2 (1–96) dissolved in 20 mM MES buffer (pH 6.1), containing 250 mM NaCl, 1 mM EDTA, and 0.1 mM TCEP, recorded at 800 MHz and 5 °C. Each [1H,15N] cross-peak corresponds to a visible (unbound) residue of syb-2 (1–96). (Side-chain amide signals of Gln are labeled by an asterisk. The spectrum is identical to the spectrum shown in Fig. 1A and is shown for comparison.) (B) Spectrum of 20 μM 2H,13C,15N syb-2 (1–116) (black) reconstituted into small liposomes constituted from a lipid mix of 5:2:2:1 DOPC/DOPS/DOPE/Chol in a buffer of 20 mM Hepes (pH 7.4 at 5 °C), 150 mM KCl, and 0.1 mM TCEP. The lipid/protein molar ratio was 100:1. For comparison, the spectrum is overlaid with the spectrum of syb-2 (1–96) (blue), identical to the spectrum shown in A. Signals from A74 to the C terminus are not visible in the spectra. (C) Spectrum of 20 μM 2H,13C,15N syb-2 (1–96) (black) in the presence of liposomes (5:2:2:1 DOPC/DOPS/DOPE/Chol), compared with the spectrum of syb-2 (1–96) without lipids (blue). In addition, soluble syb-2 signals from A74 to the C terminus disappear in the spectra, indicating lipid binding. Comparison is shown of normalized intensity ratios, , of the intensities in the 1H-15N TROSY-HSQC spectra for syb-2 (1–116) reconstituted in liposomes divided by the intensities of soluble syb-2 (1–96) (D), normalized intensity ratios of soluble syb-2 (1–96) in the presence of liposomes divided by soluble syb-2 (1–96) in the absence of liposomes (E), and normalized intensity ratio of syb-2 (1–116) reconstituted in liposomes divided by soluble syb-2 (1–96) in the presence of liposomes (F). (G) Percentage of lipid-bound syb-2 (1–116) population, as inferred from the intensity ratio shown in D via . (H) Same as in G, but for syb-2 (1–96). (I) Correlation plot between 15N R2 rate constants measured for syb-2 (1–96) and the percentage of bound population. Residues that show reduced mobility (inferred from higher R2 rate constants) in free syb-2 (without lipids) reveal a higher lipid binding affinity in the presence of liposomes. The Pearson correlation coefficient between the percentage of the bound population and R2 is ρ = 0.82.
The Syb-2 SNARE Motif and LD Interact with Lipids.
In accordance with the experiments on syb-2 (1–116) reconstituted liposomes, resonances from A74 to M96 disappeared for soluble syb-2 (1–96) in the presence of liposomes (Fig. 3C). These data show that the membrane interaction of the region between residues A74 and M96 (LD plus adjacent residues of the SNARE motif) is due to an intrinsic tendency to bind to membranes rather than being a consequence of proximity when anchored by the TMR.
In contrast to the C-terminal regions, the highly disordered and flexible N-terminal 20 residues do not appear to interact with membranes. This is evident from the residue-specific normalized intensity ratios of membrane-anchored syb-2 (1–116) divided by the intensities of soluble syb-2 (1–96) in the absence of liposomes (Fig. 3D) or from the normalized intensity ratio of syb-2 (1–96) in the presence and absence of liposomes (Fig. 3E), . A value close to 1 indicates little lipid interaction, while a smaller value indicates lipid binding and the presence of a lipid-bound conformation (discussed below); for a direct comparison of the intensities, we refer to SI Appendix, Fig. S6. In the N-terminal region, no or only very little decrease in normalized intensities is observed. In contrast, the SNARE motif shows decreasing intensities from the N to C terminus (Fig. 3 D and E), with an value of 0.55 for Q36 and 0.13 for Q71 of syb-2 (1–96). From A74 onward, the C-terminal SNARE motif and LD vanish from the spectra in the presence of liposomes (Fig. 3E). Syb-2 (1–116) embedded in liposomes shows a trend very similar to that of soluble syb-2 (1–96) in the presence of liposomes (Fig. 3 D and F). This indicates a gradually increasing lipid interaction from the N-terminal SNARE motif toward the C terminus. The decrease of intensities in the presence of lipids can be explained by a slow exchange process (millisecond time scale) between an unbound conformation (visible) and a bound conformation (invisible). Fig. 3 G and H shows the degree of bound population, as inferred from the decrease of resonance intensities in the presence of lipids, , compared with the intensities of the reference sample in solution, , assuming a simple two-state model, with the population of the bound state described by . Data indicate an N- to C-terminal gradually increasing population of the (invisible) lipid-bound conformation of syb-2, with the C-terminal SNARE motif and LD being fully bound and invisible in the presence of lipids. The increasing lipid affinity of syb-2 (1–96) from an N- to C-terminal population seems to correlate with increasing rigidity (decreasing flexibility) and increasing α-helical propensity observed for the free form of syb-2 (1–96), as inferred from the NMR relaxation and secondary chemical shift data (Figs. 1C, 2, and 3 G–I). NMR relaxation Rex data (Fig. 2D) indicate conformational exchange of the C-terminal SNARE motif and LD between an unstructured conformation and a presumably α-helical conformation, as inferred from secondary chemical shift data (Fig. 1C), pointing to an increased α-helical propensity for that region.
To investigate whether lipid binding induces a long-range structural change in syb-2 (1–96), we recorded NMR PRE data for the same mutants as described above, but now in the presence of lipids (SI Appendix, Fig. S5). We find that in the lipid-bound conformation, the syb-2 N terminus is transiently sampling the proximity of the N-terminal SNARE motif as well as that of the zero layer and beyond (SI Appendix, Fig. S5A). The data for S79C and K91C (SI Appendix, Fig. S5 B and C) consistently indicate that the N-terminal SNARE motif (apart from the very N terminus) shows little interaction with the C-terminal SNARE motif, while the transient proximity between the LD and C-terminal SNARE motif, observed already in solution (SI Appendix, Fig. S4 B and C), is also conserved for the lipid-bound conformation.
Fluorescence and Stopped-Flow Data Show Weak Lipid Binding of the Syb-2 N Terminus and LD.
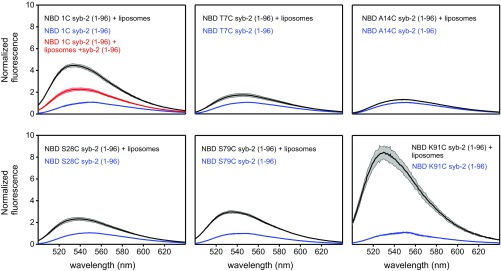
To obtain independent information on the lipid binding of the syb-2 (1–96) SNARE motif and LD (compare Fig. 3), we performed fluorescence binding studies using six single-site cysteine mutants (1C, T7C, A14C, S28C, S79C, and K91C) of syb-2 (1–96) labeled with 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD). NBD is an environment-sensitive probe that increases its fluorescence emission intensity when transferred to a hydrophobic environment and allows one to study the binding of specific residues to the membrane (32). K91C in the LD displayed the strongest increase in fluorescence, indicating strong lipid binding (Fig. 4). Surprisingly, the 1C mutant showed an NBD fluorescence increase similar to that of the K91C mutant, while the S28C and S79C mutants, with fluorescence probes located in the N-terminal SNARE motif and C-terminal SNARE motif, respectively, showed only a moderate increase in fluorescence emission intensity, implying that those residues do interact with the lipid membrane, but less strongly (Fig. 4). Similar results were observed when labeling the same mutants with 6-bromoacetyl-2-dimethylaminonaphthalene (BADAN), another environment-sensitive probe comparable to NBD (SI Appendix, Fig. S7). To exclude that the introduction of point mutants and labeling affect membrane binding of syb-2, we measured membrane binding by coflotation. There was no difference between the various mutants and unlabeled or labeled proteins used (SI Appendix, Fig. S8).
Fig. 4.
Lipid binding of syb-2 monitored by fluorescence. Fluorescence spectra of NBD-labeled cysteine mutants were recorded between 500 nm and 640 nm in 20 mM Hepes, 150 mM KCl, and 0.1 mM TCEP (pH 7.4) at 8 °C. Blue traces show spectra acquired in the absence of liposomes, and black traces shown spectra acquired in the presence of liposomes. (Left Upper) Red trace shows spectra acquired in the presence of liposomes and unlabeled syb-2 in excess. The colored area represents the SD for n = 3.
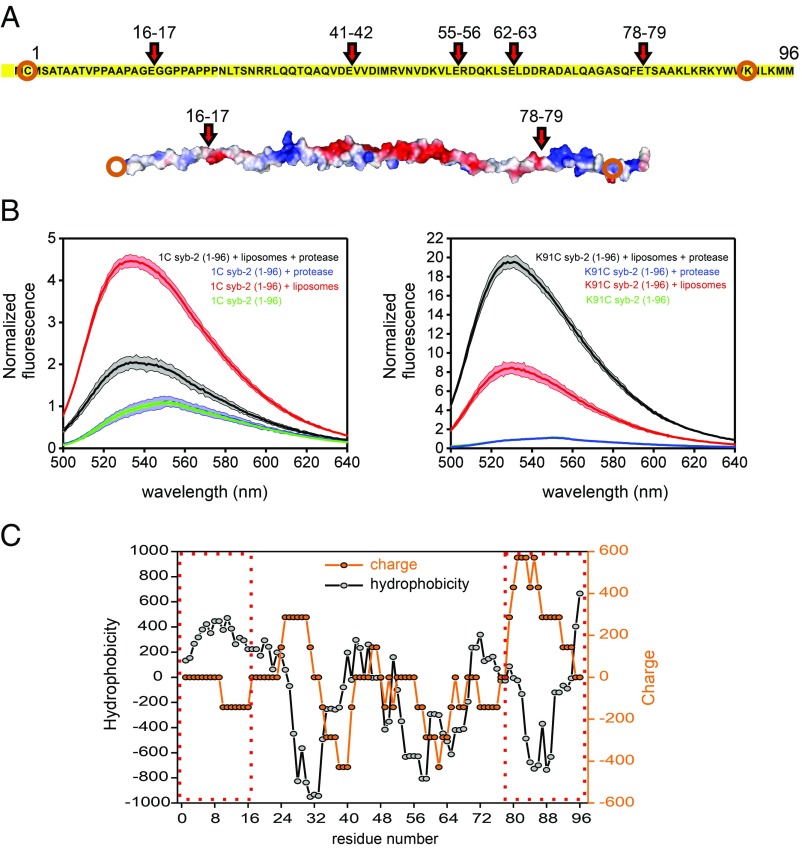
Next, we performed syb-2 membrane binding experiments following digestion by the endoproteinase Glu-C (Fig. 5A). After cleaving, membrane binding of NBD-labeled syb-2 fragments can be studied. This allows one to discriminate whether syb-2 lipid binding sites bind independently to lipids or, by contrast, bind in a cooperative manner. After digestion, the fluorescence of the K91C mutant increased, while the fluorescence of the 1C mutant decreased (Fig. 5B). This indicates cooperativity between different lipid binding sites of syb-2, probably due to the different electrostatic potential and hydrophobicity of the digested fragments (Fig. 5 A and C).
Fig. 5.
Cooperative lipid binding of syb-2. Lipid binding of labeled syb-2 fragments after cleavage of syb-2 (1–96) with endoproteinase Glu-C. (A) Predicted cleavage sites (showing the residue numbers) of endoproteinase Glu-C from Staphylococcus aureus V8 in syb-2 (1–96) and electrostatic potential (determined using PyMOL, modified from Protein Data Bank ID code 2KOG). Orange circles indicate the labeled residue in each fragment. (B) Fluorescence spectra of NBD-labeled cysteine mutants 1C and K91C were recorded before and after digestion by endoproteinase Glu-C. Green traces show spectra acquired in the absence of liposomes, red traces show spectra acquired in the presence of liposomes before digestion, and blue and black traces show spectra acquired after digestion in the presence of liposomes (black) or without (blue). The colored area represents the SD for n = 3. (C) Hydrophobicity (ExPASy, ProtScale, Eisenberg et al., https://web.expasy.org/protscale/) and charge (EMBOSS, charge, www.bioinformatics.nl/cgi-bin/emboss/charge) scores of syb-2 (1–96). A seven-residue window is used for both calculations. Dashed red boxes indicate the N-terminal and C-terminal fragments, respectively.
Binding Kinetics of Syb-2.
To study the membrane binding kinetics of syb-2 in more detail, we performed rapid mixing experiments using a stopped-flow approach. The fluorescence increase of Texas Red-labeled vesicles was monitored after rapid mixing with an equal volume of Alexa Fluor 488-labeled syb-2 (1C mutant).
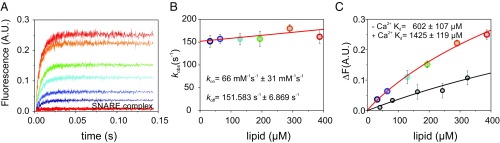
Our results show that syb-2 membrane binding reached equilibrium within the first 50 ms (Fig. 6A), revealing fast binding kinetics, in line with the NMR results. Additionally, as shown in Fig. 6B, the kinetics exhibit a fast dissociation rate (koff), resulting in low-affinity binding in the millimolar range (Kd ∼ 600 μM) (Fig. 6C). Then, we carried out the same experiment in the presence of 500 μM CaCl2. Unfortunately, due to the low signal-to-noise ratio of the fluorescence time courses, reliable association rate constant (kon) and koff values could not be calculated. Nevertheless, fluorescence increase versus lipid concentration in the presence of calcium indicated slightly lower affinities, suggesting that calcium slightly decreases the membrane binding of syb-2 (Fig. 6C). Furthermore, we repeated the experiment using Alexa Fluor 488-labeled syb-2, after forming a zippered SNARE complex consisting of 1:1:1 syx-1a/SNAP-25/syb-2. Surprisingly, no binding of the syb-2 N terminus was observed (Fig. 6A) once the soluble SNARE complex had formed, suggesting that even though the syb-2 N terminus is not involved in the SNARE complex, the zippered SNARE motif exerts negative cooperativity on the binding of the syb-2 N-terminal domain.
Fig. 6.
Lipid binding kinetics of syb-2. Stopped-flow measurements with an Alexa Fluor 488-labeled N-terminal cysteine mutant of syb-2. (A) Time course of Texas Red fluorescence emission at increasing total lipid concentrations is shown for syb-2 (lines) and at highest total lipid concentration for the SNARE complex (red symbols). (B) Observed kobs versus total lipid concentrations. The y-intercept provides the dissociation rate constant, koff, and the slope yields the association rate constant, kon. Bars represent the SEs of three to five technical repeats. The solid line shows a weighted linear fit (Eq. 2). (C) Fluorescence increase of Texas Red fluorescence emission is shown at different total lipid concentrations, in the absence (colored circles) and presence (black circles) of Ca2+, and fitted by a Hill equation (Eq. 3) with fixed n = 1. We obtained Kd = 602 ± 107 μM and Kd = 1,425 ± 119 μM, respectively. A.U., arbitrary unit.
Discussion
In this work, we have combined NMR relaxation and lipid binding studies with steady-state and stopped-flow fluorescence measurements of syb-2 in its monomeric (prefusion) form to investigate structural dynamics and lipid binding properties of syb-2 with residue-specific spatial resolution and millisecond time resolution. We find that syb-2 is intrinsically disordered but shows a gradually increasing rigidity from the N to C terminus that correlates with an increase in lipid binding affinity. The entire SNARE motif shows transient and weak lipid binding, characterized by a high off-rate and low affinity (millimolar range).
NMR relaxation experiments conducted in this study confirm that unbound monomeric syb-2 is intrinsically disordered, as previously reported by Hazzard et al. (6), but they also reveal a large variation in syb-2 internal plasticity. The N-terminal residues are extremely flexible, with flexibility gradually decreasing toward the syb-2 C-terminal LD. We suggest that this gradually decreasing flexibility progressively increases the speed of SNARE zippering, in agreement with the finding that the syb-2 N-terminal SNARE motif assembles more slowly than the syb-2 C-terminal SNARE motif due to a larger energy barrier (33, 34). Notably, on top of the general trend of increasing rigidity from N- to C-terminal LD, there is a conspicuous rigid spot around the zero layer (centered at R56). Both the LD and zero layer were identified as transition sites in optical tweezer experiments investigating SNARE zippering (33). The observed high rigidity of the LD that correlates with higher lipid binding affinity would lower the energy barrier for membrane fusion. For example, molecular dynamics experiments using coarse-grained models (35) indicate that unstructured flexible LDs (as opposed to what we find) prevent close approach of the membranes, thus interfering with fusion.
Using both soluble and membrane-anchored versions of syb-2, our data reveal that the LD and the C-terminal part of the SNARE motif interact with lipid membranes. Importantly, we also observed weak and transient binding of the N-terminal and central part of the SNARE motif suggesting that membrane binding is indeed an intrinsic property of the SNARE motif and LD, and does not arise from a proximity effect to the bilayer induced by the membrane anchor of syb-2. In addition, we observed interactions between different regions of syb-2, such as transient proximity between the LD and C-terminal SNARE motif and between the N terminus and the adjacent N-terminal region of the SNARE motif, with the latter only being seen when bound to membranes.
Interestingly, when syb-2 (1–96) is cleaved into two fragments, membrane binding of the N-terminal fragment decreases, whereas that of the C-terminal fragment increases. Evidently, there are conformational constraints by which the N- and C-terminal parts influence each other with respect to membrane binding. It is conceivable that membrane attraction by the high positive charge density at the C terminus (36) (Fig. 5) is balanced by membrane repulsion caused by negatively charged residues more upstream in the SNARE motif (Fig. 5 A and C). After cleavage, this balance is removed and the peptides can be expected to assume conformations in which exposure of hydrophobic side chains is further minimized, with charged side chains dominating membrane interactions.
In addition to positive charge clusters, the C terminus of syb-2 (LD region) is rich in hydrophobic side chains (Fig. 5) that may contribute to membrane binding. Thus, Brewer et al. (24) showed that the syb-2 (1–96) C terminus binds to 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine vesicles, while the C-terminal SNARE motif (lacking LD) showed only low lipid binding affinity. Moreover, Kweon et al. (21, 22) reported the penetration of residues W89 and W90 in membrane-reconstituted syb2 (1–116) which, however, was not observed by us when using the soluble fragment (1–96), and thus appears to be dependent on membrane anchorage by the transmembrane domain (SI Appendix, Fig. S10).
Generally, membrane binding of syb-2 (1–96) is of low affinity (in the millimolar range) due to a high dissociation rate. This finding rationalizes why syb-2 is not stably sequestered by membranes and why interactions with other SNAREs and accessory proteins are not hindered, as suggested previously (16, 24). Our findings also resolve partially contradicting conclusions of two previous studies (15, 24). Ellena et al. (15) reported that the N-terminal SNARE motif is α-helical and lipid-bound, whereas in the more recent study by Brewer et al. (24), this region was reported to be unstructured and not binding to lipids. This discrepancy has been attributed to the different lipid environments, detergent vs. lipids (24), and the resulting membrane curvature (16). As evident from SI Appendix, Table S1, our experimental conditions (buffer, pH, lipid composition) are similar to those used by Brewer et al. (24). Our data indicate the presence of two conformations: a major unstructured and unbound conformation of the N-terminal SNARE motif that is visible by NMR and a minor lipid-bound conformation that is invisible by NMR and in slow exchange with the unbound conformation. We conclude that Ellena et al. (15) have presumably observed a conformation similar to the lipid-bound conformation (with the detergent dodecylphosphocholine mimicking the lipid bilayer), while Brewer et al. (24) observed only the unbound unstructured conformation. In fact, Brewer et al. (24) observed reduced NMR signal intensities of the N-terminal SNARE motif in the presence of lipids, but the dynamic equilibrium character (slow exchange) of the N-terminal SNARE motif (which we observe) may have escaped their attention because of the sparsity of their dataset in that region. The dynamic equilibrium between a lipid-bound conformation and an unbound conformation of the N-terminal SNARE that we observe is also consistent with recent findings by EPR spectroscopy (16).
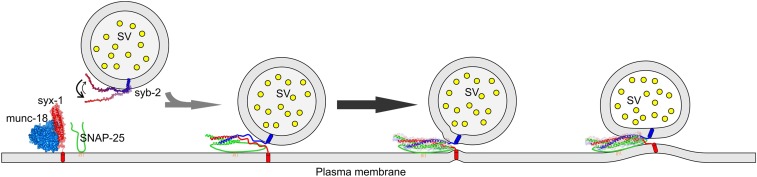
Taken together, our results suggest a picture (Fig. 7) in which syb-2 is probing the membrane in a highly dynamic and transient manner, mainly controlled by its high dissociation rate, which allows syb-2 to be accessible for SNARE zippering. Such a dynamic state would also be compatible with the proposed binding of syb-2 to Munc13-1 (37, 38) and Munc18-1 (37–39) that is thought to template SNARE assembly (38, 40). Upon SNARE complex zippering, syb-2 would dissociate from Munc13 and Munc18 (38, 39) and bind preferentially to the membrane because of the increase of the local lipid concentration near the membrane interface. Our results, together with recent results by Gao et al. (33) and Li et al. (34), suggest that the high flexibility of the syb-2 N-terminal SNARE motif retards initiation of SNARE zippering by increasing its kinetic barrier. Additionally, the gradual increase in lipid binding affinity of syb-2 may pull vesicular and plasma membranes into increasingly closer juxtaposition during SNARE zippering. Lipid binding of the C-terminal SNARE motif and the LD may lower the additional energy barrier for fusion imposed by the highly negatively charged SNARE motifs and/or the energy barrier of membrane repulsion. Thus, transient membrane binding of syb-2 may compensate for the repulsive forces between the two membranes themselves and/or the SNARE motifs and the membranes, helping to destabilize the hydrophilic-hydrophobic boundary in the bilayer. This provides a mechanistic perspective on how an IDP and its lipid interactions can lower the energy barrier for membrane fusion. Thus, we speculate that the SNARE protein sequences may be optimized to get a balance between hydrophobic and polar residues to regulate the SNARE flexibility and, at the same time, the binding between membranes and SNAREs. We suggest that the combination of syb-2 flexibility and membrane interactions plays an important role in the SNARE assembly and membrane bending during exocytosis.
Fig. 7.
Model of syb-2 dynamics and lipid binding in SNARE zippering and membrane fusion. In its precomplex state, syb-2 binds in a highly transient equilibrium to the vesicle membrane. Additionally, syb-2 shows decreasing flexibility from the N-terminal region to the C-terminal region that influences the rate of SNARE zippering. During exocytosis, intrinsic syb-2 membrane binding may help to compensate for repulsive forces between membranes and/or membranes and negatively charged side chains in the SNARE motifs, facilitating membrane fusion.
Materials and Methods
NMR Sample Preparation.
The 2H15N13C-labeled syb-2 (1–96) was expressed in deuterated M9 minimal media purified following standard protocols [protein constructs from Rattus norvegicus were expressed in Escherichia coli strain BL21 (DE3) onpET28 vectors] (41). Purified protein was dialyzed in 20 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 0.1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP). For NMR backbone assignment and 15N relaxation experiments, syb-2 (1–96) was further dialyzed in 20 mM MES (pH 6.1), 250 mM NaCl, 1 mM EDTA, and 0.1 mM TCEP to circumvent adverse amide exchange effects at higher pH.
For NMR studies on full-length syb-2 comprising the TMR, 2H15N13C-labeled syb-2 (1–116) was expressed in deuterated M9 minimal media; purified following standard protocols (41, 42); and dialyzed in 20 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.1 mM TCEP, and 1% octyl glycoside (wt/wt).
For PRE measurements, a paramagnetic (1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate (MTSL) spin-label was covalently attached to the different single-site cysteine mutants 1C, S79C, and K91C syb-2 (1–96). This was achieved by adding a five- to 10-fold excess of MTSL, dissolved in a small amount of ethanol, to a diluted sample of the respective single-site cysteine mutant of 15N syb-2 (1–96) [dissolved in 20 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA] after the ion-exchange chromatography purification step. The sample was incubated overnight at 4 °C. Excess MTSL spin-label was removed afterward by running the sample on a desalting column.
Liposome Preparation.
Small unilamellar vesicles (SUVs) composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; Avanti catalog no. 850375), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE; Avanti catalog no. 850725), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS; Avanti catalog no. 840035), and Chol (Avanti catalog no. 700000) at a molar ratio of 5:2:2:1 DOPC/DOPS/DOPE/Chol were prepared by mixing of the different lipids in chloroform stock solutions (Avanti Polar Lipids) in the desired proportions. Lipids were dried in an oxygen-free nitrogen stream, followed by at least 3 h under vacuum in a vacuum desiccator. Dried lipids were resuspended in 20 mM Hepes (pH 7.4), 150 mM KCl, 1 mM EDTA, and 0.1 mM TCEP buffer containing 3% (wt/vol) sodium cholate. Cholate was removed by size-exclusion chromatography using a self-packed column filled with 0.5 g of Sephadex G50 (Sigma–Aldrich) resuspended in 20 mM Hepes (pH 7.4), 150 mM KCl, 1 mM EDTA, and 0.1 mM TCEP.
Large unilamellar vesicles (LUVs) were prepared using the same lipid mix as described above for the SUVs. Dried phospholipids were resuspended in buffer [20 mM Hepes, 150 mM KCl, 0.1 mM TCEP (pH 7.4)] by vigorous vortexing. LUVs with a diameter of around ∼90 nm were prepared by extrusion of the rehydrated phospholipid suspensions with 21 strokes through 0.1-μm and 0.05-μm polycarbonate membranes (Millipore, Inc.) with a miniextruder (Avanti Polar Lipids). The homogeneity of liposome preparations was confirmed by dynamic light scattering measurements with a DynaPro machine (Wyatt Technology).
Sample Preparation for Fluorescence and Stopped-Flow Measurements.
Alexa Fluor 488 C5 maleimide (Thermo Fisher Scientific), iodoacetyl-N′-7-nitrobenz-2-oxa-1,3-diazol-4-yl (IANBD) amide (Thermo Fisher Scientific), and BADAN (Thermo Fisher Scientific) were used for specific labeling of the different single-cysteine mutants. Proteins were incubated with a fivefold molar excess of the dye overnight at 4 °C. Unreacted dye was removed from the labeled protein with a PD-10 desalting column (GE Healthcare) using 20 mM Hepes, 150 mM KCl, and 0.1 mM TCEP (pH 7.4).
NMR Backbone Assignment and Secondary Structure Analysis.
The 15N1H TROSY-heteronuclear single-quantum coherence (HSQC) spectra of 160 μM 2H15N13C-labeled syb-2 (1–96) dissolved in 20 mM MES (pH 6.1), 250 mM NaCl, 1 mM EDTA, and 0.1 mM TCEP were recorded on a 700-MHz Bruker Avance I spectrometer, equipped with a 5-mm room temperature probe. The experimental temperature was 5 °C to minimize adverse amide exchange effects. A 3-mm NMR tube was used to reduce adverse spectroscopic effects of higher salt concentrations. Spectral backbone resonance assignment was achieved using TROSY-based 3D backbone assignment experiments, including HNCO, HNCA, HN(CO)CA, HN(CA)CB, and HN(COCA)CB (43), implemented with gradient echo/anti-echo encoding for TROSY line selection and mixed constant-time evolution in the 15N dimension to allow for long evolution times and high resolution, very similar to the method described by Lakomek et al. (44). Spectra were analyzed, and backbone assignment was performed using the NMR Draw software package (45). Secondary chemical shifts were analyzed using TALOS+ software (46).
NMR Relaxation Experiments.
The 15N R1, R1ρ, and {1H}-15N NOE experiments using a TROSY detection scheme (47) were recorded on 160 μM 2H15N13C-labeled syb-2 (1–96) dissolved in 20 mM MES (pH 6.1), 250 mM NaCl, 1 mM EDTA, and 0.1 mM TCEP at 5 °C using a Bruker Avance III 600-MHz spectrometer equipped with a TCI cryogenic probe. Spectral dimensions were ppm 13.98 ppm, and the acquisition time was 72 ms in the dimension and 122 ms in the dimension. Relaxation delays were sampled for six different delay durations (15N R1 delays: 40, 800, 160, 640, 320, and 480 ms; 15N R1ρ delays: 5, 75, 15, 35, 55, and 45 ms) using a spin-lock radio frequency (RF) amplitude of 2 kHz for the R1ρ experiment. The 15N R2 rates were derived from R1ρ and R1 rates as described by Lakomek et al. (47). Hahn-echo–based R2 relaxation experiments were conducted at 600 MHz and 800 MHz using TROSY detection. Measurements were conducted at 5 °C, similar to the method described by Lakomek et al. (44). Relaxation data were analyzed using NMR Draw and dedicated scripts (45, 47). The fit error of the relaxation rate constants was estimated by Monte Carlo simulations using dedicated NMR Pipe scripts (45, 47).
NMR Studies of Syb-2 (1–96) and Syb-2 (1–116) Liposome Interactions.
The 15N1H TROSY-HSQC spectra on ∼20 μM 2H15N13C-labeled syb-2 (1–116) reconstituted in liposomes composed of 5:2:2:1 DOPC/DOPS/DOPE/Chol at a molar ratio [100:1 lipid/protein molar ratio; 20 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.1 mM TCEP] were recorded on a Bruker Avance I 800-MHz spectrometer equipped with a cryogenic probe; the experimental temperature was 5 °C. To study lipid interactions of the nontransmembrane part of syb-2, 20 μM 2H15N13C-labeled soluble syb-2 (1–96) [20 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.1 mM TCEP] was studied in the presence of the same liposomes (5:2:2:1 DOPC/DOPS/DOPE/Chol, 100:1 lipid/protein molar ratio). Spectral dimensions were ppm 15.02 ppm, and the acquisition time was 92 ms in the dimension and 85 ms in the dimension. Spectra were recorded with 64 transients per increment and an interscan (recovery) delay of 1 s, resulting in a total experimental time of 8.5 h. For reference purposes, 15N-1H TROSY-HSQC spectra were recorded on 200 μM unbound (free) 2H15N13C-labeled syb-2 (1–96) [20 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.1 mM TCEP] with 16 transients, resulting in a total experimental time of 2 h. For comparison of intensities (Fig. 3 and SI Appendix, Fig. S6), intensities of lipid-reconstituted 2H15N13C-labeled syb-2 (1–116) or 2H15N13C-labeled syb-2 (1–96) were scaled by a factor of 2.5 relative to the reference spectra of unbound 2H15N13C-labeled syb-2 (1–96) to account for the difference in molar concentration and number of transients recorded. Normalized intensity ratios of bound and unbound syb-2, , were calculated.
The degree of bound population was calculated from the decrease of resonance intensities in the presence of lipids, , compared with the intensities in the reference sample, , assuming a simple two-state model with the population of the bound state described by
PRE data for the different single-site cysteine mutants were also recorded in the presence of liposomes by repeating the PRE measurement as described above for ca. 20 μM syb-2 (1–96) Cys-1, S79C, and K91C in the presence of liposomes; the lipid-to-protein ratio was ∼100:1. For each increment, 64 scans were recorded, resulting in a total experimental time of 8 h for each experiment.
Fluorescence-Based Liposome Binding Assay.
IANBD amide and BADAN (Thermo Fisher Scientific) were used for specific labeling of the cysteine mutants. Proteins were incubated with a fivefold molar excess of the dye while gently shaking overnight at 4 °C. Unreacted dye was removed from the labeled protein with a PD-10 desalting column (GE Healthcare) using 20 mM Hepes, 150 mM KCl, and 0.1 mM TCEP (pH 7.4). To induce binding, 0.5 μM labeled protein was mixed with ∼500 μM total lipid in 20 mM Hepes, 150 mM KCl, and 0.1 mM TCEP (pH 7.4). The fluorescence emission was measured at 8 °C with a Fluorolog-3 spectrophotometer (model FL322; HORIBA Jobin Yvon) using a 5-nm slit width. The excitation wavelength for IANBD was set at 478 nm, and fluorescence emission was monitored from 500 to 640 nm. For BADAN, the excitation wavelength was set at 380 nm, and fluorescence emission was monitored from 420 to 600 nm. For endoproteinase Glu-C digestion (Sigma–Aldrich), 15 enzyme units were added and incubated for 1 h at room temperature.
Stopped-Flow Experiments.
Kinetic experiments were carried out with an Applied Photophysics SX.20 stopped-flow spectrophotometer at 8 °C. Alexa Fluor 488 (Thermo Fisher Scientific)-labeled protein (1 μM) was mixed with equal volumes of increasing concentrations of Texas Red-labeled LUVs (50:20:18:10:2 DOPC/DOPS/DOPE/Chol/Texas Red-phosphatidylethanolamine molar ratio), under pseudo–first-order conditions. The excitation wavelength was set on 495 nm, and a 610-nm cutoff filter was used to collect the Texas Red emission for the different vesicle concentrations tested. The resulting time courses were fit to a monoexponential function:
| [1] |
where F(t) equals the observed fluorescence at time t, F0 is the final fluorescence, Aobs represents the observed amplitude, and kobs is the observed rate constant. Observed rate constants were plotted as a function of total lipid concentration and fitted with the equation:
| [2] |
where kon represents the apparent association constant and koff represents the apparent dissociation rate constant.
The observed fluorescence change (ΔF) was plotted as a function of total lipid and fitted to the following Hill equation:
| [3] |
where ΔFmax represents the calculated maximum fluorescence change, n represents the apparent Hill constant, and Kd is the dissociation constant.
Liposome Coflotation Assay.
Protein (10 μM final concentration) was mixed with liposomes (3 mM final concentration) to reach a lipid/protein molar ratio of 300:1 and incubated for 10 min at room temperature. Then, a 50-μL sample was mixed with 50 μL of 80% (wt/vol) Nycodenz and overlaid with 50 μL of 30% (wt/vol) Nycodenz. Thirty microliters of 20 mM Hepes, 150 mM KCl, and 0.1 mM TCEP (pH 7.4) was added as a top layer. Samples were spun for 60 min at 55,000 rpm at 4 °C in an S55-S swinging bucket rotor in a Sorvall Discovery M150 SE analytical ultracentrifuge (Thermo Scientific). After centrifugation, 45-μL samples were taken from the gradient and analyzed by silver staining (Pierce Silver Stain Kit; Thermo Fisher Scientific). Liposomes appeared in the top layer of the Nycodenz gradient.
Supplementary Material
Acknowledgments
We are indebted to Ursula Welscher-Altschäffel, Sonja Pribicevic, and Delane Espinueva for protein purification assistance; Dr. Vytautas Gapsys for protein electrostatic representation assistance; and Prof. Marina V. Rodnina for access to stopped-flow facilities. We thank all members of the R.J. laboratory for helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft (DFG)/NIH Research Career Transition award program (Grant DFG LA 2724/1-1 to N.-A.L.), the European Union Marie-Curie program (Grant 627767 to N.-A.L.), NIH Grants P01 GM72694 and GB10440.155740 (to R.J.), and Max Planck Gesellschaft financial support (Grant PSBICH11200 to R.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813194116/-/DCSupplemental.
References
- 1.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 2008;27:2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fasshauer D, Otto H, Eliason WK, Jahn R, Brünger AT. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- 5.Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- 6.Hazzard J, Südhof TC, Rizo J. NMR analysis of the structure of synaptobrevin and of its interaction with syntaxin. J Biomol NMR. 1999;14:203–207. doi: 10.1023/a:1008382027065. [DOI] [PubMed] [Google Scholar]
- 7.Liang B, Kiessling V, Tamm LK. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proc Natl Acad Sci USA. 2013;110:19384–19389. doi: 10.1073/pnas.1314699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 10.Rizo J, Südhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices–Guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- 11.Kreutzberger AJ, Liang B, Kiessling V, Tamm LK. Assembly and comparison of plasma membrane SNARE acceptor complexes. Biophys J. 2016;110:2147–2150. doi: 10.1016/j.bpj.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zdanowicz R, et al. Complexin binding to membranes and acceptor t-SNAREs explains its clamping effect on fusion. Biophys J. 2017;113:1235–1250. doi: 10.1016/j.bpj.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawidowski D, Cafiso DS. Munc18-1 and the syntaxin-1 N terminus regulate open-closed states in a t-SNARE complex. Structure. 2016;24:392–400. doi: 10.1016/j.str.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakhanwal S, Lee CT, Urlaub H, Jahn R. An activated Q-SNARE/SM protein complex as a possible intermediate in SNARE assembly. EMBO J. 2017;36:1788–1802. doi: 10.15252/embj.201696270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellena JF, et al. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci USA. 2009;106:20306–20311. doi: 10.1073/pnas.0908317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang B, Dawidowski D, Ellena JF, Tamm LK, Cafiso DS. The SNARE motif of synaptobrevin exhibits an aqueous-interfacial partitioning that is modulated by membrane curvature. Biochemistry. 2014;53:1485–1494. doi: 10.1021/bi401638u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J, Pluhackova K, Böckmann RA. The multifaceted role of SNARE proteins in membrane fusion. Front Physiol. 2017;8:5. doi: 10.3389/fphys.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray DH, Tamm LK. Clustering of syntaxin-1A in model membranes is modulated by phosphatidylinositol 4,5-bisphosphate and cholesterol. Biochemistry. 2009;48:4617–4625. doi: 10.1021/bi9003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Bogaart G, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams D, Vicôgne J, Zaitseva I, McLaughlin S, Pessin JE. Evidence that electrostatic interactions between vesicle-associated membrane protein 2 and acidic phospholipids may modulate the fusion of transport vesicles with the plasma membrane. Mol Biol Cell. 2009;20:4910–4919. doi: 10.1091/mbc.E09-04-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kweon DH, Kim CS, Shin YK. Insertion of the membrane-proximal region of the neuronal SNARE coiled coil into the membrane. J Biol Chem. 2003;278:12367–12373. doi: 10.1074/jbc.M211123200. [DOI] [PubMed] [Google Scholar]
- 22.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Pluhackova K, Bruns D, Böckmann RA. Synaptobrevin transmembrane domain determines the structure and dynamics of the SNARE motif and the linker region. Biochim Biophys Acta. 2016;1858:855–865. doi: 10.1016/j.bbamem.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 24.Brewer KD, Li W, Horne BE, Rizo J. Reluctance to membrane binding enables accessibility of the synaptobrevin SNARE motif for SNARE complex formation. Proc Natl Acad Sci USA. 2011;108:12723–12728. doi: 10.1073/pnas.1105128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarzinger S, et al. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 26.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 27.Salvi N, Abyzov A, Blackledge M. Multi-timescale dynamics in intrinsically disordered proteins from NMR relaxation and molecular simulation. J Phys Chem Lett. 2016;7:2483–2489. doi: 10.1021/acs.jpclett.6b00885. [DOI] [PubMed] [Google Scholar]
- 28.Schneider R, et al. Visualizing the molecular recognition trajectory of an intrinsically disordered protein using multinuclear relaxation dispersion NMR. J Am Chem Soc. 2015;137:1220–1229. doi: 10.1021/ja511066q. [DOI] [PubMed] [Google Scholar]
- 29.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 32.Seven AB, Brewer KD, Shi L, Jiang QX, Rizo J. Prevalent mechanism of membrane bridging by synaptotagmin-1. Proc Natl Acad Sci USA. 2013;110:E3243–E3252. doi: 10.1073/pnas.1310327110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F, Tiwari N, Rothman JE, Pincet F. Kinetic barriers to SNAREpin assembly in the regulation of membrane docking/priming and fusion. Proc Natl Acad Sci USA. 2016;113:10536–10541. doi: 10.1073/pnas.1604000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risselada HJ, Kutzner C, Grubmüller H. Caught in the act: Visualization of SNARE-mediated fusion events in molecular detail. ChemBioChem. 2011;12:1049–1055. doi: 10.1002/cbic.201100020. [DOI] [PubMed] [Google Scholar]
- 36.Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25:429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- 37.Sitarska E, et al. Autoinhibition of Munc18-1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. eLife. 2017;6:e24278. doi: 10.7554/eLife.24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, et al. Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly. Nat Commun. 2019;10:69. doi: 10.1038/s41467-018-08028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Su L, Rizo J. Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry. 2010;49:1568–1576. doi: 10.1021/bi9021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker RW, et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114. doi: 10.1126/science.aac7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 42.Fasshauer D, Antonin W, Margittai M, Pabst S, Jahn R. Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J Biol Chem. 1999;274:15440–15446. doi: 10.1074/jbc.274.22.15440. [DOI] [PubMed] [Google Scholar]
- 43.Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc. 1999;121:844–848. [Google Scholar]
- 44.Lakomek NA, et al. Internal dynamics of the homotrimeric HIV-1 viral coat protein gp41 on multiple time scales. Angew Chem Int Ed Engl. 2013;52:3911–3915. doi: 10.1002/anie.201207266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lakomek NA, Ying J, Bax A. Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Jardetzky O. Investigation of the neighboring residue effects on protein chemical shifts. J Am Chem Soc. 2002;124:14075–14084. doi: 10.1021/ja026811f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.