Significance
The exclusion of immune cells from the tumor microenvironment has been associated with poor prognosis in the majority of cancers. We report that when considering 21 solid cancer types, immune cell exclusion is widely associated with the presence of a stem cell-like phenotype in tumors (“stemness”). Stemness positively correlates with higher intratumoral heterogeneity, possibly by protecting antigenic clones from elimination by the immune system. The activation of a stemness program appears to limit antitumor immune responses via tumor cell-intrinsic silencing of endogenous retrovirus expression, repression of type I interferon signaling, and up-regulation of immunosuppressive checkpoints. Our work suggests that targeting the stemness phenotype in cancer will promote T cell infiltration and render tumors more responsive to immune control.
Keywords: cancer stemness, antitumor immunity, intratumoral heterogeneity
Abstract
Regulatory programs that control the function of stem cells are active in cancer and confer properties that promote progression and therapy resistance. However, the impact of a stem cell-like tumor phenotype (“stemness”) on the immunological properties of cancer has not been systematically explored. Using gene-expression–based metrics, we evaluated the association of stemness with immune cell infiltration and genomic, transcriptomic, and clinical parameters across 21 solid cancers. We found pervasive negative associations between cancer stemness and anticancer immunity. This occurred despite high stemness cancers exhibiting increased mutation load, cancer-testis antigen expression, and intratumoral heterogeneity. Stemness was also strongly associated with cell-intrinsic suppression of endogenous retroviruses and type I IFN signaling, and increased expression of multiple therapeutically accessible immunosuppressive pathways. Thus, stemness is not only a fundamental process in cancer progression but may provide a mechanistic link between antigenicity, intratumoral heterogeneity, and immune suppression across cancers.
Tumor infiltration by T cells has been associated with improved clinical outcomes in a broad range of tumor types. Despite this, a large proportion of solid cancers appears nonpermissive to lymphocyte infiltration or nonimmunogenic (immunologically “cold”), and thus protected from cytolytic attack by lymphocytes, such as CD8+ T cells (1). The advent of immunotherapies, such as immune checkpoint inhibitors that rely on preexisting antitumor immune responses, has made an improved understanding of the mechanisms underlying the cold tumor phenotype essential.
Mounting evidence suggests that tumor cells can exhibit stem cell-like properties, ranging from characteristic gene-expression profiles to experimentally validated long-term self-renewal and repopulation capacities. The cancer stem cell (CSC) hypothesis posits that a subpopulation of tumor cells is capable of self-renewal and is responsible for the long-term maintenance of tumors (2). This hypothesis provides compelling explanations for clinical observations, such as therapeutic resistance, tumor dormancy, and metastasis (3). CSCs have been identified in a variety of human tumors, as assayed by their ability to initiate tumor growth in immunocompromised mice (4, 5). However, considerable controversy remains as to how best to define CSCs and the extent to which different tumor types exhibit a hierarchical organization. These controversies notwithstanding, there is increasing evidence that stem cell-associated molecular features, often referred to as “stemness,” are biologically important in cancer (6). It is unclear whether the stemness phenotype reflects the presence of bona fide CSCs in tumors or simply the coopting of stem cell-associated programs by non-CSC tumor cells (or both). Whatever the underlying mechanism may be, stemness has emerged as an important phenomenon due to its strong association with poor outcomes in a wide variety of cancers (7, 8). Moreover, stemness appears to be a convergent phenotype in cancer evolution (9, 10), suggesting it is a fundamentally important property of malignancy.
The evolution of transformed cells in the tumor microenvironment is shaped by diverse selective pressures, including the host immune response. Experimental work has shown that embryonic, mesenchymal, and induced pluripotent stem cells possess immune modulatory properties, while resistance to immune-mediated destruction has also recently been shown to be an intrinsic property of quiescent adult tissue stem cells (11) and CSCs (12). Similarly, immune selection has been shown to drive tumor evolution toward a stemness phenotype that inhibits cytotoxic T cell responses (13). Moreover, a recent analysis of The Cancer Genome Atlas (TCGA) revealed negative associations between stemness and some metrics of tumor leukocyte infiltration (14). Finally, CSCs have been proposed as a driver of intratumoral heterogeneity (6, 9). Consistent with this, we (15) and others (16) have reported negative associations between immune cell infiltration and intratumoral heterogeneity.
Motivated by these observations, we hypothesized that the stemness phenotype of cancer cells may confer immunosuppressive properties on tumors, resulting in immunologically cold microenvironments that both foster and maintain intratumoral heterogeneity. To address this, we performed an integrated analysis of stemness, immune response, and intratumoral heterogeneity across cancers. We recover pervasive negative associations between antitumor immunity and stemness, and strong positive associations between stemness and intratumoral heterogeneity. We further find that cancer cell lines with high stemness have cell-intrinsic immunosuppressive features, suggesting that immunologically cold microenvironments can arise due to the presence of high-stemness cancer cells. We propose that cancer stemness provides a link between tumor antigenicity, intratumoral heterogeneity, immune suppression, and the resulting evolutionary trajectories in human cancer.
Results
Derivation and Comparison of Stemness Signatures.
Recent studies have provided evidence that cancer stemness can be represented by core gene-expression programs across diverse cancer types (8, 10, 14, 17, 18). Building on this prior work, we inferred tumor stemness from cancer transcriptomes using single-sample gene set enrichment analysis (ssGSEA) with a modified version of a gene set developed by Palmer et al. (17) to measure the level of plasticity and differentiation of mesenchymal stem cells, pluripotent stem cells, terminally differentiated tissues, and human tumors across >3,200 microarray samples. Intriguingly, the authors of this gene set identified a cluster of “immune” genes that negatively loaded the principal components they used to infer stemness; however, they did not further explore this relationship. To adopt this gene set for use in ssGSEA and avoid biasing our analysis toward recovering negative associations between stemness and immunity, we omitted this immune gene cluster from our signature. We also omitted cell proliferation markers to avoid recovering a signature of proliferation rather than stemness (19).
We validated the performance of the resulting 109 gene signatures (Dataset S1A) on diverse datasets, finding that it recapitulated the expected degree of stemness in both malignant and nonmalignant cell populations (SI Appendix, Fig. S1). We further validated this signature using the stem cell-based validation dataset used by Malta et al. (14) in their recent pan-cancer stemness analysis (GSE30652), which revealed similarly high classification accuracy for our signature compared with theirs [multiclass area under the curve (AUC) 0.92 vs. 0.91, respectively] (SI Appendix, Fig. S2 A and B). We then analyzed RNA sequencing data from 8,290 primary cancers representing 21 solid cancer types (from TCGA) and found our signature showed good concordance with that of Malta et al. and two other recently published signatures (Spearman’s ρ = 0.43, 0.74, and 0.66, respectively) (SI Appendix, Fig. S2 C–F).
Stemness Varies Across Cancers and Predicts Patient Survival.
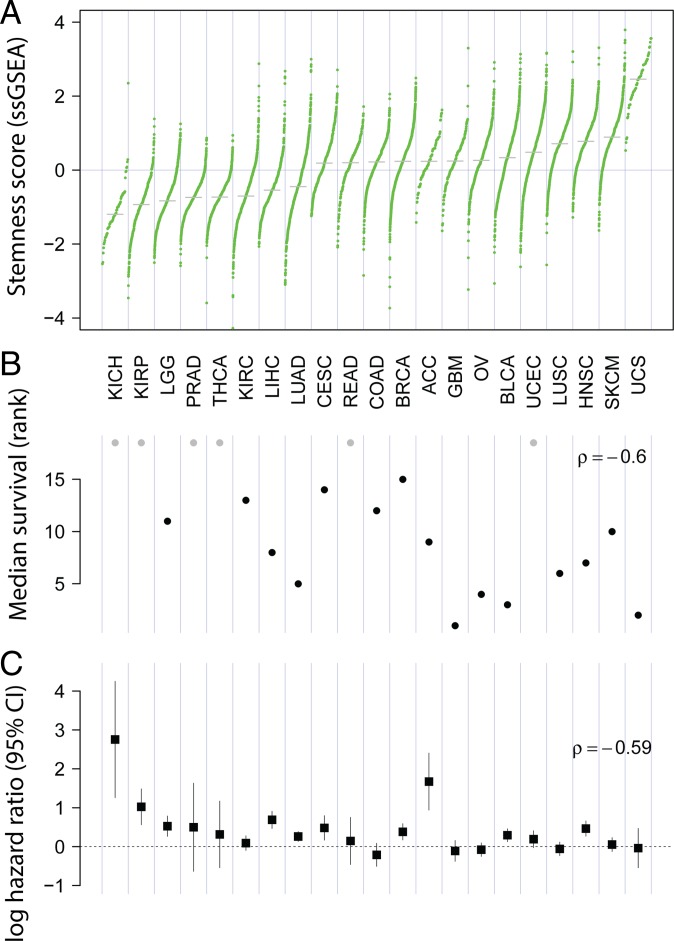
Using our signature, we found that stemness varied strongly across TCGA samples, with cancer type explaining 54% of the variation (ANOVA; adjusted R2) (Fig. 1A). Consistent with prior reports of stemness being a negative prognostic factor (7, 8), we found a strong negative relationship between median stemness and median overall survival across cancer types (Fig. 1B) (ρ = −0.60; P = 0.004; n = 21 cancers). Within cancer types, Cox regression likewise showed stemness to be significantly negatively prognostic for overall survival in the majority of cancers (Cox proportional hazards; P < 0.05), and significantly positively prognostic for none (Fig. 1C), underscoring the relevance of this signature both within and across cancers. We also noted a significant decrease in the magnitude of the hazard associated with stemness within cancers as median stemness increased (ρ = −0.59; P < 0.01) (Fig. 1C), pointing to a potential threshold effect with a saturating hazard in cancers with higher average stemness. For reference, we compared the prognostic association of our ssGSEA-based stemness with the mRNAsi signature of Malta et al. (14) derived using one-class logistic regression (OCLR), for which positive associations in some cancers were reported. Using pan-cancer Cox regressions stratified by cancer, we found ssGSEA-based stemness to be substantially more predictive of survival in this modeling framework [log hazard ratio = 0.23 ± 0.03 (coefficient ± SE); P < 10−15 vs. not significant for mRNA stemness index (mRNAsi)], demonstrating this signature uncovers the negative outcomes associated with high-stemness cancers expected from previous reports (7, 8). This relationship held when controlling for tumor purity, demonstrating that a relationship between tumor purity and patient survival is not a confounding factor (and see below).
Fig. 1.
Stemness and survival across cancers. (A) Stemness score varies widely across 21 solid cancers from TCGA. Each point represents an individual case, and cancer types are ordered by median stemness score (z-scored ssGSEA). (B) Median survival decreases with increasing median stemness (P = 0.004). Gray points represent cancers in which median overall survival times were not evaluable. (C) Stemness associates with poor outcome within cancers. Log hazard ratio (±95% CI) for the association of stemness with overall survival is shown. Hazard decreases with increasing average stemness of cancers (P = 0.008). Cox models control for patient age and tumor purity. Cancer acronyms are used as defined by TCGA (https://portal.gdc.cancer.gov).
Stemness Negatively Associates with Immune Cell Infiltration Across Solid Cancers.
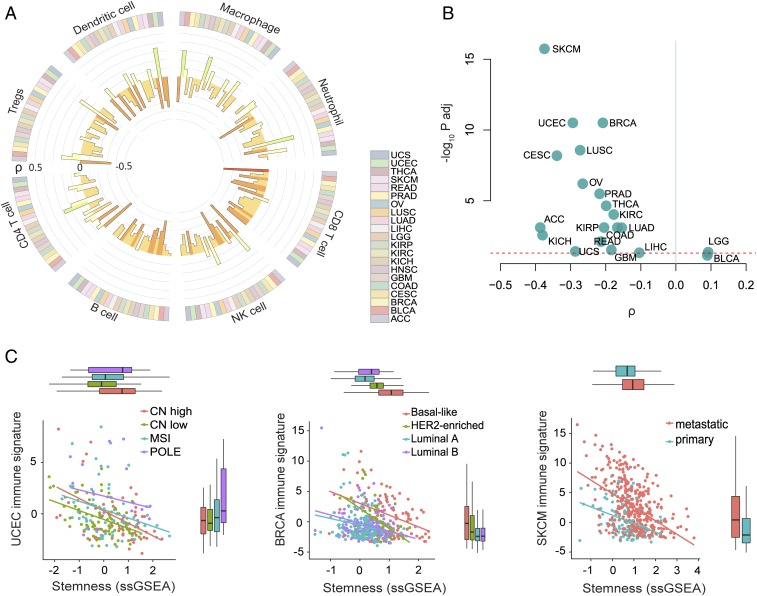
To evaluate the relationship between stemness and antitumor immunity, we generated signatures of predicted immune cell infiltration for each patient sample using xCell, an ssGSEA-based tool that infers cellular content in the tumor microenvironment (20). CD8+ T cells, which have a well-established association with favorable prognosis in a majority of solid cancers (21), showed a clear negative association with stemness for most cancers (Fig. 2A). We also considered other cell types important for antitumor immunity, including NK cells and B cells, and again observed recurrent negative associations with stemness (Fig. 2A). Additional cell types, such as CD4+ T cells, Tregs, and neutrophils, showed more variable associations with stemness, indicating this relationship does not apply to all infiltrating immune cell populations in all cancers (Fig. 2A).
Fig. 2.
Stemness negatively associates with immune cell signatures. (A) Circos plot showing the association between stemness score and the presence of 8 inferred immune cell types across 21 cancer types (colored bars in outer ring). The color and height of the inner bars represent the Spearman correlation ρ values for each cell type and cancer type. (B) Volcano plot showing the association between stemness score and immune signature (sum of z-scored signatures of CD8+ T cells, NK cells, and B cells) for each cancer. The dashed line indicates Padj = 0.05. (C) Association between stemness score and immune signature in the different molecular subtypes of endometrial (UCEC) and breast (BRCA) cancer, and within primary and metastatic melanoma (SKCM) samples (Padj < 10−7). Each point represents one case. Colors indicate the different molecular subtypes of UCEC and BRCA, or sample types for SKCM. CN, copy number; MSI, microsatellite instable; POLE, polymerase epsilon.
To generate a robust single score for antitumor immunity (hereafter referred to as “immune signature”), we aggregated xCell predictions for CD8+ T cell, NK cell, and B cell infiltration, reasoning that these cells represent important anticancer effector cells across diverse cancers (22, 23). Indeed, this immune signature was significantly associated with increased survival in the majority of cancers (SI Appendix, Fig. S3A; overall survival curves for all patients stratified by median stemness and immune signature are shown in SI Appendix, Fig. S3B). A notable exception was kidney cancer (i.e., KIRP), where a negative association was observed, in accord with prior reports for this malignancy (24). Consistent with our analyses using single immune cell-type scores (Fig. 2A), the immune signature showed a negative association with stemness within nearly all cancers (Fig. 2B). This negative association was also recovered using other published stemness gene sets (18, 19, 25) (Dataset S1B), most clearly with stemness scores reflecting NANOG, SOX2, and MYC signaling, and to a lesser extent with those reflecting embryonic stem cell programs (SI Appendix, Fig. S4). Furthermore, when we used CIBERSORT infiltration scores (26) in place of our immune signature, we found that increased stemness was associated with strong polarization of infiltrating leukocyte populations toward a macrophage-dominated, CD8+ T cell-depleted composition for most cancers (linear model controlling for cancer type; P < 10−15 for both cell fractions).
Whereas the preceding analyses were performed within cancer types, we also evaluated the relationship between stemness and immune signature across cancer types. Unexpectedly, we found no significant association between median immune signature and median stemness score across cancers (P = 0.82). This suggests that factors other than stemness control the differences in immune cell infiltration across cancer types, while the association with stemness applies within individual cancer types. It also demonstrates that our stemness metric is not recovering negative associations with immunity simply due to the lower tumor purity that is inextricably associated with the presence of infiltrating immune cells.
We next examined whether the negative association between immune signature and stemness was influenced by tumor subtype or stage. Here, we focused on breast and endometrial cancers, which have well-characterized subtypes with strong prognostic associations, and melanoma, for which both primary and metastatic samples are available within TCGA. In breast cancer, stemness varied markedly across subtypes (ANOVA; adjusted R2 = 0.26), with the basal subtype having the highest stemness, as expected (19), and the luminal-A subtype the lowest (Fig. 2C). In endometrial cancer, the highest stemness score was observed in high-copy number (CN) alteration (CN-high) and polymerase-epsilon mutant tumors, and the lowest stemness score was seen in low-CN alteration tumors (CN-low) [Tukey honest significant difference (HSD); P = 0.003; CN-high vs. CN-low tumors]. Finally, we observed a substantially higher stemness score in metastatic compared with primary melanoma lesions (Fig. 2C). In all these cancers, we observed recurrent negative associations between stemness and immune signature which remained significant when controlling for cancer subtype and tumor purity (see below; linear models; P < 10−7).
To investigate in an unbiased manner whether processes apart from antitumor immunity negatively correlate with stemness, we conducted differential expression tests to identify gene-expression patterns associated with the lowest versus highest stemness quintiles (<20th versus >80th percentiles) for each analyzed cancer type. Even with this unbiased approach, nearly all of the pathways recurrently enriched in low-stemness samples within a cancer were immune-related (SI Appendix, Fig. S5). Recognizing that the presence of nonmalignant cells can confound expression analyses of bulk-sequenced tumor samples by diluting tumor-specific expression signatures, we performed additional analyses to control for such effects. First, using recently published estimates of purity across TCGA (27), we found that the association between tumor purity and stemness was negligible and failed to reach significance in a pan-cancer linear model controlling for cancer type (P = 0.23). Second, we refit the differential expression models described above to control for tumor purity and repeated the pathway enrichment analyses, still finding that immune-associated pathways were enriched in low-stemness tumors (SI Appendix, Fig. S6).
To evaluate the relative contributions of malignant versus nonmalignant cells (e.g., stromal cells) to the stemness score, we applied our stemness metric to a single-cell RNA sequencing (scRNA-seq) dataset of lung cancer (28), which comprises a comprehensive inventory of different cell types from the tumor microenvironment (>52,000 cells from five patients). The average stemness score was much higher in cancer cells than any other cell type, including fibroblasts, myeloid cells, or lymphocytes (SI Appendix, Fig. S7) (Tukey HSD; P < 10−15), strongly supporting the notion that the stemness signature largely emanates from cancer cells rather than stromal or other cell types in the tumor microenvironment.
Stemness Associates with Immunologically Cold Cancers Measured via Immunohistochemistry.
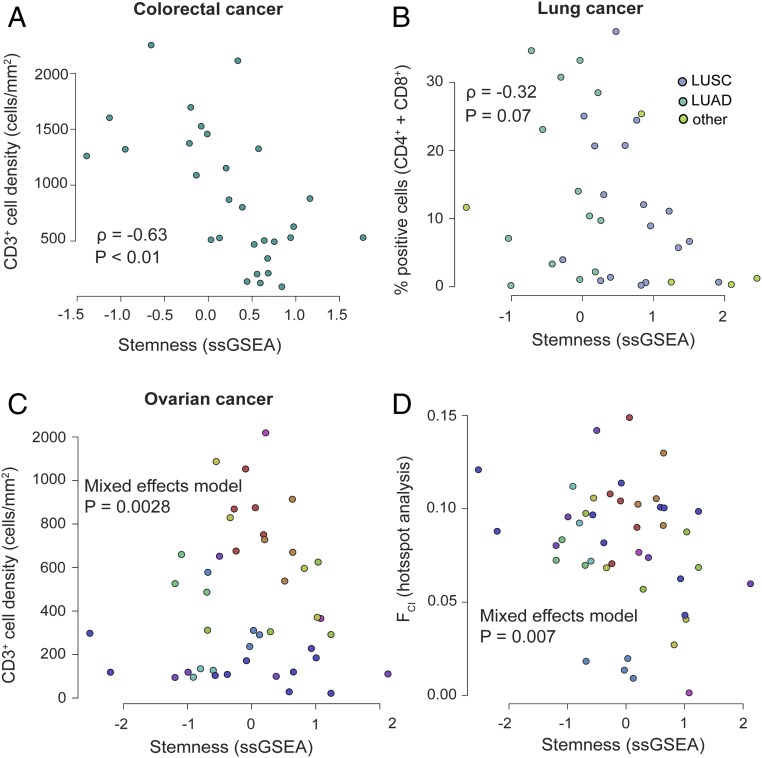
To confirm the negative association between stemness and lymphocyte infiltration, we turned to three patient cohorts with matched immunohistochemistry (IHC)-based T cell infiltration scores and gene-expression data suitable for computing a stemness score. Using a cohort of 33 colorectal cancer patients (29), we found a strong negative association between stemness and total infiltrating CD3+ T cells (ρ = −0.63; P < 0.001) (Fig. 3A). Furthermore, in this cohort the xCell-based immune signature was strongly correlated with infiltrating CD3+ cells (ρ = 0.69; P < 0.001), supporting its fidelity for measuring immune-cell infiltration. With this validation in hand, we compared the xCell-based immune signature and stemness for the total patient cohort with available microarray data (n = 585), and again observed a clear negative correlation (ρ = −0.22; P < 10−7).
Fig. 3.
Relationship between stemness and immune cell infiltrates in different cancer cohorts scored via IHC. (A) Stemness score negatively associates with tumor-infiltrating CD3+ T cells in colorectal cancer (P < 0.01; n = 33; data from ref. 29). Each point represents one patient sample. (B) Stemness negatively associates with tumor-infiltrating T cells (in this case the sum of CD4+ and CD8+ cells) in lung cancer (P = 0.07; n = 35; data from ref. 30). Colors denote adeno versus squamous cell lung cancers. (C) Stemness is significantly associated with tumor-infiltrating CD3+ T cells in a multisite dataset of high-grade ovarian cancer (P = 0.028; n = 44 samples from 12 patients; data from ref. 15). Colors represent individual patients. (D) By hotspot analysis, the fraction of tissue area occupied by colocalizing tumor and immune cells (FCI) is negatively associated with stemness in a multisite dataset from high grade ovarian cancer (P < 0.01; 44 samples from 12 patients; data from ref. 15). Colors represent individual patients.
We next evaluated this relationship in a cohort of 35 lung cancer patients with matched RNA-sequencing and IHC-based quantitation of immune cell infiltrates (30). For consistency with the above analysis, we calculated the infiltration of T cells by summing previously calculated CD4+ and CD8+ cell fractions. Although we observed a negative association between the stemness score and the percent of infiltrating T cells, this did not reach statistical significance (ρ = −0.32; P = 0.07) (Fig. 3B). Nonetheless, the additional RNA-seq data in this cohort revealed a clear negative association between the stemness and immune signature at the transcriptional level (n = 199 tumor samples; ρ = −0.19; P = 0.007).
Finally, we evaluated a small cohort of high-grade serous ovarian cancer (HGSC) cases (15) for which matched IHC-based T cell counts and microarray-based gene-expression data were available for multiple tumor sites within each patient (n = 44 samples from 12 patients). Consistent with the above findings, we found a negative association between stemness and total CD3+ T cells (negative binomial mixed effects model; P = 0.0028) (Fig. 3C). In addition, we previously subjected samples from this cohort to Getis-Ord Gi* “hotspot” analysis to quantify immune cell engagement with tumor cells (15). Intriguingly, all hotspot metrics showed a clear negative association with stemness (mixed-effects models; P < 0.01; 44 samples from 12 patients) (Fig. 3D, FCI shown) (31), suggesting that stemness negatively influences lymphocyte engagement with tumor cells.
Stemness Associates with Intratumoral Heterogeneity.
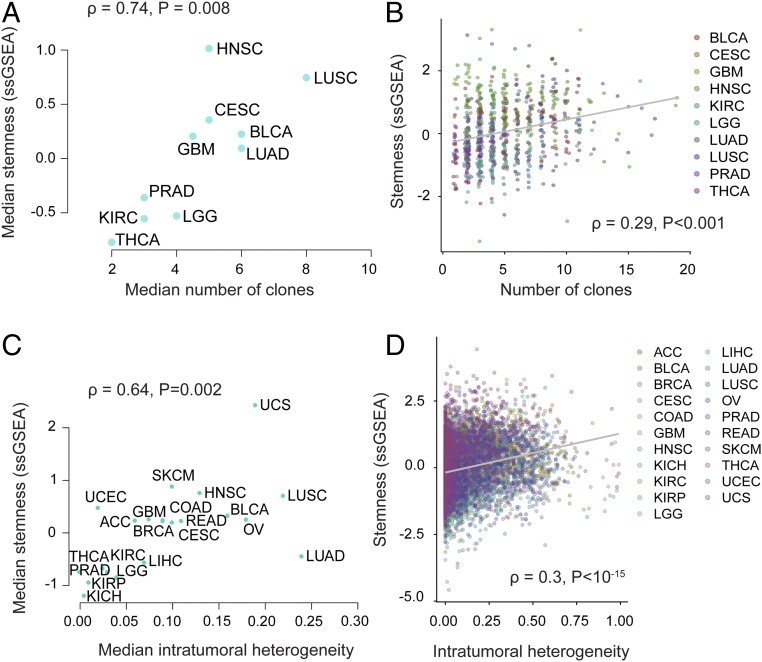
Stemness has been proposed to foster tumor clone diversity, with the replicative potential of CSCs enabling greater tumor heterogeneity (6, 9). Our hypothesis suggests that stemness could additionally promote intratumoral heterogeneity by inhibiting immune selection against new cancer clones. These predictions have not, to our knowledge, been systematically tested within or across cancers. Therefore, we compared stemness and intratumoral heterogeneity using data from two recent TCGA studies (26, 32). Using data from the first study (32), we found a dramatic positive correlation between median stemness and median number of clones in a cancer (n = 935 patients across 11 cancers; ρ = 0.75; P = 0.008) (Fig. 4A). Furthermore, we found a positive association between stemness and tumor clone count in a linear model controlling for cancer site and tumor purity, indicating that these associations are discernible both across and within cancers (P = 0.0002) (Fig. 4B). Using pan-cancer predictions of intratumoral heterogeneity from a second, larger TCGA-based study (26), we recovered a similarly strong association (ρ = 0.64; P = 0.002; n = 6,791 samples across 21 cancers) (Fig. 4C), which was again significant across all samples when controlling for cancer site and tumor purity in a linear model (P < 10−15) (Fig. 4D).
Fig. 4.
Stemness associates with intratumoral heterogeneity within and across cancers. (A) Median stemness and median clonality [inferred by Andor et al. (32)] are strongly correlated across cancers (n = 11; P = 0.008). (B) Stemness score and clonality [inferred by Andor et al. (32)] are correlated across patients while controlling for cancer type (n = 935; P = 0.0002). Colored points represent different tumor sites. (C) Median stemness score and median intratumoral heterogeneity score [inferred by Thorsson et al. (26)] are strongly correlated across cancers (n = 20; P = 0.002). (D) Stemness score and intratumoral heterogeneity [inferred by Thorsson et al. (26)] are correlated across patients while controlling for cancer type (n = 6,791; P < 10−15). Colored points represent different tumor sites. Spearman ρ values are shown.
Potential Mechanisms and Consequences of Stemness-Associated Immunosuppression.
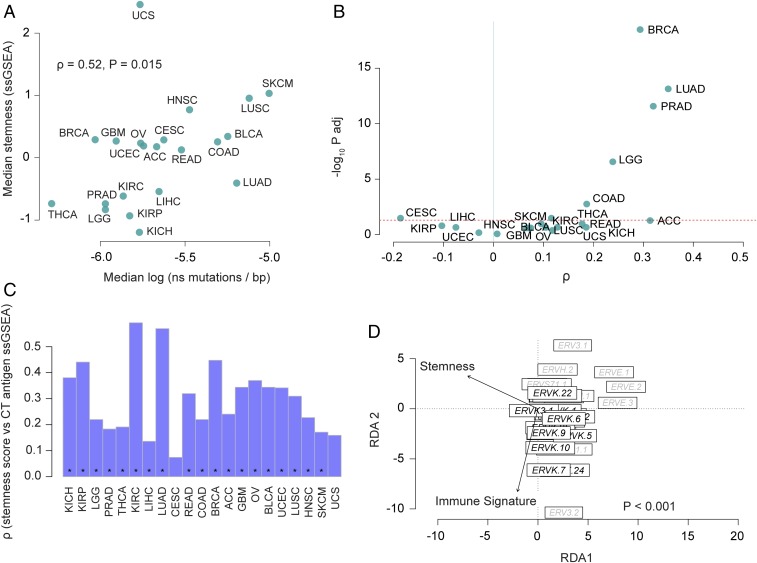
We evaluated the association between stemness and several known genetic and environmental factors that affect antitumor immunity. Antitumor immunity involves T cell recognition of neo-antigens arising from somatic mutations (33). Therefore, we examined the association between stemness, immune signature, and nonsynonymous mutation load, analyzing TCGA samples with available mutation calls (n = 6,682). While median mutation load correlated with median immune signature across cancers (ρ = 0.49; P = 0.02, n = 21), there was generally little correlation within cancers, as has been reported in other TCGA-based analyses (34) (SI Appendix, Fig. S8A). Across cancers, median mutation load showed a positive association with median stemness (ρ = 0.52; P = 0.015, n = 21) (Fig. 5A). Similarly, within cancers, we generally found positive associations between mutation load and stemness (Fig. 5B). We recovered qualitatively similar but slightly stronger associations between stemness and neoantigen load computed with NetMHCpan in the above pan-cancer analysis (26) (SI Appendix, Fig. S8 B and C).
Fig. 5.
Mutation load, CT antigen expression and ERV associations with stemness. (A) Median stemness and median mutation load are positively correlated across cancers (n = 21; P = 0.015). Mutation load is represented as log-transformed nonsynonymous mutations per base (log10 ns mutations per base pair). (B) Volcano plot reveals stemness score and mutation load correlate within some cancers (upper right quadrant). The x axis represents Spearman correlation ρ values, and the y axis represents −log10-adjusted P values (Padj). Dashed red line indicates the significance threshold, Padj value = 0.05. (C) Stemness score and CT antigen expression (ssGSEA of CT antigen gene set) positively correlate in most cancers. Bar plots show the Spearman ρ values for each cancer type, and asterisks denote Padj < 0.05. (D) Redundancy analysis triplot reveals stemness negatively associates with multivariate ERV expression (P < 0.001; 33 ERVs evaluated in 4,252 samples, analysis conditioned by cancer type).
Cancer-testis (CT) antigens are tumor antigens that normally are expressed in gametogenic tissue but become aberrantly expressed in a broad range of malignancies, often leading to an immune response (35). Using a set of 201 CT genes curated by the CTdatabase (Dataset S2) (36), we generated an ssGSEA CT antigen score and found strong positive associations with stemness within cancers (Fig. 5C). Accordingly, there was a generally negative association between CT antigen score and the immune signature, which was significant in 8 of 21 cancers (Padj < 0.05) (SI Appendix, Fig. S9). Thus, like neoantigens, CT antigens show a positive association with stemness and a negative association with immune signature.
Normal stem cells have been shown to suppress endogenous retrovirus (ERV) expression, presumably to prevent insertional mutagenesis in long-lived stem cell lineages (37). Conversely, ERV expression can be activated in cancer cells (38, 39), where it can potentially elicit antitumor immune responses by activating viral defense mechanisms and the type I IFN response (40) or by yielding immunogenic foreign epitopes (41). Despite these possibilities, associations between immunity and ERV expression were inconsistently observed in a recent pan-cancer analysis (42). To better understand this relationship, we investigated interactions between stemness, immune signature, and ERV expression. Because of the repetitive nature of ERVs, we used ERV-specific read-mappings (42) to evaluate ERV expression in 4,252 TCGA samples that overlapped with our stemness and immune signature analysis. Using redundancy analysis (a constrained extension of principal components analysis), we found that multivariate ERV expression was not clearly associated with the immune signature, consistent with prior reports (42); members of the ERVK family were an exception, showing moderate positive associations with immune signature (Fig. 5D). In contrast, ERV expression showed a pervasive negative association with stemness (P < 0.001) (Fig. 5D), consistent with the notion that suppression of ERV expression is a feature of the stem cell phenotype (43). Thus, both immune signature and ERV expression are negatively associated with stemness, but these appear to be largely orthogonal relationships.
Tumor Cell Intrinsic Mechanisms of Stemness-Mediated Immunosuppression.
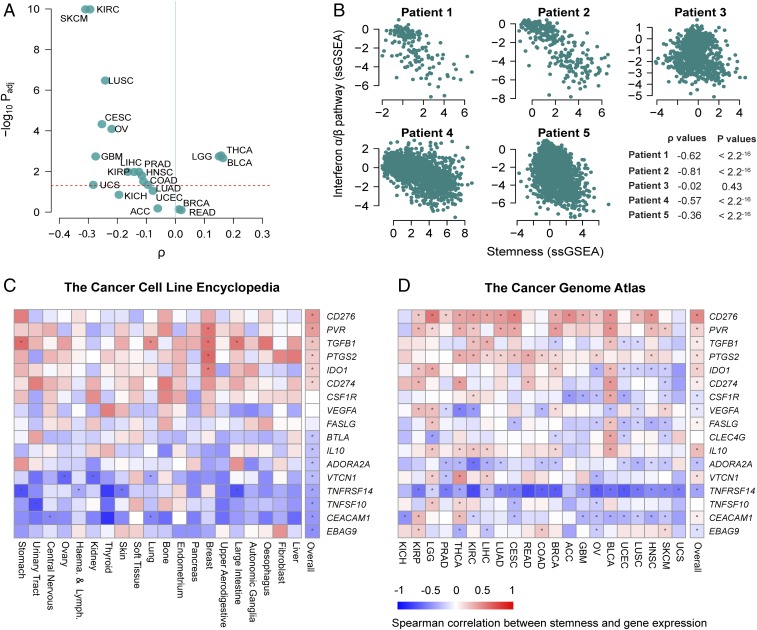
To address whether the negative association between stemness and immune signature is attributable to cancer cell-intrinsic processes, we calculated stemness scores for 1,048 cancer cell lines using gene-expression data from the Cancer Cell Line Encyclopedia (CCLE) (44), as well as the cancer cell fraction from the scRNA-seq study of lung cancer, mentioned above (28). We first assessed associations between stemness scores and the expression of 11 mapped ERVs in CCLE transcriptomes and found that three of three ERVs with nonnegligible expression levels across cell lines were significantly negatively associated with stemness, two of which remained significant when controlling for tissue of origin (linear models; Padj < 0.05) (SI Appendix, Fig. S10). We next generated an ssGSEA score for type I IFN signaling (reactome IFN α/β pathway gene set) and generally observed negative correlations with stemness within cancers in the TCGA dataset (Fig. 6A). When evaluating this association in the CCLE dataset, we observed a clear negative relationship between the IFN signature and stemness (ρ = −0.22. P < 10−10) (SI Appendix, Fig. S11A), which remained significant when controlling for tissue of origin of the cell line (linear model; P < 0.001) and when omitting cell lines derived from hematopoietic lineages (linear model; P = 0.02).
Fig. 6.
Cell-intrinsic stemness score associates with decreased type I IFN signaling and increased expression of CD276 and PVR. (A) Volcano plot reveals that stemness score and type I IFN signaling (reactome IFN α/β pathway ssGSEA) are negatively correlated in most cancers (upper left quadrant). The x axis represents Spearman correlation ρ values, and the y axis represents −log10 adjusted P values (Padj). Dashed red line indicates the significance threshold, Padj = 0.05. (B) Stemness score is negatively associated with type I IFN signaling (reactome IFN α/β pathway ssGSEA) in tumor cells from four of five lung cancer patients, based on scRNA-seq data from ref. 28. Each point represents a single tumor cell. (C and D) Heatmaps showing Spearman correlations for stemness and select immunosuppressive genes based on data from the CCLE (C) and TCGA (D). Spearman correlations were calculated within tissues represented in the CCLE by more than 10 cell lines. Genes are ranked according to the final column (“overall”), which represents the correlation across all samples, irrespective of cell line or tumor type. Red-blue intensities reflect the correlation ρ values. Asterisks denote Benjamini–Hochberg-corrected significant associations (Padj < 0.05).
Analysis of the aforementioned lung cancer scRNA-seq dataset (28) also revealed a striking negative association between cancer cell-intrinsic stemness and IFN α/β signaling in four of five patients (Fig. 6B). To further test this association in nonneoplastic lineages, we took advantage of the stem cell gene-expression dataset previously used to validate stemness signatures (GSE30652), which likewise showed a striking negative association between stemness and type I IFN signaling (ρ = −0.81; P < 10−15) (SI Appendix, Fig. S11B).
Finally, to examine other cell-intrinsic mechanisms of immunosuppression, we analyzed a curated list of immunosuppressive genes previously reported to be expressed in human cancer cells. Using both the CCLE and pan-cancer TCGA datasets, the expression of each of these genes was assessed in relation to our stemness signature (Fig. 6 C and D). This revealed positive associations between stemness and a number of immunosuppressive genes, including CD276 [B7-H3, shown to inhibit T cell activation and autoimmunity (45)], PVR [CD155, a member of the B7/CD28 superfamily, shown to exhibit potent inhibitory action in different subsets of immune cells (46)], and TGFB1 [a key player in the induction of immunological tolerance (47)]. Thus, the stemness phenotype is associated with expression of several gene products that could potentially serve as targets for immune modulation.
Discussion
Although cancer stemness, antitumor immunity, and intratumoral heterogeneity have all emerged as important features of cancer in recent years, their covariation across cancers has not been systematically investigated. Here we report that stemness is associated with suppressed immune response, higher intratumoral heterogeneity, and dramatically worse outcome for the majority of cancers. Although correlative analyses such as ours do not reveal causality, we propose that the stemness phenotype found in cancer cells, similar to that in normal stem cells, involves the expression of immunosuppressive factors that engender the formation of immune-privileged microenvironments in which tumor clone diversification can occur. The resulting heterogeneity may provide a substrate for the selection of treatment-resistant clones, resulting in inferior clinical outcomes. Thus, our findings implicate stemness as a shared therapeutic target to achieve the dual objectives of constraining tumor evolution and enhancing antitumor immunity.
A recent pan-cancer analysis reported inconsistent relationships between cancer stemness and immunity, recovering negative relationships between stemness and tumor-infiltrating lymphocytes for some cancers and positive relationships for others (14). While this work provided a valuable perspective on stemness across cancers, our results differ in that we recover much stronger and more pervasive negative relationships with immune infiltration and survival. In contrast to our ssGSEA approach, the OCLR approach used above was validated on a cohort that lacked any malignant samples. Moreover, when we reproduced their analysis, we found many components of their stemness score (i.e., positive OCLR model weights) were immunologically relevant genes; for example, among the 50 most positive gene weights were IDO1, LCK, KLRG2, PSMB9 (a component of the immunoproteasome), and multiple TNF-receptors. With the OCLR approach, higher expression of such immune genes contributes positively to the stemness score, precluding an unbiased assessment of the relationship between stemness and tumor immunity. As mentioned, it is also possible that an immune cell signature in the tumor microenvironment could negatively correlate with transcriptional signatures from cancer cells simply through dilution of cancer-specific transcripts by infiltrating immune cells. We took numerous precautions to ensure this was not driving our results. These included controlling for tumor purity in linear models throughout the analyses, showing a nonsignificant relationship between stemness and tumor purity, and identifying a negative relationship between stemness and cell-intrinsic IFN signaling in individual lung cancer cells (by scRNA-seq; Fig. 6B) and in cancer cell line transcriptomes (SI Appendix, Fig. S11A).
A contribution of cancer stemness to intratumoral heterogeneity has been postulated for some time (6, 9), but direct evidence has been lacking. We recovered a dramatic positive association between stemness and multiple metrics of intratumoral heterogeneity across cancers (Fig. 4), which is especially noteworthy given that these metrics were derived from different data types (i.e., mRNA vs. DNA). Given recent work from our group and others linking increased intratumoral heterogeneity with decreased immune cell infiltration (15, 16), one could speculate that stemness might contribute to intratumoral heterogeneity by both increasing the replicative capacities of individual tumor clones and by shielding antigenic clones from elimination by the immune system.
We found generally positive associations between stemness and mutation load within cancers (Fig. 5B), and clear evidence of this across cancers (Fig. 5A), consistent with studies demonstrating accumulation of mutations in normal adult stem cells (48). We also found strong positive associations between stemness and CT antigen expression (Fig. 5C), which is consistent with reports of CT antigen expression in mesenchymal (49), embryonic stem cells (50), and CSCs (51). It is also consistent with prior studies (42) and the present work finding nonsignificant or negative correlations between immune infiltration and CT antigen expression (Fig. 5D). Thus, the negative association between stemness and immune response is not readily attributable to low neoantigen or CT antigen load, strongly implicating the involvement of other mechanisms.
ERVs, which constitute ∼8% of the human genome, are known to be suppressed in pluripotent and embryonic stem cells (43) yet activated in human cancer (38, 39), leading us to ask which behavior would predominate in high stemness cancers. We found a strong negative association between stemness and ERV expression in both TCGA (Fig. 5D) and cancer cell line data (SI Appendix, Fig. S10), indicating that the stemness phenotype in human cancer retains this property of normal stem cells.
We identified a negative correlation between stemness and type I IFN signaling in the TCGA (Fig. 6A), CCLE (SI Appendix, Fig. S11A), and scRNA-seq datasets (Fig. 6B). Whether ERV suppression underlies the low intrinsic IFN signaling in high stemness cancer cells awaits experimental investigation. In support of our results, an attenuated innate immune response is a major characteristic of embryonic stem cells (52), and we find clear confirmation of this in nonneoplastic stem cell transcriptomes (SI Appendix, Fig. S11B). Collectively, these data suggest that activation of a stemness program in tumors could limit antitumor immune responses by silencing ERVs and repressing type I IFN signaling in a cell-intrinsic manner (53).
Using CCLE data, we found a clear association between stemness and the expression of several immunosuppressive genes, including CD276, PVR, and TGFB1 (Fig. 6C). These associations are especially intriguing given that CCLE-based gene expression profiles are independent of any ongoing influence of the immune system. CD276, a B7 family ligand, is now being clinically targeted due to expression on both cancer cells and tumor-infiltrating blood vessels (54). Intriguingly, CD276 is coexpressed with CD133, a marker that distinguishes cell populations enriched for CSCs in colorectal cancer (55). PVR is a key ligand in an emerging checkpoint pathway involving TIGIT, an inhibitory receptor expressed on T cells and other immune cells (46). Although no association between stem cells and PVR has been described so far in mammalian stem cells, expression of PVR can be activated by sonic hedgehog signaling (56), a pathway essential for self-renewal and cell fate determination in normal and CSCs (57). TGFB1 and other TGFB family members have well-documented roles in development (58) and CSC proliferation and maintenance (59, 60).
Recent work has demonstrated that tumor-intrinsic oncogenic signaling pathways have immune suppressive properties (61), but this too can be understood in the context of stemness. For example, molecular pathways involving WNT/B-catenin, MYC, PTEN, and LKB1 have been implicated in the inhibition of antitumor immunity (61), yet they also play important roles in stem cell maintenance (62–65). Thus, stemness may provide a unifying framework for understanding how various oncogenic signaling pathways engender an immunosuppressive tumor microenvironment.
Although it remains unclear if stemness metrics derived from bulk tumor samples represent rare populations of bona fide CSCs or a wholesale shift of the cancer cell population toward a higher stemness phenotype, our findings provide rationale for therapeutic targeting of the stemness phenotype itself. In particular, if stemness plays a causative role in the formation of cold tumor microenvironments, it may prove beneficial to target specific molecules or pathways that are inherent to the stemness phenotype, such as the aforementioned immunosuppressive molecules. High-stemness cancers might also be rendered more sensitive to immune control by administering drugs that induce cell differentiation to irreversibly disrupt the stemness phenotype (66). This might bring the additional benefit of constraining further tumor evolution, creating the conditions for durable clinical responses.
Materials and Methods
Data Acquisition and Processing.
All analyses were conducted with R software newer than version 3.4.2. We extracted clinical parameters and molecular subtypes for TCGA data from the pan-cancer curated clinical data of Liu et al. (67). Mutation data were downloaded from Firebrowse (www.firebrowse.org) as the number of nonsynonymous mutations per base. Tumor purity (27), mRNAsi (14), intratumoral heterogeneity (26, 32), and CIBERSORT and neoantigen scores (26) were accessed by extracting the relevant data for overlapping samples from the respective supplemental materials.
For correlations between stemness scores and IHC-based immune cell counts, immune cell infiltration data were provided by the authors of the respective studies (29). We obtained matching expression data (microarray or RNA-sequencing) from the Gene Expression Omnibus (GEO: GSE39582, GSE81089), or from the authors (15, 29, 30). Other expression data for validation was obtained from the GEO (GSE30652, GSE15192, GSE31257, GSE76009).
We accessed RNA sequencing as upper quartile-normalized fragments per kilobase of transcript per million mapped reads using the TCGAbiolinks R/Bioconductor package (68), for each cancer of interest, and expression data were merged across cancers. For genes with multiple annotated transcripts, we selected the transcript with the highest expression to represent the gene, then filtered the expression set to include only primary samples (except for melanoma, for which we included metastases), removed patients with duplicate samples, and removed any patients without a consensus purity score in Aran et al. (27) to enable purity corrections in analyses. For microarray datasets, we converted probe IDs to human gene symbols using biomaRt (69), and retained the probe with the highest expression for each gene, as above. We accessed the scRNA-seq data and tSNE embeddings in Lambrechts et al. (28) from EBI (E-MTAB-6149), while cell type annotations and anonymized patient codes were provided by the authors. Where appropriate (e.g., for linear modeling), expression data were log2(x + 1) -transformed.
We calculated stemness and other ssGSEA signatures using the GSVA package in R (gene sets in Dataset S1) (70) without normalization, and subsequently scaled values as z-scores within datasets of interest. For ssGSEA calculations on scRNA-seq, we first omitted genes below the median of average expression across samples; this left representation for 61 of 109 of the genes in the stemness signature. xCell enrichment scores were calculated in R, using the rawEnrichment analysis (20) function, which omits scaling scores to [0, 1] and correction for correlations among related cell types (20), as we sought to avoid introducing nonlinearities from these steps into analysis. To generate the immune signature, we summed z-scored signatures of cell types of interest (CD8+ T cells, NK cells, B cells). Because z-scoring of ssGSEA scores was done within each dataset, these scores should not be directly compared across datasets.
Quantification and Statistical Analysis.
We used nonparametric Spearman’s correlation to assess pairwise associations between variables of interest within cancers, or for median values across cancers, adjusting for multiple tests using the Benjamini–Hochberg method, where appropriate. For analyses across multiple cancer types or subtypes, we used linear models, controlling for site or subtype as fixed main effects, and inspecting model residuals to ensure model assumptions were reasonable. For analyses controlling for purity, purity (as consensus purity estimate from ref. 27) was included as a covariate (main effect) in linear models.
We conducted survival analyses using Cox proportional hazards models, calculating 95% CIs on log hazard ratios. We tested model assumptions using cox.zph (71). Where there were significant violations of model assumptions (P < 0.05), we inspected model Schoenfield residuals. We found that for the few instances in which model assumptions were violated, this was attributable to higher than predicted survival for some long-term survivors. For analyses across all patients and cancer types, we stratified Cox models by cancer type. For IHC data with multiple samples taken from the same patient, we modeled patient as a random effect in linear mixed-effects models implemented in lme4 in R (72) and assessed the significance of fixed effects of interest using likelihood-ratio tests on nested models.
Pathway enrichment analysis was conducted using ReactomePA (73) after testing for differential expression between quintiles using limma (74). For enrichment analysis, we selected the top 1,000 significantly down-regulated genes in high stemness cancers based on the moderated t statistic; in cancers where <1,000 genes were significantly down-regulated (Padj < 0.05), we selected all significantly down-regulated genes for downstream analysis. Significance of enrichment was evaluated at Padj < 0.05, and recurrently enriched pathways were defined as those that were significantly enriched in the greatest number of cancers. In limma analyses that included purity as a covariate, purity was log-transformed for consistency with the transformation of expression values.
To assess ERV expression, we first variance-filtered mapped ERVs to select those above the median interquartile range of expression using the genefilter R package (75). We conducted partial redundancy analysis using the vegan package (76), implementing the default distance metric (Euclidean). We conditioned the analysis by cancer type to control for cancer type-specific effects, and tested the significance of multivariate associations using permutation tests (n = 1,000).
Supplementary Material
Acknowledgments
We thank Dr. John Dick (University of Toronto) and Dr. Julian Lum (BC Cancer) for critical review of the manuscript, and Dr. Peter Watson (BC Cancer) for helpful discussions. We are grateful to The Cancer Genome Atlas and Cancer Cell Line Encyclopedia for access to data that enabled this study. Funding was provided by the BC Cancer Foundation (to B.H.N.), Canadian Cancer Society (to B.H.N.), Canada’s Networks and Centres of Excellence (to B.H.N.), Canadian Institutes of Health Research (to B.H.N.), Terry Fox Research Institute (to B.H.N.), Cancer Research Society (to B.H.N.), Vanier Canada Graduate Scholarship (to A.W.Z.), and Canadian Institutes of Health Research Postdoctoral fellowships (to P.T.H. and A. Miranda).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818210116/-/DCSupplemental.
References
- 1.Gajewski TF. The next hurdle in cancer immunotherapy: Overcoming the non-T-cell-inflamed tumor microenvironment. Semin Oncol. 2015;42:663–671. doi: 10.1053/j.seminoncol.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 3.Nassar D, Blanpain C. Cancer stem cells: Basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Ng SW, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 8.Smith BA, et al. A human adult stem cell signature marks aggressive variants across epithelial cancers. Cell Rep. 2018;24:3353–3366.e5. doi: 10.1016/j.celrep.2018.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaves M. Cancer stem cells as ‘units of selection’. Evol Appl. 2013;6:102–108. doi: 10.1111/eva.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, He X. The convergent cancer evolution toward a single cellular destination. Mol Biol Evol. 2016;33:4–12. doi: 10.1093/molbev/msv212. [DOI] [PubMed] [Google Scholar]
- 11.Agudo J, et al. Quiescent tissue stem cells evade immune surveillance. Immunity. 2018;48:271–285.e5. doi: 10.1016/j.immuni.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruttel VS, Wischhusen J. Cancer stem cell immunology: Key to understanding tumorigenesis and tumor immune escape? Front Immunol. 2014;5:360. doi: 10.3389/fimmu.2014.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noh KH, et al. Cancer vaccination drives Nanog-dependent evolution of tumor cells toward an immune-resistant and stem-like phenotype. Cancer Res. 2012;72:1717–1727. doi: 10.1158/0008-5472.CAN-11-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malta TM, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang AW, et al. Interfaces of malignant and immunologic clonal dynamics in ovarian cancer. Cell. 2018;173:1755–1769. doi: 10.1016/j.cell.2018.03.073. [DOI] [PubMed] [Google Scholar]
- 16.Safonov A, et al. Immune gene expression is associated with genomic aberrations in breast cancer. Cancer Res. 2017;77:3317–3324. doi: 10.1158/0008-5472.CAN-16-3478. [DOI] [PubMed] [Google Scholar]
- 17.Palmer NP, Schmid PR, Berger B, Kohane IS. A gene expression profile of stem cell pluripotentiality and differentiation is conserved across diverse solid and hematopoietic cancers. Genome Biol. 2012;13:R71. doi: 10.1186/gb-2012-13-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shats I, et al. Using a stem cell-based signature to guide therapeutic selection in cancer. Cancer Res. 2011;71:1772–1780. doi: 10.1158/0008-5472.CAN-10-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aran D, Hu Z, Butte AJ. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: A systematic review with meta-analysis. Br J Cancer. 2011;105:93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melero I, Rouzaut A, Motz GT, Coukos G. T-cell and NK-cell infiltration into solid tumors: A key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4:522–526. doi: 10.1158/2159-8290.CD-13-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol Immunol. 2017;14:662–674. doi: 10.1038/cmi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jochems C, Schlom J. Tumor-infiltrating immune cells and prognosis: The potential link between conventional cancer therapy and immunity. Exp Biol Med (Maywood) 2011;236:567–579. doi: 10.1258/ebm.2011.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharya B, et al. Gene expression in human embryonic stem cell lines: Unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 26.Thorsson V, et al. Cancer Genome Atlas Research Network The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 29.Becht E, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mezheyeuski A, et al. Multispectral imaging for quantitative and compartment-specific immune infiltrates reveals distinct immune profiles that classify lung cancer patients. J Pathol. 2018;244:421–431. doi: 10.1002/path.5026. [DOI] [PubMed] [Google Scholar]
- 31.Nawaz S, Heindl A, Koelble K, Yuan Y. Beyond immune density: Critical role of spatial heterogeneity in estrogen receptor-negative breast cancer. Mod Pathol. 2015;28:766–777. doi: 10.1038/modpathol.2015.37. [DOI] [PubMed] [Google Scholar]
- 32.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Şenbabaoğlu Y, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. doi: 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 36.Almeida LG, et al. CTdatabase: A knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–D819. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudet F, et al. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol. 2004;24:1640–1648. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strissel PL, et al. Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: Emergence of new molecular targets. Oncotarget. 2012;3:1204–1219. doi: 10.18632/oncotarget.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol. 2013;5:307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boller K, Janssen O, Schuldes H, Tönjes RR, Kurth R. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J Virol. 1997;71:4581–4588. doi: 10.1128/jvi.71.6.4581-4588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang BX, et al. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee YH, et al. Inhibition of the B7-H3 immune checkpoint limits tumor growth by enhancing cytotoxic lymphocyte function. Cell Res. 2017;27:1034–1045. doi: 10.1038/cr.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahnke K, Enk AH. TIGIT-CD155 interactions in melanoma: A novel co-inhibitory pathway with potential for clinical intervention. J Invest Dermatol. 2016;136:9–11. doi: 10.1016/j.jid.2015.10.048. [DOI] [PubMed] [Google Scholar]
- 47.Johnston CJ, Smyth DJ, Dresser DW, Maizels RM. TGF-β in tolerance, development and regulation of immunity. Cell Immunol. 2016;299:14–22. doi: 10.1016/j.cellimm.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blokzijl F, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature. 2016;538:260–264. doi: 10.1038/nature19768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saldanha-Araujo F, et al. Cancer/testis antigen expression on mesenchymal stem cells isolated from different tissues. Anticancer Res. 2010;30:5023–5027. [PubMed] [Google Scholar]
- 50.Lifantseva N, et al. Expression patterns of cancer-testis antigens in human embryonic stem cells and their cell derivatives indicate lineage tracks. Stem Cells Int. 2011;2011:795239. doi: 10.4061/2011/795239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada R, et al. Preferential expression of cancer/testis genes in cancer stem-like cells: Proposal of a novel sub-category, cancer/testis/stem gene. Tissue Antigens. 2013;81:428–434. doi: 10.1111/tan.12113. [DOI] [PubMed] [Google Scholar]
- 52.Guo YL, et al. Attenuated innate immunity in embryonic stem cells and its implications in developmental biology and regenerative medicine. Stem Cells. 2015;33:3165–3173. doi: 10.1002/stem.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends Immunol. 2013;34:67–73. doi: 10.1016/j.it.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seaman S, et al. Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer cell. 2017;31:501–515.e8. doi: 10.1016/j.ccell.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bin Z, et al. Overexpression of B7-H3 in CD133+ colorectal cancer cells is associated with cancer progression and survival in human patients. J Surg Res. 2014;188:396–403. doi: 10.1016/j.jss.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 56.Solecki DJ, Gromeier M, Mueller S, Bernhardt G, Wimmer E. Expression of the human poliovirus receptor/CD155 gene is activated by sonic hedgehog. J Biol Chem. 2002;277:25697–25702. doi: 10.1074/jbc.M201378200. [DOI] [PubMed] [Google Scholar]
- 57.Cochrane CR, Szczepny A, Watkins DN, Cain JE. Hedgehog signaling in the maintenance of cancer stem cells. Cancers (Basel) 2015;7:1554–1585. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watabe T, Miyazono K. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–115. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- 59.Peñuelas S, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 60.Ikushima H, et al. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 61.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18:139–147. doi: 10.1038/nrc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nusse R. Wnt signaling and stem cell control. Cell Res. 2008;18:523–527. doi: 10.1038/cr.2008.47. [DOI] [PubMed] [Google Scholar]
- 63.Murphy MJ, Wilson A, Trumpp A. More than just proliferation: Myc function in stem cells. Trends Cell Biol. 2005;15:128–137. doi: 10.1016/j.tcb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 64.Hill R, Wu H. PTEN, stem cells, and cancer stem cells. J Biol Chem. 2009;284:11755–11759. doi: 10.1074/jbc.R800071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Thé H. Differentiation therapy revisited. Nat Rev Cancer. 2018;18:117–127. doi: 10.1038/nrc.2017.103. [DOI] [PubMed] [Google Scholar]
- 67.Liu J, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Colaprico A, et al. TCGAbiolinks: An R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hänzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Therneau TM. 2018 survival: Survival Analysis. R Package Version 2.38. Available at https://CRAN.R-project.org/package=survival. Accessed on December 20, 2018.
- 72.Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- 73.Yu G, He QY. ReactomePA: An R/bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 74.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gentleman R, Carey V, Huber W, Hahne F. 2018 genefilter: Methods for Filtering Genes from High-Throughput Experiments. R Package Version 1.64.0. Available at bioconductor.org/packages/genefilter. Accessed on December 20, 2018.
- 76.Oksanen J, et al. 2015 vegan: Community Ecology Package. R Package Version 2.5-4. Available at https://CRAN.R-project.org/package=vegan. Accessed on December 20, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.